Abstract
Background
PAX3 plays an important role in mammalian embryonic development. Known mutations in PAX3 are etiologically associated with Waardenburg syndrome and syndromic neural tube defects (NTDs). Mutations in the murine homologue, Pax3, are responsible for the phenotype of Splotch mice, in which nullizygotes are 100% penetrant for NTDs.
Methods
The study sample included 74 infants with spina bifida (cases) and 87 non-malformed infant controls. The conserved paired-box domain, as well as upstream genomic region of PAX3 was subjected to re- sequencing and those identified SNPs were evaluated as haplotypes. The associations of haplotypes for selected gene regions and the risks of spina bifida were further studied.
Results
Nineteen SNPs were observed; 15 observed in controls had been submitted to National Center for Biotechnology Information (NCBI) database with allele frequencies. The PAX3 gene variant T-1186C (rs16863657) and its related haplotype, T -C-T-C-C-G-C-C-C of 9 SNPs, were found to be associated with an increased risk of spina bifida, with an odds ratio of 3.5 (95% confidence interval, 1.2–10.0) among Hispanic whites.
Conclusions
Our analyses indicated that PAX3 SNPs were not strong risk factors for human spina bifida. However, additional follow-up of the PAX3 gene variant T-1186C (rs16863657) and its related haplotype, T-C-T-C-C-G-C-C-C may be important in other populations.
Keywords: PAX3, polymorphisms, spina bifida, haplotype, association study
Introduction
Neural tube defects (NTDs) have complex etiologies that include both genetic and environmental factors. Some environmental factors such as folic acid intake have been well described in terms of their importance to NTD risk (Berry et al., 1995). However, potential genetic contributions to risk have been more resistant to identification.
The Paired-boX (PAX) gene family encodes specific DNA binding transcription factors that play important roles in embryonic development. Of the 9 PAX genes, PAX 2, 3, 5, 6, 7 and 8 are expressed during the formation and organization of the neural tube along its entire axis (Hol et al. 1996; Helwig et al. 1995; Tremblay and Gruss 1994; Balling et al. 1988). PAX genes contain a paired-box DNA binding domain, with or without an octapeptide and homeo domain (Dahl et al., 1997). The paired box domain is a highly conserved DNA-binding domain of 128 amino acids located at the amino terminus of the protein. Analysis of mouse models and human syndromes has revealed the importance of PAX genes in their role as regulators of normal development (Mansouri et al., 1999; Pani et al., 2002). Heterozygous splotch mice display pigmentary abnormalities while homozygotes die during gestation with spina bifida or exencephaly (Vogan et al. 1993; Auerbach 1954). Mutations within the human PAX3 gene have been associated with Waardenburg syndrome, a condition occasionally associated with NTDs (Baldwin et al. 1992; Hoth et al. 1993; Hol et al. 1995). In 1995, Hol and colleagues identified a 5bp deletion in exon 5 of the PAX3 gene in a patient with spina bifida and mild manifestations of Waardenburg syndrome. A subsequent study identified 2 spina bifida patients who also had small interstitial chromosomal deletions involving PAX3 (Nye et al., 1998).
In the current study, we investigated a California population to determine whether polymorphisms in PAX3 were associated with risk of human spina bifida.
Methods
Study Subjects
The study design is a population-based case-control study. Cases and controls were ascertained by the California Birth Defects Monitoring Program, a population-based active surveillance system for collecting information on infants and fetuses with congenital malformations. Diagnostic and demographic information was collected by program staff from multiple sources of medical records for all liveborn and stillborn fetuses. Nearly all major structural anomalies diagnosed within 1 year of delivery were ascertained. Overall ascertainment for major malformations has been estimated as 97% complete (Schulman J, et al., 1993). Eligible for this study were live born infants only, owing to the fact that the source of DNA was from newborn screening bloodspot.
Included for study were 74 infants with spina bifida and 87 non-malformed control infants born during the period 1983–1986 in selected counties in California. Among cases, 47.3 % were non-Hispanic Whites, 41.9 % were Hispanic Whites including U.S. born and foreign born, 4.0 % were African American and 6.8 % were other ethnic groups. Among controls, 52.9 % were non-Hispanic Whites, 34.5 % were Hispanic Whites including U.S. born and foreign born, 4.6 % were African American and 8.0 % were other ethnic groups. All newborn blood samples were obtained with approval from the State of California Health and Welfare Agency Committee for the Protection of Human Subjects. Genomic DNA was extracted from dried newborn screening blood spot on filter paper using Puregene DNA Purification kit (Gentra, Minneapolis, MN) according to the manufacturer’s instructions and was amplified using PCR (Schwartz EI, et al, 1990).
