Abstract
Predation is one of the key ecological mechanisms allowing species coexistence and influencing biological diversity. However, ecological processes are subject to contemporary evolutionary change, and the degree to which predation affects diversity ultimately depends on the interplay between evolution and ecology. Furthermore, ecological interactions that influence species coexistence can be altered by reciprocal coevolution especially in the case of antagonistic interactions such as predation or parasitism. Here we used an experimental evolution approach to test for the role of initial trait variation in the prey population and coevolutionary history of the predator in the ecological dynamics of a two-species bacterial community predated by a ciliate. We found that initial trait variation both at the bacterial and ciliate level enhanced species coexistence, and that subsequent trait evolutionary trajectories depended on the initial genetic diversity present in the population. Our findings provide further support to the notion that the ecology-centric view of diversity maintenance must be reinvestigated in light of recent findings in the field of eco-evolutionary dynamics.
Keywords: community dynamics, genetic diversity, eco-evolutionary dynamics, predator–prey interaction, Pseudomonas fluorescens SBW25, Tetrahymena thermophila
1. Introduction
Natural selection acts on the fitness variance of phenotypes [1–3], and adaptive evolution is predicted to be enhanced in populations with high genetic diversity [4]. Adaptive evolution can lead to niche divergence and thus facilitate coexistence in competitor or predator–prey communities [5–7]. Classic competition theory predicts that species coexistence is possible when intraspecific competition is stronger than interspecific competition [5]. However, owing to contemporary evolution, the impact of competitors on each other might not be constant [8]. For example, the high number of coexisting species in microbial communities [9] in even simple environments [10–13] might be explained by high levels of within-species clonal diversity [5,14,15] and result in rapid evolution to use underexploited or new ecological niches [13]. Recent work has shown that traits and species interactions in microbial food webs can be altered by evolutionary change, as de novo mutations and changes over time in the frequencies of genotypes from standing genetic variation can occur at the same temporal scale as ecological processes [16–18]. It has previously been shown that consumers such as predators or parasites can have significant indirect effects on the outcome of competition, and thus on the maintenance of species diversity [19], by compensating for differences in traits [5,20,21]. Direct interaction between competing species might also change due to rapid evolution, resulting in resource use divergence [13].
In microbial communities, competitive interactions are common [22–25], which can lead to rapid exclusion of community members under standard conditions [21]. Recent studies have proposed that coexistence and escape from competitive exclusion is facilitated by evolutionary change in between-species interactions [13,26–28], resulting in, cross-feeding [26] or a niche shift to underexploited resources [29–31]. In most bacterial experiments, monoclonal isolates are assembled [13,21,23,26,32] and any evolutionary change is based on de novo mutations [33–35], potentially imposing constraints on evolution [4]. Our aim was to study the role of initial genetic variation on coexistence as the associated phenotypic variation might affect competition [36]. Our study system consisted of two bacterial species, Escherichia coli and Pseudomonas fluorescens, competing for shared resources and consumed by a keystone predator, the ciliate Tetrahymena thermophila. To investigate if genetic diversity can promote coexistence, we compared community dynamics with monoclonal P. fluorescens populations to genetically diverse populations obtained by pooling different P. fluorescens clones. The genetic diversity is represented by differences in phenotypic traits, including growth capacity and level of defence against ciliate predation. To investigate if diversity in the predator population also has an effect, we added a population containing diverse ciliate phenotypes obtained by pooling ciliates that had coevolved with either of the two bacterial species. The reason for the two predator treatments is that we hypothesize that if predation is a key factor allowing our two bacterial species to coexist, evolutionary adaptation in the predator allowing for more efficient predation might further facilitate the coexistence [37].
In our experiment, we tracked the community dynamics and the evolutionary outcome of both bacterial species when (i) growing without a predator, (ii) with a naive ciliate predator and (iii) with coevolved predators. These predation treatments are hereafter rereferred as ‘no predation’, ‘naive’ and ‘coevolved’ treatments correspondingly. We also manipulated the genetic diversity of the P. fluorescens population in a full factorial design. We used isogenic lines of P. fluorescens (and E. coli) to inoculate the experiments with minimum standing genetic variation as control population (hereafter ‘ancestor’). Further, we increased the genetic diversity of P. fluorescens by adding evolved diverse populations from previous experiments (full-diversity), or artificially assembled a population consisting of a subset of clones (high-diversity). We assessed the variability in interaction between the two bacterial species by measuring competitive dynamics and the level of coexistence. Ecological dynamics in population size were followed for 16 days and evolutionary dynamics were estimated by testing whether the bacteria evolved defence against the ciliate (measured as prey defence level), as well as by estimating bacterial fitness (measured as growth capacity).
We found that manipulating within-species bacterial diversity affected the frequencies of the two competitors over time but this effect depended on the presence and evolutionary history of the ciliate. Notably, the highest frequency of P. fluorescens, expected to be the inferior competitor, was observed in communities with standing genetic variation in both the bacteria and the ciliates. Our results show that the relative contribution of evolution (temporal changes in growth capacity and/or defence traits) and ecology (competitive interactions and predation) to changes in the frequency of the two bacteria over time was strongly dependent on standing genetic variation.
