The developmental regulator TCP4 binds to and activates SAUR genes to facilitate cotyledon opening in the light, but high levels of the transcription factor PIF3 inhibit this binding in the dark.
Abstract
Light elicits different growth responses in different organs of plants. These organ-specific responses are prominently displayed during de-etiolation. While major light-responsive components and early signaling pathways in this process have been identified, this information has yet to explain how organ-specific light responses are achieved. Here, we report that members of the TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) transcription factor family participate in photomorphogenesis and facilitate light-induced cotyledon opening in Arabidopsis (Arabidopsis thaliana). Chromatin immunoprecipitation sequencing and RNA sequencing analyses indicated that TCP4 targets a number of SMALL AUXIN UPREGULATED RNA (SAUR) genes that have previously been shown to exhibit organ-specific, light-responsive expression. We demonstrate that TCP4-like transcription factors, which are predominantly expressed in the cotyledons of both light- and dark-grown seedlings, activate SAUR16 and SAUR50 expression in response to light. Light regulates the binding of TCP4 to the promoters of SAUR14, SAUR16, and SAUR50 through PHYTOCHROME-INTERACTING FACTORs (PIFs). PIF3, which accumulates in etiolated seedlings and its levels rapidly decline upon light exposure, also binds to the SAUR16 and SAUR50 promoters, while suppressing the binding of TCP4 to these promoters in the dark. Our study reveals that the interplay between light-responsive factors PIFs and the developmental regulator TCP4 determines the cotyledon-specific light regulation of SAUR16 and SAUR50, which contributes to cotyledon closure and opening before and after de-etiolation.
INTRODUCTION
When plants such as Arabidopsis (Arabidopsis thaliana) germinate in the dark in soil, they undergo etiolation (or skotomorphogenesis). Etiolation is characterized by the formation of an apical hook, closed cotyledons, a rapidly elongating hypocotyl, and lack of chlorophyll accumulation. When the apical region of the seedling emerges from the soil and perceives light, dramatic morphological and physiological changes occur (de-etiolation), including, among other responses, unfolding of the apical hook, opening of cotyledons, and inhibition of hypocotyl elongation to help the plant establish autotrophic growth (von Arnim and Deng, 1996; Chen et al., 2004; Kami et al., 2010). The transcription factors PHYTOCHROME-INTERACTING FACTORs (PIFs), ETHYLENE-INSENSITIVE3, and ELONGATED HYPOCOTYL5 (HY5) modulate overlapping aspects of these light responses (Shi et al., 2018). Among these, PIFs transcription factors play a central role in maintaining essentially all aspects of etiolation and appear to play a predominant role over other transcription factors in modulating cotyledon morphology. The loss of PIFs (pif1 pif3 pif4 pif5, or pifq) results in open cotyledons in the dark (Leivar et al., 2008; Shin et al., 2009; Sentandreu et al., 2011; Shi et al., 2018). When etiolated seedlings are exposed to light, PIF proteins undergo rapid phosphorylation and degradation, an event that is necessary for de-etiolation, including the timely opening of cotyledons (Al-Sady et al., 2006; Dong et al., 2017; Ni et al., 2017). Phytohormones such as gibberellin, brassinosteroid, and jasmonate affect etiolation and light responses largely via mechanisms involving CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and PIFs (Feng et al., 2008; Li and He, 2016; Zheng et al., 2017).
The light signaling pathways have been defined in a general sense, but the responses to light signals are highly tissue dependent: while rapid cell expansion halts in hypocotyl cells upon exposure to the light, cotyledon cells expand and differentiate (Blum et al., 1994; Wei et al., 1994; von Arnim and Deng, 1996). Light-responsive gene expression also tends to be tissue specific (Kohnen et al., 2016; Sun et al., 2016). This is exemplified by the identification of a group of SMALL AUXIN UP RNA (SAUR) genes whose transcriptional responses to light display extraordinary organ specificity (Sun et al., 2016). The SAUR gene family was initially defined as a set of auxin-inducible genes that regulate development, especially in hypocotyls (McClure and Guilfoyle, 1987; Ren and Gray, 2015), many of which act by promoting cell expansion (Chae et al., 2012; Spartz et al., 2012; Kong et al., 2013). In etiolated seedlings, SAUR14, SAUR16, and SAUR50 are induced by auxin in hypocotyls, but not in cotyledons; conversely, they can be induced by the light in cotyledons, but not in hypocotyls (Sun et al., 2016). These SAURs are direct target genes of the PIFs (Sun et al., 2016), but since PIFs are expressed in both cotyledons and hypocotyls (Zhang et al., 2013; Sun et al., 2016), PIF functions alone cannot explain how the light responsiveness of SAUR gene expression is differentially controlled in an organ-specific manner.
Members of the TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) family of plant-specific transcription factors regulate leaf morphology, branching, leaf senescence, petal development, and numerous other aspects of development as well as defense responses in Arabidopsis (Palatnik et al., 2003; Martín-Trillo and Cubas, 2010; Efroni et al., 2013; Huang and Irish, 2015). Within the CINCINNATA-like subgroup (CIN-like TCPs; Supplemental Figure 1), five TCPs, including TCP4-like genes (TCP3, TCP4, and TCP10), are targets of the microRNA miR319 (Palatnik et al., 2003). In situ hybridization data indicate that TCP4 is predominantly expressed in cotyledons and young leaves (Palatnik et al., 2003) and regulates the morphogenesis of these organs (Koyama et al., 2010). In addition, TCP4 activates auxin biosynthetic genes to promote hypocotyl elongation in light-grown seedlings (Challa et al., 2016). A few TCP genes are also involved in light responses. TCP17 might regulate shade-induced hypocotyl elongation (Zhou et al., 2018). TCP2 interacts with cryptochromes and participates in blue light–stimulated photomorphogenesis (He et al., 2016).
Here, we describe a cotyledon-specific, light-induced TCP4-SAUR16 and SAUR50 transcriptional regulatory module that facilitates light-induced cotyledon opening during the de-etiolation of seedlings. In this pathway, PIF3 confers light signal responsiveness by repressing TCP4-induced activation of the SAUR genes in the dark, while TCP4 provides cotyledon specificity. Our data support a model in which PIF3, which accumulates in the dark, inhibits the binding of TCP4 to the promoter of the SAUR16 and SAUR50 genes. Light-triggered degradation of PIF3 proteins allows TCP4 to activate the expression of these SAUR genes, which contributes to the changes in cotyledon morphology during de-etiolation.
RESULTS
TCP4-Like Genes Play a Positive Role in Light-Induced Cotyledon Opening
By analyzing previously published organ-specific and light-dependent transcriptome data (Sun et al., 2016), we found that the transcripts of the CIN-like clade of TCP genes, including TCP4-like genes (TCP3, TCP4, and TCP10), were predominantly expressed in cotyledons, but not in hypocotyls in etiolated or light-exposed seedlings (Supplemental Figure 1; Sun et al., 2016). We set out to study the potential involvement of TCPs in light-induced cotyledon development. Cotyledon opening and expansion are often mentioned together to describe changes in cotyledons during de-etiolation. However, cotyledon opening displays faster kinetics than cotyledon expansion (Supplemental Figure 2). When 3-d-old, dark-grown seedlings (wild-type Columbia [Col]) were transferred to white light, the cotyledons started to open from the tip, or the distal end, at ∼4 h after light treatment (DL4 h). The cotyledon opening process took place within 24 h after transfer to light for most seedlings, and the cotyledons were fully opened (opening angles to 180°) before the end of day 2 in the light. The expansion of cotyledons was a much slower process, taking several days (Supplemental Figure 2) or as many as 8 d in the light before the growth curve plateaued (Blum et al., 1994). The different kinetics of cotyledon opening and expansion suggests that the control mechanisms of the two light-induced processes might not be identical.
