Abstract
Evolutionary biologists have long sought to identify phenotypic traits whose evolution enhances an organism's performance in its environment. Diversification of traits related to resource acquisition can occur owing to spatial or temporal resource heterogeneity. We examined the ability to capture light in the Cryptophyta, a phylum of single-celled eukaryotic algae with diverse photosynthetic pigments, to better understand how acquisition of an abiotic resource may be associated with diversification. Cryptophytes originated through secondary endosymbiosis between an unknown eukaryotic host and a red algal symbiont. This merger resulted in distinctive pigment–protein complexes, the cryptophyte phycobiliproteins, which are the products of genes from both ancestors. These novel complexes may have facilitated diversification across environments where the spectrum of light available for photosynthesis varies widely. We measured light capture and pigments under controlled conditions in a phenotypically and phylogenetically diverse collection of cryptophytes. Using phylogenetic comparative methods, we found that phycobiliprotein characteristics were evolutionarily associated with diversification of light capture in cryptophytes, while non-phycobiliprotein pigments were not. Furthermore, phycobiliproteins were evolutionarily labile with repeated transitions and reversals. Thus, the endosymbiotic origin of cryptophyte phycobiliproteins provided an evolutionary spark that drove diversification of light capture, the resource that is the foundation of photosynthesis.
Keywords: photosynthesis, phycobilin, photosynthetically usable radiation, resource acquisition
1. Introduction
Ecological diversification can be driven by traits that allow for the invasion of new adaptive zones [1], increase fitness, or increase reproductive or ecological specialization [2]. In several taxa, resource acquisition has been important to taxonomic diversification. For example, in Darwin's finches, diversification is tied to the acquisition of seeds via different beak morphologies that partition the available resource pool [3]. Similarly, jaw and tooth morphology has been implicated in diversification of cichlids, host plant use in phytophagous insects and host specialization for parasites [4–10]. In these instances, consumers are partitioning a diverse biotic resource base. It is less clear whether partitioning abiotic resources will lead to diversification via evolutionary changes in resource acquisition. One abiotic resource that could be partitioned in this manner is light, which is available in a broad spectrum of wavelengths and exhibits considerable heterogeneity in aquatic environments [11]. We sought to understand whether phytoplankton diversify based on acquisition of this abiotic resource.
The intensity and spectrum of light in an aquatic ecosystem can vary substantially depending on the ambient concentrations of phytoplankton, chromophoric dissolved organic matter, and suspended sediment. Light that could be used for photosynthesis is in the visible portion of the spectrum (400–700 nm) and is referred to as photosynthetically available radiation (PAR; [11]). However, the amount of radiant energy actually absorbed by an organism is a measurable phenotype referred to as photosynthetically usable radiation (PUR; [12]). The calculation of PUR uses both the PAR value measured in the growth environment and the absorption spectrum of the organism [12]. Therefore, PUR is the portion of PAR that an organism captures and may use for photosynthesis; it is a measure of resource acquisition. The higher the PUR, the better a particular organism is at absorbing available light.
Although PUR can be considered a phenotype, it is specific to a particular light environment and thus, comparisons among taxa depend on having data from comparable environments [13]. Formal comparisons of PUR across taxa are rare because differences in light environments mean that estimates from different studies may not be comparable, and large-scale experiments examining diverse algae are generally lacking. As a consequence, we have a relatively poor overall understanding of the diversification of light capture in comparison to other types of resource acquisition. Recently, we calculated PUR for 33 strains of cryptophyte algae grown under the same light conditions (PAR) in laboratory incubators [14]. We found that PUR varied nearly threefold across the phylum, with substantial variation even within some genera (Cryptomonas, Hemiselmis and Chroomonas). This implies substantial evolutionary diversification of resource acquisition with respect to light within cryptophytes, though the mechanism is yet unclear.
The Cryptophyta are a phylum of unicellular, eukaryotic algae that originated via secondary endosymbiosis between an unknown single-celled eukaryote host and a red algal ancestor [15–19]. Endosymbiosis provides its hosts with new biochemical functions and increases genomic complexity [20–24]. Through their endosymbiosis, cryptophytes acquired a second nuclear genome (the nucleomorph) and a plastid, and thus they acquired photosynthesis. Did endosymbiosis also provide cryptophytes with the capacity to evolve new photosynthetic machinery that allowed diversification of light capture?
Overall, differences in pigment composition create a fantastic diversity of colour among cryptophyte species. All phytoplankton, including cryptophytes, have chlorophyll a as a major light-harvesting pigment [25]. Cryptophytes also use accessory pigments (α-carotene, alloxanthin, chlorophyll c2 and phycobiliproteins) for capture of wavelengths not well absorbed by chlorophyll a [26–28]. The ratio of chlorophyll a to other pigments varies among cryptophytes, and in many species, the cellular concentration of phycobiliproteins is higher than that of non-phycobiliprotein pigments [14].
Cryptophyte phycobiliproteins are composed of two α and two β protein subunits and four covalently bonded chromophores called phycobilins [16,29]. The complex of the chromophores and protein subunits makes a complete light-capturing unit. The α subunits of phycobiliproteins are encoded in the nuclear genome from the ancestral host and have no known orthologues in other organisms, including red algae [16,30–32]. The α subunit genes have duplicated and diversified in the nuclear genome of cryptophytes [31,32], whereas the β subunit gene is found in the plastid that originated from the algal endosymbiont and is similar to genes in red algae [16,18,29]. Thus, the cryptophyte phycobiliproteins are unique pigment–protein complexes that originated through secondary endosymbiosis.
Cryptophyte phycobiliproteins are categorized as either a phycoerythrin (Cr-PE) or phycocyanin (Cr-PC); no species has been shown to contain both [26]. The ‘Cr’ modifier indicates that all of the cryptophyte phycobiliproteins are red algal derivatives and not the ‘true’ phycoerythrins or phycocyanins of cyanobacteria (see [16]). Cryptophytes with Cr-PE generally appear pink to red to humans, though some are yellow, orange or brown. Cr-PC species generally appear green to blue–green. The maximum absorption peaks of different cryptophyte phycobiliproteins form a near continuum ranging from 538 to 650 nm but are divided into eight classes (table 1; [16,33]). These absorption ranges (table 1) represent the major absorption peak and its accompanying shoulders. There are six known cryptophyte phycobilin chromophores that can be bonded to the protein components in various combinations [16,29]. Four chromophores are found only in cryptophytes, while the other two are also in red algae [16]. Previous reports have proposed that Cr-PE is the ancestral cryptophyte phycobiliprotein. This is supported by both the proposed biosynthesis pathway of the Cr-PC, which includes Cr-PE as an intermediate, and the high sequence similarity of the β subunit gene to a gene in red algae [16,29,34].
Table 1.