2. Methods
(a). Model system
We constructed our communities using two bacterial species and a ciliate predator, adopting a previously used model system [21]. The two bacterial species are Escherichia coli ATCC 11303 and Pseudomonas fluorescens SBW25 cultured in 1% King's B (KB) liquid culture medium. In general, the ancestral E. coli strain seems to be the dominant competitor in co-cultures but is more limited by predation than P. fluorescens (electronic supplementary material, figure S1). As a generalist predator, capable of consuming both bacterial species, we used the ciliated protozoan Tetrahymena thermophila CCAP 1630/1U. Prior to the experiments, all bacterial stocks were kept at –80°C and ciliate stocks were cultured axenically in proteose peptone yeast extract (PPY) medium containing 20 g of proteose peptone and 2.5 g of yeast extract in 1 l of deionized water.
(b). Obtaining trait diversity
For P. fluorescens and the ciliate predator, we manipulated genetic diversity by combining samples isolated from a long-term predator selection experiment (LTPE). The LTPE was started using a single-colony P. fluorescens SBW25 and E. coli ATCC 11303, and an axenic culture of the ciliate T. thermophila 1630/1U (CCAP). Material from only these two selection lines was used in the current experiment. Each bacterial strain was cultured alone and together with the ciliate (three replicates each) in 20 ml glass vials containing 6 ml of 5% KB medium, with 1% weekly transfer to fresh medium. Cultures were kept at 28°C (±0.1°C) with shaking at 50 r.p.m. Every four transfers (28 days), bacterial and predator densities were estimated using optical density as a proxy for bacterial biomass and direct ciliate counts as described previously [38], and samples were freeze-stored with glycerol at –20°C for later analysis. This experiment had been running for 20 months when we isolated the populations for the current experiment.
We isolated coevolved ciliates from each of the six LTPE P. fluorescens and E. coli lines (three replicate lines each) and pooled them together in equal densities to obtain a diverse ciliate population referred as ‘coevolved ciliates’. This mix of ciliates was cultivated axenically in PPY until the start of the experiment. The full-diversity population of P. fluorescens was harvested by mixing all three replicate populations from samples freed from live ciliates through freeze-storage at –20°C (ciliates do not survive under these conditions). For ancestral and high-diversity populations, we isolated individual colonies of the ancestral strains and from two time points in the LTPE, after six months and after 20 months, using Tryptone Bile X-Glucuronide agar (TBX, Sigma-Aldrich) and CFC agar plates (CFC supplement: 10 mg of cetrimide and fucidin and 50 mg cephalosporin in 1 l of PPY agar). We determined the position of each clone in trait space (see below) comprising growth capacity and level of anti-predatory defence. For both ancestral bacterial strains, we isolated 10 colonies and picked one isolate representing the ancestral trait space mean. We initially isolated and characterized a pool of 200 clones and picked 20 clones among them representing a broad range of phenotypes in growth capacity and defence against predation. The high-diversity population was established by randomly combining 10 out of these 20 clones.
(c). Determining position in trait space
For individual clones from both bacterial species, we determined growth capacity and defence level. For these measurements, we used the Bioscreen C spectrophotometer (Growth Curves AB Ltd, Helsinki, Finland) to estimate the optical density of growing bacterial samples (100 wells) at 5 min intervals for 48 h. Frozen samples were revived in fresh medium and allowed to acclimatize for 24 h, after which they were pin-replicated to fresh conditions (1% KB) in Bioscreen-compatible honeycomb plates. These plates were incubated at 28°C under constant shaking in the Bioscreen device. As a proxy for biomass yield, we calculated the area under the curve to obtain growth capacity for each clone. After 48 h, ciliates were added to the samples to estimate biomass loss due to predation. Comparing change in the bacterial biomass of control treatments without ciliates with treatments containing ciliates allowed us to measure the loss of bacterial biomass due to predation. Comparing between individual clones allowed us to estimate which clones are well defended and which clones are poorly defended. Briefly, ciliates were cultivated 5 days in advance in fresh PPY medium. The medium was removed by centrifugation (2 × 8 min at 3300 r.p.m.), and the populations were starved overnight in M9 salt solution. Initial T. thermophila cell densities were enumerated directly from live subsamples (2.5 ml) using a compound microscope (Zeiss Axioskop 2 plus, Oberkochen, Germany) and diluted to obtain 1000 cells ml–1 inoculated to each microcosm. For control treatments, we filtered ciliates out from the culture vial and added ciliate-free filtrate. The optical density of the samples was tracked again for 48 h, and the loss of biomass due to predation was estimated by comparing the control with the predation treatment. This protocol allowed us to determine growth capacity and defence level in the same population. For determining evolution, we picked 10 E. coli and 10 P. fluorescens clones from each microcosm at the end of the experiment. For these clones, the evolved position in trait space (i.e. growth capacity and defence level) was estimated using the protocol described above.
(d). Estimating ciliate growth on both bacterial strains
To estimate ciliate performance, we isolated both the ciliates and the bacterial strains from the LTPE. Briefly, we isolated the bacteria by freezer-storage, which effectively killed all ciliates. Axenic ciliate populations were obtained by culturing experimental populations to high density in PPY medium containing 24, 50, 50 and 33 µg ml–1 of the antibiotics kanamycin, rifampicin, streptomycin and tetracycline, respectively. Axenicity was controlled for by plating on 50% PPY agar plates (on which all experimental bacterial strains grow). Following this, ciliates were transferred to antibiotic-free PPY medium and cultured to high density. For the growth assay, we grew both evolved bacterial strains in 5% KB to equal density based on optical density. Bacterial cells were centrifuged, and the medium was replaced with M9 salts, preventing further bacterial growth. Ciliates from both coevolved lines were grown in PPY to high density and the medium was replaced with M9 minimum medium. Ciliate populations were starved overnight and density was determined by counting live cells. We added these starved ciliates to both evolved bacterial lines. Three replicates per treatment were cultivated for 48 h after which ciliate growth rate was estimated. For these final counts, we took pictures from samples fixed with Lugol's solution using inverse light microscopy.
