Abstract
Inflammasomes are multi-protein platforms that initiate innate immunity by recruitment and activation of Caspase-1. The NLRP1B inflammasome is activated upon direct cleavage by the anthrax lethal toxin protease. However, the mechanism by which cleavage results in NLRP1B activation is unknown. Here we find that cleavage results in proteasome-mediated degradation of the N-terminal domains of NLRP1B, liberating a C-terminal fragment that is a potent Caspase-1 activator. Proteasome-mediated degradation of NLRP1B is both necessary and sufficient for NLRP1B activation. Consistent with our “functional degradation” model, we identify IpaH7.8, a Shigella flexneri ubiquitin ligase secreted effector, as an enzyme that induces NLRP1B degradation and activation. Our results provide a unified mechanism for NLRP1B activation by diverse pathogen-encoded enzymatic activities.
One-Sentence Summary:
Two distinct pathogen enzymes activate an innate immune sensor called NLRP1B by a mechanism that requires proteasome-mediated degradation of NLRP1B.
In animals, pathogens are generally recognized by germline-encoded innate immune receptors that bind directly to conserved pathogen-associated molecular patterns (PAMPs) such as bacterial lipopolysaccharide or flagellin (1). The recognition of PAMPs permits robust self-nonself discrimination, but PAMP receptors cannot readily distinguish pathogens from nonpathogens because PAMPs are found on innocuous microbes as well as pathogens. Plants also use germline-encoded receptors to detect PAMPs, but in addition, respond to infection by indirect detection of secreted pathogen enzymes called “effectors” (2). In this mode of recognition, called “effector-triggered immunity”, intracellular proteins of the nucleotide-binding domain leucine-rich repeat (NLR) superfamily sense effector-induced perturbation of host signaling pathways. Because innocuous microbes do not deliver effectors into host cells, effector-triggered immunity is inherently pathogen-specific. It has been proposed that animals may also detect pathogen-encoded activities (3–10), but there have been relatively few examples of this mode of pathogen recognition understood in molecular detail.
We sought to determine how a particular mammalian NLR protein called NLRP1 senses pathogen-encoded activities. NLRP1 is the founding member of a class of proteins that form inflammasomes (11), multi-protein platforms that initiate immune responses by recruiting and activating pro-inflammatory proteases, including Caspase-1 (CASP1) (12–14). CASP1 cleaves and activates specific cytokines (interleukin (IL)-1β and −18) and the pore-forming protein Gasdermin D, leading to a type of host cell death called pyroptosis. In certain murine strains, a specific NLRP1 family member called NLRP1B is activated via direct proteolysis of its N-terminus by the lethal factor (LF) protease secreted by Bacillus anthracis (15–18). Previous studies demonstrated that N-terminal proteolysis is sufficient to initiate NLRP1B inflammasome activation (16), but the molecular mechanisms by which proteolysis activates NLRP1B have been elusive.
Like other NLRs, NLRP1B contains a nucleotide-binding domain and leucine-rich repeats (Fig. 1A). However, NLRP1B also exhibits several unique features. First, the NLRP1B caspase activation and recruitment domain (CARD) is C-terminal instead of N-terminal, as it is in other NLRs. Second, NLRP1B is the only NLR that contains a function-to-find domain (FIIND). The FIIND constitutively undergoes an auto-proteolytic event, resulting in two separate NLRP1B polypeptides that remain noncovalently associated with each other (19–21). Mutations that abolish FIIND auto-processing block inflammasome activation (20, 21), but it remains unclear why FIIND auto-processing is essential for NLRP1B function. Lastly, proteasome inhibitors specifically block NLRP1B inflammasome activation, but do not affect other inflammasomes or inhibit LF protease, suggesting the proteasome is specifically required for activation of NLRP1B itself (22–26).
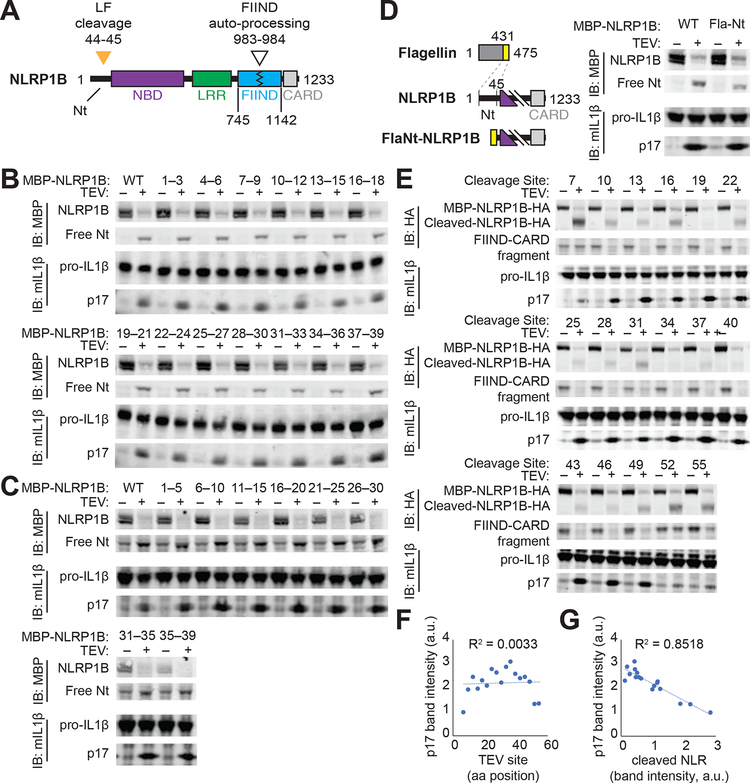
Fig. 1. The N-terminal domain of NLRP1B does not mediate auto-inhibition.