Cryptophyte phycobiliprotein classes and their associated absorption range. (Cr, cryptophytes; PE, phycoerythrin; PC, phycocyanin. Wavelengths are given in nanometres (nm). Adapted from table 1 in Glazer & Wedemayer [16]. Cryptophyte phycobiliproteins are named according to the wavelength of maximal absorption, e.g. 545 nm for Cr-PE 545.)
| phycobiliprotein class | absorption range (nm) |
|---|---|
| Cr-PE 545 | 538–551 |
| Cr-PE 555 | 553–556 |
| Cr-PE 566 | 563–567 |
| Cr-PC 569 | 568–569 |
| Cr-PC 577 | 576–578 |
| Cr-PC 615 | 612–615 |
| Cr-PC 630 | 625–630 |
| Cr-PC 645 | 641–650 |
The unusually high diversity of colour in the phylum Cryptophyta provides an opportunity to investigate how light capture abilities have diversified. Our goal was to determine which physiological and biophysical characteristics contribute to the diversification of resource acquisition (light capture) in photosynthetic cryptophytes. Using phylogenetic comparative methods, we tested whether pigment type, pigment quantity, pigment density and absorption spectra characteristics were evolutionarily associated with diversification of light capture.
2. Methods
(a). Data collection
We obtained 42 strains of cryptophytes from culture collections (electronic supplementary material, table S1). Thirty-two are named species, two are different strains of one species (Cryptomonas pyrenoidifera), eight are strains identified to a genus and two are unidentified strains; in total, these strains represent at least 12 genera (electronic supplementary material, table S1). Phenotypic characteristics (whole-cell absorption spectra, cell size measurements, pigment concentrations, phycobiliprotein absorption spectra and PUR values) for 33 of the strains in this study were previously reported in Cunningham et al. [14]. Specific characteristics of those strains plus the nine additional strains that we measured are listed in the electronic supplementary material, table S1. To make these measurements, algal cultures were grown under full spectrum irradiance of approximately 30 µmol photons m−2 s−1, nutrient replete conditions, and in media and temperature conditions established by culture collection centres (see the electronic supplementary material, table S1). Phenotypic measurements were taken during exponential phase as determined by cell counts. Detailed culture conditions, methods for all phenotypic measurements and procedures for DNA extraction, polymerase chain reaction (PCR) amplification and sequencing are listed in the electronic supplementary material, S1 and follow Cunningham et al. [14].
(b). Time-proportional phylogenies
Molecular sequence data for the 42 strains consisted of the plastid ribulose bisphosphate carboxylase, large subunit (rbcL) and nuclear small subunit (SSU) and partial large subunit (LSU) rDNA nucleotide sequences collected following Johnson et al. [35], Marin et al. [36], Hoef-Emden [37] and Cunningham et al. [14]. Methods for DNA extraction, PCR amplification and sequencing are detailed in the electronic supplementary material, S1. PCR amplification of rbcL failed in Unid. sp. CCMP1179. Plastid rbcL sequences averaged approximately 943 bp and ranged from 522 to 1068 bp; nuclear SSU sequences averaged 1698 bp and ranged from 1266 to 1823 bp; and the partial nuclear LSU sequences averaged 900 bps and ranged from 622 to 1018 bp. For an outgroup, sequences of the haptophyte Emiliania huxleyi rbcL, SSU and LSU loci were downloaded from National Center for Biotechnology Information (NCBI; GenBank accession numbers KC404516, KC404120 and KC404160).
To account for potentially differing evolutionary histories between the plastid genome (red algal ancestor) and nuclear genome (unknown single-celled eukaryote) [15–19], we constructed two separate maximum-likelihood phylogenies; one for rbcL and one for SSU/LSU. Sequences were aligned using ClustalW2 [38] and visually inspected and edited using the BioEdit Sequence Alignment Editor [39]. PartitionFinder2 [40] was used to determine the best-fitting nucleotide substitution model. Maximum-likelihood phylogenies were estimated with RAxML v. 8.2.7 [41] using the autoMRE option and a general time reversal model with a portion of invariant sites and a γ distribution (GTR + I + G). We transformed the two maximum-likelihood phylogenies (electronic supplementary material, figures S1 and S2) into time-proportional phylogenies using treePL [42]. We calibrated the phylogeny using 1.0 for the root of the tree owing to the lack of fossil or molecular calibration points (see the electronic supplementary material, S1 for details).
(c). Phylogenetic generalized least-squares analyses
Our data consist of a 41 strain rbcL time-proportional phylogeny, a 42 strain SSU/LSU time-proportional phylogeny and phenotypic measurements. These measurements include concentrations for five pigments (chlorophyll a, chlorophyll c2, alloxanthin, α-carotene and cryptophyte phycobiliprotein), PUR and phycobiliprotein maximum absorption peak wavelength (nm; table 1). The pigment concentrations are presented in per cell (pg cell−1) and per cubic micrometer (pg µm−3) to account for differing cell volumes among species. Phenotypic data for all 42 strains of cryptophytes can be found in the electronic supplementary material, table S1.
Phylogenetic generalized least-squares (PGLS) analyses were conducted in R using the package caper v. 0.5.2 and the ‘pgls’ function [43,44]. Our PGLS analyses sought to identify predictors of PUR, or, more generally, to determine which phenotypic characteristics have an evolutionary association with diversification of light capture. Collinearity of the variables was evaluated using variance inflation factors (VIF). Owing to large VIF values (greater than 10.0), when more than two pigment concentrations were evaluated together, we combined the non-phycobiliprotein pigments (chlorophyll a and chlorophyll c2, alloxanthin, and α-carotene) into a single value annotated as ‘non-phycobiliprotein pigments'. The PGLS results reported here using the non-phycobiliprotein pigments’ as a predictor are qualitatively similar to analyses performed with non-phycobiliprotein pigments individually (results not shown). We performed multi-predictor analyses with PUR as the dependent variable and the predictor variables being non-phycobiliprotein pigments, phycobiliprotein concentration and phycobiliprotein maximum absorption peak wavelength. Two separate multi-predictor analyses were conducted to evaluate the influence of pigment density using concentration per cell (pg cell−1) and biovolume (pg µm−3) for the pigments. Prior to PGLS analyses, data were log10-transformed to better fit a normal distribution. Normality and homogeneity of the residuals were evaluated using a qq-plot of residuals, a histogram of residuals and a plot of the residuals against the fitted values. Phylogenetic signal (λ) was estimated using maximum likelihood.
(d). Ancestral state estimates
Each species of cryptophyte has only one spectroscopically distinct phycobiliprotein and these phycobiliproteins are grouped into discrete classes (table 1; [16]). However, it is not known if these maximum absorption peaks have evolved in an incremental fashion or if only certain cryptophyte phycobiliprotein pigment–protein configurations are possible; therefore, we treated the data as both discrete and continuous and performed two separate ancestral state reconstructions based on character type.
We estimated discrete ancestral states for phycobiliprotein maximum absorption peak wavelength class (Cr-PE 545, Cr-PE 555, etc.) using joint stochastic character mapping [45] performed in R using the package phytools v. 0.6-00 and the function ‘make.simmap’ [46]. We used the fitDiscrete function [47] and the Akaike weights criteria to determine the best fit model (ER, equal rates; SYM, symmetric transitions are equal; ARD, all rates different) for our data and time-proportional phylogeny (see above). We found that an ER model was the best overall fit for phycobiliprotein maximum absorption peak wavelength (wi = 99.9%) for both phylogenies. We implemented the ER model and 1000 stochastic character maps that were summarized to produce posterior probabilities for each node. We estimated continuous character ancestral states using fast estimation of the maximum-likelihood ancestral states in R using phytools v. 0.6-00 and the ‘fastAnc’ function [46]. We also calculated 95% confidence intervals for continuous character state estimates. All ancestral state reconstruction figures were drawn using phytools v. 0.6-00 functions [46].
