Abstract
Recent studies have highlighted the imperatives of including diverse and under-represented individuals in human genomics research and the striking gaps in attaining that inclusion. With its multidecade experience in supporting research and policy efforts in human genomics, the National Human Genome Research Institute is committed to establishing foundational approaches to study the role of genomic variation in health and disease that include diverse populations. Large-scale efforts to understand biology and health have yielded key scientific findings, lessons and recommendations on how to increase diversity in genomic research studies and the genomic research workforce. Increased attention to diversity will increase the accuracy, utility and acceptability of using genomic information for clinical care.
ToC blurb
Including diverse populations in genomic studies has the potential to improve the use of genomic data in the clinic. Here, researchers from the National Human Genome Research Institute review the benefits of increasing diversity, the challenges to overcome and key recommendations for how to achieve this goal.
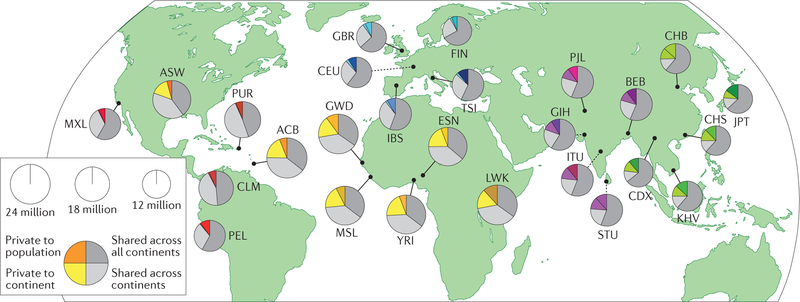
Genomic diversity, which is defined as variation in alleles and allele frequency, is inherent within and across human populations. The study of how genomic diversity arises among populations, including changes in allele frequencies across space and time, is the domain of population genetics. Understanding the contribution of genomic variation to health and disease has been a major focus of human genomics research over the past three decades. Success in sequencing the human genome has motivated a new generation of scientists to apply novel technologies to population-based studies, resulting in over 3,000 published genome-wide association studies (GWAS) to date1. More recently, population-based genome-sequencing studies, which are ideal for discovering rare genomic variants associated with disease, are becoming more common2,3. For GWAS and genome-sequencing studies, the value of a human genome reference sequence against which novel genomic variation can be identified is evident4. However, even before the launch of the Human Genome Project (HGP) in 1990, it became evident that there was no single human genome and that human genomic variation clusters in population groups separated by geography and ancestral history5. A recent analysis of genomic variation across the 1000 Genomes Project global reference sample observed that most (>70%) common genomic variation is shared across continental groups6 (FIG. 1). Ultimately, knowledge of how genomic variants and their downstream biological effects vary across populations increases our ability to understand genomic contributions to health and disease and to apply this knowledge to clinical care.
Figure 1. Shared genomic variation across global populations.
Greater than ~70% of the genomic variation in sampled populations is shared across multiple continents. The area of each pie is proportional to the number of polymorphisms within a population. Pies are divided into four slices, representing variants private to a population (darker colour unique to population), private to a continental area (lighter colour shared across continental group), shared across continental areas (light grey) and shared across all continents (dark grey). Colour families approximate continental origin (red, ancestry from the Americas; orange/yellow, African ancestry; blue, European ancestry; purple, South Asian ancestry; green, East Asian ancestry). Dashed lines indicate populations sampled outside their ancestral continental region. ACB, Barbadian; ASW, African Americans in south-west USA; BEB, Bengali; CDX, Dai Chinese; CEU, Utah residents with northern and western European ancestry; CHB, Han Chinese; CHS, southern Han Chinese; CLM, Colombian; ESN, Esan; FIN, Finnish; GBR, British; GIH, Gujarati; GWD, Gambian; IBS, Spanish; ITU, Telugu; JPT, Japanese; KHV, Kinh Vietnamese; LWK, Luhya; MSL, Mende; MXL, Mexican American; PEL, Peruvian; PJL, Punjabi; PUR, Puerto Rican; STU, Tamil; TSI, Tuscan; YRI, Yoruba. Adapted with permission from REF. 6, Macmillan Publishers Limited.
A recent analysis of the GWAS Catalog1, a curated database of genomic variants associated with human disease that is provided jointly by the National Human Genome Research Institute (NHGRI) and the European Bioinformatics Institute (EMBL-EBI), revealed a striking lack of diverse and under-represented populations in published GWAS7. In this context, the terms ‘diverse’ and ‘under-represented’ refer to the characteristics related to peoples’ ancestry and to the physical and social environments in which they live and receive health care. Non-European participants represented only 19% of individuals studied in GWAS7 even though over three-fourths of the world population live in Africa and Asia8. Other types of genetic association studies, though not as well catalogued as GWAS, are similarly heavily skewed towards non-European ancestry individuals9. Reasons commonly given for this under-representation include a desire to avoid potential population stratification10 by focusing on ancestrally homogeneous populations and reduced participation by populations of non-European ancestry due in part to mistrust arising from past misuses of their genetic and genomic data11. The net effect is that European ancestry cohorts are far larger and better characterized than non-European ancestry cohorts in extant genomics research studies.
Recent findings have highlighted the scientific need to include diverse and under-represented populations in human genomics research12–14. Genome-sequencing studies of global populations, such as the 1000 Genomes Project, have shown that despite the observation that 96–99% of a single individual’s genome is composed of common (minor allele frequency (MAF) >5% ) variants, most genomic variation is rare (MAF <0.5%) and population-specific6. Inferences drawn from studying a single ancestral group can thus be potentially incomplete or even inaccurate, which can have important implications for research15. For example, the association of PCSK9 loss-of-function mutations with low cholesterol levels and low coronary heart disease risk was identified in an African-American cohort16 and would have been missed had the studies been restricted to individuals of European ancestry. Limiting studies to a single ancestry group can also propagate misinterpretations and have clinical consequences, as illustrated by a study that identified genomic variants in African Americans that were initially classified as pathogenic for hypertrophic cardiomyopathy by use of largely European ancestry samples17. These variants were subsequently observed to be prevalent in African ancestry populations, making them unlikely to be disease-causing.
The NHGRI has a multidecade history of supporting research and community engagement efforts to increase diversity in population-based genomics research and to advance translation for improving clinical care (TABLE 1). For example, the NHGRI supports investigators in several efforts to identify disease-associated genomic variants in diverse populations, including the Population Architecture using Genomics and Epidemiology (PAGE)18, Human Heredity and Health in Africa (H3Africa)19 and Centers for Common Disease Genomics (CCDG)20 programmes. In this Perspective, we describe the challenges to achieving diversity in genomic studies . We also demonstrate how attention to diversity can enhance the use of genomic information in medical care. Finally, we outline actions that can be taken by researchers and those who fund and publish their work which ensure suitable attention is given to diversity. Although it is beyond the scope of this Perspective, we also acknowledge that the need for increased diversity in biomedical research extends far beyond genomics research and strongly endorse increased diversity across the entire research enterprise.
Table 1 |.