(A) Schematic of mouse NLRP1B domain architecture. Nt, N-terminus; NBD, nucleotide-binding domain; LRR, leucine-rich repeat; FIIND, function-to-find domain; CARD, caspase activation and recruitment domain. FIIND auto-processing (white triangle) is not complete and thus NLRP1B appears as a doublet [(B to D), upper blot]. Orange triangle, lethal factor (LF) cleavage site. (B, C) The indicated N-terminal amino acids of NLRP1 were mutated to alanine [(B), AAA] or glycine-serine-glycine [(C), GGSGG], and inflammasome activation was induced by co-expression of the tobacco-etch virus (TEV) protease which cleaves a TEV site engineered into the NLRP1B N-terminus. Activation was monitored by immunoblot (IB) for p17, generated upon CASP1 processing of pro-IL-1β. MBP, maltose binding protein tag. (D) The Nt of NLRP1B was replaced with a heterologous sequence from flagellin (FlaNt) and inflammasome activation was assessed as in (B). (E to G) A TEV site was positioned at the indicated positions along the NLRP1B Nt, and inflammasome activation was assessed as in (B). Cleaved NLRP1B was detected by immunoblotting for a C-terminal HA tag [(E), upper blot]. The relative intensity of cleaved IL-1β p17 was plotted relative to the position of the TEV site (F) or the relative level of cleaved NLRP1B protein (G). Gel images are representative of single experiment preformed once (B, C, E) or three times (D).
Cleavage of NLRP1B by LF results in a loss of 44 amino acids from the N-terminus of NLRP1B, an event that correlates with its activation (15–18). We and others have previously proposed an “auto-inhibition” model to explain NLRP1B activation (13, 27). In this model, the N-terminus of NLRP1B functions as an auto-inhibitory domain that is lost after cleavage by LF. The NLRP1B N-terminus may mediate auto-inhibition either through direct engagement with other NLRP1B domains in cis, or by binding to an inhibitory co-factor. A clear prediction of the auto-inhibition model is that sequences within the N-terminus should be required to prevent spontaneous inflammasome activation. To identify such sequences, we systematically mutated the NLRP1B N-terminus by replacing groups of three consecutive amino acids with alanines (Fig. 1B) or by replacing groups of five sequential amino acids with a flexible GGSGG motif (Fig. 1C). Each mutant was also engineered to contain an N-terminal TEV protease site to enable inducible NLRP1B cleavage and activation (16, 28). Inflammasome activity was monitored by CASP1-dependent processing of pro-IL-1β to p17 in a reconstituted inflammasome system in transfected 293T cells (16, 28). To our surprise, none of the mutants was auto-active, even though all showed full activity upon cleavage. To test if any N-terminal sequence could mediate auto-inhibition, we replaced the entire N-terminus with a heterologous alpha-helical domain from bacterial flagellin. Again, surprisingly, the hybrid Fla–NLRP1B protein was not auto-active, but was still functional after N-terminal cleavage (Fig. 1D). Together with prior experiments (20, 28, 29), these results led us to reconsider a model in which specific N-terminal sequences of NLRP1B mediate auto-inhibition.
We then asked whether the precise site of N-terminal proteolysis is a major determinant of NLRP1B activation. We generated a series of NLRP1B variants in which a TEV protease cleavage site was positioned at regular intervals from the N-terminus. We found that cleavage of as few as ten amino acids from the N-terminus was sufficient to activate NLRP1B. Furthermore, there was no significant correlation between the position of TEV cleavage and NLRP1B activity (Fig. 1E, F). In contrast, there was a striking negative correlation between the amount of TEV-cleaved NLRP1B protein and inflammasome activation (Fig. 1E, G). This correlation, as well as prior evidence that proteasome inhibitors block NLRP1B activity (22–26), led us to hypothesize that proteasome-mediated degradation of NLRP1B is an important step in its activation. Indeed, the proteasome inhibitors MG132 and Bortezomib not only abrogated NLRP1B activation in our reconstituted 293T system, but also prevented loss of the cleaved NLRP1B protein after LF treatment (Fig. 2A). By contrast, the inhibition of p97/VCP by NMS-873 had no effect on NLRP1B activation.
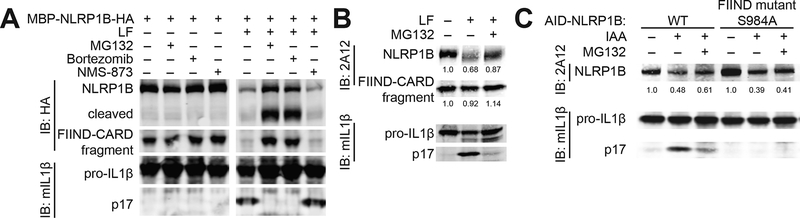
Fig. 2. Degradation of NLRP1B is necessary and sufficient for NLRP1B inflammasome activation.
