Abstract
Execution of the meiotic and mitotic cell division programs requires distinct gene expression patterns. Unlike mitotic cells, meiotic cells reduce ploidy by following one round of DNA replication with two rounds of chromosome segregation (Meiosis I and Meiosis II). However, the mechanisms by which cells prevent DNA replication between Meiosis I and Meiosis II are not fully understood. Here, we show that transcriptional repression of two essential DNA replication genes, CDC6 and SLD2, is associated with production of shorter meiosis-specific RNAs containing the 3’ end of both genes. Despite the short CDC6 RNA coding for a short protein (Cdc6short), this protein is not essential for meiosis and it does not have either a positive or negative impact on DNA replication. Production of CDC6short mRNA does not require the upstream CDC6 promoter (PCDC6) and is not a processed form of the full-length RNA. Instead, CDC6short depends on transcription initiation from within the ORF upon repression of P CDC6. Finally, using CDC6 genes from related yeast, we show that repression of full-length CDC6 mRNA is evolutionarily conserved and that this repression is consistently associated with production of unique short CDC6 RNAs. Together, these data demonstrate that meiotic cells transcriptionally repress full-length CDC6 and SLD2, and that inactivation of PCDC6 results in heterogeneous transcription initiation from within the CDC6 ORF.
Keywords: DNA replication, Meiosis, Cell Cycle, Cdc6, Sld2, Mcm2–7
Introduction
Sexual reproduction requires the generation of haploid gametes from diploid cells. To produce these gametes, cells undergo meiosis, a specialized cell division program that is designed to reduce ploidy by half (Ohkura 2015). During meiosis, a single round of DNA replication (S phase) is followed by two rounds of chromosome segregation (the meiotic divisions), Meiosis I (MI) and Meiosis II (MII). In contrast, ploidy is maintained during mitotic cell divisions which alternate between single rounds of DNA replication and chromosome segregation. A key requirement of the meiotic program is that DNA replication must be inhibited between the meiotic divisions. This inhibition targets multiple proteins acting at distinct steps during DNA replication initiation (Holt et al. 2007; Phizicky et al. 2018), but the mechanisms of inhibition are not fully understood.
Mitotic cells use oscillations of cyclin-dependent kinase (CDK) activity to temporally separate two essential steps of DNA replication, origin licensing and origin firing (Bell and Labib 2016). The low-CDK activity during G1 is permissive for origin licensing, the process of loading the Mcm2–7 helicase onto origins of replication in an inactive state. Three additional proteins catalyze Mcm2–7 loading; Cdc6, Cdt1, and the Origin Recognition Complex (ORC) (Remus et al. 2009; Evrin et al. 2009). At the G1-S transition, CDK bound to S-phase cyclins (S-CDK) oppositely regulates origin firing and origin licensing. First, S-CDK triggers origin firing by phosphorylating two essential proteins, Sld2 and Sld3. These modifications are required for helicase activation and replisome assembly, the molecular events that mediate origin firing (Masumoto et al. 2002; Tanaka et al. 2007; Zegerman and Diffley 2007). Second, S-CDK and CDK bound to M-phase cyclins (M-CDK) prevent origin licensing during S, G2, and M phases. To prevent this step of DNA replication, these kinases phosphorylate and inhibit multiple helicase-loading proteins including Cdc6 which, when phosphorylated, is targeted for proteolytic degradation (Drury et al. 1997; Calzada et al. 2000; Drury et al. 2000; Perkins et al. 2001; Arias and Walter 2007). By using CDK activity to separate origin licensing and origin firing, cells ensure that DNA replication happens exactly once per cell cycle.
During meiosis, the link between DNA replication and oscillations of CDK-activity presents a unique problem. CDK activity has been shown to decrease upon exit from MI and then increase upon entry into MII (Choi et al. 1991; Iwabuchi et al. 2000; Carlile and Amon 2008), a time when DNA replication must remain inhibited. This oscillation of CDK activity is required for MII chromosome segregation (Buonomo et al. 2003; Marston et al. 2003; Fox et al. 2017). However, an oscillation of CDK activity is also sufficient for replication of the entire genome in mitotic cells (Broek et al. 1991; Dahmann et al. 1995). Thus, there must be meiosis-specific mechanisms that prevent DNA replication between MI and MII while CDK-activity oscillates.
We recently demonstrated that meiotic cells inhibit both origin licensing and origin firing during the meiotic divisions, and that two critical targets of inhibition are Cdc6 and Sld2 (Phizicky et al. 2018). Both proteins are targeted for proteolytic degradation during meiosis, and CDC6 is also transcriptionally repressed. This co-inhibition of both proteins during meiosis is in contrast to the situation in mitotic cells wherein Cdc6 and Sld2 are inactivated during different stages of the cell cycle (Arias and Walter 2007; Bell and Labib 2016), and underscores the importance of preventing DNA replication during the meiotic divisions.
Consistent with the distinct requirements of meiosis and mitosis, meiotic cells activate genes required for meiosis and repress genes that promote mitotic cell division. In S. cerevisiae, genome-wide studies have revealed that not only do meiotic and mitotic cells transcribe and translate different genes, but also that many genes expressed in both cell types make different RNA and protein isoforms (Chu et al. 1998; Lardenois et al. 2011; Brar et al. 2012; Zhou et al. 2017; Hollerer et al. 2017; Cheng et al. 2018). In particular, thousands of meiosis-specific short-RNAs have been identified that are produced by a combination of antisense transcription of mitotically-expressed open-reading frames (ORFs), intergenic transcription, and truncated RNA isoforms containing only part of an annotated ORF (Lardenois et al. 2011; Zhou et al. 2017; Hollerer et al. 2017). In addition to their specificity for meiosis, these short RNAs constitute >10% of the translational capacity of meiotic cells (Hollerer et al. 2017), and protein products have been observed in a limited number of cases (Zhou et al. 2017; Hollerer et al. 2017). However, the molecular mechanisms resulting in meiosis-specific expression of these short RNAs, as well as the function of their protein products, have not been elucidated.
Here, we show that repression of CDC6 and SLD2 mRNAs during meiosis is associated with production of heterogeneous short RNAs corresponding to the 3’ end of both genes. Although the short CDC6 RNA codes for a short protein (Cdc6short), this protein is not essential for meiosis nor does it impact origin licensing. We found that production of Cdc6short is dependent on one or more promoter(s) inside the ORF that are activated coincident with repression of PCDC6. Examination of CDC6 genes from related yeast species suggests that repression of full-length CDC6 mRNA and production of unique short CDC6 RNAs is an evolutionarily conserved feature of these genes. Together, these data show that full-length CDC6 and SLD2 are transcriptionally repressed during meiosis, and that the transcriptional inactivation of full-length CDC6 is associated with new sites of transcription initiation within the CDC6 ORF.
Results
Repression of CDC6 and SLD2 during meiosis coincides with expression of shortened isoforms.