PAX3 Gene Re-Sequencing
The genomic region of PAX3 gene, including the paired-box domain, was re-sequenced with VariantSEQr re-sequencing system (Applied Biosystems, Foster city, CA). The primer set (RSS000013589 _02) for PAX3 included nine pairs of primers and covered a 6-kb genomic region containing 5 exons and flanking intronic sequences. PCRs (30s at 94 ÿ, 45s at 60 ÿ, 60 s at 72 ÿ for 40 cycles) were performed in a final volume of 10 ÿl containing 60 ng genomic DNA, 2.0 ÿl VariantSEQr primer mix, 250 ÿM of each dNTP, in 2.0 mM MgCl2, 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 8% Glycerol, and 1.5 U of Taq DNA polymerase on PE9700 thermalcycler. Then, PCR products were cleaned up by digestion with ExoSAP-IT enzymes (USB Corporation, Cleveland, Ohio) and 3 ÿl of final products were applied to sequencing reaction (10 s at 96ÿ, 5 s at 50 ÿ, 4 m at 60 ÿ for 25 cycles) with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster city, CA). The fragments were precipitated with isopropanol, denatured with 12 ÿl HiDi formamide solution at 95 ÿ for 5 min, and loaded onto 3730 DNA Analyzer. Sequencing results were analyzed with SeqScape v2.5 software (Applied Biosystems, Foster City, CA).
(CA) n Repeats Genotyping
Primers (forward: 5′-CCCAGATGCCTTCTTA and reverse: 5’ FAM-CAGGGAG-ATGGCAGTT-3′) were used to amplify fragments. The fragment is located ~300 bp upstream of transcription start site and contains the (CA) n dinucleotide repeats. PCR (35 s at 94 ÿ, 30s at 55.7 ÿ, 60 s at 72 ÿ for 35 cycles) was performed in a final volume of 25 ÿl containing 60 ng of genomic DNA, 6.25 pmol of each primer, 200 ÿM of each dNTP, in 1.5 mM MgCl2, 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 1 ÿg / ml bovine serum albumin and 0.4 U Taq DNA polymerase. Data were collected by 3730 DNA Analyzer (ABI, Foster city, CA) and analyzed by GeneMapper v3.5 (ABI, Foster city, CA).
Statistical analyses
Deviations from Hardy-Weinberg Equilibrium among control infants were evaluated using a χ2 test. Genotyping frequencies for detected variants within the PAX3 gene were compared between cases and controls and odds ratios and 95% confidence intervals (CIs) were estimated utilizing SAS software (version 9.1). Haplotype analysis was performed with Haploview software (version 3.2) (Barrett JC, et al, 2005) and evaluated with χ2 tests.
Results
Nineteen SNPs were observed by re-sequencing in upstream genomic region and exons 1–4 of PAX3 gene containing the paired-box domain (table 1, figure 1). Two SNPs in coding regions did not result in an amino acid substitution. The flanking sequences and SNPs are shown in Table 2. Fifteen novel SNPs observed in controls had been submitted to National Center for Biotechnology Information (NCBI) database with allele frequencies. These were rs28945085, rs16863657, rs28945086, rs28945087, rs28945088, rs28945668, rs28945089, rs28945090, rs28945091, rs12623857, rs28945092, rs28945093, rs28945094, rs28945095, rs28945096,.
Table 1.