3. Results
(a). Time-proportional phylogenies
We used a plastid genome marker (rbcL, large subunit) and two nuclear genome markers (SSU and LSU) to produce two phylogenies to account for the potentially unique and distinct evolutionary histories of cryptophytes [15–19]. These phylogenies (electronic supplementary material, figures S1 and S2) were both rooted with the haptophyte E. huxleyi.
The plastid gene (rbcL) phylogeny (electronic supplementary material, figure S1; figure 1) has strong support for a monophyletic clade consisting of the autotrophic cryptophytes with the exception of two Proteomonas species that formed a moderately supported outgroup to other cryptophytes. Within the major clade of cryptophytes, we recovered five strongly supported clades consisting of (1) Rhodomonas minuta, Geminigera crypophila and Cryptomonas erosa; (2) a Hemiselmis/Chroomonas clade; (3) a Rhodomonas/Rhinomonas/Storeatula clade; (4) Guillardia theta and Hanusia phi; and (5) a Cryptomonas clade. We found that clade 1 has moderate support for being the outgroup to clades 2–5. Furthermore, a branch with moderate support indicates that clades 4 and 5 are closely related. Interestingly, this relationship between clade 4 (Guillardia/Hanusia) and clade 5 (Cryptomonas) was also recovered with near identical support in the Hoef-Emden [37] nuclear SSU phylogeny but is not seen in other, more recent and speciose phylogenies using both the nuclear SSU and LSU markers [14,48]. Clades 2–5 are consistently recovered in more speciose cryptophyte phylogenies [14,37,48]. Uncertainty in the placement of Proteomonas species, Geminigera species and the recently named Baffinella frigidus species ([49]; formerly Unid. sp. CCMP 2293) is a persistent problem in cryptophyte phylogenetics.
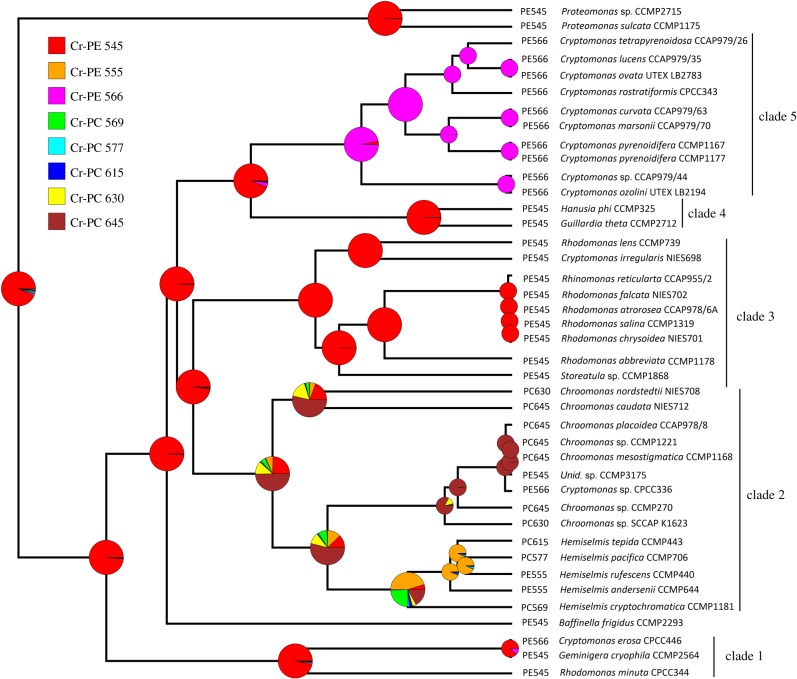
Figure 1.
Discrete character ancestral state estimates of phycobiliprotein maximum absorption peak wavelength mapped onto the rbcL phylogeny. Node labels indicate the posterior probability (PP) of each ancestral state. The size of the pie graphs are for better visualization towards the root of the phylogeny and are not indicative of PP values. Tip labels denote each species' state. Phylogenetic clade designations are derived from the rbcL maximum-likelihood bootstrap support values (see the electronic supplementary material, figure S1). Colours do not indicate the actual qualitative colours of the cultures. Wavelengths for each class are in nanometres (nm). Cr, cryptophytes; PE, phycoerythrin; PC, phycocyanin.
The SSU and LSU nuclear genome phylogeny (electronic supplementary material, figure S2) also found strong support for clades 2–5. However, in the SSU and LSU phylogeny, clade 1 consisted of only Rho. minuta and Geminigera cryophila. Cryptomonas erosa grouped with other Cryptomonas species in clade 5, suggesting a potentially interesting evolutionary history for Cr. erosa. Another interesting difference is that the Proteomonas species and Unid. sp. CCMP1179 (not in the rbcL phylogeny due to PCR amplification failure) formed a moderately supported clade with the Rhodomonas/Rhinomonas/Storeatula clade (clade 3). These marked differences suggest that some cryptophyte species may have had evolutionary histories distinct from other strains within the same genus.
A reoccurring issue with SSU and LSU cryptophyte phylogenies is that the backbone branches connecting clades 2–5 are generally short and lack strong support (electronic supplementary material, figure S2; [14,37,48]), potentially reflecting an ancient period of rapid diversification. Our rbcL phylogeny, however, has relatively long backbone branches with moderate to strong support. Therefore, in this study, we report the statistical results from the use of the plastid gene (rbcL) phylogeny as the main results.
(b). Phylogenetic generalized least-squares analyses
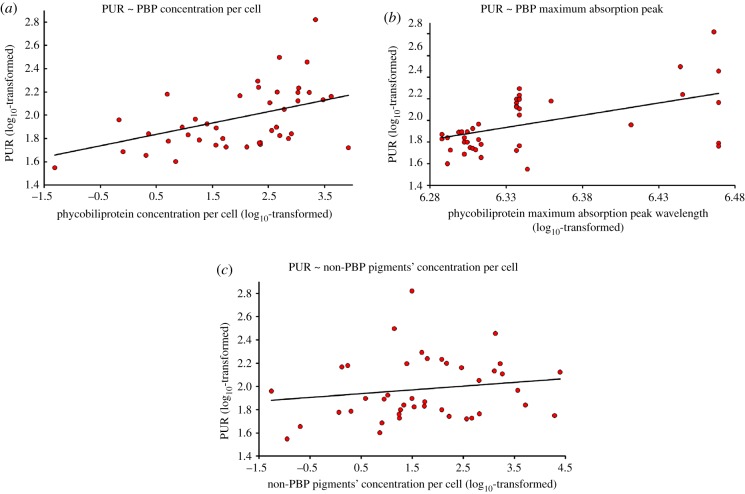
In order to identify which phenotypic traits are evolutionarily associated with cryptophyte PUR, our measure of resource acquisition ability, we performed multi-predictor PGLS analyses. We first evaluated whether the cellular (pg cell−1) concentration of the pigments and phycobiliprotein maximum absorption peak wavelength are predictors of PUR. We found that phycobiliprotein concentration per cell and phycobiliprotein maximum absorption peak wavelength were positively associated with PUR (λ = 0.512, adj. R2 = 0.452; F3,37 = 11.97, p < 0.001; table 2 and figure 2; electronic supplementary material, figure S3), while the non-phycobiliprotein pigments were not associated with PUR. These results indicate that cryptophytes with a higher absolute phycobiliprotein concentration per cell and longer phycobiliprotein maximum absorption peak wavelengths capture more light in our full-spectrum experimental environment.