(A) 293T cells transfected with constructs encoding NLRP1B, CASP1 and pro-IL-1β were assayed for inflammasome activation as in Fig. 1 in the presence (+) or absence (−) of proteasome inhibitors (MG132 (10 μM) and Bortezomib (1 μM)) or p97/VCP inhibitor (NMS-873 (0.5 μM)). (B) Immortalized 129 bone-marrow macrophages were treated with lethal factor (LF) to activate the NLRP1B inflammasome ± MG132. Endogenous NLRP1B was detected by immunoblot (IB) with 2A12. Relative band intensities are indicated. (C) The plant auxin-interacting degron (AID) was fused to the N-terminus of indicated GFP-NLRP1B variants. Specific degradation was induced with indole-3-acetic acid (IAA) in TIR1-expressing 293T cells. Inflammasome activation was assessed by immunoblot (IB) for IL-1β p17. Relative band intensities are indicated. Gel images are representative of experiments performed at least three times. S, serine. A, alanine.
To determine if endogenous NLRP1 is also lost from cells after cleavage, we derived a monoclonal antibody (2A12) against the C-terminal CARD domain of NLRP1B (Fig. S1). Using this antibody, we could track endogenous NLRP1B in 129S1/SvimJ (129) bone marrow-derived macrophages (BMMs) after treatment with LF (Fig. 2B). As in reconstituted 293T cells, LF treatment led to a loss of full-length NLRP1B, which was at least partially reversed by MG132 treatment. In contrast, proteasome inhibitors have no effect on NLRP3 (23, 26) or NAIP5/NLRC4 inflammasome activation (Fig. S2), consistent with the hypothesis that the proteasome functions uniquely in NLRP1B inflammasome activation.
A limitation of our study is that the precise mechanism by which NLRP1B is targeted to the proteasome after LF cleavage remains unknown. However, a protein quality control pathway known as the N-end rule pathway ubiquitylates cleaved proteins (30, 31), resulting in their proteasomal degradation, and N-end rule inhibitors block NLRP1B activation (25). Therefore, although our assays were not sensitive enough to detect NLRP1B ubiquitylation after LF cleavage, we propose that cleavage of NLRP1B reveals a destabilizing neo-N-terminus, which targets NLRP1B for ubiquitylation by N-end rule E3 ubiquitin ligases. Consistent with this hypothesis, in independent and parallel work, the Bachovchin laboratory has identified N-end rule pathway components that mediate NLRP1B ubiquitylation and are critical for LF-mediated NLRP1B activation [Chui, A.J. et al (2019) Science]. We note that the TEV-cleavable NLRP1B variants that we examined generate a glycine at the neo-N-terminus after TEV cleavage. According to the N-end rule, glycine is a stabilizing N-terminal amino acid (32), a prediction apparently at odds with our observation that TEV cleavage results in NLRP1B destabilization (Fig. 1). Importantly, however, aminopeptidase inhibitors block the LF-mediated killing of RAW264.7 cells (25). Thus, it appears likely that the neo-N-terminus generated by primary cleavage is further processed by aminopeptidases, resulting in the exposure of otherwise internal amino acids to N-end rule recognition. When we swapped the P2′ residues between two differentially activated TEV-cleavable NLRP1B variants, we found that the P2′ residue could also modulate activity (Fig. S3). Thus, the stability and activity of cleaved NLRP1B depends on more than just the identity of the neo-N-terminal amino acid, consistent with a growing body of evidence that multiple determinants underlie N-end rule degradation (33).
In the above experiments, the inhibition of NLRP1B activation by proteasome inhibitors may have been due to stabilization of a negative regulator of NLRP1B rather than to stabilization of NLRP1B itself. Therefore, we next asked whether specific degradation of NLRP1B is sufficient to induce its activation. To achieve selective degradation of NLRP1B, the auxin-interacting degron (AID) (34, 35) was fused to the N-terminus of NLRP1B. Upon addition of the auxin hormone indole-3-acetic acid (IAA), AID recruits a co-expressed TIR1 E3 ligase that specifically ubiquitylates AID-fusion proteins, targeting them to the proteasome. Indeed, IAA induced rapid degradation of the AID–NLRP1B fusion protein and stimulated robust IL-1β processing (Fig. 2C). Notably, FIIND auto-processing was also required for IAA-induced activation of AID–NLRP1B (Fig. 2C). Thus, the proteasomal degradation of NLRP1B itself is both necessary and sufficient for NLRP1B inflammasome activation.