NHGRI efforts relevant to enhancing diversity in human genomics research
| Programme or activity | Date of inception | Description | Refs and/or website |
|---|---|---|---|
| Ethical, Legal and Social Implications (ELSI) | 1990 | A programme established as an integral part of the Human Genome Project (HGP) to foster basic and applied research on the ethical, legal and social implications of genetic and genomic research for individuals, families and communities | https://www.genome.gov/elsi/ |
| DNA Polymorphism Discovery Resource | 1998 | A resource of DNA samples and cell lines from 450 US residents for discovering DNA sequence polymorphisms |
105 https://www.genome.gov/10001552/dna-polymorphism-discovery-resource/ |
| Diversity Action Plan (DAP) | 2002 | A training programme supporting educational activities that increase the diversity of the biomedical, behavioural and clinical research workforce in genomics | https://www.genome.gov/14514228/history-of-nhgris-minoritydiversity-action-plan/ |
| The International HapMap project (HapMap) | 2002 | An international collaboration to develop a haplotype map of the human genome, relating variations in human DNA sequences to genes associated with health |
44 https://www.genome.gov/10001688/international-hapmap-project/ |
| The Cancer Genome Atlas (TCGA) | 2006 | An effort to generate comprehensive, multidimensional maps of the key genomic changes in 33 types of cancer | https://cancergenome.nih.gov/ |
| Genome Reference Consortium (GRC) | 2006 | A consortium working to create assemblies that better represent the complex allelic diversity of the human genome and provide more robust substrates for genome analysis | https://www.ncbi.nlm.nih.gov/grc |
| Electronic Medical Records and Genomics (eMERGE) | 2007 | A consortium devoted to developing, disseminating and applying approaches to research that combine biorepositories with electronic medical record systems for genomic discovery and genomic medicine implementation research |
106 https://www.genome.gov/27540473/electronic-medical-records-and-genomics-emerge-network/ |
| 1000 Genomes Project | 2008 | A collaboration between research groups around the world to produce an extensive catalogue of human genomic variation to support medical research studies |
6 https://www.genome.gov/27528684/1000-genomes-project/ |
| GWAS Catalog | 2008 | A quality-controlled, manually curated, literature-derived collection of all published genome-wide association studies (GWAS) | https://www.ebi.ac.uk/gwas/ |
| Population Architecture using Genomics and Epidemiology (PAGE) | 2008 | A consortium of US studies that focuses on analysing the relationship between genomic variants and a range of common diseases and traits in ancestrally diverse populations |
107 http://pagestudy.org/ |
| Human Heredity and Health in Africa (H3Africa) Initiative | 2012 | An initiative aiming to facilitate a contemporary research approach to the study of genomics and environmental determinants of common diseases with the goal of improving the health of African populations by contributing to the development of the necessary expertise among African scientists and to the establishment of networks of African investigators | http://h3africa.org/ |
| Implementing Genomics in Practice (IGNITE) | 2013 | A consortium created to increase the use of genomic medicine by supporting the development of methods for incorporating genomic information into clinical care and the exploration of methods for effective implementation, diffusion and sustainability in diverse clinical settings |
80 https://www.genome.gov/27554264/implementing-genomics-in-practice-ignite/ |
| Centers for Common Disease Genomics (CCDG) | 2016 | A national, collaborative large-scale genome-sequencing effort to identify rare risk and protective variants contributing to multiple common disease phenotypes | https://www.genome.gov/27563570/ |
| High-quality reference genomes programme | 2016 | An initiative to increase the population diversity represented in high-quality assemblies so that references are more useful for analyses of individual genomes from diverse populations | https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-15–027.html |
| Roundtable on inclusion and engagement of under-represented populations in genomics | 2016 | A 2015 meeting convened to discuss the opportunities and challenges associated with the inclusion and engagement of under-represented populations in genomics research | https://www.genome.gov/pages/about/nachgr/february2016agenda-documents/2015_09_16_roundtable_report_final.pdf |
| Clinical Sequencing Evidence-Generating Research (CSER2) | 2017 | A national consortium conducting interdisciplinary research to evaluate the clinical utility of integrating genome sequencing into clinical care in diverse populations and settings | https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-16–011.html |
| NHGRI Community Engagement in Genomics Working Group (CEGWG) | 2017 | A working group of the National Advisory Council for Human Genome Research. The mission of the CEGWG is to engage communities to ensure that genomics and genomic medicine benefits all | https://www.genome.gov/27568486/community-engagement-in-genomics-working-group/ |
NHGRI, National Human Genome Research Institute.
Lessons learnt on diversity
Consideration of diversity improves the research process at all levels from study design to translation of research findings. Currently, the primary policy effort to ensure diversity in studies funded by the National Institutes of Health (NIH) rests with the NIH policy on inclusion of women and minorities as participants in human subjects research21. Inclusion monitoring is the process by which NIH-funded researchers report the numbers of enrolled individuals who are women and/or belong to an ethnic or racial minority and is performed regularly (typically annually) throughout the grant period. The reporting categories are not intended to reflect genetic ancestry, though overall concordance with self-reported race is high for individuals of African and European ancestry (albeit less so for non-African, non-European individuals)22. In the experiences of the NHGRI, monitoring inclusion is necessary but not sufficient for ensuring an adequate emphasis on diversity in accordance with institute priorities13. While there are no formal requirements for analysing or disseminating diversity-focused scientific findings, our emphasis on funding diversity-related grants and programmes has led to peer-reviewed publications from NHGRI-supported investigators demonstrating the shortcomings of population under-representation and the importance of increasing diversity in genomic studies (TABLE 2). These scientific accomplishments are also shared publicly through lecture series23 and workshops24. Recognizing that inclusion monitoring is a means to an end and that formal requirements for analysing and disseminating diverse data are absent, we describe lessons that point to effective ways to increase diversity in human genomics research. These lessons are organized according to various steps in the research cycle (TABLE 3): formulating research questions; providing funding opportunities; recruiting diverse participants; analysing and interpreting results; applying results; and ensuring opportunities for diverse researchers.
Table 2 |.
Key scientific findings of NHGRI-funded programmes related to diversity
| Programme | Publication date | Finding | Implications for future studies | Refs |
|---|---|---|---|---|
| 1000 Genomes Project | 2008 | ● 86% of genomic variants in a global reference population were restricted to a single continental group ● 35% of disease-associated variants have no proxy shared across continental groups |
Multiethnic populations will be needed for comprehensive discovery studies and for follow-up studies refining initial signals that are discovered | 6 |
| Electronic Medical Records and Genomics (eMERGE) | 2012, 2017 | ● A GWAS examining venous thromboembolism in African Americans identified three novel genomic variants unique to this population ● A GWAS in a diverse population identified an African ancestry-specific novel white blood count associated variant |
GWAS and discovery studies that include diverse populations are needed to identify population-specific genomic variants associated with disease |
72 108 |
| Human Heredity and Health in Africa (H3Africa) Initiative | 2014 | In sub-Saharan African participants, carrier frequencies of variants related to spinal muscular atrophy are much lower than those in European and Asian populations | Conditions described as panethnic may be based on incomplete allele frequency data from global populations. Risk assessment and genetic counselling should be tailored accordingly | 73 |
| Clinical Sequencing Exploratory Research (CSER) | 2015 | The yield of identifiable actionable secondary findings from exome sequencing in unselected populations is greater in those of European ancestry (2.0%) than in those of African ancestry (1.1%) | Given greater genomic diversity in African populations, databases and literature from which actionable findings are currently identified are under-represented for diverse ancestry. The utility of these resources will increase as the diversity of the underlying data increases | 77 |
| The Cancer Genome Atlas (TCGA) | 2015 | Analysis of tumour sequence, gene expression and proteomic data from multiple subtypes of breast cancer suggests race-specific gene signatures in women of African-American ancestry compared with those of European ancestry. Over 40% of the differences in subtype were explained by germline variants | Subtype-specific therapies could be developed to target subtypes that disproportionately affect African-American women. Additional research is needed to better understand the underlying pathogenesis and implications for treatment | 35 |
| GWAS Catalog | 2016 | In 2009, 96% of participants from GWAS publications were of European ancestry. In 2016, this proportion was 81%. | Non-European populations are greatly under-represented in genomic discovery studies and the resources that derive from them | 7 |
| Implementing Genomics in Practice (IGNITE) | 2017 | In interviews with African-American patients before and after receiving APOL1 genotyping results, themes of empowerment, accountability, promise and risk emerged. Patients viewed these genomic testing results as holding more promise than peril | Communication strategies tailored for diverse patients may bring the value of genomic testing to diverse patients. Assessing genomic risk may motivate providers to overcome clinical inertia in addition to hypertension management | 50 |
| Population Architecture using Genomics and Epidemiology (PAGE) | 2017 | In five different ancestry groups, associations between 36 established loci and obesity in European ancestry populations were assessed. Most loci generalized across ancestral groups, but several novel signals were also identified | Multiethnic populations are essential for replicating and refining initial signals from GWAS and for identifying causal variants | 75 |
GWAS, genome-wide association studies; NHGRI, National Human Genome Research Institute.