Primer Sequences for Resequencing PAX3 Gene
| Primer ID | Sequences (5′ → 3′) | Fragment length (bp) |
|---|---|---|
| RSA000021427 | Fw: TGTAAAACGACGOCCAGTGGTGCCAGCACICTAAGAACCCA | 595 |
| Re: CAGGAAACAGCIATGACCGGTGATCTGACGGCAGCCAA | ||
| RSA000021430 | Fw: TGTAAAACGACGGCCAGTAAGAAGTGTCCAGGGCGCGT | 571 |
| Re: CAGGAAACAGCIAIGACCGGTCIGGGTCIGGGAGTCCG | ||
| RSA000021432 | Fw: TGTAAAACGACGGCCAGTTATCCGGAGCGTGGAGAGCC | 578 |
| Re: CAGGAAACAGCTATGACCTTCTCCGCCCTCAGCAACTG | ||
| RSA000021433 | Fw: TGTAAAACGACGGCCAGTCGATTGGCCGACGGGTAGAC | 554 |
| Re: CAGGAAACAGCTATGACCGGGCACTGACCACAGGAAGG | ||
| RSA000579700 | Fw: TGTAAAACGACGGCCAGTAATGGCAACAGAGTGAGAGCTTCC | 587 |
| Re: CAGGAAACAGCTATGACCAGGAGACACCCGCGAGCAGT | ||
| RSA000579702 | Fw: TGTAAAACGACGGCCAGTTAAACGCTCTGCCTCCGCCT | 600 |
| Re: CAGGAAACAGCTATGACCGGGATGTGTTCTGGTCTGCCC | ||
| RSA001307840 | Fw: TGTAAAACGACGGCCAGTCAGTTGCTGAGGGCGGAGAA | 596 |
| Re: CAGGAAACAGCTATGACCAGAACCGCAGCTTGCCAGAG | ||
| RSA001307844 | Fw: TGTAAAACGACGGCCAGTAGAGCAGCGCGCTCCATTTG | 596 |
| Re: CAGGAAACAGCTATGACCGCTCGCCGTGGCTCTCTGA | ||
| RSA001307846 | Fw: AACGACGGCCAGTCTGGGAGGAGTCCAGGGTGC | 461 |
| Re: AAACAGCTATGACCAGGCGAGTTCGTGGCACTT |
The 5′ end was flanked with 18 bp of universal M13 sequence.
Figure 1.
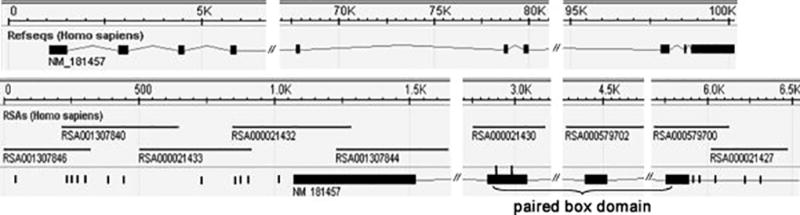
Table 2.
SNPs Observed in Genomic Region of PAX3 Gene Containing Paired-Box Domain
| Location | AA position |
Submitter ID* | Submitted SNP (NCBI) |
Reference SNP (NCBI) |
Relevant sequence |
|---|---|---|---|---|---|
| Genomic region | PAX3_C-1219G | ss38343224 | rs28945085 | GCTTGCCAGAG(C/G)GGTTTTTACA | |
| Genomic region | PAX3_T-1186C | ss38343225 | rs16863657 | GCCAGGTTTT(T/C)TYCTGCACCCTCC | |
| Genomic region | PAX3_T-1184C | ss38343226 | rs28945086 | GCCAGGTTTTYT(T/C)CTGCACCCTCC | |
| Genomic region | PAX3_C-1166T | ss38343227 | rs28945087 | CCCTCCCCCATA(C/T)CCCCAGCCT | |
| Genomic region | PAX3_C-1072G | ss38343228 | rs28945088 | CAAAAGAACAT(C/G)AGCGCACCCT | |
| Genomic region | PAX3_G-900A | N/A | N/A | GCCAAGAGAAATGAGAGC (G/A) AGACCTACAG | |
| Genomic region | PAX3_C-749T | ss38343229 | rs28945668 | GAGAAAGACACACA(C/T)ACACACACACACACA | |
| Genomic region | PAX3_C-693G | ss38343230 | rs10193524 rs28945089 | CACACACACACAGAGTGACA(C/G)AGACAGAGAGACAGAGACAGAGA | |
| Genomic region | PAX3_G-576A | ss38343231 | rs28945090 | CCTGGGCAAGGGGC(G/A)CAGCGCGGGT | |
| Genomic region | PAX3_G-569C | N/A | N/A | AGGGGCGCAGCGC(G/C)GGTCCCCCTCGGGGCCAGC | |
| Genomic region | PAX3_C-565A | ss38343232 | rs28945091 | CAGCGCGGGT(C/A)CCCCTCGGGGCCAGCA | |
| Genomic region | PAX3_G-396C | N/A | N/A | AAGAACTAATAAATGCTCCC(G/C)AGCCCGGATCCCCGCACTCG | |
| Exon 3 | 43 | PAX3_T129C | ss38343233 | rs12623857 | CGTCAACCAGCTCGGCGG(T/C)GTTTTTATCAACGGCAGG |
| Exon 3 | 52 | PAX3_C156G | ss38343234 | rs28945092 | GGCAGGCCGCTGCC(C/G)AACACATCCGCC |