To explain the seemingly paradoxical observation that NLRP1B degradation leads to its activation, we propose the following “functional degradation” model (Fig. 3A). This proposal relies on the prior observation that FIIND auto-processing is required for activation of NLRP1B. The FIIND domain comprises two separate sub-domains, termed ZU5 and UPA, with auto-processing occurring near the C-terminal end of the ZU5 domain (at F983|S984) (19–21). After auto-processing, the C-terminal fragment of NLRP1B consists of an UPA domain fused to the CARD that is required for CASP1 recruitment and activation. Prior to activation, the FIIND(UPA)–CARD fragment is non-covalently associated with the rest of NLRP1B. After cleavage by LF, NLRP1B is targeted to the proteasome, a processive protease that degrades polypeptides by feeding them through a central barrel (36). Critically, however, directional (N-to-C-terminus) and processive degradation of NLRP1B by the proteasome would be interrupted by the covalent break within the auto-processed FIIND domain. We thus propose that the C-terminal FIIND(UPA)–CARD fragment is released and is able to seed inflammasome assembly (Fig. 3A).
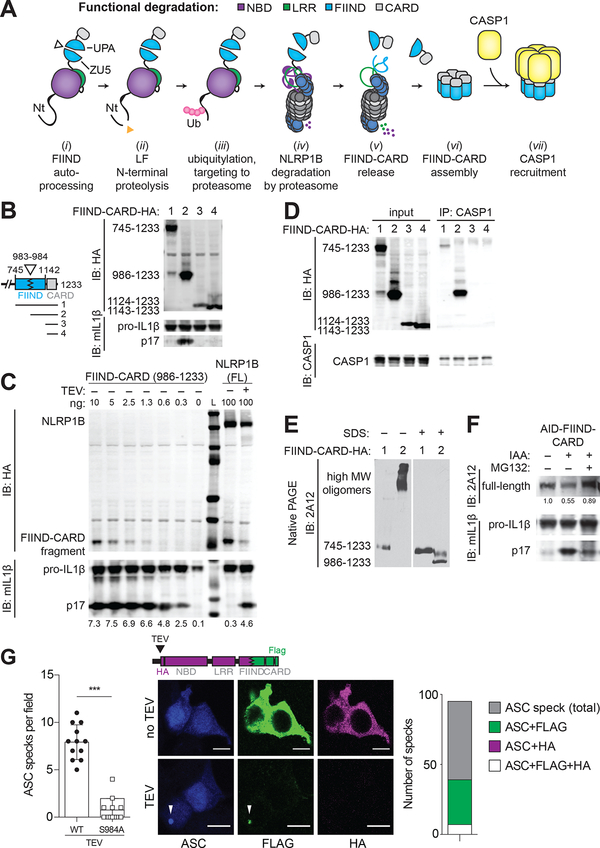
Fig. 3. NLRP1B is activated by “functional degradation”.
(A) A model for NLRP1B activation via “functional degradation”: (i) constitutive auto-processing of the NLRP1B FIIND domain results in two non-covalently associated polypeptides: NBD–LRR–FIIND(ZU5) and FIIND(UPA)–CARD; (ii) Lethal factor (LF) protease cleavage of the NLRP1B Nt exposes a neo-Nt; (iii) N-end rule factor recognition of the neo-Nt results in ubiquitylation of NLRP1B; (iv) NLRP1B is degraded by the proteasome, resulting in (v) release of the FIIND(UPA)–CARD fragment; (vi) The FIIND(UPA)–CARD fragment self-assembles into a high molecular weight oligomer which (vii) serves as a platform for CASP1 maturation and downstream inflammasome signaling. (B) The ability of the indicated C-terminal HA-tagged expression constructs to induce inflammasome activity was tested in 293T cells as in Fig. 1. (C) 293T cells were transfected with the indicated amounts (ng) of plasmid encoding HA-tagged FIIND(UPA)–CARD fragment (left) or full-length (FL) NLRP1B (± co-transfected TEV protease) and inflammasome activation was measured as in Fig. 1. L, Ladder of protein molecular weight standards. (D) 293T cells were transfected with CASP1 and HA-tagged constructs as depicted in (B). Lysates were immunoprecipitated (IP) with anti-CASP1 and immunoblotted (IB) as indicated. (E) 293T cells were transfected with CASP1 and HA-tagged constructs as depicted in (B) and analyzed by native PAGE and immunoblot with anti-NLRP1B antibody (2A12). Proteins were native or denatured with sodium dodecyl sulfate (SDS) as indicated. (F) 293T cells were transfected with an expression construct encoding AID-FIIND(ZU5+UPA)–CARD and treated with IAA or MG132 as indicated. Relative band intensities are shown. Gel images are representative of experiments performed at least three times for (B, D, E, F) or a single (C) experiment is shown. (G) 293T cells were transfected with expression constructs for ASC (blue) and TEV-cleavable NLRP1B encoding a C-terminal FLAG (green) tag and HA (magenta) tag (inserted after the TEV site, as shown). The number of ASC specks per field (+/− SD) was quantified for TEV-treated samples, compared to cells expressing a S984A FIIND auto-processing NLRP1B mutant. Representative images depict cytosolic FLAG and HA signal in untreated samples, with FLAG colocalization with ASC specks (white arrows) and concomitant loss of the HA signal in TEV-expressing cells. The total number of ASC specks, or ASC specks positive for both FLAG and HA (n=7) or only FLAG (n=32) or HA (n=0) in TEV-treated samples is quantified from 12 fields from three independent experiments. Scale bar, 10 microns. Significance was determined by student’s t-test ***, P < 0.001.