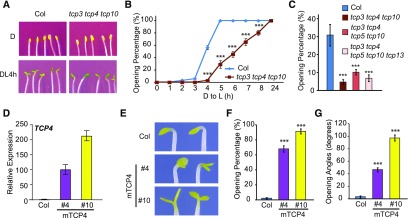
In this study, we focused specifically on light-induced cotyledon opening and examined this phenotype in tcp mutants and TCP4 overexpression lines. In the dark, tcp3 tcp4 tcp10 triple mutant seedlings exhibited closed cotyledons, similar to those of Col (Figure 1A). When scored for the percentage of seedlings with open cotyledons, we found that most cotyledons of Col seedlings opened within 5 h after being transferred to light, while tcp3 tcp4 tcp10 mutant seedlings showed slower kinetics of cotyledon opening, suggesting that these three TCP4-like genes are necessary for a timely response to light during cotyledon opening (Figures 1A and 1B). Higher order mutants of CIN-like TCPs also showed retarded cotyledon opening after light exposure, but the phenotypes were no more severe than those of tcp3 tcp4 tcp10 (Figure 1C). These results suggest that the TCP4-like genes likely have greater functional involvement in light-induced cotyledon opening than other TCP genes.
Figure 1.
TCP4-Like Genes Promote Light-Induced Cotyledon Opening.
(A) to (C) The tcp3 tcp4 tcp10 triple mutant shows delayed light-induced cotyledon opening. Four-day-old dark-grown Col and tcp3 tcp4 tcp10 seedlings were illuminated with continuous white light (at 40 µmol/m2/s). (A) Photographs of 4-d-old dark-grown seedlings (D) or after DL4 h. (B) Cotyledon opening percentages at the indicated time points after light exposure. (C) Relative cotyledon opening percentages of Col and tcp mutants at DL4 h. Data are shown as the mean ± se.
(D) to (G) Overexpression of TCP4 results in cotyledon opening in the dark. (D) The expression levels of TCP4 mRNA in 4-d-old dark-grown Col and 35S:Myc-mTCP4#4 and #10 (mTCP4 #4 and #10) seedlings were determined by RT-qPCR analysis. Data are shown as the mean ± sd of three biological replicates (see “Methods”). (E) Images of mTCP4#4 and mTCP4#10 dark-grown seedlings (4 d) showing open cotyledons. Cotyledon opening percentages (F) and cotyledon opening angles (G) of Col, mTCP4#4, and mTCP4#10 seedlings after 4 d of growth in the dark. Data are shown as the mean ± se.
Statistical analysis was performed via two-tailed Student’s t test compared with Col: ***, P < 0.001.
We examined two TCP4 overexpression lines (35S:Myc-mTCP4), mTCP4#4 and mTCP4#10, in which the Cauliflower mosaic virus 35S promoter was used to drive a mutated form of the TCP4 (mTCP4) coding sequence that is resistant to miR319-mediated downregulation (Tao et al., 2013). The mTCP4#10 line showed higher expression of TCP4 than mTCP4#4 (Figure 1D). In contrast to the closed cotyledons of Col seedlings, most mTCP4#4 and mTCP4#10 seedlings developed open cotyledons in the dark (Figures 1E and 1F). The percentage of seedlings with open cotyledons and the angles of cotyledon separation appeared to be positively correlated with the expression levels of TCP4 (Figures 1E to 1G). Taken together, these results indicate that the TCP4-like genes are not required for etiolation but are necessary for rapid, light-induced cotyledon opening during de-etiolation and that ectopic overexpression of TCP4 can cause cotyledon opening in the dark.
SAUR Genes Are Potential Targets of TCP4
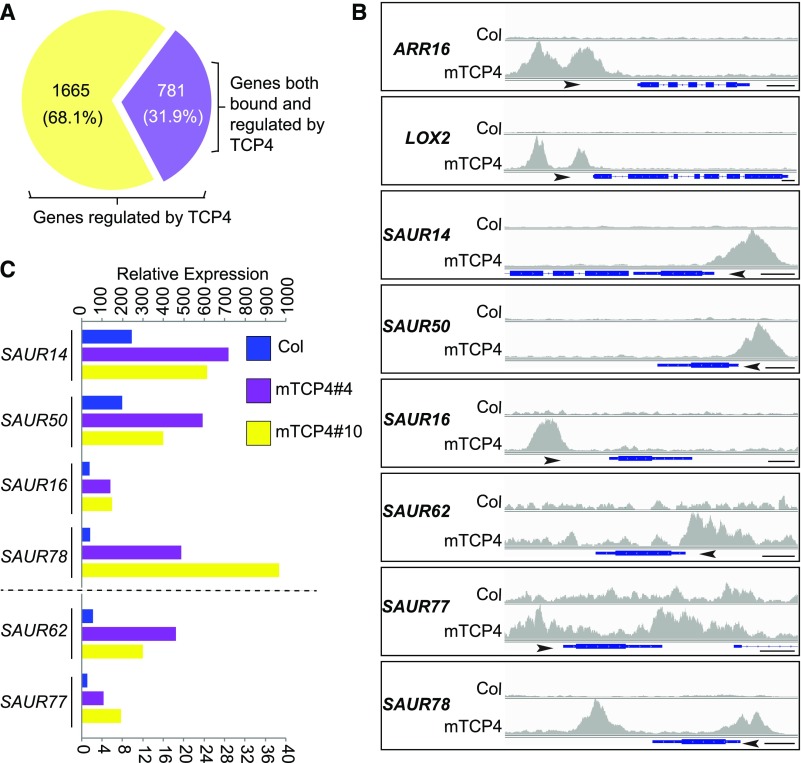
In an effort to elucidate the potential mechanism(s) by which TCP4 promotes cotyledon opening, we performed chromatin immunoprecipitation sequencing (ChIP-seq) and whole transcriptomic RNA sequencing (RNA-seq). ChIP-seq of 4-d-old, dark-grown mTCP4#10 seedlings identified ∼6000 TCP4-bound genes (Supplemental Figure 3A; Supplemental Data Set 1). Most of the binding sites of these genes were located at promoter regions (Supplemental Figure 3B), with strong enrichment of the GGACCA motif in the promoters (Supplemental Figure 3C). We performed RNA-seq analysis using cotyledon tissues of dark-grown mTCP4#4 and mTCP4#10 lines, thereby identifying 4000 and 4042 genes, respectively, that were differentially expressed compared with Col. Among these, 2446 genes were coregulated in both lines (Supplemental Figures 4A and 4B; Supplemental Data Set 2), suggesting that the expression of these genes in cotyledons is likely affected by TCP4 overexpression. Among the 2446 TCP4-regulated genes in cotyledons, 781 genes whose promoters were also bound by TCP4 in the ChIP-seq experiment were identified (Figure 2A; Supplemental Data Set 3). This group of genes includes known TCP4 targets such as ARABIDOPSIS RESPONSE REGULATOR16, LIPOXYGENASE2, CONSTANS, and YUCCA5 (highlighted in Supplemental Data Set 3; Efroni et al., 2013; Challa et al., 2016; Kubota et al., 2017; Liu et al., 2017). Also found in this group were six SAUR genes, including SAUR14, SAUR50, SAUR16, SAUR62, SAUR77, and SAUR78, whose promoters were bound by TCP4 and whose expression levels in cotyledons were elevated in dark-grown mTCP4#4 and mTCP4#10 plants (Figures 2B and 2C). The results for these SAUR genes were verified by ChIP-quantitative (q)PCR and RT-qPCR (Supplemental Figures 3D and 4C).
Figure 2.
A Group of SAURs Are Potential Targets of TCP4.
(A) Among the 2446 differentially expressed genes in mTCP4 cotyledons, 781 genes were also bound by mTCP4 in the ChIP-seq analyses.
(B) Peak graphs showing the ChIP-seq raw reads at the indicated gene loci in mTCP4 and Col samples. The arrows indicate the directions of transcription, and the blue bars indicate the transcripts of each gene. Bars = 250 bp. AAR16, ARABIDOPSIS RESPONSE REGULATOR16; LOX2, LIPOXYGENASE2.
(C) Lines mTCP4#4 and mTCP4#10 show increased expression of SAUR genes in cotyledons of dark-grown seedlings. The graph shows the total counts based on the RNA-seq data. Note that different axis labels are used for SAUR14, SAUR50, SAUR16, and SAUR78 versus SAUR62 and SAUR77.