| 3′-UTR | PAX3_A672C | ss38343235 | rs28945093 | ATCTGACGGCAGCCA(A/C)GCCCAGCTCGGATCAAGG | |
| 3′-UTR | PAX3_C701A | N/A | N/A | CTCGGATCAAGGTCCCTTCATG(C/A)GCGGTGTCTCTGCGCCTGA | |
| 3′-UTR | PAX3_T722C | ss38343236 | rs28945094 | GTGTCTCTGCGCCTGAG(T/C)AACGACATGTCC | |
| 3′-UTR | PAX3_C1020T | ss38343237 | rs28945095 | GGTCTCCGGAGTTT(C/T)CTCGCATTAAAGGAG | |
| 3′-UTR | PAX3_T1087C | ss38343238 | rs28945096 | CAAAATTGCCCCCACA(T/C)TGGCTGCCTTA |
Nucleotide +1 is the A of the ATG-translation initiation codon; the nucleotide 5′ to +1 is numbered −1.
N/A: not applicable.
Deviations from Hardy-Weinberg equilibrium were evaluated in controls for each SNP. Statistical evidence (P <.05) for deviations was observed for rs28945089 and rs28945094. Stratified by maternal race/ethnicity, deviations of those 2 SNPs remained significant in non-Hispanic Whites.
Analyses focused on (CA) n repeats, single SNPs, and haplotypes. We did not observe significant associations between (CA) n repeats polymorphism and risk of spina bifida (data not shown). For SNP analyses, Table 3 shows allele frequencies by case and control status and by maternal race/ethnic background. Overall, substantial differences between cases and controls in allele frequencies were not observed. One exception, however, was rs16863657. The major allele (T) for this SNP in Hispanic white case infants was substantially less frequent than in control infants.
Table 3.
Single SNP Associations
| Allele frequencies: case, control (X2, P value)
|
|||||
|---|---|---|---|---|---|
| SNP | Major allele | Hispanic Whites | Non-Hispanic Whites | African-American | Others |
| rs28945096 | T | 0.98, 0.98 (0.00, .98) | 1.00, 0.99 (0.78, .38) | 0 | 0 |
| rs28945095 | C | 1.00, 0.98 (1.01, .32) | 0 | 0 | 0 |
| rs28945094 | T | 1.00, 0.98 (0.98, .32) | 1.00, 0.95 (3.17, .07) | 0 | 0.90, 1.00 (1.05, .30) |
| PAX3_C701A | C | 0 | 0.99, 1.00 (1.24, .26) | 0 | 0 |
| rs28945093 | A | 0 | 0.99, 0.99 (0.02, .88) | 0 | 0 |
| rs28945092 | C | 0 | 0 | 1.00, 0.88 (0.81, 0.37) | 0 |
| rs12623857 | T | 0.97, 0.90 (2.07, .15) | 0.85, 0.91 (1.04, .31) | 0.67, 0.88 (0.88 , 0.35) | 1.00, 0.88 (1.32, .25) |
| PAX3_G-396C | G | 0 | 0 | 0.83, 1.00 (1.44, 0.23) | 0 |
| rs28945091 | C | 0.95, 0.98 (1.03, .31) | 0 | 0 | 0 |
| PAX3_G-569C | G | 0 | 0.99, 1.00 (1.07, .30) | 0 | 0 |
| rs28945090 | G | 1.00, 0.98 (1.01, .32) | 0 | 0 | 0 |
| rs28945089 | C | 0.59, 0.58 (2.67, .10) | 0 | 0 | 0.80, 0.50 (1.8, .18) |
| rs28945668 | C | 0 | 1.00, 0.99 (0.80, .37) | 0 | 0 |
| rs28945088 | C | 0 | 1.00, 0.99 (0.69, .41) | 0 | 0 |
| rs28945087 | C | 0 | 1.00, 0.99 (0.74, .39) | 0 | 0 |
| rs28945086 | T | 0 | 1.00, 0.99 (0.82, .37) | 0 | 0 |
| rs16863657 | T | 0.60, 0.82 (6.82, .01*) | 0.81, 0.82 (0.02, .90) | 0 | 0.80, 0.80 (0.39, .53) |
| rs28945085 | C | 0.98, 0.93 (2.11, .15) | 0.92, 0.94 (0.31, .58) | 0.83, 1.00 (1.44, 0.23) | 0 |
P < .01.