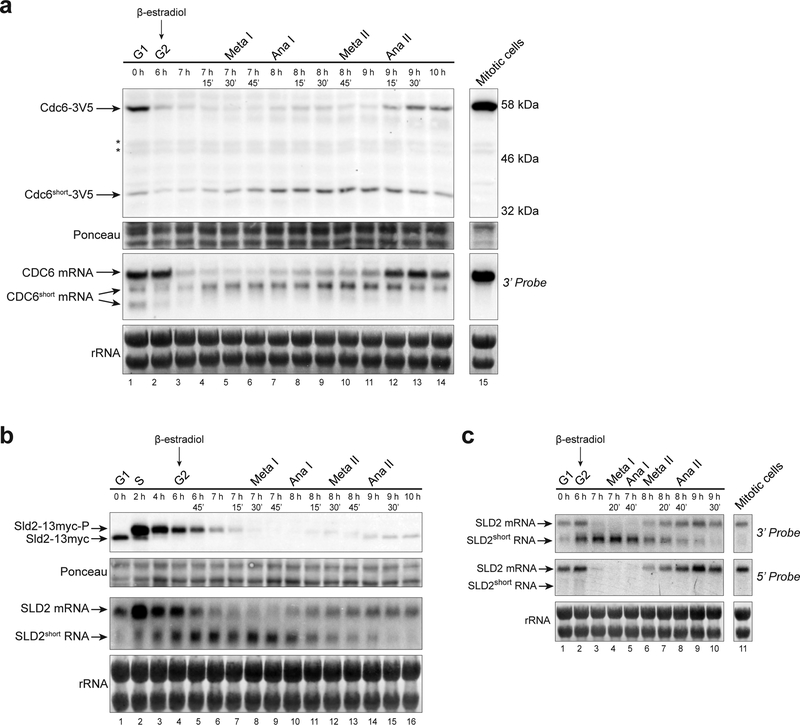
To understand how meiotic cells repress expression of Cdc6 and Sld2 proteins during the meiotic divisions, we analyzed the protein and RNA expression of both genes using a previously described meiotic-synchronization system (Benjamin et al. 2003; Carlile and Amon 2008). In this system, meiotic cells undergo G1 and S phases before accumulating at a G2- arrest. Release from this arrest allows cells to progress synchronously through both meiotic divisions (Fig. S1). While examining Cdc6 protein levels, we identified a short isoform of Cdc6 (Cdc6short) that is 20 kDa smaller than the full-length protein and was expressed throughout the meiotic divisions (Fig. 1a, top). Cdc6short protein expression coincided with expression of a shorter CDC6 mRNA isoform (Fig. 1a, bottom). Both the short RNA and short protein isoforms were anti-correlated with the corresponding full-length products. Cdc6short protein was detected via a C-terminal tag indicating that Cdc6short included the C-terminus of the ORF. Consistent with this conclusion, we detected the CDC6short mRNA using a 3’-end specific probe by northern blot (Fig. 1a). Neither the CDC6short mRNA nor protein were detectable in asynchronous mitotic cells (Fig. 1a). Thus, a meiosis-specific mechanism produces CDC6short.
Fig. 1. Short isoforms of CDC6 and SLD2 are specifically expressed during meiosis.
(a) - (c) The time after transfer into sporulation medium and the peak of each cell-cycle stage are denoted above each lane. β-estradiol was added to the culture medium after taking samples for the 6-hour time point to release cells from the G2-arrest. For quantification of cell-cycle stages, see Fig. S1a, S1b, and S1c, respectively.
(a) A short protein and RNA isoform of CDC6 is produced during meiosis but not in asynchronous mitotic cells (yDP71). Top: Cdc6–3V5 immunoblots from meiotic and mitotic cells. Asterisks (*) denote bands observed in untagged control samples on the same blot. Bottom: CDC6 RNA species were detected by northern blot using a 3’-end specific probe.
(b) A short RNA isoform of SLD2 is produced during meiosis (yDP336). Top: Sld2–13myc immunoblot during meiosis. Bottom: SLD2 RNA species were detected by northern blot with a probe to the whole ORF.
(c) SLD2short RNA is specific to meiotic cells and is enriched for the 3’-end of the gene (yDP336). Top: SLD2 northern blot with a 3’-end specific probe detects both the full-length SLD2 mRNA and SLD2short RNA. SLD2short RNA is not strongly detected in asynchronous mitotic cells.
Bottom: SLD2 northern blot with a 5’-end specific probe detects full-length SLD2 mRNA but not SLD2short RNA.
Similar to CDC6, we found that full-length SLD2 mRNA was reduced during the meiotic divisions, and that this decrease correlated with expression of a short SLD2 RNA species (Fig. 1b). We did not, however, detect a short Sld2 protein isoform (Fig. S1b). To determine which end of the full-length RNA was included in SLD2short, we performed northern blots using probes specific for the 5’- and 3’-end of the gene. Although both probes detected full-length SLD2 mRNA, only the 3’-probe detected the SLD2short RNA (Fig. 1c). Like CDC6short, SLD2short RNA was not detectable in mitotically-dividing cells (Fig. 1c). Together, these data show that both CDC6 and SLD2 produce a shortened RNA isoform during meiosis while the full-length mRNA of both genes is repressed.
Cdc6short is produced using an internal start codon and contains the C-terminal portion of CDC6
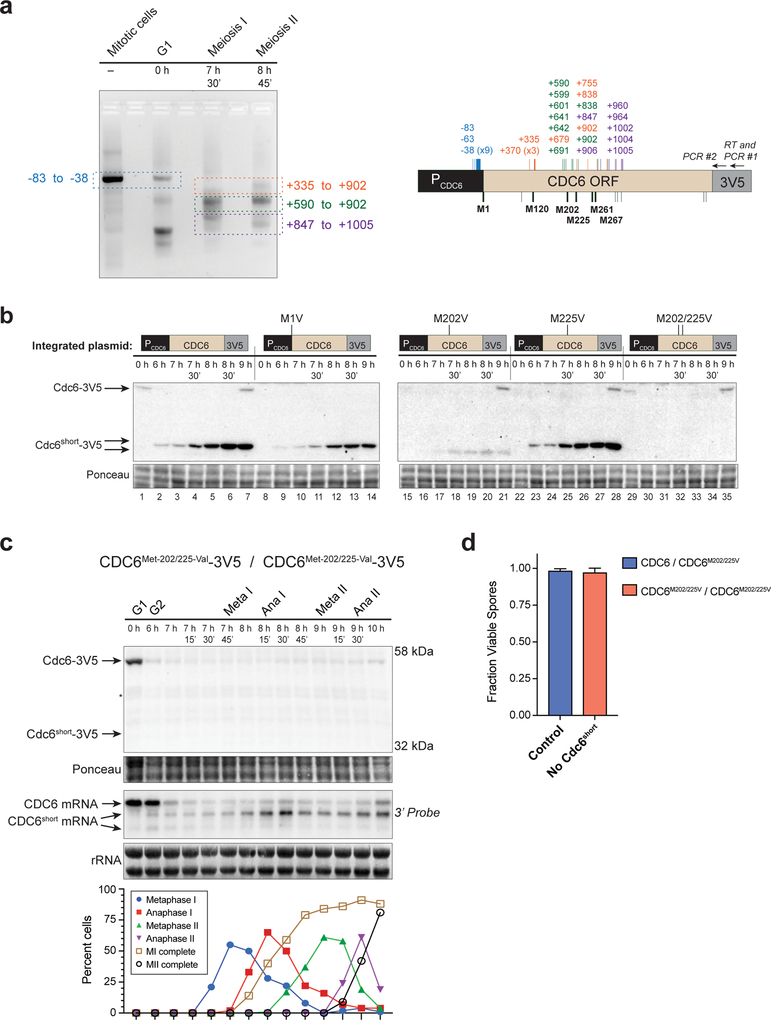
We focused our studies on CDC6short because the production of a short protein product in addition to the short mRNA suggested a potential function. To elucidate any potential functions of Cdc6short, we wanted to identify which portion of Cdc6 was expressed in this isoform. To determine the expressed region, we performed 5’ RACE on CDC6 mRNAs from different cell-cycle stages (Fig. 2a). Full-length CDC6 was detectable in asynchronous mitotic and pre-meiotic G1 cells, and the 5’ end of these products mapped primarily to the previously- defined start site of transcription (nucleotide −38, Fig. 2a) (Pelechano et al. 2013). cDNA from mitotic cells was present almost entirely as the full-length band, consistent with our northern blots (Fig. 1a and 2a). In contrast, during both meiosis I and II, multiple shorter versions of CDC6 cDNA were identified but none of the full-length product (Fig. 2a). The 5’ ends were located throughout the ORF, ranging from nucleotides +335 to +1,005, and none of the clones contained spice-site junctions. The robust detection of full-length CDC6 cDNA from asynchronous mitotic cells shows that these shorter products are not due to RNA secondary structures blocking reverse transcription. SLD2 cDNA showed similar 5’-end heterogeneity during meiosis and a lack of splice-site junctions (Fig. S2a). Along with our northern blots, these data demonstrate that the CDC6short and SLD2short RNAs have many different sub-species, but that all of these forms are missing part of the 5’ end.