(e). Microcosm experiments with manipulated community structure
Microcosms for experimental lines were set up in deep 96-well plates filled with 500 µl medium containing M9 salts and 1% KB (0.2 g l–1 Peptone number 3 and 0.1 ml–1 glycerol). Communities consisting of P. fluorescens and E. coli were assembled and either (i) no ciliates (control), (ii) naive ciliates or (iii) coevolved ciliates were added to the microcosms. For P. fluorescens, we initiated populations with three different levels of initial genetic diversity: (i) minimal level, obtained by culturing a population from a single ancestral clone; (ii) full-diversity established from populations obtained from the LTPE; and (iii) high-diversity around ancestral trait mean. For E. coli, we used only one ancestral clone to set up the populations. For ancestral P. fluorescens populations, we added one of four clones isolated from the ancestor to each replicate. For the full-diversity populations, bacteria that had evolved in medium either alone or together with a ciliate, both from the LTPE, were used. For the high-diversity populations, we randomly combined 10 out of 20 clones representing the trait space with respect to growth capacity and defence level (electronic supplementary material, figure S2). Both species were added in even densities based on optical density. The nine different treatments, consisting of three ciliates and three genetic diversity levels, were replicated four times. Plates were incubated at 28°C under constant shaking (50 r.p.m.). Every 48 h, 10% of the community was transferred to fresh medium. We recorded biomass by measuring optical density at 600 nm (Tecan Infinite M200 plate reader) and stored samples at –80°C after each round to archive the time series. The experimental period was 16 days, representing approximately 50 bacterial and ciliate generations. After reviving the archived samples from days 0, 2, 6 and 16, we determined the ratio between E. coli and P. fluorescens using selective TBX and CFC agar plates, respectively. With these selective media and culture conditions, we were able to clearly distinguish and enumerate both bacterial species from mixed samples. From the last time point, we also isolated 10 individual colonies (clones) from both species that were stored at –80°C for later analysis.
(f). Data analysis
All analyses were performed in R [39]. We used generalized estimating equation models (geeGLMs) to compare the proportion of P. fluorescens, accounting for the time-series structure following individual microcosms. We modelled the proportion of P. fluorescens using ‘genetic diversity’ and ‘predation’ both in interaction as explanatory variables together with ‘time’ as continuous variable. To account for the temporal replication, ‘microcosm ID’ was included as a random effect. We used a model of the binomial family and included an ‘ar1’ correlation structure (continuous-time first-order autoregressive correlation structure) to account for temporal correlation. We used the function geeglm from the package geepack [40] with the family ‘binomial’ with a logit link. To model the bacterial biomass data, we followed a similar approach and used estimated equation models based on the Gaussian family. Again, the model investigated the main effects ‘genetic diversity’ and ‘predation’ over ‘time’, including ‘ID’ and an ‘ar1’ correlation structure. We simplified the model by dropping non-significant terms. For analysis of ciliate densities, we compared only the treatments containing ciliates using a model based on the Gaussian family, after log10 transformation of the data. For the main effects model, ‘genetic diversity’ and ‘predation’ were included together with ‘time’, ‘ID’ to account for temporal replication and the ‘ar1’ correlation structure. Again, the model was simplified to remove non-significant terms. For statistical analysis, we removed day 0 measurements from all data, as these represent diluted, not maximum densities of established populations represented by all the other sampling points. For comparing performance of the two coevolved ciliate lines growing on the two bacteria from the LTPE, we used ANOVA with ciliate ID (ciliates coevolved with P. fluorescens or with E. coli) and bacterial ID (P. fluorescens and E. coli) as explanatory variables. We applied a model selection process in which the interaction between both variables was dropped. To test for the effect of the treatments on individual traits (defence or growth), we used generalized least-squares (gls) models, as implemented in the nlme [41] package, assuming a residual variance structure dependent on the experimental treatments. Multiple comparisons for gls models were performed using the package emmeans [42], with p-value adjustment according to the Tukey post hoc method and significance level α = 0.05.