The risk associated with rs16863657 for spina bifida was further explored (table 4). The odds ratio associated with rs16863657 was 3.5 (95% CI: 1.2–10.0), indicating that SNP, rs16863657, was associated with a substantial increased risk of spina bifida in Hispanic Whites. Haplotype analyses were performed for various race/ethnic groups. Since all observed SNPs were in linkage disequilibrium, these SNPs were defined as whole block for these analyses. Results of these analyses are shown in Table 5. For African Americans, 5 haplotypes were investigated, including C-C-G-C, G-C-G-C, C-T-G-C, C -T-C-C, and C-T-G-G, which were represented by four SNPs, rs28945092 (C/G), rs12623857 (T/C), PAX3_G-396C, and rs28945085 (C/G). For Hispanic whites, 7 haplotypes were investigated, T-C-T-C-C-G -G-T-C, T-C-T-C-C-G- C-C-C, T-C-T-C-C-G-C -T-C, T-C-T-T-C-G-C- T-G, T-C-T-C-A-G-C-C -C, T-C-T-T-C-G-C-T- C, and C-C-T-C-C-G-C-T-C, corresponding to rs28945096 (T/C), rs28945095 (C/T), rs28945094 (T/C), rs12623857 (T/C), rs28945091 (C/A), rs28945090 (G/A), rs28945089 (C/G), rs16863657 (T/C) and rs28945085 (C/G). For non-Hispanic white, 5 haplotypes were investigated, T-T-C-A-C-G-C-C-C-T-T-C, T-T-C-A -C-G-C-C-C-T-C-C, T-T-C-A-T-G-C-C-C-T-T-G, T- T-C-A-T-G-C-C-C-T-T-C, and T-C-C-A-C-G-C-C-C-T-T-C, corresponding to rs28945096 (T/C), rs28945094 (T/C), PAX3_C701A, rs28945093 (A/C), rs12623857 (T/C), PAX3_G-569C, rs28945668 (C/T), rs28945088 (C/G), rs28945087 (C/T), rs28945086 (T/C), rs16863657 (T/C), and rs28945085 (C/G). For the other race/ethnic groups, 6 haplotypes were investigated, T-C-G-T, T-C-C-T, T-C-C-C, C-C-G-T, T-T-G-C, and T-T-C-C, corresponding to rs28945094 (T/C), rs12623857 (T/C), rs28945089 (C/G), and rs28945085 (T/C). For these various comparisons, only 1 haplotype appeared to be more frequent among infants with spina bifida than control infants. That is, the haplotype frequency of T-C-T-C-C-G-C-C- C in Hispanic Whites was 0.35 and 0.15 in cases and controls respectively, including the SNP, rs16863657. Because of the numbers of analytic comparisons made in this study, we cannot exclude the possibility that this latter result is also consistent with expected random variation.
Table 4.
PAX3_T-1186C (rs28945085) Genotype in Hispanic White Population
| Control (%) | Case (%) | OR | 95% CI | |
|---|---|---|---|---|
| Overall | ||||
| TT | 19 (63.3) | 10 (33.3) | Reference | |
| CC, TC | 11 (36.7) | 20 (66.7) | 3.5 | 1.2–10.0 |
Table 5.