Fig. 2. Cdc6short is produced from a 5’-end truncated mRNA via the internal start codon, Met202.
(a) 5’ RACE of CDC6 shows heterogeneous 5’ ends during the meiotic divisions. Left: Agarose gel of PCR products. Portions that were gel purified for ligation into a cloning vector are denoted by colored boxes, along with the range of 5’ ends detected for each species. Right: Diagram of CDC6 promoter, ORF, and 3V5 tag with the relative location of 3V5-specific primers for the RT reaction and PCR reactions. Colored vertical lines and numbers indicate the 5’ ends of individually sequenced clones from specific gel-purified sections. Potential translation start codons of Cdc6short are indicated on the bottom of the CDC6 ORF. Those that were mutated are denoted by thick black lines and labeled by amino acid number, and other in-frame ATGs are depicted as thin black lines.
(b) The primary start codon of Cdc6short protein is Met202. Plasmids containing PCDC6-Cdc6–3V5 with either WT CDC6 or Met→Val start codon mutations were integrated into yeast, and the subsequent strains were assayed for the ability to produce Cdc6short during meiosis. WT CDC6 (yDP198, lanes 1–7); CDC6Met−1-Val (yDP213, lanes 8–14); CDC6Met−202-Val (yDP231, lanes 15–21); CDC6Met−225-Val (yDP232, lanes 22–28); CDC6Met−202/225-Val (yDP227, lanes 29–35). The time after transfer into sporulation medium is indicated above each lane. Protein was detected by V5- immunoblot.
(c) Endogenous CDC6Met−202/225-Val (yDP259) prevents expression of Cdc6short protein but not CDC6short mRNA. Top: Immunoblot of Cdc6Met−202/225-Val-3V5 during meiosis. Asterisk (*) denotes a band observed in untagged control samples on the same blot. Middle: CDC6 RNA species were detected by northern blot using a 3’-end specific probe. The time after transfer into sporulation medium and the peak of each cell-cycle stage are denoted above each lane. Bottom: Quantification of cell-cycle stages.
(d) Spore viability of tetrads resulting from meiosis that occurred with or without Cdc6short protein. Control tetrads were from diploids that were heterozygous null for Cdc6short (CDC6–3V5 × CDC6Met−202/225-Val-3V5; yDP33 × yDP245), and compared to diploids that were homozygous null for Cdc6short (CDC6Met−202/225-Val-3V5 × CDC6Met−202/225-Val-3V5; yDP246 × yDP245). The graph shows the mean and standard deviation from three independent experiments.
Due to the large number of 5’ ends detected for CDC6short, it was unclear which AUG was used as the translation start codon for the Cdc6short protein. To address this question, we integrated a second copy of CDC6 at the LEU2 locus with mutations in potential translation start codons (ATG (M) → GTG (V)). Unlike the full-length protein which relied entirely on Met1, Cdc6short depended mostly on Met202, but also used Met225 at low efficiency (Fig. 2b). None of the other ATG codons tested prevented either full-length Cdc6 or Cdc6short protein expression (Fig. 2b, S2b). In combination with the 5’ RACE results, these data lead us to conclude that Cdc6short contains the Cdc6 sequence from Met202 to the C-terminus.
To determine whether Cdc6short is essential for meiosis, we replaced the endogenous copy of CDC6 with the Met-202/225-Val mutant. This mutant prevented expression of Cdc6short protein but not CDC6short mRNA (Fig. 2c). Eliminating Cdc6short protein did not prevent completion of meiosis I or meiosis II (Fig. 2c) nor did it cause decreased viability of the resulting spores (Fig. 2d). From these data, we conclude that Cdc6short protein is not essential for meiosis or spore formation.
Cdc6short cannot participate in or inhibit origin licensing
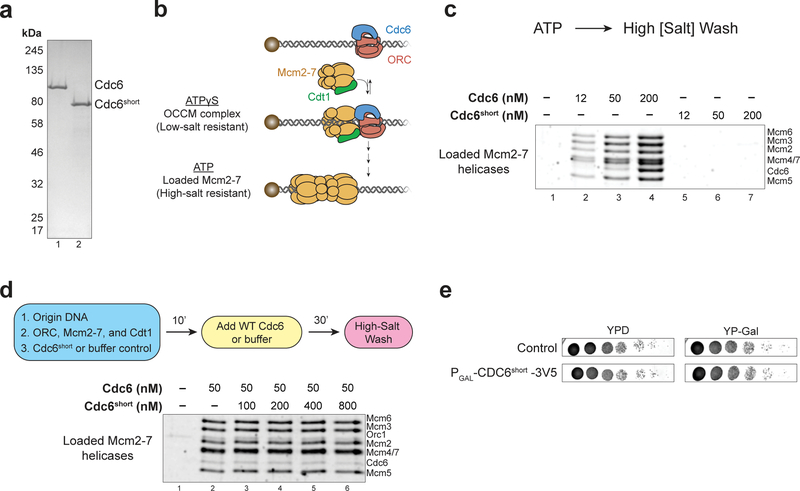
We hypothesized that Cdc6short could act as a dominant negative protein isoform to inhibit origin licensing during the meiotic divisions. To determine whether Cdc6short impacted origin licensing, we purified Cdc6 and Cdc6short (Fig. 3a) and tested their function using a reconstituted Mcm2–7 loading assay (Remus et al. 2009; Evrin et al. 2009). In this assay, origin-containing DNA attached to a magnetic bead was incubated with the four proteins required for Mcm2–7 loading: Cdc6, Cdt1, ORC, and Mcm2–7. After this incubation, the DNA-beads were washed and the proteins that remained associated with DNA were detected (diagram in Fig. 3b). Although wild-type (WT) Cdc6 efficiently loaded Mcm2–7 complexes onto origin DNA, Cdc6short did not facilitate this event (Fig. 3c). To test whether Cdc6short acted as a dominant-negative, we pre-incubated Cdc6short with ORC, Mcm2–7, Cdt1, and origin-containing DNA before adding WT Cdc6. Even when present in 16-fold excess compared to the WT Cdc6, Cdc6short did not inhibit Mcm2–7 loading (Fig. 3d). To determine whether Cdc6short could form initial physical interactions with helicase-loading proteins, we repeated the same experiment in the presence of ATPγS, a slowly-hydrolyzable ATP-analog (diagram in Fig. 3b). ATPγS stalls Mcm2–7 loading at an intermediate stage of assembly that contains one copy each of ORC, Cdt1, Cdc6, and Mcm2–7 bound to the origin DNA (the OCCM complex) (Randell et al. 2006; Sun et al. 2013; Ticau et al. 2015; Zhai et al. 2017; Ticau et al. 2017; Yuan et al. 2017). If Cdc6short could interact with these proteins, we would expect them to associate with DNA in a Cdc6short-dependent manner. Unlike WT Cdc6, Cdc6short did not form the OCCM complex in ATPγS, suggesting that it cannot physically interact with at least one of these proteins (Fig. S3a). Finally, to test whether Cdc6short acts as a dominant-negative inhibitor in vivo, we overexpressed this protein in mitotically diving cells. Consistent with our in vitro data, Cdc6short overexpression did not cause a significant growth defect compared to control cells, as would have been expected if it functioned as a dominant-negative isoform (Fig. 3e). Thus, Cdc6short cannot load Mcm2–7 onto origins of replication nor can it inhibit Mcm2–7 loading by WT Cdc6 in vitro or in vivo.