SAUR Genes Are Required for mTCP4-Induced Cotyledon Opening in the Dark
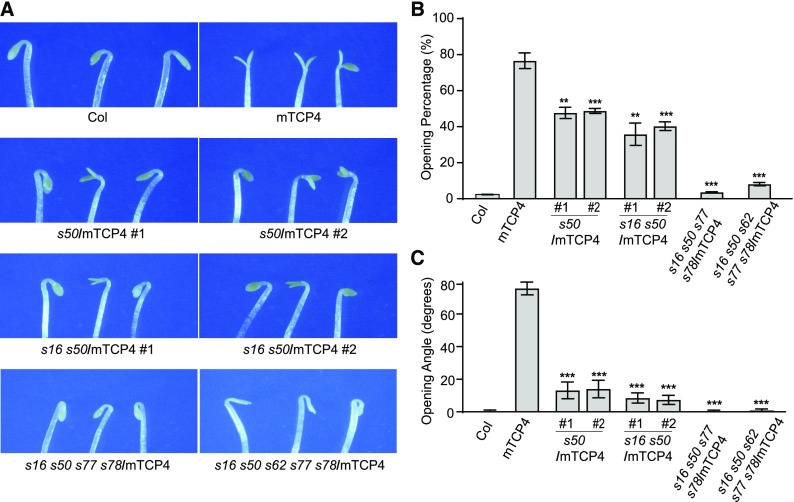
Similar to the tcp3 tcp4 tcp10 triple mutant, we found that the saur16 saur50 double mutant exhibited slower kinetics during light-induced cotyledon opening than Col (Supplemental Figure 5; Sun et al., 2016), while seedlings with high levels of SAUR50 overexpression resembled mTCP4 seedlings, with open cotyledons in the dark (Sun et al., 2016). These observations led us to hypothesize that TCP4 might activate SAUR genes to cause cotyledons to open in the dark. To investigate whether the open-cotyledon phenotype of dark-grown mTCP4 seedlings could be attributed to the increased expression of SAUR genes, we generated mutations in multiple SAUR loci in the mTCP4#10 background using a clustered regularly interspaced short palindromic repeat (CRISPR)/ CRISPR associated protein 9 (Cas9) strategy (Supplemental Figure 6A). As shown in Figure 3, the percentages of seedlings showing open cotyledons, as well as the opening angles of cotyledons in dark-grown mTCP4#10, were significantly suppressed in the saur50 single mutant and saur16 saur50 double mutant backgrounds. The cotyledon phenotypes of dark-grown mTCP4 overexpression lines were almost completely rescued in the saur16 saur50 saur77 saur78 and saur16 saur50 saur62 saur77 saur78 higher order mutants (Figure 3). These genetic data suggest that the elevated expression of SAUR genes may largely account for the premature cotyledon opening phenotype of dark-grown mTCP4.
Figure 3.
Mutations in the SAUR Genes Suppress the Premature Cotyledon Opening Phenotype of mTCP4 Plants.
(A) Photographs showing cotyledons of 4-d-old dark-grown seedlings of Col, mTCP4, saur50/mTCP4 (s50/mTCP4), saur16 saur50/mTCP4 (s16 s50/mTCP4), saur16 saur50 saur77 saur78/mTCP4 (s16 s50 s77 s78/mTCP4), and saur16 saur50 saur62 saur77 saur78/mTCP4 (s16 s50 s62 s77 s78/mTCP4).
(B) Cotyledon opening percentages of the indicated lines, showing the reduced occurrence of premature cotyledon opening in the dark when multiple indicated SAUR genes were mutated.
(C) Cotyledon opening angles of the indicated lines, showing the reduced severity of the phenotype caused by multiple SAUR gene mutations in mTCP4.
Data are shown as the mean ± se. Statistical significance was calculated using two-tailed Student’s t test against mTCP4: **, P < 0.01; ***, P < 0.001.
TCP Transcriptionally Activates SAUR50 and SAUR16 to Promote Light-Induced Cotyledon Opening
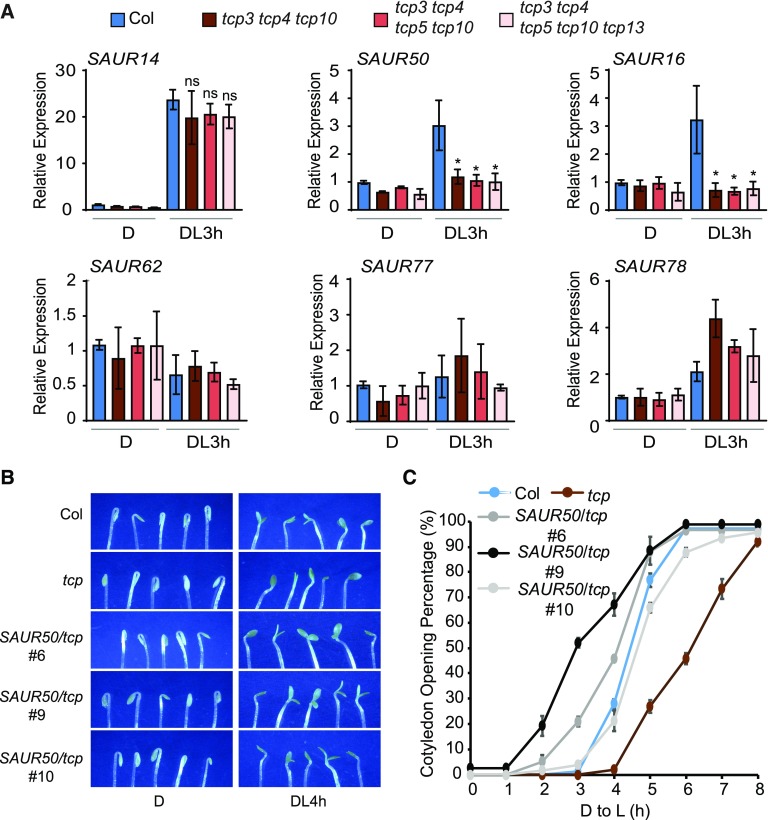
We then examined whether the expression of these SAUR genes in cotyledons is dependent on the TCPs and responsive to light. As shown in Figure 4A, the expression of SAUR14, SAUR50, and SAUR16 in cotyledons was robustly induced by 3 h of light irradiation. Remarkably, the light-induced expression of SAUR50 and SAUR16 was abolished in the tcp3 tcp4 tcp10 triple mutant (Figure 4A). The expression of other SAUR genes did not appear to be drastically affected by the tcp mutants tested. Among these was SAUR14, whose expression was light induced, but was apparently not specifically dependent on the TCP genes tested (Figure 4A). With regard to SAUR50 and SAUR16 expression in cotyledons, the data unequivocally showed that their light induction, or simply transcriptional activation, was entirely dependent on TCP4-like genes.
Figure 4.
SAUR50 and SAUR16 Are Activated by the Expression of TCP4-like Genes to Promote Cotyledon Opening.
(A) Expression of SAUR16 and SAUR50 in cotyledons is light induced and is dependent on the presence of TCP4-like genes. Expression levels of the indicated SAURs in cotyledons of Col and the indicated tcp mutants grown in the dark (D) or after light treatment for 3 h were determined by RT-qPCR. Data are shown as the mean ± sd of three biological replicates. Statistical analysis was performed using two-tailed Student’s t test against Col (DL3 h): ns, P > 0.05; *, P < 0.05. DL3 h, 3 h after light exposure.
(B) and (C) Overexpression of SAUR50 rescues the delayed cotyledon opening phenotypes of tcp3 tcp4 tcp10 (tcp) mutants during the dark-to-light transition. (B) Photographs showing cotyledons of the indicated lines taken in the dark or 4 h after light exposure. (C) Four-day-old dark-grown seedlings were illuminated with white light, and cotyledon opening percentages at the indicated time after light exposure were scored. Data are shown as the mean ± se. D, dark; D to L, dark to light.