Table 2.
Multi-predictor PGLS results of PUR as the dependent variable and phycobiliprotein maximum absorption peak wavelength (phycobiliprotein max. abs. peak), phycobiliprotein concentration per cell (pg cell−1), and the additive value of the absolute concentrations per cell (pg cell−1) of alloxanthin, α-carotene, chlorophyll a and chlorophyll c2 (non-phycobiliprotein pigments) as predictors. (All values were log10 transformed before analysis (log). *Significant at p < 0.05; ***significant at p < 0.001.)
| estimate | std. error | t-value | p-value | |
|---|---|---|---|---|
| (intercept) | −13.504 | 3.631 | −3.695 | <0.001*** |
| log phycobiliprotein conc. (pg cell−1) | 0.099 | 0.041 | 2.425 | 0.020* |
| log phycobiliprotein max. abs. peak | 2.411 | 0.582 | 4.141 | <0.001*** |
| log non-phycobiliprotein pigments (pg cell−1) | −0.001 | 0.037 | −0.024 | 0.981 |
Figure 2.
Log10-transformed data are plotted with a trend line indicating the relationships between photosynthetically usable radiant energy (PUR) and phycobiliprotein concentration per cell (a), PUR and phycobiliprotein maximum absorption peak wavelength (b), and PUR and the additive value of the concentrations per cell of the non-phycobiliprotein pigments (c; alloxanthin, α-carotene, chlorophyll a and chlorophyll c2). (Online version in colour.)
The results from our primary PGLS analysis (table 2) indicate that specific traits of phycobiliproteins, per cell quantity of cryptophyte phycobiliproteins and maximum absorption peak wavelength allow some species to have increased ability to capture light. To account for differences in cell volume among strains, we tested whether the density of pigments per cubic micrometre (biovolume concentration) are also evolutionarily associated with PUR in a second PGLS analysis. In contrast with the first PGLS analysis (table 2), we did not find that the biovolume concentration of phycobiliprotein was significant (table 3). However, as in the first PGLS analysis, we found phycobiliprotein maximum absorption peak wavelength was positively associated with PUR and the non-phycobiliprotein pigments were again not associated with PUR (λ = 0.499, adj. R2 = 0.26; F3,38 = 5.69, p = 0.003; table 3). Collectively, these PGLS results indicate that the phycobiliprotein concentration per cell is a significant predictor of PUR but that cell volume is seemingly unrelated to differences in PUR among cryptophytes. Furthermore, the phycobiliprotein class (table 1) retains statistical significance in both analyses and is clearly an important trait governing the differential ability of light capture in cryptophytes.
Table 3.
Multi-predictor PGLS results of PUR as the dependent variable and phycobiliprotein maximum absorption peak wavelength (phycobiliprotein max. abs. peak), phycobiliprotein concentration per cubic micrometre (pg µm−3), and the additive value of the relative concentrations per cubic micrometer (pg µm−3) of alloxanthin, α-carotene, chlorophyll a and chlorophyll c2 (non-phycobiliprotein pigments) as predictors. (All values were log10 transformed before analysis (log). **Significant at p < 0.01.)
| estimate | std. error | t-value | p-value | |
|---|---|---|---|---|
| (intercept) | −12.863 | 4.416 | −2.913 | 0.006** |
| log phycobiliprotein conc. (pg µm−3) | 0.038 | 0.055 | 0.683 | 0.499 |
| log phycobiliprotein max. abs. peak | 2.363 | 0.683 | 3.460 | 0.001** |
| log non-phycobiliprotein pigments (pg µm−3) | 0.003 | 0.052 | 0.063 | 0.950 |
For both the primary and secondary PGLS analyses, we found qualitatively similar results using the nuclear SSU and LSU time-proportional phylogeny (electronic supplementary material, figure S2 and tables S2 and S3) to those found with the rbcL phylogeny, indicating that these PGLS results are robust to differing evolutionary histories and phylogenetic uncertainty.
(c). Ancestral state estimates
The evolutionary association of phycobiliprotein maximum absorption peak wavelength and resource acquisition ability prompted us to investigate the evolutionary history of phycobiliproteins. Did the evolution of cryptophytes involve a transition from a phycobiliprotein with a shorter maximum absorption peak (the phycoerythrins; table 1) to one able to absorb longer wavelengths (the phycocyanins; table 1)? We addressed this question by reconstructing the ancestral states of phycobiliprotein maximum absorption peak wavelength. Here, we report ancestral state estimates for this trait as discrete (figure 1) and continuous (electronic supplementary material, figure S4) characters.
We first wanted to test the hypothesis that Cr-PE was the ancestral state of cryptophytes and that Cr-PC evolved in cryptophytes secondarily [16,29,34]. Furthermore, we sought to determine if there is evidence suggesting that cryptophyte phycobiliprotein maximum absorption peak evolved from shorter to longer wavelengths. Using the phycobiliprotein maximum absorption peak wavelengths as a discrete character (figure 1), ancestral state estimates found the strongest support for Cr-PE 545 (0.932 posterior probability (PP)) followed by Cr-PE 566 (0.016 PP) and Cr-PC 569 (0.013 PP) with all other phycobiliprotein classes (table 1) having effectively no support. Our continuous character ancestral state estimate (electronic supplementary material, figure S4) for the root of the phylogeny was approximately 558 with a 95% confidence interval of 299.9–816.6, which covers the entirety of the visible spectrum (400–700 nm).
The 1000 trees with mapped discrete character states had an average of approximately 14 state changes with the only state changes occurring approximately once being from Cr-PE 545 to Cr-PE 566, Cr-PC 645 to Cr-PE 545 and Cr-PC 645 to Cr-PC 630. Interestingly, the only state change from a Cr-PE type to a Cr-PC type occurring on a basal branch leading to a phylogenetically significant clade occurred on the branch leading to the Chroomonas/Hemiselmis clade. Furthermore, several state reversals (PC to PE and PC to PE) appear to have occurred within the Hemiselmis subclade (figure 1). However, the other major clades (clade 1 and clades 3–5; electronic supplementary material, figure S4) have no shifts between pigment types (Cr-PE to Cr-PC) indicating that evolutionary lability is unevenly distributed across the phylogeny. Together, these results indicate that we should accept the hypothesis that phycocyanin evolved from phycoerythrin. Furthermore, the discrete and continuous ancestral state estimates suggest a trend towards longer maximum absorption peaks that enable enhanced light capture ability.
The nuclear SSU and LSU ancestral state reconstruction of the cryptophyte phycobiliprotein maximum absorption peak also found strong support for a Cr-PE 545 ancestral state for cryptophytes (results not shown).