Fig. 3. Cdc6short cannot participate in nor inhibit Mcm2–7 loading.
(a) Colloidal-stained protein gel of purified Cdc6 and Cdc6short. See methods for purification details.
(b) Diagram of Mcm2–7 loading on origin-containing DNA coupled to a magnetic bead. The OCCM (ORC-Cdc6-Cdt1-Mcm2–7) complex is detectable when the reaction contains ATPγS and is washed in low-salt buffer, whereas loaded Mcm2–7 helicases are detectable when the reaction contains ATP and is washed in high-salt buffer.
(c) Cdc6 can catalyze Mcm2–7 loading, whereas Cdc6short cannot. Titrations of Cdc6 (lanes 2–4) and Cdc6short (lanes 5–7) are shown, and a reaction lacking Cdc6 (lane 1) shows that detection of Mcm2–7 complexes depends on the helicase‑loading reaction.
(d) Cdc6short does not have a dominant negative effect on Mcm2–7 loading in vitro. Top: Flowchart of experiment. Bottom: Pre-incubation of Cdc6short at the indicated concentration with the other helicase-loading proteins and origin DNA does not prevent WT Cdc6 from promoting Mcm2–7 loading (lanes 2–6). A reaction lacking Cdc6 (lane 1) shows that detection of Mcm2–7 complexes depends on the helicase‑loading reaction.
(e) Overexpression of Cdc6short-3V5 does not cause growth defects in mitotic cells. Strain containing PGAL-Cdc6short-3V5 overexpresses Cdc6short-3V5 on YP-Gal plates but not YPD plates. The control strain does not overexpress Cdc6short-3V5 on either medium.
Production of Cdc6short during meiosis does not require activity from the upstream CDC6 promoter
Given the apparent lack of function for Cdc6short protein, we asked why cells produce CDC6short mRNA during meiosis. Perhaps CDC6short expression is associated with repression of full-length CDC6, and is not important for making a protein product. We hypothesized two possible mechanisms that would cause the observed anti-correlation between full-length CDC6 and CDC6short levels (Fig. 1a): (1) the full-length mRNA is cleaved or partially degraded from the 5’-end to make CDC6short mRNA, which is then translated by cap-independent mechanisms to produce Cdc6short protein; (2) the upstream CDC6 promoter (PCDC6) is mutually exclusive with a cryptic promoter within the CDC6 ORF. A key distinction between these models is that the first model would require PCDC6 for Cdc6short production but the second would not.
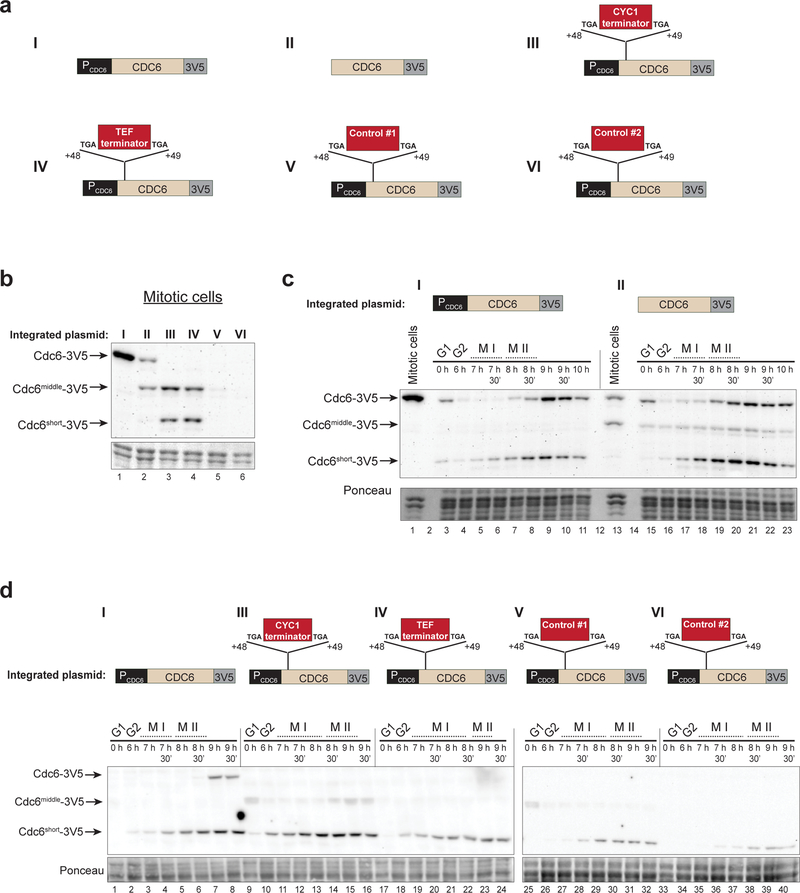
To address these two possibilities, we inserted a second copy of the CDC6 gene with multiple promoter alterations and examined the production of Cdc6 isoforms from this locus using the 3V5 tag (Fig. 4a). We found that shorter Cdc6 isoforms are produced under any condition during which PCDC6 is inactivated, supporting a model in which expression of the full- length and short CDC6 RNAs depends on mutually-exclusive promoters (Fig. 4b–4d). Deletion of nucleotides −550 to −1 strongly reduced PCDC6 activity resulting in dramatic reduction of full- length Cdc6 protein in asynchronous mitotic cells. Interestingly, these cells produced an intermediate length Cdc6 isoform (Cdc6Middle, discussed further below) (Fig. 4b). During meiosis, the promoter deletion did not affect full-length Cdc6 production, but did increase levels of Cdc6middle and Cdc6short (Fig. 4c, panel II). It is noteworthy that the decreased production of full-length Cdc6 using this deletion in mitotic cells is similar to the amount produced by meiotic cells (Fig. 4c). These data suggest that meiotic cells produce less Cdc6 than mitotic cells due to reduced activity of PCDC6 during meiosis. The basal Cdc6 expression in the Pcdc6 deletion construct is likely to be the result of a promoter in the LEU2-integrating vector that is juxtaposed with the CDC6 ORF upon deletion of PCDC6.