Table 3 |.
How greater diversity accelerates discovery and translation efforts
| Step in research cycle | Potential benefits |
|---|---|
| Formulate research questions investigating genomic and environmental contributors to health disparities | ● Improved study design ● More precise assessment of genetic and environmental risk factors |
| Provide dedicated funding support | ● Increased ability to address challenges to recruitment ● Continuity and stability of research teams |
| Recruit diverse participants and communities | ● Adequate sample sizes for analysis ● Enrolment reflective of population disease burden ● More equitable distribution of benefits of genomic research ● Consensus building and shared oversight |
| Improve analysis and interpretation by use of foundational genomic data resources | ● Higher-quality reference sequences, yielding more accurate variant calls in diverse participants ● Expanded availability of population-specific allele frequencies ● Better imputation and facile data integration, yielding larger sample sizes for analysis ● Fine mapping to identify causal variants ● More accurate identification of clinically relevant variants ● Identification of novel variants |
| Apply knowledge to health care systems | ● Identification of implementation opportunities applicable to all types of health delivery systems ● Implementation of interventions that might otherwise be missed |
| Increase diversity among researchers and clinicians | ● Facilitation of enrolment of diverse participants ● Improved workforce diversity |
Formulate research questions that investigate genomic and environmental contributors to health disparities.
The importance of social and environmental factors that interact with genetic variants to influence health outcomes is increasingly being recognized in genomics research. The extent to which genomics can inform our understanding of disparities in disease incidence and outcomes has been widely debated25–31. Some have posited that the role of genomics has been overstated relative to social determinants and environmental factors26,29, and others have emphasized the need to continue to study genomic contributions28,31. Although the primary role of social and environmental factors on health disparities is well documented — with low income and low education consistently associated with a wide range of poor health outcomes32 — such factors have received insufficient emphasis in genomics research. The ability of genomic studies to understand disease aetiology and health disparities may be greatly improved if such studies are designed to incorporate and analyse data on social and physical environment29. Because social, environmental and genomic factors are not mutually exclusive and most human diseases and traits arise from a combination of these factors, attention to robust study design and inclusion of these factors when formulating research questions will maximize opportunities to better understand the often-complicated dynamic among them33,34. For example, markedly poorer survival from breast cancer in women of African-American compared with women of European ancestry led researchers to hypothesize that this disparity might be due in part to genomic factors. A recent analysis of publicly available tumour and germline data from The Cancer Genome Atlas (TCGA) collected at multiple stages of breast cancer development revealed significantly increased survival in women of European ancestry compared with those of African-American ancestry, with approximately 40% of the differences in cancer subtype attributable to germline genomic factors35. As only limited environmental factors were available in TCGA, the environmental contribution to understanding cancer health disparities could not be assessed, providing future opportunities for large-scale efforts to address, for example, by the International Cancer Genome Consortium for Medicine36 or other National Cancer Institute (NCI) cancer genomics efforts such as the Early Onset Malignancies Initiative37.
Provide dedicated funding support.
Studies that include under-represented populations in research are often more time-intensive in recruiting participants or collecting complete outcome data and more resource-intensive, requiring more personnel and research expenses, than those with less-inclusive designs38. Challenges faced by individuals belonging to under-represented populations that constrain their ability to participate in research studies may include, for example, socioeconomic factors (such as lack of discretionary time, loss of income owing to time away from work or lack of access to transportation and information resources), cultural factors (such as language differences) or health research-specific factors (such as poorer overall health status and greater mistrust of research or health care systems). In addition, in settings of limited access to basic health care, participants are less likely to prioritize research participation if no clinically actionable information is received38–40. Practically, this can translate into lower enrolment rates and higher attrition, with the resulting need for additional study resources to compensate and meet intended study enrolment targets. These challenges to enrolment and retention should be factored into the study design by providing realistic enrolment projections; however, high attrition rates can be a formidable obstacle to receiving favourable evaluations of the proposed studies during scientific peer review before funding. Data analysis of diverse populations can also be complicated, albeit only modestly, by the need to account for ancestry-related variables and interactions with environmental factors41,42.
These obstacles may create disincentives to invest in establishing newer, more diverse research cohorts, thereby perpetuating existing disparities and gaps in scientific knowledge. It is thus critical that funding agencies consider opportunities to establish diversity-emphasizing programmes (particularly large-scale efforts) as a complement to existing efforts in populations fully participating in genomics research. Such programmes should prioritize the inclusion of diverse participants and tailor scientific questions accordingly. They should also commit resources with awareness of the substantial challenges to recruiting and retaining diverse participants as well as to analysing and interpreting their data. In addition to addressing these recruitment challenges, the benefits of dedicated support include facilitating the continuity and stability of research teams.
Recruit diverse participants and communities.
Adequate recruitment of diverse participants assures sample sizes that are well powered to evaluate study aims and guides enrolment of participants reflective of the disease burden in the population at large. Inclusion of diverse participants in genomic studies can be increased by community engagement designed to meet specific cultural expectations, maximizing the possibility that the research outcomes will benefit individuals from these populations43. The Polymorphism Discovery Resource and HapMap44 projects were early efforts incorporating community consultations that highlighted the need for deliberate consideration of ethical, legal and social implications (ELSI) as part of the study design. As NHGRI-supported efforts translate findings from foundational resources to the biology of disease and science of medicine, community-centred and participant-centred advisory boards and workshops as well as the use of ‘embedded ELSI’ research in major research programmes continue to have a prominent role45–48. For example, one study in the Implementing Genomics in Practice (IGNITE) 49 network established community consultations to guide research questions and strategies in implementing genomic medicine related to hypertension-related kidney disease in inner-city African Americans. In this study, participant and researcher feedback was used to develop intuitive and useful communication strategies50. Embedding genomic expertise and resources directly in the community that hopes to benefit from the research facilitates consensus building and shared oversight. It also creates a collaborative environment in which shared priorities can guide the research, including its aims, implementation, dissemination and evaluation. This in turn can accelerate the accumulation of expertise within the scientific and participant communities and the generation of new scientific insights regarding the human genome and its role in health and disease.
Recent NIH efforts have intentionally sought the advice of patients and community members to improve precision medicine and clinical research51,52. While recruitment of diverse populations has been improved by established approaches such as including researchers from these communities and working within trusted social networks38,39, novel approaches to communication and data sharing (for example, using social media or online platforms to communicate with participants and obtain their input) are also promising53. Data sharing among scientific investigators has traditionally occurred through centralized NIH databases, such as dbGaP54 and ClinVar55. Populations that have limited familiarity with or access to technology or are especially distrustful because of historical experiences often view data sharing cautiously. Alternative approaches to data sharing that are more participant-focused may be needed to engage these populations , as data-sharing requirements have been and continue to be major impediments to participation among some populations56,57,58. Models in which study participants have greater control over what information is shared (and when) are gaining traction57,59 and may help to address this barrier.
Improve analysis and interpretation by use of foundational genomic data resources.