Our new “functional degradation” model of NLRP1B inflammasome activation has several virtues. First, it explains how N-terminal cleavage results in proteasome-dependent NLRP1B activation without a requirement for specific N-terminal “auto-inhibitory” sequences. Second, the model accounts for why the NLRP1B CARD is C-terminal, rather than N-terminal, as only the C-terminus of NLRP1B remains after proteasome-mediated degradation. Lastly, the model explains why FIIND domain auto-processing is required for NLRP1B activity: an unprocessed FIIND mutant would be fully degraded and would not release a C-terminal CARD-containing fragment.
A strong prediction of the “functional degradation” model is that the C-terminal FIIND(UPA)–CARD fragment possesses inflammasome activity. Consistent with prior work (21, 37), we found that the FIIND(UPA)–CARD fragment was indeed sufficient to promote robust CASP1 activity in our 293T reconstituted inflammasome assay, whereas the full-length FIIND(ZU5+UPA)–CARD appeared inactive despite auto-processing (Fig. 3B). Likewise, the isolated CARD domain (lacking any portion of the FIIND) also appeared inactive, implying that the FIIND(UPA) domain contributes to inflammasome formation. This pattern was observed across a range of expression levels (Fig. S4). Importantly, the FIIND(UPA)–CARD fragment appears to be a highly potent activator of inflammatory signaling. By titrating the amount of expression construct, we found that the FIIND(UPA)–CARD fragment appeared to be up to ~150 times more potent than TEV-cleavable full-length NLRP1B (Fig. 3C). These results imply that only a tiny fraction of the total NLRP1B in a cell may need to be degraded to liberate sufficient amounts of the FIIND(UPA)–CARD fragment for robust inflammasome activation. Thus, even if NLRP1B is only degraded a fraction of the time in the “productive” N-to-C-terminal direction, this would still likely be sufficient for robust inflammasome activation.
The “functional degradation” model predicts that the mature/assembled NLRP1B inflammasome consists of the FIIND(UPA)–CARD fragment and CASP1. In support of this hypothesis, only the FIIND(UPA)–CARD fragment co-immunoprecipitated robustly with CASP1 (Fig. 3D) and assembled into higher-order oligomers when assessed via non-denaturing PAGE (Fig. 3E). These results indicate that the NBD and LRR domains of NLRP1B are dispensable for inflammasome activation. Indeed, IAA-induced degradation of an AID–FIIND(ZU5+UPA)–CARD fragment is sufficient to induce CASP1 activity (Fig. 3F). Thus, the FIIND–CARD module appears to be sufficient to impart both auto-inhibition and proteasome-induced activation to NLRP1B. Prior studies found that NBD and LRR mutants of NLRP1B are constitutively active (27, 28, 37, 38), a result that could be explained by functional degradation if the mutations destabilize NLRP1B. Moreover, Although the role of the NBD and LRRs of NLRP1B remains to be determined, we hypothesize that these domains may contribute to recognition of pathogen-encoded effectors.
To visualize the simultaneous degradation of the N-terminal domains and the release of the FIIND(UPA)–CARD fragment upon NLRP1B activation, we engineered a variant of NLRP1B with a C-terminal FLAG tag and an HA-tag following the N-terminal TEV cleavage site. In unstimulated cells expressing this variant, FLAG and HA co-stained within the cytosol (Fig. 3G). In contrast, cells co-transfected with TEV protease showed inflammasome activation as indicated by formation of “specks” containing the ASC adaptor protein. Importantly, we found that the FLAG-tagged UPA–CARD fragment, but not the HA-tagged N-terminal domains, robustly formed puncta co-localized at the ASC speck (Fig. 3G). Moreover, we also observed a near complete loss of the HA signal, consistent with N-terminal degradation upon TEV cleavage. The FLAG signal was also lost upon TEV cleavage when FIIND processing was disrupted (Fig. S5). Taken together, these results indicate that the C-terminal UPA–CARD is released to seed inflammasome formation upon proteolytic cleavage and the subsequent proteasomal degradation of the NLRP1B N-terminal domains.
A major implication of our “functional degradation” model is that in addition to detecting pathogen-encoded proteases such as LF, NLRP1B can also potentially sense any enzymatic activity that results in NLRP1B degradation. Several pathogens encode E3 ubiquitin ligases that promote virulence through degradation of host target proteins (39). We therefore tested whether the type III secretion system (T3SS)-secreted IpaH family of E3 ubiquitin ligases, encoded by the intracellular bacterial pathogen Shigella flexneri (40–42), are detected by NLRP1B. Using our reconstituted 293T cell system, we found that IpaH7.8, but not IpaH1.4, 4.5 or 9.8, markedly reduced NLRP1B protein levels and induced IL-1β processing in an NLRP1B-dependent manner (Fig. 4A). IpaH4.5 also reduced IL-1β levels in cells, but we did not pursue this observation. IpaH7.8 selectively activated the 129 but not the C57BL/6 (B6) allele of NLRP1B (Fig. 4B). As expected, FIIND auto-processing was required for IpaH7.8-induced NLRP1B activation (Fig. 4C). Truncation of either the IpaH7.8 LRR or E3 domains, as well as mutation of the catalytic cysteine residue (CA) required for E3 ligase activity, also abolished IpaH7.8-mediated inflammasome activation (Fig. 4D). We observed that IpaH7.8, but not IpaH9.8 or an IpaH7.8 catalytic mutant (IpaH7.8CA), was able to directly ubiquitylate the 129 allele, but not the B6 allele, of NLRP1B (Fig. 4E) in a reconstituted ubiquitylation assay. These results show that IpaH7.8/ubiquitylation-dependent degradation of NLRP1B can result in its activation.