Fig. 4. Inactivation PCDC6 is sufficient for Cdc6short expression in mitotic cells and does not prevent Cdc6short expression during meiosis.
(a) Diagram of CDC6-derived constructs integrated at the LEU2 locus. The control sequences for constructs V and VI are similar in length to the CYC1 and TEF terminator sequences, and are derived from regions of the GFP ORF (V) and GST ORF (VI).
(b) Inactivation of PCDC6 is sufficient for production of shorter Cdc6 protein isoforms (immunoblot) in asynchronous mitotic cells, either as Cdc6short (constructs III and IV) or as an intermediate length isoform which we denote as Cdc6middle (constructs II, III, and IV). Protein was detected by V5-immunoblot. Lanes 1–6: yDP198, yDP242, yDP249, yDP250, yDP251, yDP252.
(c) Deletion of PCDC6 does not prevent expression of Cdc6short during meiosis but causes mitotic cells to have reduced levels of full-length Cdc6. Construct I (yDP198, lanes 1, 3–11); Construct II (yDP242, lanes 13, 15–23). Protein was detected by V5-immunoblot. The time after transfer into sporulation medium and the completion of MI and MII are indicated. For quantification of MI and MII completion, see Fig. S4a.
(d) Preventing transcription elongation from PCDC6 not prevent expression of Cdc6short during meiosis. Construct I (yDP198, lanes 1–8); Construct III (yDP249, lanes 9–16); Construct IV (yDP250, lanes 17–24); Construct V (yDP251, lanes 25–32); Construct VI (yDP252, lanes 33–40). Protein was detected by V5-immunoblot. The time after transfer into sporulation medium and the completion of MI and MII are indicated. For quantification of MI and MII completion, see Fig. S4b.
To eliminate any transcription elongation originating from within or before PCDC6, we made constructs with transcription terminators or similarly sized control sequences inserted between nucleotides +48 and +49 of the CDC6 ORF (Fig. 4a). These sequences were flanked by stop codons to prevent translation of full-length Cdc6, but these insertions do not affect the Cdc6short ORF. We found that both the CYC1 and TEF terminator sequences were sufficient to induce Cdc6short and Cdc6middle expression in asynchronous mitotic cells, whereas control sequences did not have this effect (Fig. 4b). All of these constructs still expressed Cdc6short during meiosis (Fig. 4d). Together, these data demonstrate that Cdc6short does not require PCDC6 activity, and that inactivation of PCDC6 is sufficient for production of shorter Cdc6 isoforms. Furthermore, Cdc6short production during meiosis cannot rely on degradation of full-length mRNA because neither the promoter deletion nor the transcription terminators prevent Cdc6short accumulation during meiosis.
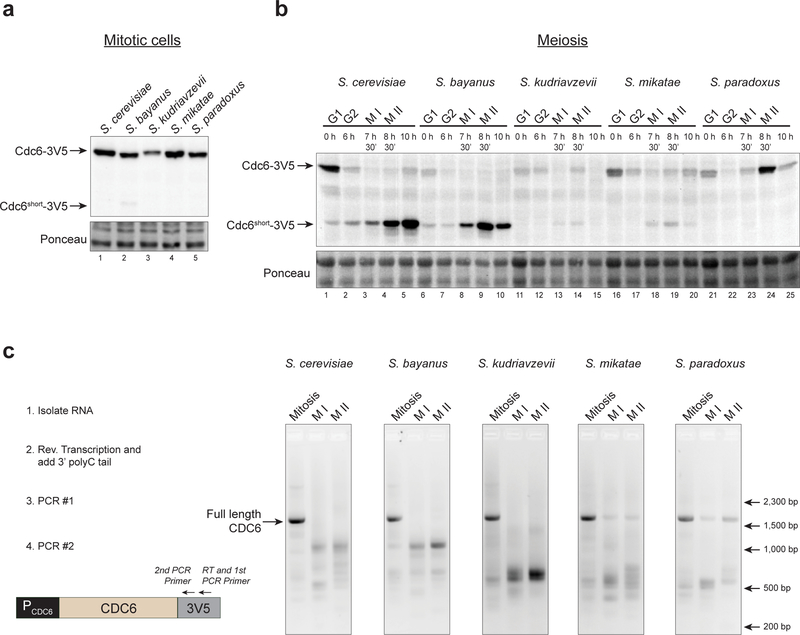
Evolutionarily-related CDC6 genes repress full-length CDC6 expression but produce unique short RNAs
The strong repression of full-length CDC6 transcription during meiosis suggests that repressing CDC6 expression is critical to prevent origin licensing (Fig. 1a) (Phizicky et al. 2018). To determine whether the mechanism to repress full-length CDC6 expression is conserved in other organisms, we cloned the CDC6 gene from four yeast species (S. bayanus, S. kudriavzevii, S. mikatae, and S. paradoxus) into plasmids and tested for RNA and protein expression. These species are estimated to be between 5 – 20 million years diverged from S. cerevisiae (Kellis et al. 2003; Scannell et al. 2011). Each gene tested included the entire CDC6 ORF, ~550 nucleotides upstream of the coding region, and a 3V5 tag at the C-terminus for detection of the protein and RNA produced. The primary Cdc6short start site (Met202) is conserved in all of these species.
We analyzed expression of CDC6 from each species during mitosis and meiosis. Each of the genes expressed full-length Cdc6 in mitotic cells at similar levels to S. cerevisiae (Fig. 5a). During meiosis, S. bayanus CDC6 showed complete repression of full-length CDC6 mRNA and expressed CDC6short protein and mRNA to similar levels as the S. cerevisiae CDC6 gene (Fig. 5b–5c). Interestingly, although the genes from both S. kudriavzevii and S. mikatae also repressed full-length CDC6 during MI and MII, only a small amount of Cdc6short protein was detected (Fig. 5b–5c). Instead, both of these genes showed meiosis-specific enrichments of much shorter RNA isoforms than those observed from S. cerevisiae. In contrast to the other CDC6 genes, S. paradoxus CDC6 did not produce detectable Cdc6short protein and had only limited enrichment of short RNA isoforms during meiosis, despite it having decreased expression of full-length CDC6 during MI (Fig. 5b–5c). Together, these data suggest that transcriptional repression of full-length CDC6 during meiosis is associated with aberrant transcription initiation from within the CDC6 ORF, and that these features are evolutionarily conserved in related yeast species.
Fig. 5. CDC6 genes from related yeast repress full-length CDC6 but produce unique short RNAs during meiosis.