3. Results
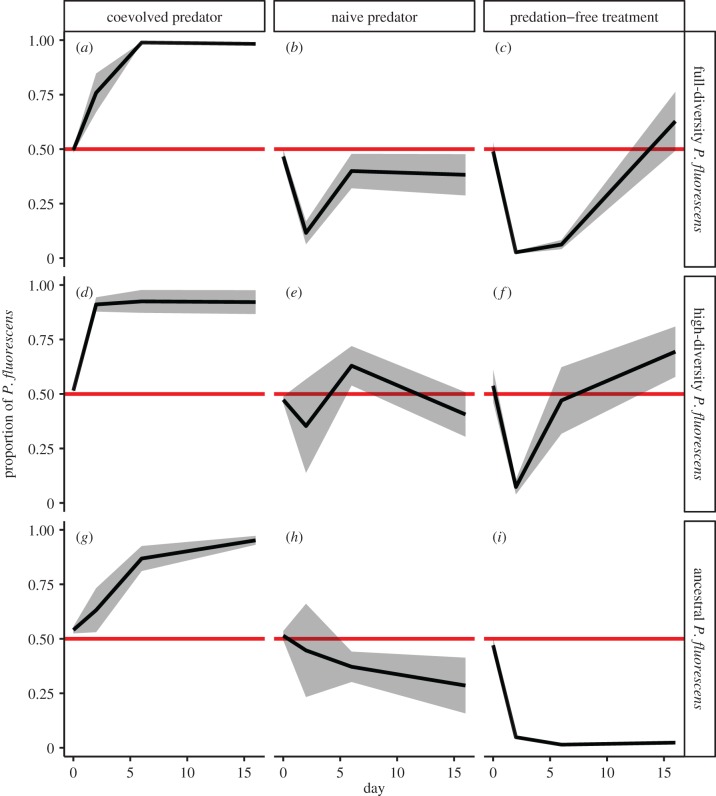
Our experiments suggested that the coexistence of Escherichia coli and P. fluorescens depended on both ciliate predation and genetic diversity (figure 1). Ciliate predation was an important factor increasing P. fluorescens proportions in general (table 1). Genetic diversity enhanced the competitive ability of P. fluorescens mainly in interaction with predation. While the ancestral P. fluorescens was almost completely excluded under competition, P. fluorescens dominated when genetic diversity and predation acted together. The effect of diversity was time-dependent suggesting that competitive ability successively increased over the course of the experiment. The proportional changes over time revealed interesting temporal dynamics congruent with the findings described above. The ancestral clone was inferior to E. coli and experienced rapid competitive exclusion over time without ciliate predation. By contrast, in the full-diversity treatment with coevolved ciliates, P. fluorescens was the superior competitor and almost excluded E. coli. Adding naive ciliates instead of coevolved ciliates decreased the competitive ability of P. fluorescens, and both competitors seemed equal. Without ciliates, P. fluorescens initially decreased in proportion but was able to recover when full-diversity was present in the population. Interestingly, the coevolved ciliate population shifted competitive balance towards P. fluorescens even in the absence of diversity.
Figure 1.
Proportion of the P. fluorescens population over time. The rows represent the different bacterial population structures (rows in figure) for P. fluorescens: (a–c) full-diversity, (d–f) high-diversity and (g–i) ancestral P. fluorescens strain. The columns represent the different predator treatments that were also applied: (a,d,g) ciliates coevolved with bacteria, (b,e,h) naive ciliate and (c,f,i) no ciliates. The black line represents the proportion of P. fluorescens (mean ± s.e.), and the red line shows the equal proportion line as reference.
Table 1.
A generalized estimated equation model showed that the main effects ‘predation’ and ‘diversity’ both had significant effects on Pseudomonas fluorescens proportions. ***p < 0.001; **p < 0.01; *p < 0.05; .p < 0.1.
| d.f. | X2 | p(>|χ|) | |
|---|---|---|---|
| time | 1 | 5 | 0.0259* |
| diversity | 2 | 5.4 | 0.0689. |
| predation | 2 | 142 | 0.0000*** |
| time × diversity | 2 | 6.1 | 0.0481* |
| time × predation | 2 | 13 | 0.0015** |
| diversity × predation | 4 | 25.4 | 0.0000*** |
| time × diversity × predation | 4 | 22.7 | 0.0001*** |
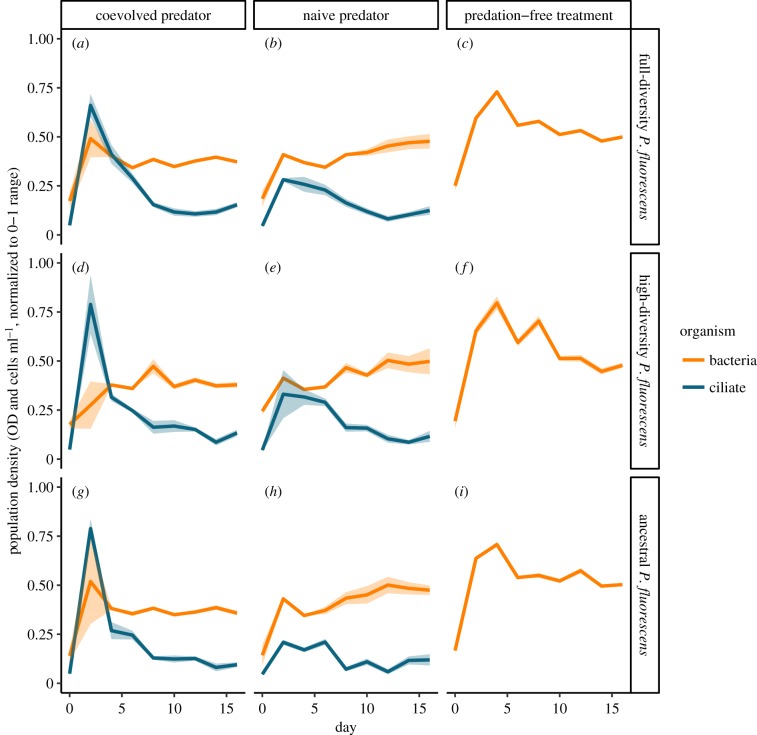
The total bacterial biomass was independent from the diversity of P. fluorescens (figure 2), but was affected by predation (electronic supplementary material, table S1). As expected, without predation, bacterial biomass was increased with no difference between P. fluorescens diversity treatments. While there was no obvious change over time under coevolved predation, bacterial biomass slightly increased over time with naive predators. In general, however, bacterial biomass seemed relatively stable over the duration of the experiment.