Haplotype Association
| Haplotype | Frequency | Case, control frequencies | X2 | P |
|---|---|---|---|---|
| Hispanic Whites: rs28945096 (T/C), rs28945095 (C/T), rs28945094 (T/C), rs12623857 (T/C), rs28945091 (C/A), rs28945090 (G/A), rs28945089 (C/G), rs16863657 (T/C), and rs28945085 (C/G) | ||||
| TCTCCGGTC | 0.52 | 0.46, 0.58 | 1.78 | 0.18 |
| TCTCCGCCC | 0.25 | 0.35, 0.15 | 6.37 | 0.01* |
| TCTCCGCTC | 0.09 | 0.10, 0.10 | 0.00 | 0.98 |
| TCTTCGCTG | 0.03 | 0.02, 0.05 | 1.03 | 0.31 |
| TCTCAGCCC | 0.03 | 0.05, 0.02 | 1.03 | 0.31 |
| TCTTCGCTC | 0.03 | 0.02, 0.05 | 0.86 | 0.35 |
| CCTCCGCTC | 0.02 | 0.02, 0.02 | 0.00 | 1.00 |
| Non-Hispanic whites: rs28945096 (T/C), rs28945094 (T/C), PAX3_C701A, rs28945093 (A/C), rs12623857 (T/C), PAX3_G-569C, rs28945668 (C/T), rs28945088 (C/G), rs28945087 (C/T), rs28945086 (T/C), rs16863657 (T/C), and rs28945085 (C/G) | ||||
| TTCACGCCCTTC | 0.65 | 0.64, 0.66 | 0.10 | 0.76 |
| TTCACGCCCTCC | 0.17 | 0.20, 0.16 | 0.23 | 0.63 |
| TTCATGCCCTTG | 0.06 | 0.06, 0.06 | 0.00 | 0.95 |
| TTCATGCCCTTC | 0.04 | 0.06, 0.20 | 2.16 | 0.14 |
| TCCACGCCCTTC | 0.02 | 0.00, 0.04 | 2.48 | 0.12 |
| African Americans: rs28945092 (C/G), rs12623857 (T/C), PAX3_G-396C, and rs28945035 (C/G) | ||||
| CCGC | 0.71 | 0.67, 0.75 | 0.12 | 0.73 |
| GCGC | 0.07 | 0.00, 0.13 | 0.81 | 0.37 |
| CTGC | 0.07 | 0.00, 0.13 | 0.81 | 0.37 |
| CTCC | 0.07 | 0.17, 0.00 | 1.44 | 0.23 |
| CTGG | 0.07 | 0.17, 0.00 | 1.44 | 0.23 |
| Others: rs28945094 (T/C), rs12623857 (T/C), rs28945089 (C/G), and rs16863657 (T/C) | ||||
| TCGT | 0.59 | 0.70, 0.47 | 1.06 | 0.30 |
| TCCT | 0.21 | 0.10, 0.33 | 1.53 | 0.22 |
| TCCC | 0.10 | 0.10, 0.10 | 0.00 | 1.00 |
| CCGT | 0.05 | 0.10, 0.00 | 1.05 | 0.30 |
| TTGC | 0.03 | 0.00, 0.05 | 0.51 | 0.47 |
| TTCC | 0.03 | 0.00, 0.05 | 0.51 | 0.47 |
P < .05.
Discussion
PAX3 belongs to the family of paired domain proteins that bind DNA and regulate gene expression. Previous studies in mouse and chick embryos demonstrate that PAX3 play an important role in neural tube development. Expression of PAX3 is detected in gestation day (GD) 8.5 embryos, with peaks of expression from day 9 to 12 followed by declining levels until no expression is evident in GD 17 embryos. The time window of PAX3 high expression is coincident with the critical time for neural tube closure. Homozygotes (splotch mice) have 100% penetrance of neural tube defects. Our study focused on conserved, paired-box domain of PAX3 gene. Nineteen SNPs were found and 16 novel ones were identified and submitted to the NCBI database. Haplotypes including these SNPs were investigated in a study population of California infants. Overall, our analyses indicated that PAX3 SNPs were not strong risk factors for human spina bifida. Despite the fact that our study is strengthened by its population-based design, our lack of observing a modest role for PAX3 SNPs and spina bifida risk may be explicable to limited sample sizes and the exclusion of spina bifida-affected pregnancies that were terminated. Nevertheless, additional follow-up of PAX3 gene variant T-1186C (rs16863657) and its related haplotype, T-C-T-C-C-G-C-C-C may be important in other populations.
Acknowledgments
The authors are indebted to Dr. George Cunningham, Dr. Fred Lorey, and Terry Kennedy for making it possible to access newborn blood specimens. We also appreciate the technical support of Ms. Consuelo Valdes, Ms. Dia R. Gentile, Ms. Sarah Seth, and Mr. James Ebot Enaw.
This work was partially supported, by funds from the Centers of Disease Control and Prevention, Centers of Excellence Award No. U50/CCU913241 and by the NIH/NINDS R01 NS050249.