(a) CDC6 genes from related yeast all produce full-length Cdc6 protein (V5 immunoblot) in asynchronous mitotic cells.
(b) Related yeast CDC6 genes produce different amounts of Cdc6short protein during meiosis. S. cerevisiae CDC6 (yDP198, lanes 1–5); S. bayanus CDC6 (yDP668, lanes 6–10); S. kudriavzevii CDC6 (yDP670, lanes 11–15); S. mikatae CDC6 (yDP671, lanes 16–20); S. paradoxus CDC6 (yDP673, lanes 21–25). Protein was detected by V5-immunoblot.
(c) CDC6 genes from related yeast repress full-length CDC6 mRNA expression during meiosis and produce distinct short CDC6 RNA isoforms. Left: Workflow for analysis of RNA products by Reverse Transcription (RT)-PCR. Right: Agarose gel of PCR products.
Discussion
Preventing DNA replication between MI and MII is critical for reducing ploidy during meiosis, but the mechanisms cells use to repress specific steps of replication initiation have not been fully elucidated. In this study, we found that two essential DNA replication, CDC6 and SLD2, are transcriptionally repressed during the meiotic divisions. For each gene, as the full- length mRNA is lost, expression of shorter RNAs that display a high degree of 5’-end heterogeneity increases. The CDC6short mRNA encodes a short form of Cdc6, but this protein does not have an obvious function during origin licensing. We provide evidence that the short RNAs are from internal promoters rather than processing of the full-length mRNA. Finally, we demonstrate that meiotic repression of full-length CDC6 and production of shorter RNA isoforms is evolutionarily conserved in genes from related yeast species. These findings provide additional insight into how DNA replication is inhibited during MI and MII and suggest that production of CDC6short is a byproduct of intragenic transcription upon repression of the full- length mRNA. In addition, our findings suggest that a subset of novel meiotic RNA and protein isoforms are a byproduct of inhibition of the associated full-length genes.
How is the CDC6short mRNA produced? The accumulation of CDC6short mRNA upon loss of the full-length mRNA (and vice versa) suggests that activation of one or more promoters inside the ORF is mutually exclusive with activation of the upstream promoter, PCDC6. Our finding that PCDC6 inactivation results in mitotic cells producing shorter Cdc6 isoforms (Fig. 4) suggests that transcription from PCDC6 represses the internal promoter(s). Consistent with this hypothesis, transcription elongation results in repressive chromatin marks being placed within gene bodies, and the lack of these marks allows transcription initiation from within ORFs (Carrozza et al. 2005; Kouzarides 2007; Wagner and Carpenter 2012; Smolle et al. 2013; Neri et al. 2017). Further support for this model, stems from the observation that the distribution of CDC6short 5’ ends spans >600 nucleotides (Fig. 2a). The elimination of repressive marks within a transcribed region would be consistent with the activation of multiple promoters within the CDC6 ORF. Although less likely, it is also possible that activation of the internal promoter(s) causes repression of PCDC6, thereby limiting full-length CDC6 expression. Our results are not consistent with CDC6short being produced as a result partial decay or processing of full-length CDC6 mRNA. These models would predict that Cdc6short would depend on PCDC6 activity, which it does not (Fig. 4). In addition, it is unlikely that partially-degraded transcripts would have the necessary requirements (e.g. a 5’ cap) for translational initiation that the short transcripts direct. Finally, we detected no evidence of splice junctions in our analysis of the CDC6short RNAs.
Despite observing many 5’ ends for CDC6short mRNA, Met202 is by far the most highly-used translation start codon during meiosis in unperturbed meiotic cells (Fig. 2). There are multiple reasons for this preference. First, we note that protein products produced from out-of-frame start codons would not be detectable by our C-terminal tag. Second, we detected very few RNAs with 5’-ends that are far enough upstream to allow translation initiation from either of the two upstream, in-frame start codons, Met89 and Met120 (Fig. 2a). Third, the sequence context of Met202 is more favorable for translation initiation than that of nearby, downstream AUGs due to its enrichment of upstream adenosines (Hinnebusch 2011; Dvir et al. 2013). Of the eleven nucleotides upstream of each start codon, Met202 has nine adenosines whereas Met225, Met261, and Met267 have two, two, and four adenosines, respectively. Yeast 5’-UTRs that are adenosine-rich, especially in the −1 to −10 region relative to the start codon, promote efficient translation initiation, likely by keeping the 5’-UTR unstructured and thus accessible to the ribosome (Hinnebusch 2011; Dvir et al. 2013). Thus, it is likely a combination of transcriptional and translational control that selects Met202 as the start codon for Cdc6short.
Although our data are consistent with PCDC6 and an internal promoter(s) being mutually-exclusive, this model may not be predictive under all conditions. Full-length CDC6 is transcriptionally repressed in mitotic cells at least from late-S phase until the end of M phase (Zwerschke et al. 1994; Piatti et al. 1995). A simple mutually-exclusive promoter model would predict that the shorter CDC6 mRNA isoforms would be observed at these stages of mitosis, yet we did not detect short CDC6 RNA or protein isoforms in asynchronous mitotic cells (Fig. 1a). Short CDC6 isoforms may not have been detected in mitotic cells either because PCDC6 is not as fully repressed as it is in meiotic cells, or because PCDC6 must be shut off for a longer time to allow chromatin changes that promote intragenic transcription initiation. Considering that Cdc6short protein is observed in mitotic cells containing transcription terminators after PCDC6 (Fig. 4b), mitotic cells most likely have all the factors required for internal initiation.
There are likely to be other transcriptional or translational regulatory mechanisms that affect which Cdc6 protein isoforms are produced. The detection of a distinct isoform, Cdc6middle, from constructs either lacking PCDC6 or those containing internal transcription-terminators suggests that different Cdc6 protein isoforms are expressed as a function of the mechanism preventing transcription elongation into the ORF. These methods of inhibiting transcription elongation into the CDC6 ORF are different than the regulated transcriptional shutoff of full-length CDC6 observed during meiosis, during which Cdc6middle is not detected above background levels (Fig. 1a and 4c). It is possible that different methods of inactivating PCDC6 cause distinct chromatin changes, thereby permitting transcription initiation from different parts of the ORF. There is also clear evidence of cell-cycle-dependent regulation of Cdc6 isoforms. All of our constructs, either with WT CDC6 or the promoter perturbations, produce different ratios of Cdc6:Cdc6middle:Cdc6short in mitotic and meiotic cells (Fig. 4).
Determining the mechanisms that direct expression of meiosis-specific RNAs will be important for assessing their function, and should allow perturbation of many RNAs at the same time. We note that neither CDC6short nor SLD2short depended on the meiotic transcription factor Ndt80, as Ndt80 is not expressed from 0–6 hours during which both short RNAs are detectable (Fig. 1). The lack of dependence on Ndt80 for CDC6short and SLD2short is in contrast to many other meiosis-specific RNA isoforms, suggesting that different mechanisms are used to produce meiosis-specific transcripts (Zhou et al. 2017; Cheng et al. 2018). Finally, it is also clear that the transcriptional regulation of CDC6 and SLD2 activity is but one of many ways that cells ensure that these proteins are eliminated at the appropriate times in the cell cycle (Drury et al. 2000; Masumoto et al. 2002; Zegerman and Diffley 2007; Reusswig et al. 2016; Phizicky et al. 2018).