The HGP used samples predominately representing European ancestry. However, subsequent efforts expanded the ancestral diversity of foundational resources of human genomic variation. The availability of high-quality reference genome sequences, such as those generated by the Genome Reference Consortium (GRC) and the High Quality Human Reference Sequence Program60,61, has been essential in aligning and analysing genome sequences from participants in human disease studies. These and other programmes are continuing to improve the quality of reference genome sequences by incorporating samples from multiple continents of origin (the Americas, Africa and Asia in addition to Europe), which will serve as better standards for clinical genome sequencing4. Estimates of allele frequency and haplotype structure in human populations began being generated shortly after the HGP, for example, by the HapMap44 and the 1000 Genomes6 projects, followed by the development of the Exome Variant Server (EVS) under the auspices of the National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project62, and more recent databases developed by the Exome Aggregation Consortium (ExAC) or the Genome Aggregation Database (gnomAD) consortium63. Together, these projects and consortia have greatly expanded the number of populations for which allele frequencies have been characterized. These comprehensive catalogues of genomic variation highlight the value of going beyond socially and politically defined racial and ethnic groups64 and demonstrate how alleles differ within and between populations. They also show that existing population labels can be arbitrary and inexact owing to gradients and admixture among groups. For example, although genome sequence data in the 1000 Genomes data set can distinguish among continental populations (African, Asian, European, American), subcontinental patterns of genomic variation are also seen within each continental population6 (FIG. 1).
The foundational genomic variation resources and methods are immediately applicable to population-based association studies. Dense and diverse haplotype information is useful for imputation, a statistical approach that infers genomic variants that are not directly genotyped from nearby variants identified in appropriate reference populations65. Imputation facilitates the combination of analyses of genotype data from different studies to maximize sample size and generalizability of findings, benefits that are magnified when applying imputation to publicly accessible reference populations66–68. Data from diverse populations are also useful for replicating initial findings, identifying novel genomic variants more common to particular ancestral groups and assessing functional impact69–73. To follow up initial GWAS signals found in populations of European ancestry, trans-ethnic fine mapping of nearby genomic regions can be used to leverage differences in the degree of linkage disequilibrium among populations. This approach is particularly useful in enhancing scientific discovery when it includes individuals from admixed populations such as those of African or Hispanic ancestry, in whom shorter blocks of linkage disequilibrium may narrow the genomic region presumed to contain a true causal variant74,75.
Appropriate diversity of reference populations is also crucial in assessing the prevalence of a genomic variant of potential clinical relevance in population databases, one of several consensus criteria for inferring pathogenicity of genomic sequence variants76. A recent analysis of 6,903 exome sequences found fewer clinically actionable findings in participants of African ancestry compared with those of European ancestry. This result is inconsistent with the known greater genomic diversity in the former population and further highlights the under-representation of diverse ancestry individuals in population databases and genomic research77. A similar under-representation of Hispanic or Latino populations is also likely given that existing reference populations capture an incomplete fraction of the expected genomic variation in the broader Hispanic or Latino population78.
Although the genomic variation yet to be identified in global populations is challenging to estimate, the expansion of existing catalogues to include sequencing of genomes from more ancestrally diverse individuals is more likely to identify novel genomic variants in the rare-to-common frequency range compared with sequencing genomes from existing individuals at deeper coverage, which will likely identify novel variants that are observed only once or are very rare6. These resources need to be complemented by the use (and refinement where necessary), of computational methods, such as principal component analyses or mixed models66, that better account and adjust for diversity during analysis. These methods were once thought to be too specialized, too computationally intensive or too reliant on manual curation for routine use, but recent developments have shown that not to be the case. Instead of excluding non-European participants to focus analyses on ancestrally homogenous samples, improved analytical tools allow multiethnic data to be routinely incorporated into analyses79.
Apply knowledge to health care systems.
As genomics becomes useful in clinical care, it becomes increasingly important to study the integration of genomic medicine in all types of health delivery systems, not just those at the forefront of implementation research. This includes systems providing care in resource-limited settings, such as federally qualified health centres and rural hospitals where under-represented and underserved populations are disproportionately likely to receive their care. Lack of support or resources for genomic medicine is a challenge for providers seeking to adopt genomic medicine in these settings, which focus more heavily on addressing pressing problems than on early adoption. The IGNITE network and Meharry Medical College site within the Electronic Medical Records and Genomics (eMERGE) network are adapting genomic medicine efforts from highly specialized centres for use in low-resourced primary care settings, for example, by streamlining approaches for genetic counselling and the return of actionable genomic information or by providing genome-sequencing capacity80,81.
The provision of high quality, equitable and appropriate care to all patients in all health care settings, whether or not that care is directly related to genomics, is a societal obligation and will guard against the potential for genomic medicine to widen existing health care disparities. Where disparities exist, lack of access to established and widely accepted approaches to preventing, diagnosing and treating disease irrespective of newer genomic approaches can be a major contributor to worse outcomes. In such settings, novel genomic medicine approaches have a fairly small impact. Conversely, because most accepted medical interventions were developed and tested in European ancestry populations, genomic variation that alters treatment response and predisposes to adverse effects more commonly in non-European ancestry populations may be missed, such as in the case of G6PD-inactivating variants in African Americans that led to massive haemolysis on exposure to quinine82 or in the case of the 100-fold increased risk of carbamazepine-induced Stevens–Johnson syndrome and toxic epidermal necrolysis in carriers of HLAB*15:02, an allele found almost exclusively in persons of Asian ancestry83. With careful attention to potential between-population differences, the integration of genomic, clinical, environmental and socioeconomic data from health care systems serving diverse populations can identify examples of genomic medicine that will collectively benefit patients of all ancestral or socioeconomic backgrounds.
Increase diversity among researchers and clinicians.
Increased diversity of scientists in all disciplines and at all levels has been shown to lead to more efficient and creative approaches to addressing complex problems84,85, yet racial and ethnic minorities remain under-represented at every level of scientific and medical training86. It is also recognized that hiring research staff with similar race or ethnicity as the study participants to be recruited facilitates enrolment of diverse participants38. The NIH are committed to enhancing the diversity of its workforce87; however, under-representation is still evident at the faculty level88 and in the NIH granting process89. For example, black or African-American investigators who submitted NIH grants were 10% less likely to receive NIH funding than white investigators, even after controlling for educational background, publication record and other social and educational characteristics89. In 2001, the NHGRI established and implemented a Diversity Action Plan (DAP) to increase the number of individuals from under-represented groups in the scientific workforce trained in genomics research90. To date, research opportunities for over 1,400 participants from under-represented groups in genomics research have been supported. About 70% of the alumni of the DAP remain in a science, technology, engineering or math field91. Successful characteristics among institutions with DAPs include enhancing academic areas in which students have limited skills, providing graduate school examination preparation classes and connecting trainees with meaningful research.
Prioritizing diversity in genomic medicine
Building upon the learnt lessons described above, the NHGRI will continue to fund, conduct and encourage diversity in biomedical research to build a strong foundation for genomic medicine. However, fulfilling our obligation to bring genomics research from bench to bedside for all will require the efforts of the entire scientific community. Broader adoption of genomic medicine for all populations can begin with existing resources, such as databases of clinically relevant genomic variants, education opportunities for providers and laboratory personnel and patient access to genetic counselling (BOX 1). Opportunities to further the research agenda for genomic medicine to benefit all individuals, but with an emphasis on diverse and underserved individuals92, are evident as well. Components of this research agenda might include recruiting diverse individuals for research studies, addressing gaps in evidence for clinical utility, facilitating the integration of data from diverse populations in commonly used resources and databases, addressing laboratory-specific and provider-specific challenges, improving provider–patient approaches to communication, researching patient-centred and family-centred measures of utility and facilitating the implementation of genomic medicine in under-resourced and underserved settings.