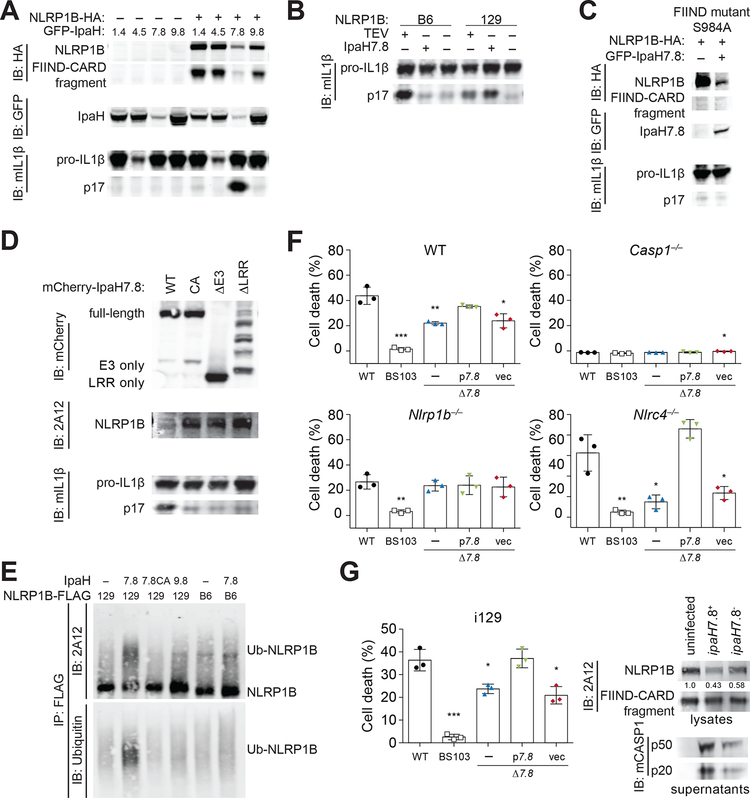
Fig. 4. The secreted Shigella flexneri IpaH7.8 E3 ubiquitin ligase activates NLRP1B.
(A, B) 293T cells were transfected with expression constructs for the 129 (A, B) or B6 (B) alleles of NLRP1B, plus CASP1, pro-IL-1β, and GFP-tagged IpaH1.4, 4.5, 7.8 or 9.8. NLRP1B expression and inflammasome activation was assessed as in Fig. 1. (C) As in (A), but cells were transfected with a serine (S) 984 to alanine (A) mutant of NLRP1B. (D) As in (A) but cells were transfected with expression constructs for mutant IpaH7.8: CA, catalytic mutant; ΔE3, deletion of Ub ligase domain; ΔLRR, deletion of LRR. (E) An in vitro ubiquitylation assay (see Methods) was used to assess the ability of IpaH7.8, IpaH7.8 catalytic mutant (7.8CA) or IpaH9.8 to ubiquitylate the 129 or B6 alleles of NLRP1B-FLAG. Reactions were immunopreciptiated with anti-FLAG before immunoblotting with anti-ubiquitin or with anti-NLRP1B (2A12). Images are representative of at least three independent experiments (C, D, E), except for (A, B), which were performed twice. (F) WT, Casp1–/–, Nlrp1b–/– or Nlrc4–/– RAW264.7 cells were infected (MOI 30) with WT Shigella flexneri strain 2457T (black circle), BS103 (virulence plasmid-cured, white box), Δ7.8 (ipaH7.8 deletion, blue triangle), p7.8 (Δ7.8 strain complemented with pCMD136 ipaH7.8, green inverted triangle), or vec (Δ7.8 strain complemented with pCMD136 empty vector, red diamond). Inflammasome-induced pyroptotic cell death was monitored by assaying for lactate dehydrogenase (LDH) activity in culture supernatants 30 minutes post-infection (+/− SD). (G) Immortalized 129 (i129) bone-marrow-derived macrophages were infected with S. flexneri strains as in (F). Cell lysates were immunoblotted with anti-NLRP1B (2A12) or anti-mouse CASP1, or cell death was measured by LDH as in (F) 2 hours post-infection (+/− SD). Data in (F, G) is representative of at least three independent experiments. Data sets were analyzed using one-way ANOVA. P-values were determined by Dunnet’s multiple comparison post-hoc test. *, P < 0.05; ** P < 0.01; ***P < 0.001.