Methods
Yeast strains and plasmids
All yeast strains and their genotypes are described in Table S1. All strains used for in vivo experiments were derived from the SK1 background with the following genotype; ho::LYS2/ho::LYS2, lys2/lys2, ura3/ura3, leu2::hisG/leu2::hisG, his3:hisG/ his3:hisG, trp1::hisG/trp1::hisG. CDC6 genes from S. bayanus, S. kudriavzevii, S. mikatae, and S. paradoxus were cloned out of the isolates given in the strain list. Plasmids used for protein expression are described in Table S2.
Meiotic time-courses
All strains put through meiotic time-courses contained PGPD–GAL4–ER, PGAL–NDT80 for controlled block-release from meiotic G2 phase. Meiotic time courses were done as described in (Berchowitz et al. 2013). Briefly, cells were diluted to an OD600 = 0.25 in BYTA medium from saturated YPD cultures. BYTA cultures were grown for 18–20 hours while shaking at 30°C. After growth in BYTA, cells were washed with water and resuspended to OD600 = 1.9 in Sporulation medium. The 0–hour sample was taken immediately after transfer to Sporulation medium. Cultures were shaken at 30°C for 6 hours to allow cells to accumulate at the NDT80 block. To release cells from the block, 1 mM β-estradiol (5 mM stock in ethanol [Sigma, E2758]) was added to the culture medium. Protein, RNA, and immunofluorescence samples were harvested in parallel at the indicated time points.
Immunofluorescence
Tubulin immunofluorescence was performed as previously described (Berchowitz et al. 2013). 100 cells were counted per time point for both spindle and nuclei scoring. Metaphase–I and Metaphase-II cells were defined as having, respectively, one or two short, thick, bipolar spindle(s); Anaphase–I and Anaphase-II cells as having, respectively, one or two long, bipolar spindle(s); Nuclei were counted as the number of distinct DNA masses.
Immunoblots and RNA Isolation
Immunoblots and RNA isolation were done as previously described (Phizicky et al. 2018).
Northern blots
Equal amounts of total RNA (between 12–16 μg) were loaded in each lane of a formaldehyde gel and transferred to a nylon membrane (GE HyBond N+, catalog number RPN3050B). rRNA was detected by methylene blue and used as a loading control, and specific RNAs were detected using gene-specific probes. 32P-labeled probes were made by Klenow extension (GE Healthcare, RPN1605) of gene-specific PCR products containing the indicated regions. Primers for Klenow extensions were random hexamers. Templates: The CDC6 template was specific to the 3’ half of the ORF (nucleotides +923 to +1488). The three SLD2 templates used covered either the entire ORF (nucleotides +1 to +1359), or were specific to the 5’ half (nucleotides +1 to +547) or the 3’ half (nucleotides +805 to +1349) of the ORF.
5’ RACE
cDNA of CDC6 or SLD2 was made from 4 μg of total RNA using Superscript IV Reverse Transcriptase (Life Technologies, catalog number 18090010) and gene-specific primers according to manufacturer’s instructions. After cDNA synthesis and purification, a poly- deoxycytosine tail was added to the 3’ end of the cDNA using Terminal Transferase with a 19:1 ratio of dCTP:ddCTP (so as to limit the length of the tail). Poly-dC-tailed cDNA was then amplified by PCR using a poly-G primer and the same gene-specific primer as was used for the RT-step. A second PCR was performed using the resulting PCR product as a template, and a different gene-specific primer was used to increase the specificity of the product. This second- round PCR product was digested with SacI and SalI restriction enzymes, sites for which were encoded on the primers in the PCR reaction. The resulting products were run on an agarose gel (shown). Sections of this gel were purified and ligated into a similarly digested vector, so as to prevent ligation reaction bias for short products. Ligation reactions were transformed and DNA from individual colonies was miniprepped. The entire ligated region was sequenced, and 5’- ends were called based on the last nucleotide that was successfully aligned before the poly- Cytosine region was reached.
Helicase loading assays
Helicase loading assays were done as described in (Kang et al. 2014). Briefly, 50 nM ORC, 150 nM Mcm2–7/Cdt1, and the indicated concentration of Cdc6 or Cdc6short were combined in a 40 μL reaction with 25 nM bead–bound 1.3 kB DNA containing the ARS1 origin. After combining the proteins and DNA, reactions were incubated at 25°C for 30 min while shaking at 1,250 rpm (Thermo-mixer, Eppendorf). Beads were then washed three times in buffer. For helicase– loading reactions, the three washes had buffer containing 300 mM KGlut, 500 mM NaCl, and 300 mM KGlut respectively. Proteins were eluted from DNA with DNase, run on SDS–PAGE gels, and detected with Krypton fluorescent stain (Fisher, PI–46629). To assay OCCM formation, ATP was replaced with 5 mM ATPγS, and beads were washed three times with buffer containing 300 mM KGlut.
Protein Purifications
Cdc6 (plasmid pDP69; MBP-FLAG-Cdc6–6xHIS) and Cdc6short (pDP70; MBP-FLAG-Cdc6short- 6xHIS, containing CDC6 amino acids 202–513) were purified from E. Coli Rosetta 2 cells grown to log phase and induced overnight with 0.5mM IPTG and shake at 18°C for 20 hours. Cells were resuspended in Buffer G (50 mM Potassium Phosphate [pH = 7.6], 5 mM Magnesium Chloride, 1% Triton, and 1 mM DTT) with 400 mM Potassium Acetate (KAc), 2 mM ATP, 1 mM PMSF, and 1× cOmplete Protease Inhibitors (Roche), and then flash frozen in liquid nitrogen. Cells were thawed and lysed with lysozyme and sonication. After centrifugation at 12,000 rpm, 10 mM imidazole was added to the supernatant which was flowed over Nickel resin equilibrated in the same buffer. The resin was washed with 20 bed volumes of buffer. Protein was eluted using Buffer G, 400 mM KAc, 2 mM ATP, and 350 mM imidazole. Peak elusions were pooled and incubated with equilibrated amylose resin, which was then washed with 20 bed volumes of Buffer G with 400 mM KAc. Resin was then washed with 5 bed volumes of Buffer G, 400 mM KAc, and 15% glycerol, and protein was eluted using the same buffer with the addition of 10 mM amylose. Peak fractions were pooled, aliquoted, flash frozen, and stored at −80°C. ORC (ySDORC) and Mcm2–7–Cdt1 (yST144) were purified as previously described (Kang et al. 2014).
Supplementary Material
Acknowledgements
We thank Luke Berchowitz for comments on the manuscript. We thank all members of the Bell Laboratory for helpful discussions. We thank John Diffley, Maitreya Dunham, and Mark Johnston for strains. DVP was supported in part by a NIH Pre-Doctoral Training Grant (GM007287). SPB is an investigator with the Howard Hughes Medical Institute.