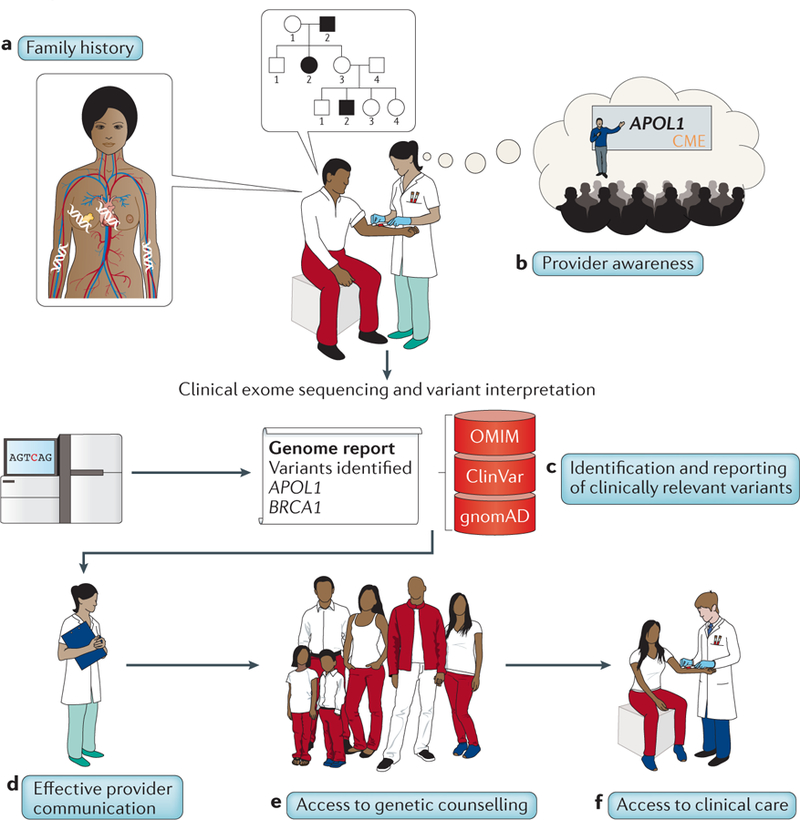
Box 1 |. Contributions of diversity to the implementation of genomic medicine: an illustrative example.
A vision of how diversity may benefit genomic medicine in the future is depicted for a hypothetical self-identified African-American individual, Mr Smith, and his primary care provider, Dr Jones. At a routine clinical visit, Dr Jones notes Mr Smith’s hypertensive status and updates Mr Smith’s family history (see figure, part a), which reveals a family history of hypertension with end-stage renal disease. The family history also reveals cases of dilated cardiomyopathy99, breast cancer100 and chronic anaemia despite negative haemoglobin (sickle cell) testing101 — events for which genomic testing could be considered. Dr Jones has recently received continuing medical education (CME) credit related to genomic medicine and is aware of a test for an APOL1 haplotype more common in individuals of African descent that increases the risk of kidney disease in patients with hypertension such as Mr Smith102 (see figure, part b). CME builds awareness of genomic medicine, including the need to integrate ancestral diversity into research and care. Dr Jones orders a clinical exome-sequencing test (the cost of which has dropped drastically compared with multiple single genomic tests), which provides information beyond APOL1 status that may be valuable in care. Mr Smith receives counselling about the risks and benefits of genomic testing with special emphasis on findings common in individuals of African ancestry but can also potentially receive additional findings in a limited set of clinically relevant variants regardless of his ancestry103. The clinical laboratory sequences and interprets Mr Smith’s DNA by use of genomic sequence resources (databases such as OMIM, ClinVar and gnomAD) from ancestrally diverse populations, enabling it to provide definitive classifications of a high proportion of variants104 and minimizing the possibility of inaccurate or uncertain results (see figure, part c). A pathogenic variant in the APOL1 gene that is disproportionately but not uniquely present in individuals of African ancestry is identified, potentially leading to more intensive hypertension management. Mr Smith also receives information about a finding in the BRCA1 gene that has implications for breast, ovarian and prostate cancer in his relatives. Dr Jones is aware of the need to consider diversity in communicating genomic results and is able to confidently and accurately communicate Mr Smith’s results (see figure, part d). Because their health care facility is focused on ensuring that patients of all ancestral and socioeconomic backgrounds have access to appropriate counselling and care, Mr Smith and his family members receive genetic counselling that is appropriate for and responsive to their needs (see figure, part e) and have access to clinical services needed for the appropriate follow-up of results. Mr Smith’s BRCA1 results prompt female relatives to undergo further testing to determine whether they carry the variant (see figure, part f).
Increased consideration of diversity in genomic medicine will likely continue to yield scientific and clinical benefit. For example, the accuracy of genomic testing will likely improve as diverse individuals are included in reference populations and participate in clinical studies of genomics-informed therapies. Research into the utility of genomic testing requires clinical research in diverse and underserved populations. For diseases in which the disease burden is disproportionately high in understudied populations, increasing participant diversity in clinical genome-sequencing studies may have greater impact. Utility will also likely improve if genomic and environmental factors are integrated into clinical decision-making regarding disease prevention or management.
Finally, the acceptability of genomic testing will likely increase as providers and laboratory personnel become more aware of the importance of understanding and accounting for diversity, thus improving their ability to interpret and return genomic results in a way that reflects patients’ genomic and sociocultural makeup.
What can be done now?
In this Perspective and elsewhere7,15, the need for greater attention to increasing the diversity of participants in human genomics research is highlighted. Although the challenges of incorporating diversity are deep-rooted, require long-term attention and extend beyond the mission of the NHGRI, much can be done in the short term.
For researchers in human genomics, increasing inclusion of research participants from diverse populations in studies and analyses is essential for providing critical information on potential differences in the impact of genomics and other factors across diverse populations. Diversity should be kept at the forefront of planning for large-scale efforts, similar to its role as a core principle of the All of Us research programme, a key element of the Precision Medicine Initiative (PMI) that aims to gather data from 1 million or more people living in the United States of America93. A focus on building respectful and collaborative partnerships with diverse and under-represented populations who will participate in studies is crucial, as is developing equitable community-engaged research that builds community trust and capacity.
For funding agencies, raising expectations for diversity in research design and diversifying the biomedical research workforce will incentivize researchers, peer reviewers, programme staff and advisers to prioritize diversity in strategic planning and funding decisions. Inviting participants and communities to advise and engage in research is ideal52. Increasing the acceptability of community participation in data-sharing plans and providing guidance for peer reviewers on the value of and reasonable expectations for inclusion of diverse participants and for community and participant engagement should also be considered.
For journal editors, stronger publication standards can emphasize attention to diversity in research design and execution. The existing recommendations from the International Committee of Medical Journal Editors94 could be broadened to require descriptions of how diverse the participants in the study were or explaining any lack of diversity.
Collaborative efforts among funding agencies are needed to ensure the inclusion of diverse populations given the broad benefits of multiethnic translational research and the need for large-scale efforts. The NHGRI currently collaborates with the National Institute for Minority Health and Health Disparities, the NHLBI, the NCI and the All of Us research effort on diversity-related programmes. We have also recently led or co-led diversity-focused sessions at national and international meetings95,96. NHGRI-led workshops have identified existing barriers that limit under-represented populations from participating in genomics research and formulated strategies to address those barriers97,24. As these efforts come to fruition, they will complement international projects on genomic medicine implementation in establishing a global knowledge base to improve human health98. Many of these efforts will include participants who are not diverse by local standards but will produce information that will be useful in regions of the world in which such locals are a minority. In continuing to advance the research agenda for genomic medicine, the NHGRI will persist in seeking long-term and sustainable collaborations with other NIH institutes and centres, other national and international funding agencies and other research programmes and organizations. Such alliances may facilitate continuity of efforts in times of scarce resources and promote strategic planning in areas of mutual interest.