Since the tcp3 tcp4 tcp10 mutant showed reduced expression of SAUR16 and SAUR50, we next asked whether the slow cotyledon opening phenotype of tcp3 tcp4 tcp10 could be rescued by overexpression of the SAUR genes. Assuming there might be functional redundancy between SAUR16 and SAUR50, we transgenically expressed only SAUR50 in the tcp3 tcp4 tcp10 background (Supplemental Figure 6B). We selected three lines with moderate overexpression of SAUR50 (<10-fold) compared with previously published strongly SAUR50-overexpressing transgenic lines (>20-fold) for phenotypic observation. Among these lines, very few seedlings exhibited open cotyledons in the dark, which allowed us to quantify the cotyledon opening kinetics of these lines during de-etiolation (Figure 4C). We found that SAUR50 overexpression efficiently accelerated the cotyledon opening process of tcp3 tcp4 tcp10 seedlings in all three lines (Figures 4B and 4C). Taken together, these data suggest that TCP4-like transcription factors are required for light-induced activation of SAUR50 and SAUR16 and that the failure to activate SAUR50/SAUR16 expression contributes to the slow cotyledon opening phenotypes of tcp3 tcp4 tcp10 during the dark-to-light transition.
Light Signals and Loss of PIFs Promote the Binding of TCP4 to the Promoters of the SAUR Genes
Our data have thus far indicated that TCP4 activates the transcription of SAUR16 and SAUR50 in cotyledons during de-etiolation. The key question that follows is how light signals regulate TCP4-induced activation of the SAUR genes. While SAUR16 and SAUR50 were robustly induced in response to light, the mRNA expression of TCP4 and most CIN-like TCPs was unresponsive or only mildly responsive to light, except for TCP10, according to the RT-qPCR data (Supplemental Figure 7A) and the RNA-seq data (Supplemental Figure 1A).
We monitored the effects of light on TCP4 proteins. The levels of Myc-mTCP4 proteins slightly rose after dark-grown plants were transferred to light, and their levels slightly declined when light-grown seedlings were transferred to the dark, suggesting that light enhances Myc-mTCP4 protein abundance (Supplemental Figure 7B). To test whether TCP4 protein stability is regulated by COP1 and DE-ETIOLATED1 (DET1), we introduced the Myc-mTCP4 transgene into the respective mutants by crossing. Myc-mTCP4 protein levels were unchanged in cop1-4 and det1-1 (Supplemental Figure 7C), suggesting that Myc-mTCP4 is unlikely to be a substrate of COP1 or DET1. The moderate, gradual increase in TCP4 protein levels in the light is consistent with its known function as a regulator of leaf development, which occurs naturally under light conditions. Nevertheless, the upregulation of TCP4 by light appeared to be too slow to account for the faster kinetics during light induction of the SAUR genes. Moreover, TCP4 and other CIN-like TCPs are fairly abundant in etiolated cotyledons, where they obviously do not activate SAUR16, SAUR50, or SAUR14 transcription as efficiently as they do after light exposure. Thus, there must be a mechanism that suppresses TCP4-induced activation of these SAUR genes in the dark.
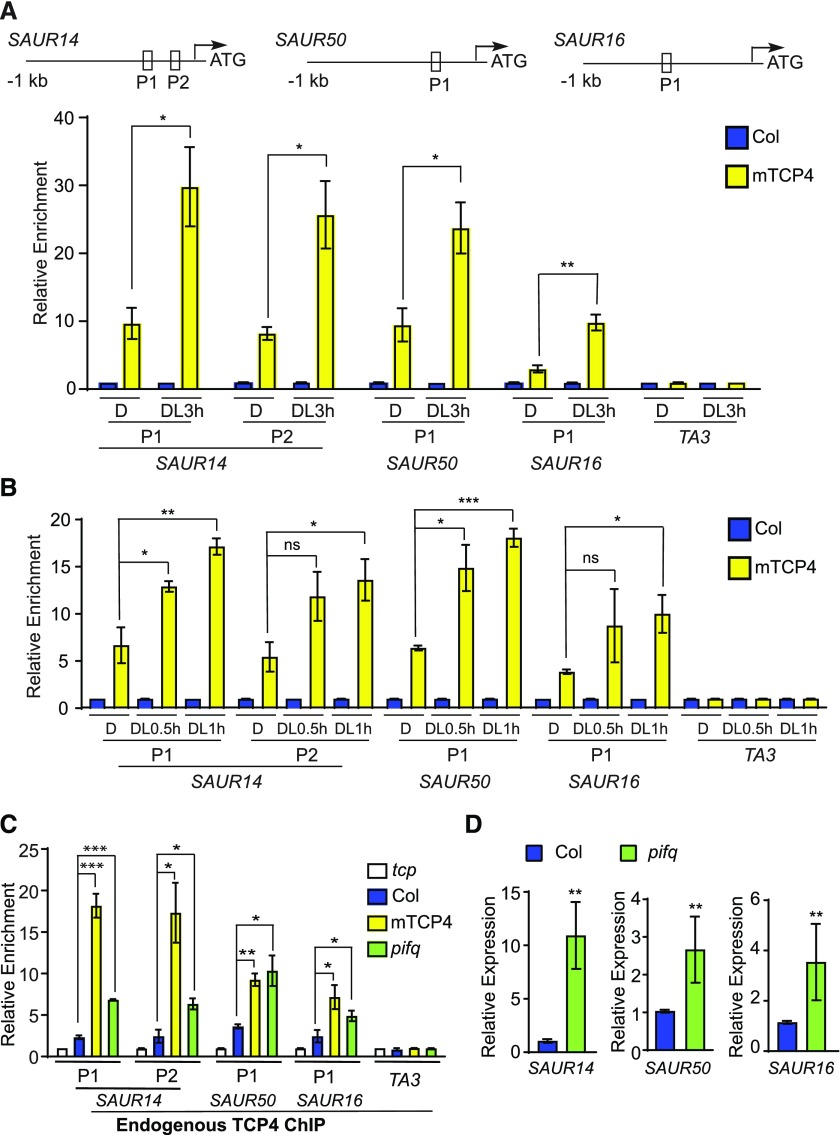
We next asked whether the binding of TCP4 to the promoters of the SAUR genes is regulated by light. By ChIP-qPCR of mTCP4, we found that the association of mTCP4 to the promoters of light-inducible SAURs (SAUR14, SAUR50, and SAUR16) was clearly increased after 3 h of light exposure (Figure 5A). Notably, the light-induced increase in TCP4 binding occurred as soon as within 0.5 or 1 h of light exposure (Figure 5B), when the mTCP4 protein level was not significantly altered (Supplemental Figure 7B). These results indicate that light signals sharply increase the binding capacity of TCP4 to the promoters of these genes, a process that appears to be distinct from the gradually rising levels of TCP proteins. More precisely, given the abundance of TCP4 in etiolated seedlings, our results imply that the binding of TCP4 proteins to these light-inducible SAUR gene promoters is inhibited in the dark.
Figure 5.
Binding of TCP4-Like Proteins to the SAUR Promoters Is Enhanced by Light Signal or the Lack of PIFs.
(A) and (B) Light increases the binding of TCP4 to the promoter regions of the SAUR genes. ChIP was performed using whole Col and mTCP4 seedlings grown in the dark or after transfer to the light for 3, 1, or 0.5 h. The relative enrichment of Col was set to 1. The TA3 transposon locus was used as a negative control. The top image shows the locations of the PCR fragments in the SAUR gene promoters. Data are shown as the mean ± se of three biological replicates. D, dark; DL0.5 h, after transfer to the light for 0.5 h; DL1 h, after transfer to the light for 1 h; DL3 h, after transfer to the light for 3 h.
(C) The PIF-deficient mutant (pifq) exhibits increased DNA binding of TCP4 to the SAUR promoters in the dark. Endogenous anti-TCP4 antibody was used in the ChIP assay in 4-d-old dark-grown seedlings. Relative enrichment value in the tcp3 tcp4 tcp10 (tcp) mutant negative control was set to 1. TA3 was used as an internal control. Data are shown as the mean ± se of three biological replicates.
(D) Expression levels of SAUR14, SAUR50, and SAUR16 are elevated in the cotyledons of the PIF-deficient mutant (pifq). Total RNA extracted from cotyledons of 4-d-old dark-grown Col and pifq seedlings was used for RT-PCR. Data are shown as the mean ± sd of three biological replicates.