4. Discussion
Evolutionary biologists have advanced at least two complementary ideas to explain diversification of ecologically critical traits. One view has long held that diversification is driven by environmental factors, and that populations diverge through adaptation in response to changes in a species' ecological circumstances [50,51]. By contrast, others have argued that key innovations arise that open a new range of habitats to a taxon, leading to diversification as it spreads across the new habitats (examples: feather coloration, [52]; nectar spurs in flowers, [53]). Although these views share an emphasis on adaptive differentiation, they differ in whether the initial step leading to diversification is the origin of a new function or the invasion of a new habitat [54]. Our results support the view that the cryptophyte phycobiliproteins, pigment–protein complexes that originated from the marriage of proteins coming from each partner in an ancient endosymbiosis, are the driving evolutionary spark behind functional diversification of light capture in these algae. This has allowed them to specialize in, and diversify within, low-light environments of varying spectral quality, where every photon captured counts [48].
We found that the concentration of phycobiliprotein per cell was a significant predictor of PUR but that the biovolumetric concentration of phycobiliprotein was not. These results suggest that cell volume itself is not important, i.e. larger cells do not necessarily have more phycobiliprotein to enhance their light capture ability. Cryptophytes have a single bilobed plastid connected by a pyrenoid or in some cases two plastids where a pyrenoid connection appears absent [48,55–57]. Therefore, a larger cell would not produce more plastids or necessarily have a higher amount of phycobiliprotein. Cryptophyte phycobiliproteins are localized to the thylakoid lumen and are not attached to the thylakoid membrane, unlike other organisms (cyanobacteria, glaucophytes and red algae) that possess phycobilin type light-harvesting pigments [58–61]. The width of the thylakoid lumen can vary but is relatively uniform across genera [55]. These cellular features combined with our results indicate that the total amount of phycobiliprotein in a cryptophyte is not determined by cell volume, number of plastids or thylakoid lumen size, but by other genetic and/or environmental factors. While environmental factors like light or nutrient availability will result in some degree of variation in the cellular concentrations of both phycobiliprotein and non-phycobiliprotein pigments [62,63], we minimized this variability to the extent possible by growing all cultures under full spectrum irradiance of approximately the same intensity (approx. 30 µmol photons m−2 s−1), nutrient replete conditions, and, for phenotypic measurements, all cells were harvested during exponential growth.
The cryptophyte phycobiliprotein maximum absorption peak was found to be positively associated with PUR regardless of which other dependent variables were included in our PGLS analyses. Generally, the results show that longer wavelength absorption peaks result in an enhanced ability to capture light while cryptophytes with shorter wavelength maximum absorption peaks have a reduced ability to capture light. Furthermore, our ancestral state reconstruction found that cryptophyte phycobiliproteins evolved from a shorter to a longer wavelength. In fact, the cryptophyte strain with the highest PUR (Chroomonas caudata NIES 712) also has the longest phycobiliprotein maximum absorption peak (Cr-PC 645; electronic supplementary material, table S1). The trend of a higher PUR for Cr-PC 645 strains probably relates to the 645 peak becoming continuous with and broadening the chlorophyll a and c2 absorption peaks in the red and far red regions of the spectrum, thus enabling a greater portion of the visible light spectrum to be absorbed (see the electronic supplementary material, figure S6; [14]). Energy transfer dynamics have been studied extensively for two cryptophyte strains (Chroomonas sp. CCMP270 and Rhodomonas sp. CS24) that have Cr-PC 645 and Cr-PE 545 phycobiliprotein classes, respectively [64–68]. These studies found that the efficiency of energy transfer from Cr-PC 645 in the thylakoid lumen to the thylakoid membrane-bound photosystems is greater than 99%, while the Cr-PE 545 of Rhodomonas sp. efficiency was approximately 98%. These differences in efficiency are attributed to small differences in the structure of the phycobiliproteins [65]. Chroomonas sp. (CCMP270) was one of our 42 strains and had the third highest PUR value, while our Rhodomonas species (all with Cr-PE 545) had PUR values that were approximately 50% of the Cr-PC 645 Chroomonas strains (electronic supplementary material, table S1). Together, these data indicate that the evolution of cryptophyte phycobiliproteins has resulted in species with not only a greater ability to capture light across the spectrum as measured by PUR, but also a greater efficiency [65–69] at transferring this energy to the photosystems.
We found that not only are the diversity of phycobiliprotein class and type evolutionarily associated with resource acquisition, but also that phycobiliprotein diversification rate is not equal across the phylogeny. The exceptional phycobiliprotein diversity found among the Hemiselmis and Chroomonas species has been well characterized in several studies [14,36,48,69]. The phycobiliprotein diversity among the 42 species used in this study indicates that six of the eight phycobiliprotein classes (table 1) are only found in the Chroomonas/Hemiselmis clade (figure 1; electronic supplementary material, figure S4). The Hemiselmis genus forms a monophyletic clade and has at least four different phycobiliprotein classes (Cr-PE 555, Cr-PC 569, Cr-PC 577 and Cr-PC 615; [14]). Our ancestral state reconstruction supports previous hypotheses suggesting multiple phycobiliprotein state changes in the Hemiselmis clade [36,48,69]. Cr-PE 555 is unique to Hemiselmis species and has evolved at least once in the genus. Our time-proportional phylogeny and ancestral state reconstruction finds that the subclade comprising Hemiselmis species probably had an ancestral character state of Cr-PE 555 (0.453 PP) and that a state change to Cr-PC 615 occurred on the branch leading to He. tepida (figure 1). The unusually high amount of phycobiliprotein variation among Hemiselmis species may relate to the open-conformation physical structure of the phycobiliprotein that appears to be unique to this genus [48,70,71]. The open-conformation may allow for the attachment of different phycobilins without additional changes in the sequences of the protein subunits. Therefore, only alterations in the biosynthesis pathways of phycobilins need to occur for adaptation to novel light environments. However, the open-conformation may come at a cost of lower excitonic coupling of the phycobiliprotein phycobilins that could reduce the efficiency of light harvesting [71].
We measured characteristics of all cryptophyte species under a standard full-spectrum light environment (electronic supplementary material, figure S5) that provided the opportunity for cells to capture photons of any wavelength. We did not attempt to mimic the specific light environments that individual species experience in the wild, but it may be important to consider whether the cells could exhibit phenotypic plasticity of photosynthetic characteristics in response to more constrained, but realistic, light environments. Chromatic adaptation (as defined in the literature on phytoplankton physiology) is a type of phenotypic plasticity that entails the non-evolutionary capability of algae to change photosynthetic pigment content and concentrations in response to variation of light spectrum [72]. Cryptophytes have been reported to display weak chromatic adaptation, largely based on species favouring one light environment over another light environment [73]. For example, Wall & Briand [74] and Ojala [73] found that five different species of cryptophytes grew better in blue light, while Kamiya & Miyachi [75] found that a Cryptomonas species grew better in green light over blue or red light. These studies, which provide evidence for chromatic adaptation, did not measure pigment concentrations directly. Instead, Ojala [73] and Vesk & Jeffrey [76] used a relative measure of phycobiliprotein/chlorophyll concentration (fluorescence quotient) and found up to a 25% increase in relative phycobiliprotein content in blue/green light environments. Our work shows that cryptophyte species’ phycobiliprotein concentration per cell is positively associated with resource acquisition ability in one light environment; it would provide further insight to test for shifts in pigment composition and concentration induced by variation in the spectral composition of the light environment.