Conclusions
The long-standing efforts of the NHGRI to promote diversity have yielded many lessons, but much additional work remains. We will continue to support human genomics research to benefit the health of individuals of all ancestries and backgrounds. Our education and outreach efforts will promote awareness of the importance of diverse, under-represented and underserved individuals in the entire research process. These efforts require a long-term commitment of the NHGRI , including measurable milestones to increase the ancestral diversity of the human genomics studies that it funds and a research agenda that addresses scientific questions of importance to diverse populations. In our view, issues of diversity must be raised in proposing research questions, developing and awarding funding opportunities, implementing studies and recruiting participants, engaging participants and their communities, evaluating and interpreting the results and disseminating and applying the resulting knowledge. The mission of the NHGRI is to understand the structural, functional and clinical implications of the human genome and the way that it interacts with the environment. We have a scientific and institutional commitment to include all populations, including populations that have been historically under-represented, in genomics research13. Until population diversity is recognized as a critical driver of scientific success, we will not fully realize the benefits of genomics for understanding disease and improving human health.
Key points.
Knowledge of how genomic variants vary by population increases our ability to understand genomic contributions to health and disease and to apply this knowledge to clinical care.
In addition to producing more robust science, studies involving diverse participants facilitate a more equitable distribution of resulting benefits.
Existing obstacles related to study enrolment and analysis can be overcome by rigorous attention to community engagement and analytic strategies, although this may come at the expense of expediency and convenience.
Researchers, funding agencies and journal editors have roles to play in increasing the inclusion of diverse participants and populations, prioritizing diversity-related research and raising publication standards, respectively.
Acknowledgements
The authors thank L. Brooks, A. Felsenfeld, T. Gatlin, G. Ginsburg, B. Graham, M. Hahn, G. Jarvik, D. Kaufman, R. Li, N. Lockhart, E. Madden, J. McEwen, J. Mulvihill, G. Petersen, D. Roden, L. Rodriguez, C. Rotimi, H. Sofia, J. Troyer, M. Williams and A. Wise for valuable discussion and feedback. The authors are grateful to the investigators supported by the US National Human Genome Research Institute (NHGRI) and the individuals who have participated in NHGRI-supported research for their contributions to further diversity-related efforts in genomics.
Glossary
- Allele frequency
A measure of the frequency of a particular allele relative to all alleles in a population; typically expressed as a percentage.
- Genome-wide association studies
(GWAS). An approach used to associate specific genomic variants with particular diseases by scanning the genomes from many different people and looking for genomic markers that can be used to predict the presence of a disease.
- Reference sequence
A genomic sequence representative of a particular species’ sequence, often used to align and analyse genome sequences from participants in human genomic studies.
- Population stratification
Differences in allele frequencies between cases and controls due to systematic differences in ancestry rather than association of genes with disease.
- Pathogenic
Pathogenicity classification for a genomic alteration that increases an individual’s susceptibility or predisposition to a certain disease or disorder.
- Haplotype structure
A pattern or block-like structure comprising a set of DNA variations, or polymorphisms, that tend to be inherited together. A haplotype can refer to a combination of alleles or to a set of single nucleotide polymorphisms found on the same chromosome.
- Admixture
The interbreeding of individuals from two isolated populations; often used in the context of ancestry arising from two or more continents of origin (for example, admixed populations).
- Imputation
A statistical approach to predicting unobserved genotypes in a study population by use of known genotypes from a reference population.
- Trans-ethnic fine mapping
An approach to refine initial GWAS results by leveraging differences in the degree of linkage disequilibrium among multiethnic populations, narrowing the genomic region in which a causal variant may reside.
- Linkage disequilibrium
The nonrandom association of alleles at different loci; a sensitive indicator of the population genetic forces that structure a genome.
- Secondary findings
Genomic test results that do not pertain to the primary diagnostic question or reason for testing; also referred to as incidental or additional findings.
Author biographies
Lucia A. Hindorff is an epidemiologist and Program Director in the Division of Genomic Medicine at the National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, USA. Her scientific interests include the integration of genetic tests into clinical care, enhancing diversity in genomic studies and practical issues related to large epidemiological studies.
Vence L. Bonham Jr is an associate investigator within the Social and Behavioral Research Branch of the Division of Intramural Research at the National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, USA, and leads the Health Disparities Genomics Unit there . His research focuses primarily on the social influences of new genomic knowledge, particularly in communities of colour.
Lawrence C. Brody is Head of the Molecular Pathogenesis Section at the National Human Genome Research Institute (NHGRI), National Institutes of Health, Bethesda, Maryland, USA, where his lab studies genetic mutations that lead to perturbations in normal metabolic pathways. As Director of the Division of Genomics and Society, he also oversees NHGRI matters related to the ethical, legal and social implications of genomic research.
Margaret E.C. Ginoza is a scientific programme analyst at the National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, USA, in the Divisions of Genomic Medicine and Genomes and Society. Her interests include the ethical and social implications of science, technology and medicine.
Carolyn M. Hutter is the acting Division Director for the Division of Genome Sciences at the National Human Genome Research Institute (NHGRI), National Institutes of Health, Bethesda, Maryland, USA,. She is responsible for NHGRI matters related to the structure and function of genomes in health and disease and is the NHGRI lead for The Cancer Genome Atlas.
Teri A. Manolio is a physician and epidemiologist and the Director of the Division of Genomic Medicine at the National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, USA,. She has a deep interest in discovering genetic changes associated with diseases by conducting biomedical research on large groups of people and leads efforts to support research translating those discoveries into diagnoses, preventive measures, treatments and prognoses of health conditions.
Eric D. Green is the Director of the National Human Genome Research Institute (NHGRI), National Institutes of Health, Bethesda, Maryland, USA, and is responsible for providing overall leadership on all institute research matters and other initiatives. His work has also included major start-to-finish involvement in the Human Genome Project and broadening the mission of the NHGRI to accelerate the application of genomics to medical care.
Footnotes
Competing interests statement
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
DATABASES
GWAS Catalog: https://www.ebi.ac.uk/gwas/
Exome Variant Server: http://evs.gs.washington.edu/EVS
Genome Aggregation Database (gnomAD): http://gnomad.broadinstitute.org/
Exome Aggregation Consortium (ExAC) Database: http://exac.broadinstitute.org/
FURTHER INFORMATION
1000 Genomes Project: https://www.genome.gov/27528684/1000-genomes-project/
The Cancer Genome Atlas (TCGA): https://cancergenome.nih.gov/
Centers for Common Disease Genomics (CCDG): https://www.genome.gov/27563570/
Clinical Sequencing Evidence-Generating Research (CSER2): https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-16–011.html
Diversity Action Plan: https://www.genome.gov/14514228/history-of-nhgris-minoritydiversity-action-plan/
DNA Polymorphism Discovery Resource: https://www.genome.gov/10001552/dna-polymorphism-discovery-resource/
Electronic Medical Records and Genomics (eMERGE): https://www.genome.gov/27540473/electronic-medical-records-and-genomics-emerge-network/
Ethical, Legal and Social Implications (ELSI): https://www.genome.gov/elsi/
Genome Reference Consortium (GRC): https://www.ncbi.nlm.nih.gov/grc
GWAS Catalog: https://www.ebi.ac.uk/gwas/
High-quality reference genomes programme: https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-15–027.html
Human Heredity and Health in Africa (H3Africa) Initiative: http://h3africa.org/
Implementing Genomics in Practice (IGNITE): https://www.genome.gov/27554264/implementing-genomics-in-practice-ignite/
The International HapMap Project (HapMap): https://www.genome.gov/10001688/international-hapmap-project/
NHGRI Community Engagement in Genomics Working Group (CEGWG): https://www.genome.gov/27568486/community-engagement-in-genomics-working-group/
Population Architecture using Genomics and Epidemiology (PAGE): http://pagestudy.org/
Roundtable on inclusion and engagement of under-represented populations in genomics: https://www.genome.gov/pages/about/nachgr/february2016agendadocuments/2015_09_16_roundtable_report_final.pdf
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Figure permission information
[FIG. 1: Adapted with permission from REF. 6, Macmillan Publishers Limited.]