S. flexneri robustly activates multiple inflammasomes (43) and can cause macrophage cell death in an IpaH7.8- and NLRP1B-dependent manner (44–46). However, a connection between IpaH7.8 and NLRP1B has not been established. Consistent with prior studies (44, 46), we found that wild-type S. flexneri induces robust LDH release from infected RAW264.7 macrophages, which is reduced in infections with a ΔipaH7.8 mutant (Fig. 4F). Cell killing by the ΔipaH7.8 strain was complemented with a plasmid expressing ipaH7.8. IpaH7.8-dependent cell death was markedly reduced in cells lacking CASP1 or NLRP1B (Fig. 4F) (47). The NLRC4 inflammasome also recognizes S. flexneri (48–50). In Nlrc4–/– RAW cells, inflammasome activation was almost entirely IpaH7.8-dependent (Fig. 4F). Although the N-end rule ubiquitin ligase Ubr2 is required for LF-mediated NLRP1B activation (Chui, A.J. et al (2019) Science, this issue), as expected, this requirement was circumvented by the direct ubiquitylation of NLRP1B by S. flexneri (Fig. S6). Immortalized 129 macrophages were also sensitive to IpaH7.8-dependent killing, which correlated with decreased levels of endogenous NLRP1B and the induction of CASP1 maturation (Fig. 4G). S. flexneri is not a natural pathogen of mice; this may be due in part to species-specific NLRP1B effector recognition, as human NLRP1 does not appear to detect IpaH7.8 (Fig. S6D). Thus, a mechanistic understanding of NLRP1B has led us to identify ubiquitin ligases as an additional category of pathogen-encoded enzymes that activates NLRP1B.
Prior work in Arabidopsis has shown that an NLR called RPS5 detects proteolytic cleavage of the host PBS1 kinase by the translocated Pseudomonas syringae effector AvrPphB (51). In this system, RPS5 appears to detect the cleavage products of PBS1 (52). Although NLRP1B also appears to detect a pathogen-encoded protease, our results suggest that the underlying mechanism is very different than that of RPS5. Instead of proteolysis generating a specific ligand, it appears that NLRP1B is itself the target of proteolysis, leading to its proteasomal degradation, and release of a functional inflammasome fragment. Thus, NLRP1B is in essence a sensor of its own stability, permitting detection of diverse pathogen-encoded enzymes, potentially including those of viruses or parasites (53–55). Although IpaH7.8 and LF protease both activate NLRP1B, we favor a scenario in which the “intended” targets of these pathogen-encoded enzymes are other host proteins, and that NLRP1B has evolved as a decoy target of these proteins. Although decoy sensors are widely deployed in plant immunity (56), such sensors have not been previously described in animals.
NLRP1 was the first protein shown to form an inflammasome (11). Our results provide a long-sought mechanism that explains how NLRP1 is activated. Our proposed mechanism may also apply to other FIIND-death domain fold containing proteins, including PIDD1 and CARD8 (19). In addition to explaining how NLRP1 senses pathogens, “functional degradation” likely provides an explanation for why naturally occurring mutations that destabilize the N-terminal pyrin domain in human NLRP1 (27) also result in its activation. It has previously been suggested that NLRP1B is activated upon ATP depletion in cells (38, 45). Our results are not incongruous with this model; indeed, ATP depletion may also indirectly affect NLRP1B stability. Thus, our results lay a foundation for identifying pathogen-encoded activators of human NLRP1 and provide a conceptual basis for designing therapeutic interventions targeting NLRP1.
Supplementary Material
Acknowledgments:
We are grateful to J. Chavarría-Smith for discussions and for laying the experimental foundations for this work. We thank P.R. Beatty and UC Berkeley undergraduates in the MCB150L course for help in generating the 2A12 monoclonal, the Rape Lab for guidance with ubiquitylation assays, J. Mogridge for the AKR/J Nlrp1b allele construct (21), A. Holland for the AID and TIR1 constructs, the Bachovchin Lab for the HEK and RAW cell lines and for sharing results prior to submission, G. Barton, J. Chavarría-Smith, H. Darwin, M. Dorrington, and J. Tenthorey for comments on the manuscript, and members of the Vance and Barton Labs for discussions.
Funding: R.E.V. is an HHMI Investigator and is supported by NIH AI075039 and AI063302; P.S.M. is supported by a Jane Coffin Childs Memorial Fund postdoctoral fellowship. C.F.L. is a Brit d’Arbeloff MGH Research Scholar and supported by NIH AI064285.
Footnotes
Competing interests: A patent related to this work has been submitted by R.E.V., A.S. and P.S.M.; and R.E.V. is a scientific advisory board member for Metchnikoff Therapeutics, Inc.
Data and materials availability: All data are available in the main text or the supplementary materials.