Figure 2.
Total densities for bacteria and ciliates. Rows represent data from the three different population structures: (a–c) full-diversity, (d–f) high-diversity and (g–i) ancestral P. fluorescens strain without diversity. Columns represent the three different predation treatments: (a,d,g) ciliates coevolved with bacteria, (b,e,h) naive ciliate and (c,f,i) no ciliates. Orange lines and points (mean ± s.e.) show total bacterial density measured by absorbance, and blue lines and squares represent ciliate densities (mean ± s.e.). Bacterial density is shown as optical density at 600 nm, and ciliate density as cells ml–1, both normalized to 0–1 range.
The densities of the predatory ciliates depended on total bacterial density but changed independent from P. fluorescens diversity (figure 2). The ciliate densities peaked initially but decreased thereafter over time resulting in low final densities (electronic supplementary material, table S2). The naive ciliate growing on ancestral P. fluorescens and E. coli had the lowest densities; however, unlike the other treatments, there was no decrease over time.
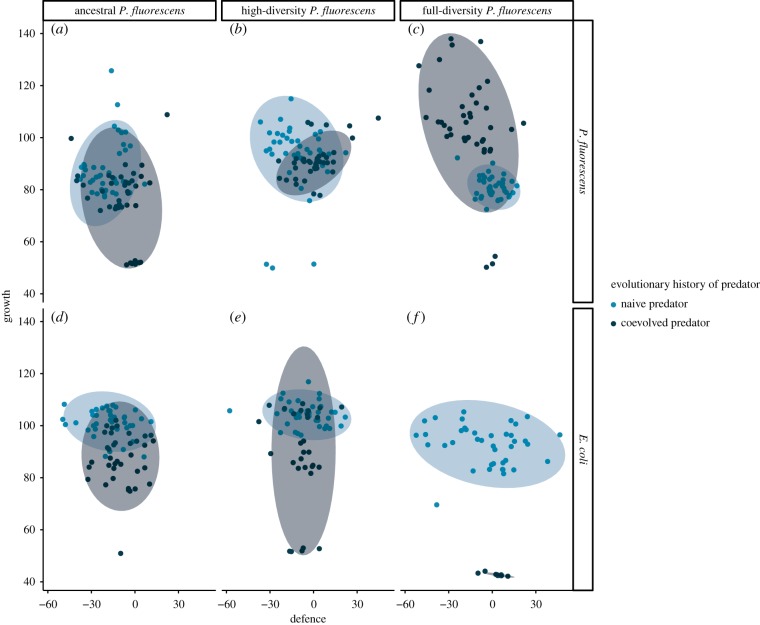
In the bacterial clones isolated from the endpoint of the experiment, there was an overall negative correlation between growth and anti-predatory defence level (Pearson r = −0.175, t = −3.7012, d.f. = 435, p = 0.0002) in line with a fitness trade-off between the two traits. Initial variability in the genetic diversity of P. fluorescens and the evolutionary state of the predator together drove the E. coli competitor (full-diversity P. fluorescens combined with coevolved predator), but not P. fluorescens itself, to diverge in trait space during the experiment (figure 3). In the treatment with the ancestral P. fluorescens strain, the presence of coevolved compared to naive predators caused increased selection for anti-predatory defence with a corresponding decrease in growth ability, while the opposite occurred in the full-diversity treatment (for gls model results for growth and defence, and multiple contrasts, see electronic supplementary material, tables S3 and S4). This is consistent with prior coevolution in the respective predator and prey populations. In turn, E. coli divergence in trait space was driven by increased defence and decreased growth with more diverse competitors or, similar to ancestral P. fluorescens, coevolved predators (for gls model results for growth and defence, and multiple contrasts, see electronic supplementary material, tables S5 and S6). Therefore, both predator coevolution and the genetic diversity of an otherwise inferior competitor resulted in decreased resource use evolution and an increased fitness advantage of anti-predatory defence.
Figure 3.
Divergence in trait space caused by the genetic diversity of P. fluorescens or predator evolutionary history (ellipses depict 95% confidence levels). Growth is the biomass yield in the absence of predation and defence is the effect of predation (0 = predation has no effect) on the biomass yield (both in optical density area units). (a–c) P. fluorescens and (d–f) E. coli clones isolated from the endpoint of a microcosm experiment.
As there was an effect of the different predation treatments, we also investigated whether the two initial coevolved ciliate populations had different grazing capacities on the evolved bacterial lines. In all conditions, we found ciliates had increased growth on E. coli, explaining why the inferior competitor P. fluorescens can coexist under predation. Ciliates coevolved with E. coli had higher growth rates compared with ciliates coevolved with P. fluorescens (ANOVA, F = 7.995, d.f. = 1,9, p = 0.0198; electronic supplementary material, figure S1 and table S7). In turn, the ciliates were able to grow marginally better on the evolved E. coli bacteria compared to the performance on P. fluorescens bacteria (ANOVA, F = 5.772, d.f. = 1,9, p = 0.0397).