Statistical significance was calculated using two-tailed Student’s t test: ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The lack of PIFs (pifq) results in open cotyledons in the dark (Al-Sady et al., 2006; Leivar et al., 2008; Shin et al., 2009), while overexpression of any of the PIF1, PIF3, PIF4, or PIF5 products in pifq can each independently work to maintain closed cotyledons in the dark (Shi et al., 2018). Since SAUR14, SAUR50, and SAUR16 are among the genes that are bound and repressed by PIFs in etiolated cotyledons (Sun et al., 2016), we examined the role of PIFs in the light-dependent regulation of TCP4 binding to the promoters of SAUR genes. If PIFs are needed to inhibit the binding of TCP4 to the SAURs promoter in the dark, the pifq mutant would show abnormally increased TCP4 binding to these sites. We tested this hypothesis by ChIP-qPCR using a native TCP4 antibody that could readily detect overexpressed Myc-mTCP4 in protein gel blots (Supplemental Figure 8). TCP4 ChIP showed strong signals on the promoter regions of SAUR14, SAUR50, and SAUR16 in mTCP4 compared with the tcp (tcp3 tcp4 tcp 10) mutant (Figure 5C), indicating that the anti-TCP4 antibody was capable of immunoprecipitating TCP4 protein in the ChIP assays. Importantly, TCP4 ChIP showed significant enrichment of these SAUR promoter regions in the pifq mutant compared with Col in dark-grown seedlings (Figure 5C). This result indicates that the SAUR14, SAUR50, and SAUR16 promoters were bound by endogenous TCP4 proteins in pifq in the dark at much higher levels than in Col. Given that TCP4 expression levels are comparable between Col and the pifq mutant (Supplemental Figure 9), our data strongly suggest that PIFs are genetically required to inhibit the binding of TCP4 to the SAUR promoters in the dark. As a result, the expression of SAUR genes was significantly derepressed in the dark-grown pifq mutant (Figure 5D), which is consistent with the increased binding of TCP4 to their promoters (Figure 5B) as well as the corresponding open cotyledon phenotype of pifq in the dark (Leivar et al., 2008; Shin et al., 2009).
The transcription factor HY5 and HY5-HOMOLOG (HYH) promotes seedling photomorphogenesis and potentially binds to the SAUR50 promoter (Lee et al., 2007). We considered the possibility that HY5 might facilitate the light-induced binding activity of TCP4 to SAUR promoters. However, the endogenous TCP4 ChIP assays showed that TCP4 was recruited to the SAUR promoters in hy5 hyh mutants in the light (Supplemental Figure 10), suggesting that HY5/HYH might not be required for the binding of TCP4 to the promoter regions of SAUR50 or SAUR16 in vivo. Taken together, the data indicate that light regulates the DNA binding activity of TCP4 to the SAUR promoters and that PIFs are required to minimize these binding activities in the dark.
PIF3 Inhibits the Binding of TCP4 to the Promoters of SAUR14/16/50
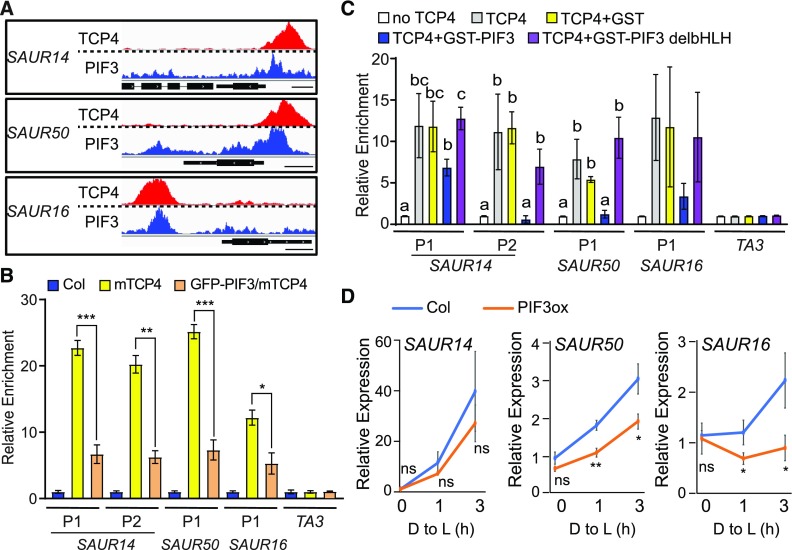
Although PIFs and TCP4 bind to different DNA motifs (Zhang et al., 2013; Supplemental Figure 3B), we noticed that the binding areas of PIF3 and those of mTCP4 (obtained in separate studies) overlap on the promoter regions of these SAUR genes (Figure 6A), hinting at the possibility of interference between transcription factors on these promoters. We tested whether PIF proteins directly inhibit the binding of TCP4 to SAUR promoters using both in vivo and in vitro experiments. First, we overexpressed the green fluorescent protein (GFP)-PIF3 transgene in mTCP4 (35S:GFP-PIF3/mTCP4; Supplemental Figure 11) and monitored the effect of PIF3 overexpression on the binding affinity of mTCP4 to the SAUR promoters by ChIP. As shown in Figure 6B, PIF3 overexpression significantly decreased the binding of TCP4 to the SAUR promoters.
Figure 6.
PIF3 Inhibits the Binding of TCP4 to the Promoters of SAUR Genes and Represses Their Expression.
(A) The binding area of PIF3 overlaps with that of TCP4 at the SAUR promoter regions. The raw reads of PIF3 ChIP-seq (Zhang et al., 2013) and TCP4 ChIP-seq (this study) at the SAUR14, SAUR50, and SAUR16 loci are shown. Bars = 250 bp.
(B) PIF3 overexpression inhibits the binding of TCP4 to the SAUR promoters in the dark. Four-day-old dark-grown seedlings of the indicated genotype were used for ChIP assays. Relative enrichment value in Col was set to 1. TA3 was used as an internal control. Data are shown as the mean ± se of three biological replicates. Statistical analysis was performed by two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
(C) PIF3 inhibits the binding of TCP4 to the SAUR promoters in vitro, and this inhibition requires the PIF3 DNA binding domain. In vitro–translated TCP4-Myc proteins captured on anti-Myc agarose were incubated with fragmented genomic DNA in the presence or absence of recombinant GST, GST-PIF3, or GST-PIF3 without the bHLH domain (GST-PIF3 delbHLH). Bound DNA was analyzed by qPCR. Data are shown as the mean ± se of three biological replicates. Different letters represent significant differences (P < 0.05 by one-way analysis of variance).
(D) Four-day-old dark-grown (D) Col and 35S:PIF3-Myc (PIF3ox) seedlings were transferred to the light for 1 or 3 h, and RNA from cotyledon tissues was analyzed to measure the expression of the indicated SAUR genes. Data are the mean ± sd of three biological replicates. Statistical analysis was performed by two-tailed Student’s t test: ns, P > 0.05; *, P < 0.05; **, P < 0.01. D to L: dark to light; PIF3ox, 35S:PIF3-Myc seedlings.
To investigate whether the inhibition by PIF3 is direct, we performed an in vitro DNA affinity purification (DAP) assay, which has been used to generate the genome-wide binding landscapes of transcription factors in Arabidopsis (Bartlett et al., 2017). In the DAP assay, in vitro–translated TCP4 proteins were incubated with genomic DNA preparation, and TCP4-associated DNAs were found to be enriched with corresponding promoter regions of SAUR14, SAUR50, and SAUR16, but not in the TA3 (transposable element) negative control (Figure 6C). Remarkably, the addition of GST (GLUTATHIONE S-TRANSFERASE)–tagged PIF3, but not the GST control, decreased the binding of TCP4 to these promoters (Figure 6C). Moreover, GST-PIF3 without the basic helix-loop-helix (bHLH) domain (GST-PIF3 deleting bHLH, GST-PIF3 delbHLH) was unable to inhibit TCP4 binding (Figure 6C), indicating that the DNA binding domain of PIF3 is required for this activity. These results suggest that PIF3 diminishes the binding of TCP4 to the promoters of SAUR14, SAUR50, and SAUR16 and that this transcription factor interference likely occurs on the DNA. These data imply that repression of the TCP4-mediated transcription of the SAURs can be achieved by increasing the level of PIF proteins, as is the case in etiolated seedlings and that light-triggered depletion of PIF can alleviate this repression and result in the activation of SAURs by TCP4.