Broad phylogenetic comparisons typically take one of two distinct strategies that involve an important experimental trade-off. In one, traits are measured on wild organisms in their natural habitats. In the other, living organisms are brought into a controlled environment and measured in a common garden experiment. The former strategy has the advantage of relying on traits as they are expressed in the habitats where selection has shaped them. However, if the traits in question have some degree of phenotypic plasticity, it comes at the cost that traits may not be directly comparable. The latter strategy, which we adopted in this study, has the advantage of ensuring traits are comparable, and thus, conclusions drawn are phylogenetically valid. However, it comes at a cost of a weaker connection to the habitats in which organisms evolved. Of course, if the traits in question are fixed rather than plastic, the environment where they are measured does not matter.
Many of the traits in our study are impossible to measure in the field, so we were constrained to using laboratory-based measurements. As described above, we opted to make measurements in a single standard light environment and harvested cells during exponential phase under nutrient replete conditions. Standardizing these conditions allows reasonable confidence that our phylogenetic comparisons are based on homologous traits. Furthermore, because we used a broad light spectrum and PUR is scaled to the amount of light available at each wavelength, we can be confident that all of our strains had available to them the light that they could capture, and that PUR measurements were not phylogenetically biased. Because phenotypic plasticity of cellular concentrations of photopigments with respect to light and nutrients is known [62,63,77], future work on the relationship between pigment characteristics and light capture as resource acquisition would benefit from a reaction norm approach that considers more environments. Such a strategy potentially resolves the trade-off between environment-specific and common-garden phylogenetic comparisons. Unfortunately, culture collections generally lack information on environmental characteristics under which cryptophyte strains were isolated, making it difficult to recreate the specific light and nutrient environments to which individual strains were adapted.
5. Conclusion
We evaluated resource acquisition in a phylum of algae, Cryptophyta, by measuring the amount of light absorbed (PUR) in the same full spectrum environment (PAR). Variation in light capture ability as measured by PUR indicates that different species have substantially different abilities to acquire light. We determined that characteristics of phycobiliproteins are evolutionarily associated with diversification of light capture ability, but other pigments, including chlorophyll, are not. Our results support the hypothesis that Cr-PE was the ancestral pigment type of cryptophyte phycobiliproteins and that the evolutionary shift to Cr-PC first occurred on the branch leading to the Chroomonas/Hemiselmis clade (figure 1). Thus, the endosymbiotic origin of the cryptophyte phycobiliproteins probably provided an evolutionary spark that led to the great diversity of cryptophyte phycobiliprotein classes (table 1) which resulted in the diversification of light capture ability and an increase in the efficiency of photosynthesis [64–71] in cryptophytes.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
DNA sequences: Genbank accessions MK818447–MK818487, MK828443–MK828483, and MK828402–MK828442.
Authors' contributions
M.J.G., T.L.R. and J.L.D. conceived the study. M.J.G. wrote the manuscript, carried out molecular work and conducted statistical analyses. M.J.G., T.L.R. and J.L.D. revised the manuscript. B.R.C. and T.L.R. performed phenotype measurements and calculations. J.M.P. performed rbcL gene amplification. E.M.L. maintained algal cultures.
Competing interests
We have no competing interests.
Funding
This material is based upon work supported by the National Science Foundation (NSF) Dimensions of Biodiversity program under grant no. 1542555.
References
- 1.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 2.Heard SB, Hauser DL. 1995. Key evolutionary innovations and their ecological mechanisms. Hist. Biol. 10, 151–173. ( 10.1080/10292389509380518) [DOI] [Google Scholar]
- 3.Grant BR, Grant PR. 1993. Evolution of Darwin's finches caused by a rare climatic event. Proc. R. Soc. Lond. B 253, 111–117. ( 10.1098/rspb.1993.0016) [DOI] [Google Scholar]
- 4.Jaenike J. 1990. Host specialization in phytophagous insects. Annu. Rev. Ecol. Syst. 21, 243–273. ( 10.1146/annurev.es.21.110190.001331) [DOI] [Google Scholar]
- 5.Kawecki TJ. 1998. Red queen meets Santa Rosalia: arms races and the evolution of host specialization in organisms with parasitic lifestyles. Am. Nat. 152, 635–651. ( 10.1086/286195) [DOI] [PubMed] [Google Scholar]
- 6.Ricklefs RE, Fallon SM. 2002. Diversification and host switching in avian malaria parasites. Proc. R. Soc. Lond. B 269, 885–892. ( 10.1098/rspb.2001.1940) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kocher TD. 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet. 5, 288–298. ( 10.1038/nrg1316) [DOI] [PubMed] [Google Scholar]
- 8.Janson EM, Stireman JO, Singer MS, Abbot P. 2008. Phytophagous insect-microbe mutualisms and adaptive evolutionary diversification. Evolution 62, 997–1012. ( 10.1111/j.1558-5646.2008.00348.x) [DOI] [PubMed] [Google Scholar]
- 9.Burress ED. 2015. Cichlid fishes as models of ecological diversification: patterns, mechanisms, and consequences. Hydrobiologia 748, 7–27. ( 10.1007/s10750-014-1960-z) [DOI] [Google Scholar]
- 10.Sudakaran S, Retz F, Kikuchi Y, Kost C, Kaltenpoth M. 2015. Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J. 9, 2587–2604. ( 10.1038/ismej.2015.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirk JT. 1994. Light and photosynthesis in aquatic ecosystems, 2nd edn Cambridge, UK: Cambridge University Press. [Google Scholar]
- 12.Morel A. 1978. Available, usable and stored radiant energy in relation to marine photosynthesis. Deep-Sea Res. 25, 673–688. ( 10.1016/0146-6291(78)90623-9) [DOI] [Google Scholar]
- 13.Moran NA. 1992. The evolutionary maintenance of alternative phenotypes. Am. Nat. 139, 971–989. ( 10.1086/285369) [DOI] [Google Scholar]