4. Discussion
Predation mediated coexistence of competitors is a widely accepted phenomenon in the field of ecology. However, very little is known about how contemporary evolution and coevolution may alter the operation of this mechanism. Our data provide compelling evidence for the role of genetic diversity in species coexistence. While monoclonal P. fluorescens is rapidly outcompeted by E. coli, it will stably coexist if the P. fluorescens population is genetically diverse (figure 1). The ensuing reduction in the population size of the competitor might also alter its evolutionary dynamics, constraining resource use evolution and making anti-predatory defence critical for population survival. As a result, coexistence is promoted, and the genetically diverse population dominates the bacterial community. This is congruent with recent theory, which predicts coexistence of diverse communities under sufficiently high trait adaptation [7], and helps to explain why natural food webs contain many co-occurring species [43,44]. Interestingly, total biomass production seems to be independent from underlying population structure. While there is obvious change in the proportions of bacterial species (figure 1), total bacterial and ciliate production is not affected (figure 2). Taken together, these results indicate that the success of species in communities depends on genetic variation in the traits under selection, although overall production might remain unaltered, which is in line with previous findings [45]. Higher biomass production would be plausible, especially if both competitors only share a small resource pool. Pseudomonas fluorescens and E. coli should both at least slightly differ in resource use, and thus they are expected to introduce additional ecological functions when both are found together [25]. However, it is possible that these functions are redundant in the experimental conditions used where competitive interactions can be strong (rapid outcompetition of P. fluorescens by E. coli), indicating exploitation of a similar set of resources [21,46].
We found predation to be highly important as an ecological force affecting coexistence of the two bacteria. This result is in line with previous studies showing the effect of predation on the coexistence of species [21,37,43,44,47]. A naive predator equalizes species proportions [21], although this also depends on growth–defence trade-offs [19,45]. While E. coli seems to grow faster in our experiment, it also experiences higher loss due to predation (electronic supplementary material, figure S1), which might explain how coexistence is possible, as P. fluorescens seems better defended against predation loss. However, a coevolved predator population which was previously exposed to different bacterial species promotes coexistence, giving an advantage to P. fluorescens. This finding might be partly explained by the fact that E. coli is more affected by predation and more efficient coevolved predators might enhance this. When P. fluorescens diversity and a coevolved predator both come together, this seems to have the strongest effect. Our findings are in line with recent studies using the same experimental system, without the species competition aspect, which have shown that the role of predator–prey coevolution can be an important factor determining intraspecific prey diversity and eco-evolutionary feedback loops [19,48]. In addition, the relative importance of ecology and evolution for coexistence has been observed to depend on the community structure and the type of consumers [21].
In our present study, we manipulated the initial trait variation in the inferior competitor (P. fluorescens). By doing this we aimed to manipulate the strength of the eco-evolutionary feedback and the speed of the trait evolution by providing different amounts of initial genetic variability. We hypothesized that with more within-species trait variation, it is possible that evolution is facilitated by selection acting on standing genetic variation already present in the population [4,18] and population trait means can quickly shift as predicted by theory [3], ultimately even changing the competitive ranking between the species. Our findings support this idea, and when looking at the traits at the end of the experiment, we observed that not only the coexistence of our prey species but also the trait evolution of the competitor was affected by the P. fluorescens pre-adaptation treatment (figure 3d,e). Furthermore, the coevolutionary history of the predator also altered the final traits in E. coli, indicating that eco-evolutionary mechanisms altering the coexistence of competitors constitute a process functioning in different trophic levels.
Taken together, initial trait variability, ecological dynamics and further trait evolution are interconnected processes which need to be investigated together to fully understand the role of evolution in species coexistence. It is not mechanistically completely clear how traits at the end of the experiment and ecological dynamic are connected in our study since links are potentially very complex. We propose that understanding these feedbacks between ecological dynamics and potentially reciprocal trait changes between competitors is important for our understanding of the contribution of evolution to species coexistence as the focus has traditionally been mostly on ecological factors. Furthermore, we need to address the role of evolution in competitors as well as the role of coevolution in consumers. When inspecting the coexistence of microbes, a recent study proposes that assembly rules in microbial communities can be predicted from two- and three-way interactions for more diverse communities [24]. In such a model, ecological interaction is the driving force, which seems applicable as long as these interactions are stable and do not change. Here we find standing genetic variation to be important at both the prey and the predator level and show how it can contribute to completely shifting the coexistence ratio between competitors, in turn, altering further trait change. Our study proposes that evolutionary aspects cannot be neglected as they might affect interactions and therefore alter coexistence. If genetic variation drives evolution, the initially estimated interaction might rapidly change and depend on further interactions in a more complex community. Finally, we argue that further experimental studies are needed to understand eco-evolutionary community dynamics in more species-rich systems such as the one we present here. Findings from these relatively simple and ‘unnatural’ systems are still vital for providing mechanistic understanding on how ecological and evolutionary dynamics interact in more complex natural systems.
Supplementary Material
Acknowledgements
We thank Rashmi Shrestha and Roosa Jokela for help during the laboratory experiments. Additionally, Veera Partanen helped to maintain the LTPE lines. T.S. finalized this manuscript during his paternity leave supported by the ZBFS.
Data accessibility
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.4b866r7 [49].
Authors' contributions
T.H., L.B. and T.S. designed research; T.S. performed the experiments; J.C. managed maintenance of LTPE lines; T.S., J.C. and T.H. analysed data; T.S. and T.H. wrote the first draft of the manuscript; all authors contributed to the final version of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
The study was funded by the Academy of Finland to T.H. (project no. 106993).