To confirm that the PIF-induced modulation of TCP4 binding to promoter regions can ultimately affect SAUR gene expression, we examined SAUR14, SAUR16, and SAUR50 mRNA levels in PIF3 overexpression plants both in the dark and during the transition from dark to light. Compared with Col, higher levels of PIF3 (PIF3 overexpression [PIF3ox]) resulted in lower expression and retarded elevation of SAUR14/16/50 expression during the dark-to-light transition (Figure 6D). It should be mentioned that, unlike SAUR16 and SAUR50, SAUR14 transcription in cotyledons was not dependent on TCP4-like factors (Figure 4A). There might be a wider range of redundancy in how SAUR14 transcription is activated. Regardless of the specificity of TCP activation, our data indicate that the mechanism of PIF-dependent dark–light transcriptional switching applies to all three light-induced SAUR genes (Figures 5 and 6).
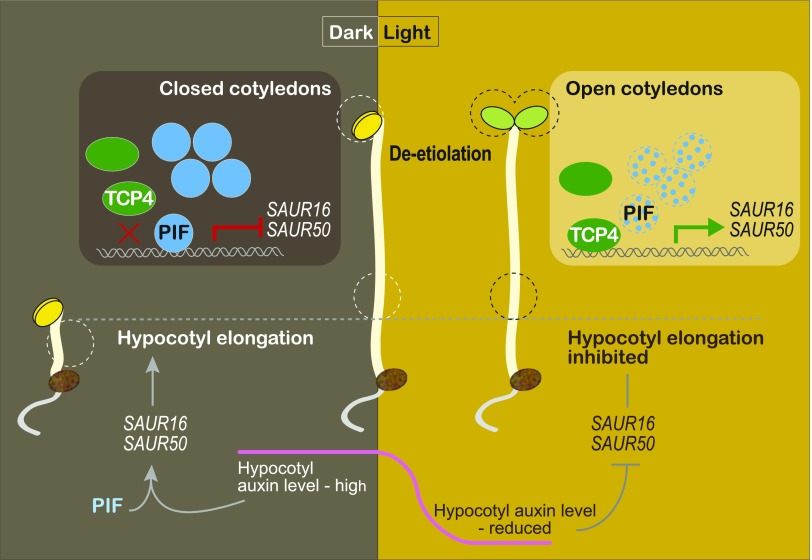
DISCUSSION
Photomorphogenesis is a light-triggered developmental process involving all parts of a plant, with different organs and tissues exhibiting different responses to the same light signal (von Arnim and Deng, 1996). Our understanding of the early light-signaling components and pathways has allowed us to address the organ specificity of light responses. To this end, we report that cotyledon-specific light activation of SAUR16 and SAUR50 is achieved by the antagonistic action of the light-signaling regulator PIF3 and the cotyledon developmental regulators TCP4-like factors, which ultimately contributes to light-induced cotyledon opening during de-etiolation of Arabidopsis (Figure 7). This model also illustrates how the regulatory strategies of SAUR16 and SAUR50 genes in cotyledons are different from that observed in hypocotyls (Figure 7). In cotyledons, SAUR16 and SAUR50 are activated by the TCP4-like transcription factors but repressed by PIFs. In the dark, the heightened accumulation of PIF3 proteins (and likely other PIFs) in the nucleus results in their binding to SAUR gene promoters where they inhibit the binding of TCP4-like factors to these proximal regions. As a result, the ability of TCP4-like transcription factors to activate SAUR16 and SAUR50 is suppressed in the dark. Upon light irradiation, PIF protein levels drastically decline, relieving the inhibition of TCP4. Consequently, more TCP4 proteins bind to the promoters of SAUR16 and SAUR50 and activate their transcription in the light. The SAUR16 and SAUR50 gene products then facilitate the opening of cotyledons. In hypocotyls, on the other hand, SAUR16 and SAUR50 are activated in the dark by a different mechanism that depends on auxin and the PIFs (Sun et al., 2016). Their distinct organ-specific activation strategies allow SAUR16 and SAUR50 to display contrasting responses to light signals: transcriptional increases in cotyledons and decreases in hypocotyls. Ultimately, the organ-specific expression of SAUR16 and SAUR50 contributes to different cellular responses to light during de-etiolation.
Figure 7.
A Model Illustrating Organ-Specific Regulation of SAUR16/50 During De-Etiolation.
In etiolated cotyledons, PIF proteins accumulate to interfere with the binding of TCP4-like transcription factors to the SAUR16/50 promoter regions, inhibiting TCP4-mediated activation of the SAUR genes in the dark. The inhibition of TCP4-SAUR16/50 transcription by PIFs contributes to the typically closed cotyledon phenotype of etiolated seedlings. During de-etiolation, PIF protein levels sharply decline, resulting in increased binding of TCP4 to the promoters of the SAURs and their elevated expression, which facilitates cotyledon opening. In hypocotyls, SAUR16/50 genes are regulated by a different mechanism involving auxin. These genes are highly expressed in the dark, and their expression declines upon the transition from dark to light, which is correlated with changes in auxin levels in the hypocotyl (Sun et al., 2016).
The TCP4-SAUR16/50 Module Is Cotyledon Specific
Cotyledons and the apical region of etiolated seedlings exhibit dramatic morphological changes during de-etiolation, including the unfolding of the apical hook and opening of the two cotyledons, followed by cotyledon expansion (von Arnim and Deng, 1996). Unfolding of the apical hook, a process that has been extensively studied, involves spatial and temporal regulation of the ethylene and auxin pathways (Zádníková et al., 2010; Béziat et al., 2017; Shi et al., 2018). Much less is known about the mechanisms of light-induced cotyledon opening. Here, we identified TCP4-SAUR16 and SAUR50 as an important transcriptional regulatory module involved in cotyledon opening. There are likely other factors and other SAUR genes that also promote cotyledon opening. For example, MISEXPRESSED IN THE DARK1 (HYDROXYSTEROID DEHYDROGENASE1), a PIF3 regulatory gene, has been implicated in cotyledon separation (Sentandreu et al., 2011). However, the functional relationship of this gene with SAUR genes is unclear.
Based on in situ hybridization and organ-specific transcriptome analyses (Supplemental Figure 1A; Palatnik et al., 2003), the TCP4-like factors are predominantly expressed in cotyledons (rather than hypocotyls) in early seedlings. This expression pattern would ensure that TCP4-induced activation of SAUR16 and SAUR50 occurs primarily in cotyledons. Curiously, despite the observations that TCP4 can activate auxin biosynthesis genes (Challa et al., 2016) and that SAUR16 and SAUR50 are capable of responding to auxin (in hypocotyl cells), the transcriptional activation of SAUR16 and SAUR50 by TCP4 in cotyledons appears to occur directly rather than via auxin. The following lines of evidence support this idea. First, the expression of SAUR16 and SAUR50 in etiolated cotyledons is insensitive to auxin (Sun et al., 2016). Second, the binding of TCP4 to the SAUR16 and SAUR50 promoters in response to light, as well as PIF levels, is tightly correlated with the expression of the SAUR genes (Figures 5 and 6).
In contrast to cotyledons, SAUR16 and SAUR50 in hypocotyl cells are highly sensitive to auxin levels. In fact, light downregulates SAUR16 and SAUR50 expression in hypocotyls in part by decreasing auxin levels (Sun et al., 2016). In light-grown seedlings, members of the CIN-like TCP transcription factor family promote hypocotyl elongation (TCP4; Challa et al., 2016) or in response to shade (TCP17; Zhou et al., 2018). In both cases, TCP4 or TCP17 functions by inducing auxin biosynthetic genes. Perhaps the transport of auxin from the cotyledons to hypocotyl cells ultimately results in the hypocotyl phenotype. Regardless, our work highlights the notion that, as a result of the TCP4-regulated mechanism in cotyledons versus an auxin-dependent mechanism in hypocotyls, the same SAUR16 and SAUR50 genes with the same promoter DNA sequences are able to respond to light signals in opposite ways in different organs.