References and Notes:
- 1.Janeway CA Jr., Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54 Pt 1, 1–13 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL, The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Blander JM, Sander LE, Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol 12, 215–225 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Chung LK et al. , The Yersinia Virulence Factor YopM Hijacks Host Kinases to Inhibit Type III Effector-Triggered Activation of the Pyrin Inflammasome. Cell Host Microbe 20, 296–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keestra AM et al. , Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature 496, 233–237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keestra-Gounder AM et al. , NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratner D et al. , The Yersinia pestis Effector YopM Inhibits Pyrin Inflammasome Activation. PLoS Pathog 12, e1006035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vance RE, Isberg RR, Portnoy DA, Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6, 10–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H et al. , Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237–241 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Fontana MF et al. , Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog 7, e1001289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinon F, Burns K, Tschopp J, The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Broz P, Dixit VM, Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Chavarria-Smith J, Vance RE, The NLRP1 inflammasomes. Immunol. Rev 265, 22–34 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Rathinam VA, Fitzgerald KA, Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell 165, 792–800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyden ED, Dietrich WF, Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet 38, 240–244 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Chavarria-Smith J, Vance RE, Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog 9, e1003452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellmich KA et al. , Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PLoS One 7, e49741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levinsohn JL et al. , Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog 8, e1002638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Osualdo A et al. , CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One 6, e27396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finger JN et al. , Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem 287, 25030–25037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frew BC, Joag VR, Mogridge J, Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog 8, e1002659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fink SL, Bergsbaken T, Cookson BT, Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci U S A 105, 4312–4317 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Squires RC, Muehlbauer SM, Brojatsch J, Proteasomes control caspase-1 activation in anthrax lethal toxin-mediated cell killing. J. Biol. Chem 282, 34260–34267 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Tang G, Leppla SH, Proteasome activity is required for anthrax lethal toxin to kill macrophages. Infect. Immun 67, 3055–3060 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickliffe KE, Leppla SH, Moayeri M, Killing of macrophages by anthrax lethal toxin: involvement of the N-end rule pathway. Cell. Microbiol 10, 1352–1362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickliffe KE, Leppla SH, Moayeri M, Anthrax lethal toxin-induced inflammasome formation and caspase-1 activation are late events dependent on ion fluxes and the proteasome. Cell. Microbiol 10, 332–343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong FL et al. , Germline NLRP1 Mutations Cause Skin Inflammatory and Cancer Susceptibility Syndromes via Inflammasome Activation. Cell 167, 187–202 e117 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Chavarria-Smith J, Mitchell PS, Ho AM, Daugherty MD, Vance RE, Functional and Evolutionary Analyses Identify Proteolysis as a General Mechanism for NLRP1 Inflammasome Activation. PLoS Pathog 12, e1006052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neiman-Zenevich J, Liao KC, Mogridge J, Distinct regions of NLRP1B are required to respond to anthrax lethal toxin and metabolic inhibition. Infect. Immun 82, 3697–3703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachmair A, Finley D, Varshavsky A, In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986). [DOI] [PubMed] [Google Scholar]
- 31.Lucas X, Ciulli A, Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr. Opin. Struct. Biol 44, 101–110 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Gonda DK et al. , Universality and structure of the N-end rule. J. Biol. Chem 264, 16700–16712 (1989). [PubMed] [Google Scholar]
- 33.Varshavsky A, The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland AJ, Fachinetti D, Han JS, Cleveland DW, Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci U S A 109, E3350–3357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M, An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Nyquist K, Martin A, Marching to the beat of the ring: polypeptide translocation by AAA+ proteases. Trends Biochem. Sci 39, 53–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao KC, Mogridge J, Expression of Nlrp1b inflammasome components in human fibroblasts confers susceptibility to anthrax lethal toxin. Infect. Immun 77, 4455–4462 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao KC, Mogridge J, Activation of the Nlrp1b inflammasome by reduction of cytosolic ATP. Infect. Immun 81, 570–579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maculins T, Fiskin E, Bhogaraju S, Dikic I, Bacteria-host relationship: ubiquitin ligases as weapons of invasion. Cell Res. 26, 499–510 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C, Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1, 77–83 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Singer AU et al. , Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat. Struct. Mol. Biol 15, 1293–1301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y et al. , Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat. Struct. Mol. Biol 15, 1302–1308 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Hermansson AK, Paciello I, Bernardini ML, The Orchestra and Its Maestro: Shigella’s Fine-Tuning of the Inflammasome Platforms. Curr. Top. Microbiol. Immunol 397, 91–115 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Prada CM et al. , Shigella flexneri IpaH(7.8) facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun 68, 3608–3619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neiman-Zenevich J, Stuart S, Abdel-Nour M, Girardin SE, Mogridge J, Listeria monocytogenes and Shigella flexneri Activate the NLRP1B Inflammasome. Infect. Immun 85, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki S et al. , Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proc Natl Acad Sci U S A 111, E4254–4263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okondo MC et al. , Inhibition of Dpp8/9 Activates the Nlrp1b Inflammasome. Cell Chem Biol 25, 262–267 e265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyes Ruiz VM et al. , Broad detection of bacterial type III secretion system and flagellin proteins by the human NAIP/NLRC4 inflammasome. Proc Natl Acad Sci U S A 114, 13242–13247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki S et al. , Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcdelta. PLoS Pathog 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Zhao Y, Shi J, Shao F, Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A 110, 14408–14413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao F et al. , Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230–1233 (2003). [DOI] [PubMed] [Google Scholar]
- 52.DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW, Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell. Microbiol 14, 1071–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cirelli KM et al. , Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathog 10, e1003927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ewald SE, Chavarria-Smith J, Boothroyd JC, NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect. Immun 82, 460–468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorfu G et al. , Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. MBio 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones JD, Vance RE, Dangl JL, Intracellular innate immune surveillance devices in plants and animals. Science 354, (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.