References
- 1.Darwin C. 1859. On the origin of species, 1st edn New York, NY: John Murray. [Google Scholar]
- 2.Fisher RA. 1930. The genetical theory of natural selection. Oxford, UK: Clarendon Press. [Google Scholar]
- 3.Lande R, Arnold S. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226. ( 10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- 4.Barrett R, Schluter D. 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44. ( 10.1016/j.tree.2007.09.008) [DOI] [PubMed] [Google Scholar]
- 5.Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. ( 10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 6.Ehrlich E, Becks L, Gaedke U. 2017. Trait-fitness relationships determine how trade-off shapes affect species coexistence. Ecology 98, 3188–3198. ( 10.1002/ecy.2047) [DOI] [PubMed] [Google Scholar]
- 7.Klauschies T, Vasseur DA, Gaedke U. 2016. Trait adaptation promotes species coexistence in diverse predator and prey communities. Ecol. Evol. 6, 4141–4159. ( 10.1002/ece3.2172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart SP, Turcotte MM, Levine JM. 2019. Effects of rapid evolution on species coexistence. Proc. Natl Acad. Sci. USA 116, 2112–2117. ( 10.1073/pnas.1816298116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivett DW, Bell T. 2018. Abundance determines the functional role of bacterial phylotypes in complex communities. Nat. Microbiol. 3, 767–772. ( 10.1038/s41564-018-0180-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gause GF. 1934. Experimental analysis of Vito Volterra's mathematical theory of the struggle for existence. Science 79, 16–17. ( 10.1126/science.79.2036.16-a) [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson GE. 1961. The paradox of the plankton. Am. Nat. 95, 137–145. ( 10.1086/282171) [DOI] [Google Scholar]
- 12.Hart SP, Usinowicz J, Levine JM. 2017. The spatial scales of species coexistence. Nat. Ecol. Evol. 1, 1066–1073. ( 10.1038/s41559-017-0230-7) [DOI] [PubMed] [Google Scholar]
- 13.Fiegna F, Scheuerl T, Moreno-Letelier A, Bell T, Barraclough TG. 2015. Saturating effects of species diversity on life-history evolution in bacteria. Proc. R. Soc. B 282, 20151794 ( 10.1098/rspb.2015.1794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendall ML, et al. 2016. Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. 10, 1589–1601. ( 10.1038/ismej.2015.241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn MW, Scheuerl T, Jezberová J, Koll U, Jezbera J, Šimek K, Vannini C, Petroni G, Wu QL. 2012. The passive yet successful way of planktonic life: genomic and experimental analysis of the ecology of a free-living Polynucleobacter population. PLoS ONE 7, e32772 ( 10.1371/journal.pone.0032772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hairston NG Jr, Ellner SP, Geber MA, Yoshida T, Fox JA. 2005. Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127. ( 10.1111/j.1461-0248.2005.00812.x) [DOI] [Google Scholar]
- 17.Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG Jr. 2003. Rapid evolution drives ecological dynamics in a predator-prey system. Nature 424, 303–306. ( 10.1038/nature01767) [DOI] [PubMed] [Google Scholar]
- 18.Scheuerl T, Stelzer C-P. 2017. Sex initiates adaptive evolution by recombination between beneficial loci. PLoS ONE 12, e0177895 ( 10.1371/journal.pone.0177895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Traulsen A, Werner B, Hiltunen T, Becks L. 2017. Dynamical trade-offs arise from antagonistic coevolution and decrease intraspecific diversity. Nat. Commun. 8, 2059 ( 10.1038/s41467-017-01957-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chase JM, Abrams PA, Grover JP, Diehl S, Chesson P, Holt RD, Richards SA, Nisbet RM, Case TJ. 2002. The interaction between predation and competition: a review and synthesis. Ecol. Lett. 5, 302–315. ( 10.1046/j.1461-0248.2002.00315.x) [DOI] [Google Scholar]
- 21.Hiltunen T, Kaitala V, Laakso J, Becks L. 2017. Evolutionary contribution to coexistence of competitors in microbial food webs. Proc. R. Soc. B 284, 20170415 ( 10.1098/rspb.2017.0415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster KR, Bell T. 2012. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850. ( 10.1016/j.cub.2012.08.005) [DOI] [PubMed] [Google Scholar]
- 23.Rivett DW, Scheuerl T, Culbert CT, Mombrikotb SB, Johnstone E, Barraclough TG, Bell T. 2016. Resource-dependent attenuation of species interactions during bacterial succession. ISME J. 10, 2259–2268. ( 10.1038/ismej.2016.11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman J, Higgins LM, Gore J. 2017. Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 109 ( 10.1038/s41559-017-0109) [DOI] [PubMed] [Google Scholar]
- 25.Langenheder S, Bulling MT, Prosser JI, Solan M. 2012. Role of functionally dominant species in varying environmental regimes: evidence for the performance-enhancing effect of biodiversity. BMC Ecol. 12, 14 ( 10.1186/1472-6785-12-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, Barraclough TG. 2012. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 10, e1001330 ( 10.1371/journal.pbio.1001330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence D, Barraclough TG. 2016. Evolution of resource use along a gradient of stress leads to increased facilitation. Oikos 125, 1284–1295. ( 10.1111/oik.02989) [DOI] [Google Scholar]