PIFs Confer the Dependency of TCP4-Induced Transcriptional Activation of SAURs on Light Signals during De-Etiolation
PIF transcription factors are required for seedlings to undergo etiolation when grown in the dark (Leivar et al., 2008; Shin et al., 2009). SAUR14, SAUR16, and SAUR50 are among the genes that PIFs repress in cotyledon cells (Sun et al., 2016). Our data suggest that in etiolated cotyledon cells, highly abundant PIF proteins in the nucleus suppress the binding of TCP4-like activators to the promoters of SAUR14, SAUR16, and SAUR50, thereby inhibiting their expression. Nevertheless, we detected the binding of TCP4 to the SAUR promoter sites in the dark, albeit at very low levels (Figure 5C, compare Col to the tcp mutant). However, in dark-grown mTCP4 seedlings, overexpressed mTCP4 appeared to have escaped inhibition by PIFs, presumably by outcompeting PIF levels. The increased binding of mTCP4 to the promoter explains the elevated expression of the SAUR genes (Figures 2C and 5C) and the open-cotyledon phenotype in the dark (Figure 1E).
Our data suggest that light stimulates SAUR14/16/50 expression in two phases: at the onset of the dark-to-light transition (de-etiolation) and during sustained light irradiation. During extended light irradiation, light increases TCP10 transcription and might slightly increase mTCP4 protein abundance (Supplemental Figure 7), which may contribute to the higher expression of the SAUR genes in the light. However, there is evidence indicating that TCP4 levels are not strongly regulated by light or light-responsive factors. TCP4 transcription is not regulated by light (Supplemental Figure 1), nor by PIFs (Supplemental Figure 9). mTCP4 protein levels are not affected by PIF3 overexpression (Supplemental Figure 11), nor by cop1 or det1 mutations in the dark (Supplemental Figure 7). Thus, TCP4 per se is probably not a light-responsive regulator.
We propose that the increased TCP4-induced activation of SAUR14, SAUR16, and SAUR50 during the de-etiolation phase directly results from the changes in PIF levels. Besides the genetic and molecular data, the kinetics of the increase in TCP4 binding to the SAUR promoters roughly matches the sharp decline in PIF protein levels after the dark-to-light transition (Figure 5). It should be mentioned that although PIF3 was used as a representative PIFs in all of the gain-of-function experiments, we do not have evidence to suggest that this type of regulation is specific to PIF3. In fact, given the functional redundancy of the PIFs in maintaining closed cotyledons in the dark (Shi et al., 2018), it appears likely that other PIFs work in a manner similar to that of PIF3 in this pathway.
HY5 and HYH are established positive regulators of photomorphogenesis (von Arnim and Deng, 1996; Chen et al., 2004; Kami et al., 2010), primarily for their functions in inhibiting hypocotyl elongation in response to light. However, unlike PIFs that regulate essentially all facets of photomorphogenesis, including both hypocotyl and cotyledon development, the role of HY5 could be demonstrated for hypocotyl development, but not for cotyledon opening or expansion (Shi et al., 2018). In agreement with these observations, our data suggest that HY5/HYH might not facilitate light-induced increases in TCP4 binding to the SAUR promoters (Supplemental Figure 10).
Mechanism of the Inhibitory Effect of PIF3 on the Binding of TCP4 to the SAUR Promoter Sites
PIFs activate or repress the expression of many light-responsive genes by binding to their promoter regions (Zhang et al., 2013), but the mechanisms of PIF-mediated transcriptional repression remain poorly understood. In some cases, PIF3 interacts with HISTONE DEACETYLASE15 to modulate histone acetylation of targeted genes (Liu et al., 2013). Here, we present an example in which PIFs repress SAUR gene expression by inhibiting the binding of the transcriptional activators (TCP4-like) to the promoters of their target genes.
How PIF3 inhibits TCP4 binding to the SAUR promoter sites remains speculative at present. Several key nuclear factors modulate light responses by regulating PIF DNA binding activity. The gibberellin inhibitors, DELLA proteins, interact with the DNA binding domain of PIFs, thereby inhibiting their ability to bind to the promoters of their target genes (de Lucas et al., 2008; Feng et al., 2008). A similar strategy is utilized by LONG HYPOCOTYL IN FAR-RED1, which interacts with PIF1 to prevent it from binding to DNA during seed germination (Shi et al., 2013). Other transcription factors such as ABSCISIC ACID INSENSITIVE5 interact with PIF1 while binding to sites on its target promoters, thereby helping PIF1 bind to its target sites (Kim et al., 2016). Little is known about whether PIFs inhibit the binding of other transcription factors to their target promoters. We have so far been unable to demonstrate a positive interaction between PIF3 and full-length TCP4 in yeast two-hybrid, in vitro pull-down, or bimolecular fluorescence complementation assays (Supplemental Figure 12). Under the same experimental conditions, we have detected interactions between PIF3 and REPRESSOR OF GA1-3 (RGA), PIF3 with itself, and TCP4 and FLOWERING BHLH1 (FBH1; Supplemental Figure 12; Liu et al., 2017). In a yeast two-hybrid test, we did detect an interaction between PIF3 and a C-terminal truncation of TCP4 lacking the DNA binding domain (Supplemental Figure 12). Perhaps the interaction between PIF3 and TCP4 is too weak to be detected using these approaches, or perhaps the interaction occurs only under certain circumstances. Nevertheless, the data do not support a model in which PIF3 binds to TCP4 and prevents it from binding to DNA via sequestration, as sequestration normally requires strong protein interactions or binding at the DNA binding domain. Still, we cannot rule out this possibility, or the possibility that other PIFs might interact with TCPs during certain developmental processes.
Instead of sequestration, our in vivo and in vitro data reveal a strict correlation between the relative level of TCP4 to PIF3 and how much TCP4 associates with the promoter sites: more TCP4 binding was observed in the light (low levels of PIFs) than in the dark (high levels of PIFs), more in pifq than in Col (Figure 5C), more in mTCP4 than in Col, and more in mTCP4 than PIF3ox/mTCP4 (Figure 6B). It appears that these two transcription factors compete with each other for binding to the promoters of their target genes. We favor the hypothesis that PIF3 inhibits the DNA binding ability of TCP4 via a mechanism that does not involve direct protein–protein interactions. The binding sites of PIF3 and TCP4 on the promoter regions of SAUR14, SAUR50, and SAUR16 are 115, 73, and 47 bases apart, respectively (Supplemental Figure 13), which are quite close to each other. Moreover, the DNA binding ability of PIF3 is critical for inhibiting the binding of TCP4 to promoters (Figure 6C), indicating that inhibition, or competition, occurs on the DNA. These observations support the hypothesis that the DNA-bound PIF protein complexes interfere with or repel TCP4 complexes. Alternatively, perhaps PIF binding induces conformational changes in DNA that make the TCP4 binding sites less accessible to TCP4.
Taken together, our findings demonstrate that TCP4 and PIF3 antagonistically regulate the expression of SAUR16 and SAUR50, which contributes to cotyledon opening during de-etiolation in Arabidopsis. This mechanism requires TCP4-like transcription factors to activate the expression of SAUR16 and SAUR50 in cotyledons and the light-responsive repressor PIF3 or PIFs to confer light responsiveness. The resulting cotyledon-specific light activation of SAUR16 and SAUR50 represents one of many transcriptional regulatory mechanisms that together underlie an organ-specific response to light signals during plant photomorphogenesis.
METHODS
Plant Materials and Growth Conditions
All the Arabidopsis (Arabidopsis thaliana) materials used in this study are in the Col ecotype background. Adult Arabidopsis plants were grown under long-day conditions with a 16-h white light (at 90 µmol/m2/s)/8-h dark cycle at 22°C. The tcp3 tcp4 tcp10, tcp3 tcp4 tcp5 tcp10, and tcp3 tcp4 tcp5 tcp10 tcp13 seeds were kindly provided by Tomotsugu Koyama (Kyoto University; Koyama et al., 2010). The hy5 hyh seeds were kindly provided by Liumin Fan (Peking University; Yang et al., 2018).