- 14.Cunningham BR, Greenwold MJ, Lachenmyer EM, Heidenreich KM, Davis AC, Dudycha JL, Richardson TL. In press. Light capture and pigment diversity in marine and freshwater cryptophytes. J. Phycol. ( 10.1111/jpy.12816 [DOI] [PubMed] [Google Scholar]
- 15.Douglas SE. 1992. Eukaryote–eukaryote endosymbiosis: insights from studies of a Cryptomonad alga. Biosystems 28, 57–68. ( 10.1016/0303-2647(92)90008-M) [DOI] [PubMed] [Google Scholar]
- 16.Glazer AN, Wedemayer GJ. 1995. Cryptomonad biliproteins—an evolutionary perspective. Photosynth. Res. 46, 93–105. ( 10.1007/BF00020420) [DOI] [PubMed] [Google Scholar]
- 17.Douglas SE, Penny SL. 1999. The plastid genome from the Cryptomonad alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J. Mol. Evol. 8, 236–244. ( 10.1007/PL00006462) [DOI] [PubMed] [Google Scholar]
- 18.Douglas SE, et al. 2001. The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091–1096. ( 10.1038/35074092) [DOI] [PubMed] [Google Scholar]
- 19.Kim JI, Yoon HS, Yi G, Kim HS, Yih W, Shin W. 2015. The plastid genome of the Cryptomonad Teleaulaz amphioxeia. PLoS ONE 10, e0129284 ( 10.1371/journal.pone.0129284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. ( 10.1038/nrg1271) [DOI] [PubMed] [Google Scholar]
- 21.Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467, 929–934. ( 10.1038/nature09486) [DOI] [PubMed] [Google Scholar]
- 22.Nowack ECM, Melkonian M. 2010. Endosymbiotic associations within protists. Phil. Trans. R. Soc. B 365, 695–712. ( 10.1098/rstb.2009.0188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagat P, Bodył A, Mackiewicz P, Stiller JW. 2013. Tertiary plastid endosymbioses in dinoflagellates. In Endosymbiosis (ed. W Löffelhardt), pp. 233–290. Vienna, Austria: Springer. [Google Scholar]
- 24.Stiller JW, Schreiber J, Yue J, Guo H, Ding Q, Huang J. 2014. The evolution of photosynthesis in chromist algae through serial endosymbiosis. Nat. Commun. 5, 5764 ( 10.1038/ncomms6764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy S, Llewellyn CA, Egeland ES, Johnsen G. 2011. Phytoplankton pigments: characterization, chemotaxonomy and applications in oceanography. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 26.Hill DRA, Rowan KS. 1989. The biliproteins of the Cryptophyceae. Phycologia 28, 455–463. ( 10.2216/i0031-8884-28-4-455.1) [DOI] [Google Scholar]
- 27.Jeffrey SW, Mantoura RFC, Wright SW, International Council of Scientific Unions, and Unesco. 1997. Phytoplankton pigments in oceanography: guidelines to modern methods. Paris, France: UNESCO Publications. [Google Scholar]
- 28.Schagerl M, Donabaum K. 2003. Patterns of major photosynthetic pigments in freshwater algae. 1. Cyanoprokaryota, Rhodophyta and Cryptophyta. Ann. Limnol. – Int. J. Limnol. 39, 35–47. ( 10.1051/limn/2003003) [DOI] [Google Scholar]
- 29.Apt KE, Collier JL, Grossman AR. 1995. Evolution of the phycobiliproteins. J. Mol. Biol. 248, 79–96. ( 10.1006/jmbi.1995.0203) [DOI] [PubMed] [Google Scholar]
- 30.Jenkins J, Hiller RG, Speirs J, Godovac-Zimmermann J. 1990. A genomic clone encoding a cryptophyte α-subunit: evidence for three α-subunits and an N-terminal membrane transit sequence. FEBS 273, 191–194. ( 10.1016/0014-5793(90)81082-Y) [DOI] [PubMed] [Google Scholar]
- 31.Gould SB, Fan E, Hempel F, Maier U, Klösgen RB. 2007. Translocation of a phycoerythrin α subunit across five biological membranes. J. Biol. Chem. 282, 30 295–30 302. ( 10.1074/jbc.M701869200) [DOI] [PubMed] [Google Scholar]
- 32.Kieselbach T, Cheregi O, Green BR, Funk C. 2018. Proteomic analysis of the phycobiliprotein antenna of the cryptophyte alga Guillardia theta cultured under different light intensities. Photosynth. Res. 135, 149–163. ( 10.1007/s11120-017-0400-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gantt E. 1979. Phycobiliproteins of cryptophyceae. In Biochemistry and physiology of protozoa (eds M Levendowsky, SH Hunter), pp. 121–137. New York, NY: Academic Press, Inc. [Google Scholar]
- 34.Beale SI. 1993. Biosynthesis of phycobilins. Chem. Rev. 93, 785–802. ( 10.1021/cr00018a0) [DOI] [Google Scholar]
- 35.Johnson MD, et al. 2016. The genetic diversity of Mesodinium and associated cryptophytes. Front. Microbiol. 7, 2017 ( 10.3389/fmicb.2016.02017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marin B, Klingberg M, Melkonian M. 1998. Phylogenetic relationships among the cryptophyta: analyses of nuclear-encoded SSU rRNA sequences support the monophyly of extant plastid-containing lineages. Protist 149, 265–276. ( 10.1016/S1434-4610(98)70033-1) [DOI] [PubMed] [Google Scholar]
- 37.Hoef-Emden K. 2008. Molecular phylogeny of phycocyanin-containing cryptophytes: evolution of biliproteins and geographical distribution. J. Phycol. 44, 985–993. ( 10.1111/j.1529-8817.2008.00530.x) [DOI] [PubMed] [Google Scholar]
- 38.Larkin MA, et al. 2007. ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948. ( 10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- 39.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 40.Lanfear R, Frandsen PB, Wright AM, Senfield T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. ( 10.1093/molbev/msw260) [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith SA, O'Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690. ( 10.1093/bioinformatics/bts492) [DOI] [PubMed] [Google Scholar]
- 43.Freckleton RP, Harvey PH, Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 160, 712–726. ( 10.1086/343873) [DOI] [PubMed] [Google Scholar]
- 44.Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N, Pearse W.. 2013. Caper: comparative analyses of phylogenetics and evolution in R. See http://cran.r-project.org/web/packages/caper/vignettes/caper.pdf
- 45.Huelsenbeck JP, Neilsen R, Bollback JP. 2003. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–138. ( 10.1080/10635150390192780) [DOI] [PubMed] [Google Scholar]
- 46.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 47.Yang Z. 2006. Computational molecular evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 48.Hoef-Emden K, Archibald JM. 2016. Cryptophyta (Cryptomonads). In Handbook of the protists (eds JM Archibald, AGB Simpson, CH Slamovits, L Margulis, M Melkonian, DJ Chapman, JO Corliss), pp. 1–41. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 49.Daugbjerg N, Norlin A, Lovejoy C. 2018. Baffinella frigidus gen. et sp. nov. (Baffinellaceae fam. nov., Cryptophyceae) from Baffin Bay: morphology, pigment profile, phylogeny, and growth rate response to three abiotic factors. J. Phycol. 54, 665–680. ( 10.1111/jpy.12766) [DOI] [PubMed] [Google Scholar]