References
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes & Development 21:497–518. doi: 10.1101/gad.1508907 [DOI] [PubMed] [Google Scholar]
- Bell SP, Labib K (2016) Chromosome Duplication in Saccharomyces cerevisiae. Genetics 203:1027–1067. doi: 10.1534/genetics.115.186452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I (2003) Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes & Development 17:1524–1539. doi: 10.1101/gad.1101503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Gajadhar AS, van Werven FJ, et al. (2013) A developmentally regulated translational control pathway establishes the meiotic chromosome segregation pattern. Genes & Development 27:2147–2163. doi: 10.1101/gad.224253.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GAG, Yassour MM, Friedman NN, et al. (2012) High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335:552–557. doi: 10.1126/science.1215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D, Bartlett R, Crawford K, Nurse P (1991) Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature 349:388–393. doi: 10.1038/349388a0 [DOI] [PubMed] [Google Scholar]
- Buonomo SBC, Rabitsch KP, Fuchs J, et al. (2003) Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Developmental Cell 4:727–739. [DOI] [PubMed] [Google Scholar]
- Calzada A, Sánchez M, Sánchez E, Bueno A (2000) The Stability of the Cdc6 Protein Is Regulated by Cyclin-dependent Kinase/Cyclin B Complexes inSaccharomyces cerevisiae. Journal of Biological Chemistry 275:9734–9741. [DOI] [PubMed] [Google Scholar]
- Carlile TM, Amon A (2008) Meiosis I is established through division-specific translational control of a cyclin. Cell 133:280–291. doi: 10.1016/j.cell.2008.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, et al. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592. doi: 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Otto GM, Powers EN, et al. (2018) Pervasive, Coordinated Protein-Level Changes Driven by Transcript Isoform Switching during Meiosis. Cell 172:910–923.e16. doi: 10.1016/j.cell.2018.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi T, Aoki F, Mori M, et al. (1991) Activation of p34(cdc2) protein kinase activity in meiotic and mitotic cell cycles in mouse oocytes and embryos. Development 113:789–795. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, et al. (1998) The transcriptional program of sporulation in budding yeast. Science 282:699–705. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JFX, Nasmyth KA (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Current Biology 5:1257–1269. doi: 10.1016/S0960-9822(95)00252-1 [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF (1997) The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J 16:5966–5976. doi: 10.1093/emboj/16.19.5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF (2000) The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Current Biology 10:231–240. [DOI] [PubMed] [Google Scholar]
- Dvir S, Velten L, Sharon E, et al. (2013) Deciphering the rules by which 5’-UTR sequences affect protein expression in yeast. Proc Natl Acad Sci USA 110:E2792–801. doi: 10.1073/pnas.1222534110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, et al. (2009) A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proceedings of the National Academy of Sciences 106:20240–20245. doi: 10.1073/pnas.0911500106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C, Zou J, Rappsilber J, Marston AL (2017) Cdc14 phosphatase directs centrosome re-duplication at the meiosis I to meiosis II transition in budding yeast. Wellcome Open Res 2:2. doi: 10.12688/wellcomeopenres.10507.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75:434–67– first page of table of contents. doi: 10.1128/MMBR.00008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollerer I, Higdon A, Brar GA (2017) Strategies and Challenges in Identifying Function for Thousands of sORF-Encoded Peptides in Meiosis. Proteomics 70:1700274. doi: 10.1002/pmic.201700274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Hutti JE, Cantley LC, Morgan DO (2007) Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Molecular Cell 25:689–702. doi: 10.1016/j.molcel.2007.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi MM, Ohsumi KK, Yamamoto TMT, et al. (2000) Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M-M transition in Xenopus oocyte extracts. EMBO J 19:4513–4523. doi: 10.1093/emboj/19.17.4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Warner MD, Bell SP (2014) Multiple Functions for Mcm2–7 ATPase Motifs during Replication Initiation. Molecular Cell. doi: 10.1016/j.molcel.2014.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, et al. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241–254. doi: 10.1038/nature01644 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Lardenois A, Liu Y, Walther T, et al. (2011) Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc Natl Acad Sci USA 108:1058–1063. doi: 10.1073/pnas.1016459108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Lee BH, Amon A (2003) The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Developmental Cell 4:711–726. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H (2002) S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415:651–655. doi: 10.1038/nature713 [DOI] [PubMed] [Google Scholar]
- Neri F, Rapelli S, Krepelova A, et al. (2017) Intragenic DNA methylation prevents spurious transcription initiation. Nature 543:72–77. doi: 10.1038/nature21373 [DOI] [PubMed] [Google Scholar]
- Ohkura H (2015) Meiosis: an overview of key differences from mitosis. Cold Spring Harbor Perspectives in Biology 7:1–15. doi: 10.1101/cshperspect.a015859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM (2013) Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497:127–131. doi: 10.1038/nature12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Drury LS, Diffley JF (2001) Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J 20:4836–4845. doi: 10.1093/emboj/20.17.4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky DV, Berchowitz LE, Bell SP (2018) Multiple kinases inhibit origin licensing and helicase activation to ensure reductive cell division during meiosis. Elife 7:497. doi: 10.7554/eLife.33309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J 14:3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JCW, Bowers JL, Rodríguez HK, Bell SP (2006) Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Molecular Cell 21:29–39. doi: 10.1016/j.molcel.2005.11.023 [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, et al. (2009) Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139:719–730. doi: 10.1016/j.cell.2009.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusswig K-U, Zimmermann F, Galanti L, Pfander B (2016) Robust Replication Control Is Generated by Temporal Gaps between Licensing and Firing Phases and Depends on Degradation of Firing Factor Sld2. Cell Rep 17:556–569. doi: 10.1016/j.celrep.2016.09.013 [DOI] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, et al. (2011) The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3 (Bethesda) 1:11–25. doi: 10.1534/g3.111.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, Workman JL, Venkatesh S (2013) reSETting chromatin during transcription elongation. Epigenetics 8:10–15. doi: 10.4161/epi.23333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Evrin C, Samel SA, et al. (2013) Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2–7 bound to DNA. Nat Struct Mol Biol. doi: 10.1038/nsmb.2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, et al. (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445:328–332. doi: 10.1038/nature05465 [DOI] [PubMed] [Google Scholar]
- Ticau S, Friedman LJ, Champasa K, et al. (2017) Mechanism and timing of Mcm2–7 ring closure during DNA replication origin licensing. Nat Struct Mol Biol 24:309–315. doi: 10.1038/nsmb.3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticau S, Friedman LJ, Ivica NA, et al. (2015) Single-Molecule Studies of Origin Licensing Reveal Mechanisms Ensuring Bidirectional Helicase Loading. Cell. doi: 10.1016/j.cell.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13:115–126. doi: 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Riera A, Bai L, et al. (2017) Structural basis of Mcm2–7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol 24:316–324. doi: 10.1038/nsmb.3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445:281–285. doi: 10.1038/nature05432 [DOI] [PubMed] [Google Scholar]
- Zhai Y, Cheng E, Wu H, et al. (2017) Open-ringed structure of the Cdt1-Mcm2–7 complex as a precursor of the MCM double hexamer. Nat Struct Mol Biol. doi: 10.1038/nsmb.3374 [DOI] [PubMed] [Google Scholar]
- Zhou S, Sternglanz R, Neiman AM (2017) Developmentally regulated internal transcription initiation during meiosis in budding yeast. PLoS ONE 12:e0188001. doi: 10.1371/journal.pone.0188001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke W, Rottjakob HW, Küntzel H (1994) The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J Biol Chem 269:23351–23356. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.