Subject categories
Biological sciences / Genetics / Genomics / Medical genomics [URI /631/208/212/2301]
Biological sciences / Genetics / Clinical genetics [URI /631/208/2489] Scientific community and society [URI /706]
Biological sciences / Genetics / Genomics / Personalized medicine [URI /631/208/212/2166]
Biological sciences / Genetics / Sequencing / Next-generation sequencing [URI /631/208/514/2254]
Biological sciences / Genetics / Population genetics [URI /631/208/457]
References
- 1.MacArthur J et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewey FE et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354, aaf6814 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Chaisson MJ, Wilson RK & Eichler EE Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet 16, 627–640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalli-Sforza LL, Menozzi P & Piazza A The History and Geography of Human Genes (Princeton Univ. Press, 1994). [Google Scholar]
- 6.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popejoy AB & Fullerton SM Genomics is failing on diversity. Nature 538, 161–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.United Nations Department of Economic and Social Affairs. World population prospects: the 2015 revision (UN, 2015). [Google Scholar]
- 9.Collins FS & Manolio TA Merging and emerging cohorts: necessary but not sufficient. Nature 445, 259 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Knowler WC, Williams RC, Pettitt DJ & Steinberg AG Gm3;5,13,14 and type 2 diabetes mellitus: an association in American Indians with genetic admixture. Am. J. Hum. Genet 43, 520–526 (1988). [PMC free article] [PubMed] [Google Scholar]
- 11.[No authors listed.] After Havasupai litigation, Native Americans wary of genetic research. Am. J. Med. Genet 152A, fm ix (2010). [DOI] [PubMed] [Google Scholar]
- 12.Khoury MJ et al. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet. Med 9, 665–674 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Green ED, Guyer MS & National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature 470, 204–213 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Manolio TA et al. Bedside back to bench: building bridges between basic and clinical genomic research. Cell 169, 6–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentley AR, Callier S & Rotimi CN Diversity and inclusion in genomic research: why the uneven progress? J. Commun. Genet 8, 255–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen J et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet 37, 161–165 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Manrai AK et al. Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med 375, 655–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Institutes of Health. Population Architecture Using Genomics and Epidemiology (PAGE), phase II — study investigators (U01). National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-12-010.html (2012). [Google Scholar]
- 19.National Institutes of Health. Human Heredity and Health in Africa (H3Africa): research projects (U01). National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/rfa-files/RFA-RM-16-015.html (2016). [Google Scholar]
- 20.National institutes of Health. Centers for common disease genomics (UM1). National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-15-001.html (2014). [Google Scholar]
- 21.National Institutes of Health. Inclusion of women and minorities as participants in research involving human subjects — policy implementation page. National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/funding/women_min/women_min.htm (2016). [Google Scholar]
- 22.Banda Y et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 200, 1285–1295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Human Genome Research Institute. Genomics and health disparities lecture series. National Human Genome Research Institute http://www.genome.gov/27561525/genomics-and-health-disparities-lecture-series/ (2017). [Google Scholar]
- 24.National Human Genome Research Institute & National Institute of Minority Health and Health Disparities. Workshop on the use of race and ethnicity in genomics and biomedical research (Rockville, 2016). [Google Scholar]
- 25.Kahn J Misreading race and genomics after BiDil. Nat. Genet 37, 655–656 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Sankar P et al. Genetic research and health disparities. JAMA 291, 2985–2989 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shields AE et al. The use of race variables in genetic studies of complex traits and the goal of reducing health disparities: a transdisciplinary perspective. Am. Psychol 60, 77–103 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Rotimi C, Shriner D & Adeyemo A Genome science and health disparities: a growing success story? Genome Med 5, 61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West KM, Blacksher E & Burke W Genomics, health disparities, and missed opportunities for the nation’s research agenda. JAMA 317, 1831–1832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt LM, Truesdell ND & Kreiner MJ Genes, race, and culture in clinical care: racial profiling in the management of chronic illness. Med. Anthropol. Q 27, 253–271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bustamante CD, Burchard EG & De la Vega FM Genomics for the world. Nature 475, 163–165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adler NE, Glymour MM & Fielding J Addressing social determinants of health and health inequalities. JAMA 316, 1641–1642 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Braveman P & Gottlieb L The social determinants of health: it’s time to consider the causes of the causes. Public Health Rep 129 (Suppl. 2), 19–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams DR, Mohammed SA, Leavell J & Collins C Race, socioeconomic status, and health: complexities, ongoing challenges, and research opportunities. Ann. NY Acad. Sci 1186, 69–101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huo D et al. Comparison of breast cancer molecular features and survival by African and European ancestry in the Cancer Genome Atlas. JAMA Oncol 10.1001/jamaoncol.2017.0595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International Cancer Genome Consortium for Medicine. Linkinggenomics to clinical information. International Cancer Genome Consortium for Medicine https://icgcmed.org/ (2017). [Google Scholar]
- 37.National Cancer Institute. Early onset malignancies initiative. National Cancer Institute https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/early-onset-disparities (2017). [Google Scholar]
- 38.George S, Duran N & Norris K A systematic review of barriers and facilitators to minority research participation among African Americans, Latinos, Asian Americans, and Pacific Islanders. Am. J. Public Health 104, e16–e31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yancey AK, Ortega AN & Kumanyika SK Effective recruitment and retention of minority research participants. Annu. Rev. Public Health 27, 1–28 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Akinleye I et al. Differences between African American and White research volunteers in their attitudes, beliefs and knowledge regarding genetic testing for Alzheimer’s disease. J. Genet. Couns 20, 650–659 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Thomas D Gene—environment-wide association studies: emerging approaches. Nat. Rev. Genet 11, 259–272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Institutes of Health. Diversity Matters. National Institutes of Health: Diversity in Extramural Programs https://extramural-diversity.nih.gov/diversity-matters (2017). [Google Scholar]
- 44.International HapMap Consortium. Integrating ethics and science in the International HapMap Project. Nat. Rev. Genet 5, 467–475 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Institutes of Health. The Electronic Medical Records and Genomics (eMERGE) network, phase III – study investigators (U01). National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-14-025.html (2014). [Google Scholar]
- 46.National Institutes of Health. Genomic sequencing and newborn screening disorders (U19). National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/rfa-files/RFA-HD-13-010.html (2012). [Google Scholar]
- 47.National Institutes of Health. Clinical sequencing exploratory research (UM1). National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-12-009.html (2012). [Google Scholar]
- 48.National Institute of Minority Health and Health Disparities. Transdisciplinary Collaborative Centers for Health Disparities Research Program (TCC). National Institute of Minority Health and Health Disparities https://www.nimhd.nih.gov/programs/extramural/transdisciplinary-collab-centers.html#precision-research (2017). [Google Scholar]
- 49.Sperber NR et al. Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med. Genom 10, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horowitz CR et al. Race, genomics and chronic disease: what patients with African ancestry have to say. J. Health Care Poor Underserved 28, 248–260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terry SF The study is open: Participants are now recruiting investigators. Sci. Transl Med 9, eaaf1001 (2017). [DOI] [PubMed] [Google Scholar]
- 52.National Human Genome Research Institute. Community Engagement in Genomics Working Group. National Human Genome Research Institute https://www.genome.gov/27568486/community-engagement-in-genomics-working-group/ (2017). [Google Scholar]