- 28.Toju H, Yamamichi M, Guimarães PR, Olesen JM, Mougi A, Yoshida T, Thompson JN. 2017. Species-rich networks and eco-evolutionary synthesis at the metacommunity level. Nat. Ecol. Evol. 1, 24 ( 10.1038/s41559-016-0024) [DOI] [PubMed] [Google Scholar]
- 29.Jousset A, Eisenhauer N, Merker M, Mouquet N, Scheu S. 2016. High functional diversity stimulates diversification in experimental microbial communities. Sci. Adv. 2, e1600124 ( 10.1126/sciadv.1600124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldford JE, Lu N, Bajic D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segre D, Mehta P, Sanchez A.2017. Emergent simplicity in microbial community assembly. bioRxiv, 205831. (doi:10.1101/205831) [DOI] [PMC free article] [PubMed]
- 31.Brockhurst MA, Colegrave N, Hodgson DJ, Buckling A. 2007. Niche occupation limits adaptive radiation in experimental microcosms. PLoS ONE 2, e193 ( 10.1371/journal.pone.0000193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friman V-P, Buckling A. 2013. Effects of predation on real-time host-parasite coevolutionary dynamics. Ecol. Lett. 16, 39–46. ( 10.1111/ele.12010) [DOI] [PubMed] [Google Scholar]
- 33.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42°C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol. 31, 2647–2662. ( 10.1093/molbev/msu209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogbunugafor CB, Eppstein MJ. 2016. Competition along trajectories governs adaptation rates towards antimicrobial resistance. Nat. Ecol. Evol. 1, 7 ( 10.1038/s41559-016-0007) [DOI] [PubMed] [Google Scholar]
- 35.Yosef I, Edgar R, Levy A, Amitai G, Sorek R, Munitz A, Qimron U. 2016. Natural selection underlies apparent stress-induced mutagenesis in a bacteriophage infection model. Nat. Microbiol. 1, 16047 ( 10.1038/nmicrobiol.2016.47) [DOI] [PubMed] [Google Scholar]
- 36.Gibert JP, DeLong JP. 2015. Individual variation decreases interference competition but increases species persistence. In Advances in ecological research (eds Pawar S, Woodward G, Dell AI), pp. 45–64. New York, NY: Academic Press. [Google Scholar]
- 37.Gibert JP, Brassil CE. 2014. Individual phenotypic variation reduces interaction strengths in a consumer-resource system. Ecol. Evol. 4, 3703–3713. ( 10.1002/ece3.1212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cairns J, Jalasvuori M, Ojala V, Brockhurst M, Hiltunen T. 2016. Conjugation is necessary for a bacterial plasmid to survive under protozoan predation. Biol. Lett. 12, 20150953 ( 10.1098/rsbl.2015.0953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team. 2010. The R project for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.r-project.org/ (accessed on 25 July 2012). [Google Scholar]
- 40.Højsgaard S, Halekoh U, Yan J. 2016. geepack: generalized estimating equation package. See https://cran.r-project.org/web/packages/geepack/index.html.
- 41.Pinheiro J, Bates D, DebRoy S, Sarkar D. 2012. Linear and nonlinear mixed effects models CRAN—Package nlme. See http://cran.r-project.org/web/packages/nlme/index.html (accessed on 12 September 2012).
- 42.Lenth R, Singmann H, Love J, Buerkner P, Herve M.. 2018. emmeans: estimated marginal means, aka least-squares means. See https://CRAN.R-project.org/package=emmeans.
- 43.Gibert JP, DeLong JP. 2017. Phenotypic variation explains food web structural patterns. Proc. Natl Acad. Sci. USA 114, 11 187–11 192. ( 10.1073/pnas.1703864114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbour MA, Fortuna MA, Bascompte J, Nicholson JR, Julkunen-Tiitto R, Jules ES, Crutsinger GM. 2016. Genetic specificity of a plant-insect food web: Implications for linking genetic variation to network complexity. Proc. Natl Acad. Sci. USA 113, 2128–2133. ( 10.1073/pnas.1513633113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barabás G, D'Andrea R. 2016. The effect of intraspecific variation and heritability on community pattern and robustness. Ecol. Lett. 19, 977–986. ( 10.1111/ele.12636) [DOI] [PubMed] [Google Scholar]
- 46.Gamfeldt L, Roger F. 2017. Revisiting the biodiversity-ecosystem multifunctionality relationship. Nat. Ecol. Evol. 1, 168 ( 10.1038/s41559-017-0168) [DOI] [PubMed] [Google Scholar]
- 47.Scheuerl T, Stelzer C-P. 2019. Asexual reproduction changes predator population dynamics in a life predator–prey system. Popul. Ecol. 61, 210–216. ( 10.1002/1438-390X.1017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiltunen T, Becks L. 2014. Consumer co-evolution as an important component of the eco-evolutionary feedback. Nat. Commun. 5, 5226 ( 10.1038/ncomms6226) [DOI] [PubMed] [Google Scholar]
- 49.Scheuerl T, Cairns J, Becks L, Hiltunen T. 2019. Data from: Predator coevolution and prey trait variability determine species coexistence Dryad Digital Repository. ( 10.5061/dryad.4b866r7) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Scheuerl T, Cairns J, Becks L, Hiltunen T. 2019. Data from: Predator coevolution and prey trait variability determine species coexistence Dryad Digital Repository. ( 10.5061/dryad.4b866r7) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.4b866r7 [49].