References
- Auerbach R. Analysis of the developmental effects of a lethal mutation in the house mouse. Exp Zool. 1954;127:305–329. [Google Scholar]
- Baldwin CT, Hoth CF, Amos JA, da-Silva EO, Milunsky A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg’s syndrome. Nature. 1992;355:637–638. doi: 10.1038/355637a0. [DOI] [PubMed] [Google Scholar]
- Baldwin CT, Hoth CF, Amos JA, da-Silva EO, Milunsky A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg’s syndrome. Nature. 1992;355(6361):637–8. doi: 10.1038/355637a0. 13. [DOI] [PubMed] [Google Scholar]
- Balling R, Deutsch U, Gruss P. Undulated, a mutation affecting the development of the mouse skeleton, has a point mutation in the paired box of Pax-1. Cell. 1988;55:531–535. doi: 10.1016/0092-8674(88)90039-6. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berry RJ, Li Z, Erickson JD, Li S, Moore CA, Wang H, Mulinare J, Zhao P, Wong LY, Gindler J, Hong SX, Correa A. Prevention of neural-tube defects with folic acid in China. China-U.S. Collaborative Project for Neural Tube Defect Prevention. N Engl J Med. 1999 Nov 11;341(20):1485–90. doi: 10.1056/NEJM199911113412001. [DOI] [PubMed] [Google Scholar]
- Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997 Sep;19(9):755–65. doi: 10.1002/bies.950190905. 1997. [DOI] [PubMed] [Google Scholar]
- Helwig U, Imai K, Schmahl W, Thomas BE, Varnum DS, Nadeau JH, Balling R. Interactions between undulated and Patch leads to an extreme form of spina bifida in double-mutant mice. Nature Genet. 1995;11:60–63. doi: 10.1038/ng0995-60. [DOI] [PubMed] [Google Scholar]
- Hol FA, Geurds MPA, Chatkupt S, Shugart YY, Balling R, Schrander-Stumpel CTRM, Johnson WG, Hamel BCJ, Mariman ECM. PAX genes and human neural tube defects: an amino acid substitutioin in PAX1 in a patient with spina bifida. J Med Genet. 1996;33:655–660. doi: 10.1136/jmg.33.8.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol FA, Hamel BC, Geurds MP, Mullaart RA, Barr FG, Macina RA, Mariman EC. A frameshift mutation in the gene for PAX3 in a girl with spina bifida and mild signs of Waardenburg syndrome. J Med Genet. 1995;32(1):52–6. doi: 10.1136/jmg.32.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth CF, Milunski A, Lipsky N, Sheffer R, Clarren SK, Baldwin CT. Mutations in the paired domain of the human PAX3 gene cause Klein-Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I) Am J Hum Genet. 1993;52:455–462. [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Goudreau G, Gruss P. Pax genes and their role in organogenesis. Cancer Research. 1999;59:1707s–1710s. [PubMed] [Google Scholar]
- Nye JS, Balkin N, Lucas H, Knepper PA, McLone DG, Charro wJ. Myelomeningocele and Waardenburg syndrome (Type 3) in patients with interstitial deletions of 2q35 and the PAX3 gene: possible digenic inheritance of a neural tube defect. Am J Med Genet. 1998;75:401–408. [PubMed] [Google Scholar]
- Pani L, Horal M, Loeken MR. Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function: implications for Pax-3- dependent development and tumorigenesis. Genes Dev. 2002;16(6):676–80. doi: 10.1101/gad.969302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman J, Edmonds LD, McClearn AB, Jensvold N, Shaw GM. Surveillance for and comparison of birth defect prevalences in two geographic areas--United States, 1983–88. MMWR CDC Surveill Summ. 1993;42:1–7. (1993) [PubMed] [Google Scholar]
- Schwartz EI, Khalchitsky SE, Eisensmith RC, Woo SL. Polymerase chain reaction amplification from dried blood spots on Guthrie cards. Lancet. 1990;336(8715):639–40. doi: 10.1016/0140-6736(90)93446-v. [DOI] [PubMed] [Google Scholar]
- Tremblay P, Gruss P. Pax: genes for mice and men. Pharmacol Ther. 1994;61(1–2):205–226. doi: 10.1016/0163-7258(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Vogan KJ, Epstein DJ, Trasler DG, Gros P. The Splotch-Delayed (Spd) mouse mutant carries a point mutation within the paired box of the Pax-3 gene. Genomics. 1993;17:364–369. doi: 10.1006/geno.1993.1333. [DOI] [PubMed] [Google Scholar]