- 53.Topolovec-Vranic J & Natarajan K The use of social media in recruitment for medical research studies: a scoping review. J. Med. Internet Res 18, e286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.National Center for Biotechnology Information. Database of Genotypes and Phenotypes (dbGaP). National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/gap (2017). [Google Scholar]
- 55.National Center for Biotechnology Information. ClinVar. National Center for Biotechnology Information https://www.ncbi.nlm.nih.gov/clinvar/ (2017). [Google Scholar]
- 56.Aronson SJ & Rehm HL Building the foundation for genomics in precision medicine. Nature 526, 336–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James R et al. Exploring pathways to trust: a tribal perspective on data sharing. Genet. Med 16, 820–826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CP et al. Facilitating health data sharing across diverse practices and communities. AMIA Jt. Summits Transl Sci. Proc 2010, 16–20 (2010). [PMC free article] [PubMed] [Google Scholar]
- 59.Hudson KL & Collins FS Biospecimen policy: family matters. Nature 500, 141–142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The Genome Reference Consortium. The Genome Reference Consortium. National Center for Biotechnology Information https://www.ncbi.nlm.nih.gov/grc (2017). [Google Scholar]
- 61.National Institutes of Health. High quality human and non-human primate genome sequences (U24). National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-15-027.html (2017). [Google Scholar]
- 62.NHLBI GO Exome Sequencing Project (ESP). Exome Variant Server. University of Washington http://evs.gs.washington.edu/EVS/ (2017). [Google Scholar]
- 63.Karczewski KJ et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 45, D840–D845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Institutes of Health. NIH policy on reporting race and ethnicity data: subjects in clinical research. National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/notice-files/NOT-OD-01-053.html (2001). [Google Scholar]
- 65.Howie B, Fuchsberger C, Stephens M, Marchini J & Abecasis GR Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet 44, 955–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price AL, Zaitlen NA, Reich D & Patterson N New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet 11, 459–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seldin MF, Pasaniuc B & Price AL New approaches to disease mapping in admixed populations. Nat. Rev. Genet 12, 523–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shriner D, Adeyemo A, Ramos E, Chen G & Rotimi CN Mapping of disease-associated variants in admixed populations. Genome Biol 12, 223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li YR & Keating BJ Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med 6, 91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenberg NA et al. Genome-wide association studies in diverse populations. Nat. Rev. Genet 11, 356–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Auer PL et al. Guidelines for large-scale sequence-based complex trait association studies: lessons learned from the NHLBI Exome Sequencing Project. Am. J. Hum. Genet 99, 791–801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heit JA et al. Identification of unique venous thromboembolism-susceptibility variants in African-Americans. Thromb. Haemost 117, 758–768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sangare M et al. Genetics of low spinal muscular atrophy carrier frequency in sub-Saharan Africa. Ann. Neurol 75, 525–532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steinthorsdottir V et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet 39, 770–775 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Fernandez-Rhodes L et al. Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the Population Architecture using Genomics and Epidemiology (PAGE) Study reveals evidence for multiple signals at established loci. Hum. Genet 136, 771–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richards S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amendola LM et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res 25, 305–315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moreno-Estrada A et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 344, 1280–1285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tian C, Gregersen PK & Seldin MF Accounting for ancestry: population substructure and genome-wide association studies. Hum. Mol. Genet 17, R143–R150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weitzel KW et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med. Genom 9, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.eMERGE Network. Collaborate. eMERGE Network https://emerge.mc.vanderbilt.edu/collaborate/ (2017). [Google Scholar]
- 82.Beutler E Glucose-6-phosphate dehydrogenase deficiency. Diagnosis, clinical and genetic implications. Am. J. Clin. Pathol 47, 303–311 (1967). [DOI] [PubMed] [Google Scholar]
- 83.Yip VL, Marson AG, Jorgensen AL, Pirmohamed M & Alfirevic A HLA genotype and carbamazepine-induced cutaneous adverse drug reactions: a systematic review. Clin. Pharmacol. Ther 92, 757–765 (2012). [DOI] [PubMed] [Google Scholar]
- 84.Berg J, Hrabowski F & Zerhouni E Training the workforce for 21st-century science. JAMA 316, 1675–1676 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Valantine HA & Collins FS National Institutes of Health addresses the science of diversity. Proc. Natl Acad. Sci. USA 112, 12240–12242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGee R Jr., Saran S & Krulwich TA Diversity in the biomedical research workforce: developing talent. Mt. Sinai J. Med 79, 397–411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.National Institutes of Health. Notice of NIH’s interest in diversity. National Institutes of Health: Grants & Funding https://grants.nih.gov/grants/guide/notice-files/NOT-OD-15-053.html (2017). [Google Scholar]
- 88.National Science Foundation, National Center for Science and Engineering Statistics. Women, minorities, and persons with disabilities in science and engineering (National Science Foundation, 2017). [Google Scholar]
- 89.Ginther DK et al. Race, ethnicity, and NIH research awards. Science 333, 1015–1019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.National Human Genome Research Institute. Plan for increasing the number of underrepresented minorities trained in genomics and ELSI research. National Human Genome Research Institute https://www.genome.gov/10001707/plan-for-increasing-the-number-of-underrepresented-minorities-trained-in-genomics-research/ (2008). [Google Scholar]
- 91.National Human Genome Research Institute. NHGRI’s 11th annual meeting of DAP and T32 training programs. National Human Genome Research Institute https://www.genome.gov/pages/about/irminorities/eleventhannualdapmeeting.pdf (2014). [Google Scholar]
- 92.National Human Genome Research Institute. Genomic medicine VIII: NHGRI’s genomic medicine portfolio. Executive summary. National Human Genome Research Institute https://www.genome.gov/multimedia/slides/gm8/gm8executivesummary_gm8exec_odgisj.pdf (2015). [Google Scholar]
- 93.Collins FS & Varmus H A new initiative on precision medicine. N. Engl. J. Med 372, 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.International Committee of Medical Journal Editors. Recommendations for the conduct, reporting, editing, and publication of scholarly work in medical journals. International Committee of Medical Journal Editors http://www.icmje.org/icmje-recommendations.pdf (2016). [PubMed] [Google Scholar]
- 95.Bonham V, Cornel M, Kaariainen H, Rotimi C & Robinson HM in European Society of Human Genetics 2017 Conference (Copenhagen, 2017). [Google Scholar]
- 96.Hindorff L et al. in ACMG Annual Clinical Genetics Meeting 2017 (Phoenix, 2017). [Google Scholar]
- 97.National Human Genome Research Institute. Roundtable on inclusion and engagement of underrepresented populations in genomics (NHGRI, 2015). [Google Scholar]
- 98.Manolio TA et al. Global implementation of genomic medicine: we are not alone. Sci. Transl Med 7, 290ps13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cirino AL & Ho C in GeneReviews® (eds Pagon RA et al.) (University of Washington, Seattle, 1993). [PubMed] [Google Scholar]
- 100.Petrucelli N, Daly MB & Pal T in GeneReviews® (eds Pagon RA et al.) (University of Washington, Seattle, 1993). [PubMed] [Google Scholar]
- 101.Bender MA & Douthitt Seibel G in GeneReviews® (eds Pagon RA et al.) (University of Washington, Seattle, 1993). [Google Scholar]
- 102.Nadkarni GN & Horowitz CR Genomics in CKD: is this the path forward? Adv. Chron. Kidney Dis 23, 120–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kalia SS et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med 19, 249–255 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Petrovski S & Goldstein DB Unequal representation of genetic variation across ancestry groups creates healthcare inequality in the application of precision medicine. Genome Biol 17, 157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collins FS, Brooks LD & Chakravarti AA DNA polymorphism discovery resource for research on human genetic variation. Genome Res 8, 1229–1231 (1998). [DOI] [PubMed] [Google Scholar]
- 106.Gottesman O et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med 15, 761–771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matise TC et al. The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am. J. Epidemiol 174, 849–859 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crosslin DR et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum. Genet 131, 639–652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]