- 50.Yoder JB, et al. 2010. Ecological opportunity and the origin of adaptive radiations. J. Evol. Biol. 23, 1581–1596. ( 10.1111/j.1420-9101.2010.02029.x) [DOI] [PubMed] [Google Scholar]
- 51.Wellborn GA, Langerhans RB. 2015. Ecological opportunity and the adaptive diversification of lineages. Ecol. Evol. 5, 176–195. ( 10.1002/ece3.1347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maia R, Rubenstein DR, Shawkey MD. 2013. Key ornamental innovations facilitate diversification in an avian radiation. Proc. Natl Acad. Sci. USA 110, 10 687–10 692. ( 10.1073/pnas.1220784110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hodges SA. 1997. Floral nectar spurs and diversification. Int. J. Plant Sci. 158, S81–S88. ( 10.1086/297508) [DOI] [Google Scholar]
- 54.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 55.Gantt E, Edwards MR, Provasoli L. 1971. Chloroplast structure of the Cryptophyceae: evidence for phycobiliproteins within intrathylakoidal spaces. J. Cell Biol. 48, 280–290. ( 10.1083/jcb.48.2.280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hill DRA. 1991. A revised circumscription of Cryptomonas (Cryptophyceae) based on examination of Australian strains. Phycologia 30, 170–188. ( 10.2216/i0031-8884-30-2-170.1) [DOI] [Google Scholar]
- 57.Hoef-Emden K, Melkonian M. 2003. Revision of the genus Cryptomonas (Cryptophyceae): a combination of molecular phylogeny and morphology provides insights into a long-hidden dimorphism. Protist 154, 371–409. ( 10.1078/143446103322454130) [DOI] [PubMed] [Google Scholar]
- 58.Mckay RML, Lichtlé C, Gibbs SP. 1992. Immunocytochemical characterization of the intrapyrenoid thylakoids of Cryptomonads. J. Phycol. 26, 64–68. ( 10.1111/j.0022-3646.1992.00064.x) [DOI] [Google Scholar]
- 59.Spear-Bernstein L, Miller KR. 1989. Unique location of the phycobiliprotein light-harvesting pigment in the Cryptophyceae. J. Phycol. 25, 412–419. ( 10.1111/j.1529-8817.1989.tb00245.x) [DOI] [Google Scholar]
- 60.MacColl R, Guard-Friar D. 1987. Phycobiliproteins. Boca Raton, FL: CRC Press. [Google Scholar]
- 61.Vesk M, Dwarte D, Fowler S, Hiller RG. 1992. Freeze fracture immunocytochemistry of light-harvesting pigment complexes in a cryptophyte. Protoplasma 170, 166–176. ( 10.1007/BF01378791) [DOI] [Google Scholar]
- 62.Lewitus AJ, Caron DA. 1990. Relative effects of nitrogen or phosphorus depletion and light intensity on the pigmentation, chemical composition, and volume of Pyrenomonas salina (Cryptophyceae). Mar. Ecol. Prog. Ser. 61, 171–181. ( 10.3354/meps061171) [DOI] [Google Scholar]
- 63.Lawrenz E, Richardson TL. 2017. Differential effects of changes in spectral irradiance on photoacclimation, primary productivity and growth in Rhodomonas salina (Crytpophyceae) and Skeletonema costatum (Bacillariophyceae) in simulated blackwater environments. J. Phycol. 53, 1241–1254. ( 10.1111/jpy.12578) [DOI] [PubMed] [Google Scholar]
- 64.Mirkovic T, Wilk KE, Curmi PMG, Scholes GD. 2009. Phycobiliprotein diffusion in chloroplasts of cryptophyte Rhodomonas CS24. Photosynth. Res. 100, 7–17. ( 10.1007/s11120-009-9412-8) [DOI] [PubMed] [Google Scholar]
- 65.Doust AB, Wilk KE, Curmi PMG, Scholes GD. 2006. The photophysics of cryptophyte light-harvesting. J. Photochem. Photobiol. A 184, 1–7. ( 10.1016/j.jphotochem.2006.06.006) [DOI] [Google Scholar]
- 66.Mirkovic T, Doust AB, Kim J, Wilk KE, Curutchet C, Mennucci B, Cammi R, Curmi PMG, Scholes GD. 2007. Ultrafast light havesting dynamics in the cryptophyte phycocyanin 645. Photochem. Photobiol. Sci. 6, 964–975. ( 10.1039/B704962E) [DOI] [PubMed] [Google Scholar]
- 67.Van der Weij-De Wit CD, Doust AB, van Stokkum IHM, Dekker JP, Wilk KE, Curmi PMG, Scholes GD, van Grondelle R.. 2006. How energy funnels from the phycoerythrin antenna complex to photosystem I and photosystem II in cryptophyte Rhodomonas CS24 cells. J. Phys. Chem. B 110, 2423–2433. ( 10.1021/jp061546w) [DOI] [PubMed] [Google Scholar]
- 68.Van der Weij-De Wit CD, Doust AB, van Stokkum IHM, Dekker JP, Wilk KE, Curmi PMG, van Grondelle R.. 2008. Phycocyanin sensitizes both photosystem I and photosystem II in cryptophyte Chroomonas CCMP270 cells. Biophys. J. 94, 2423–2433. ( 10.1529/biophysj.107.113993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lane CE, Archibald JM. 2008. New marine members of the genus Hemiselmis (Cryptomonadales, Cryptophyceae). J. Phycol. 44, 439–450. ( 10.1111/j.1529-8817.2008.00486.x) [DOI] [PubMed] [Google Scholar]
- 70.Collini E, Wong CY, Wilk KE, Curmi PMG, Brumer P, Scholes GD. 2010. Coherently wired light-harvesting in photosynthetic marine algae at ambient temperature. Nature 463, 644–648. ( 10.1038/nature08811) [DOI] [PubMed] [Google Scholar]
- 71.Harrop SJ, et al. 2014. Single residue insertion switches the quaternary structure and exciton states of cryptophyte light harvesting proteins. Proc. Natl Acad. Sci. USA 111, E2666–E2675. ( 10.1073/pnas.1402538111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Engelmann TW. 1902. Ueber experimentelle erzeugung zweckmässiger aenderungen der färbung pflanzlicher chromophylle durch farbiges licht. Arch. Anat. Physiol. (Physiol. Abt.) 1902, 333–335. [Google Scholar]
- 73.Ojala A. 1993. The influence of light quality on growth and phycobiliprotein/chlorophyll a fluorescence quotients of some species of freshwater algae in culture. Phycologia 32, 22–28. ( 10.2216/i0031-8884-32-1-22.1) [DOI] [Google Scholar]
- 74.Wall D, Briand F. 1979. Response of lake phytoplankton communities to in situ manipulation of light intensity and colour. J. Plankton Res. 1, 103–112. ( 10.1093/plankt/1.1.103) [DOI] [Google Scholar]
- 75.Kamiya A, Miyachi S. 1984. Effects of light quality on formation of 5-aminolevulinic acid, phycoerythrin and chlorophyll in Cryptomonas sp. cells collected from the subsurface chlorophyll layer. Plant Cell Physiol. 25, 831–839. ( 10.1093/oxfordjournals.pcp.a076778) [DOI] [Google Scholar]
- 76.Vesk M, Jeffrey SW. 1977. Effect of blue-green light on photosynthetic pigments and chloroplast structure in unicellular marine algae from six classes. J. Phycol. 13, 280–288. ( 10.1111/j.1529-8817.1977.tb02928.x) [DOI] [Google Scholar]
- 77.Faulkner ST, Rekully CM, Lachenmyer EM, Kara E, Richardson TL, Shaw TJ, Myrick ML. 2018. Single-cell and bulk fluorescence excitation signatures of seven phytoplankton species during nitrogen depletion and resupply. Appl. Spectrosc. 73, 304–312. ( 10.1177/0003702818812090) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: Genbank accessions MK818447–MK818487, MK828443–MK828483, and MK828402–MK828442.