Seeds were surface sterilized with 15% (w/v) sodium hypochlorite for 10 min and washed at least four times with sterile double-distilled water before sowing on Murashige and Skoog plates containing Suc (4.4 g/L Murashige and Skoog powder, 10 g/L Suc, and 8 g/L agar, pH 5.7). After stratification at 4°C for 3 d, the seeds were treated with white light (at 40 µmol/m2/s) for 3 to 4 h (to synchronize germination) and incubated in the dark for 4 d. For ChIP-seq, 4-d-old dark-grown seedlings were used to prepare the DNA samples. For ChIP-qPCR, 4-d-old dark-grown seedlings were treated with 40 µmol/m2/s white light for 3 h and used to prepare the DNA samples. For gene expression and phenotypic analysis in the dark, the cotyledons of 4-d-old seedlings were used for RNA extraction and phenotypic observation, respectively. For gene expression and phenotypic analysis during the dark-to-light transition, 4-d-old dark-grown seedlings were transferred to 40 µmol/m2/s white light for the indicated time, and cotyledons were used for RNA extraction and phenotypic observation. For protein gel blot analyses, 4-d-old dark-grown seedlings were exposed to 40 µmol/m2/s white light or 4-d-old seedlings grown under 40 µmol/m2/s white light were transferred to the dark for the indicated time for total protein extraction.
Generation of Transgenic Plants
To generate pBA-MYC-mTCP4, the Gateway recombinant cassette (attR1-ccdB-attR2) was amplified from the pK2GW7 plasmid (Ghent University) using primer pair attR1 F-MluI and attR2 R-SacI. The PCR fragments were digested with MluI and SacI and cloned into the MluI and SacI sites of binary vector myc-pBA (Zhou et al., 2005) to generate myc-pBA-GW. The full-length coding region of TCP4 was amplified from Arabidopsis cDNA by RT-PCR using primer pair TCP4-1 and TCP4-2 and cloned into pENTRY/D-TOPO (Invitrogen) to generate pENTR-TCP4. The point mutation resistant to miR319 was introduced using primer pair TCP4m-1 and TCP4m-2 to generate pENTR-mTCP4. pBA-MYC-mTCP4 was generated by LR reaction with myc-pBA-GW and pENTR-mTCP4. The construct was transferred into Agrobacterium tumefaciens GV3101 and then transformed into Col using the floral dip method (Clough and Bent, 1998). The primers are listed in Supplemental Table.
To generate 35S:GFP-PIF3/mTCP4, the GFP coding sequence without the stop codon was amplified from pJim19bar-sGFP (Sun et al., 2016) using XbaIGFPf/XhoIGFPr and inserted into pJim19 hyg empty vector using XbaI and XhoI, yielding pJim19 hygNGFP. The PIF3 coding sequence was amplified from pT7CFE1-PIF3-CMyc (Dong et al., 2017) using XhoIPIF3f/SacIPIF3r and inserted into pJim19 hygNGFP using XhoI/SacI, yielding pJim19 hygNGFP-PIF3. The construct was delivered into 35S:Myc-mTCP4 10-2 via Agrobacterium GV3101 transformation using the floral dip method.
Generation of CRISPR Mutants
To generate saur mutations in 35S:Myc-mTCP4, the guide sequences targeting SAUR14, SAUR50, SAUR16, SAUR62, SAUR77, and SAUR78 were inserted into the pAtU6-26-SK vector (Feng et al., 2013), yielding pAtU6-SAUR14, pAtU6-SAUR50, pAtU6-SAUR16, pAtU6-SAUR62, pAtU6-SAUR77, and pAtU6-SAUR78, respectively (Sun et al., 2016). The pAtU6-SAUR50 expression cassette was digested with SpeI/SalI and inserted into the NheI/SalI sites of pAtU6-SAUR14, yielding pAtU6-SAUR14 and pAtU6-SAUR50. The pAtU6-SAUR16, pAtU6-SAUR62, pAtU6-SAUR77, and pAtU6-SAUR78 expression cassettes were inserted one by one using the same strategy, and ultimately generating pAtU6-SAUR14, pAtU6-SAUR50, pAtU6-SAUR16, pAtU6-SAUR62, pAtU6-SAUR77, and pAtU6-SAUR78. The expression cassette was digested with KpnI/SalI and ligated to pEC-Cas9 (Wang et al., 2015) to generate pEC-Ca9-AtU6-SAUR14, pAtU6-SAUR50, pAtU6-SAUR16, pAtU6-SAUR62, pAtU6-SAUR77, and pAtU6-SAUR78. Finally, the expression cassette for Cas9 and guide RNA was digested with KpnI/EcoRI and ligated to the pCAMBIA1300 vector, yielding pCAMBIA1300-EC-Cas9-AtU6-SAUR14, AtU6-SAUR50, AtU6-SAUR16, AtU6-SAUR62, AtU6-SAUR77, and AtU6-SAUR78. The construct was transformed into 35S:Myc-mTCP4 10-2 via Agrobacterium GV3101 using the floral dip method. Mutations were genotyped by PCR-based sequencing of targeted gene loci. The primers used for PCR of each locus are listed in Supplemental Table.
Chromatin Immunoprecipitation
The methods used for ChIP were described previously (Sun et al., 2016). Briefly, 4-d-old seedlings were cross-linked for 20 min by vacuum filtration in 1% formaldehyde solution. The chromatin solution was incubated with 10 μL of anti-Myc (C3956, Sigma-Aldrich) or anti-TCP4 antibody (this study) overnight in 1 mL of ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, and 167 mM NaCl) and captured by 80 μL of ChIP dilution buffer prewashed protein A beads for 1 h. After washing, the immune complex captured by protein A was eluted in 500 μL of elution buffer (1% SDS and 0.1 M NaHCO3). Next, 20 μL of 5 M NaCl was added for reverse cross-linking at 65°C overnight, and DNA was purified by phenol/chloroform/isoamyl alcohol extraction and used for ChIP-qPCR or ChIP-seq. The primers used for ChIP-qPCR are listed in Supplemental Table 1.
For ChIP-seq, DNA samples (20 ng) pooled from three independent batches of 4-d-old dark-grown seedling samples were used for library construction and deep sequencing. The raw sequence data were aligned to the Arabidopsis genome The Arabidopsis Information Resource 10 (TAIR10) using Bowtie version 0.12.9 (Langmead et al., 2009). Only reads uniquely mapped to the reference genome were used for binding-peak identification. We performed the peak calling process in a comparison between 35S:mTCP4 and Col-0 with MACS2 (Zhang et al., 2008) version 2.1.1 and then annotated with TAIR10 annotation to find the specific target genes of TCP4 (Feng et al., 2012). We performed motif discovery on the +/−200 bp of the 500 most enriched peaks using the online MEME program (Bailey and Elkan, 1994) to identify the motifs to which TCP4 bound.
Whole Transcriptomic RNA-Seq
The cotyledons of 4-d-old dark-grown Col, mTCP#4, and mTCP#10 were used for total RNA extraction following the instructions of the RNeasy plant mini kit (74,903; Qiagen). RNA-seq and data analyses were performed as described previously (Sun et al., 2016), with minor modifications. After removing adapters and trimming low-quality bases, pair-end reads were mapped to the Arabidopsis TAIR10 genome using the graph-based alignment tool HISAT2 with default parameters. The alignment files were used as input for HTSeq (Anders et al., 2015) to count aligned reads per gene. Counts generated by HTSeq were corrected for library size using DESeq2, and differentially expressed genes were identified using criterion fold change > 1.5. Since we had two overexpression lines to compare with each other, we only used one repeat for RNA-seq.
RT-Quantitative PCR
Total RNA was extracted from cotyledons of 4-d-old seedlings using the RNeasy plant mini kit (74,903; Qiagen). Total RNA (1 μg) was used for reverse transcription using SuperScript III Reverse Transcriptase (18,080,044; Invitrogen). The qPCR was performed in a CFX96 real-time system using iQ SYBR Green Super Mix (1,708,880; Bio-Rad). ACTIN2 was used as an internal control. The primers used for RT-qPCR are listed in Supplemental Table.
Protein Gel Blot Analysis
Four-day-old seedlings were ground into a powder in liquid nitrogen. Total proteins were extracted using denaturing buffer (8 M urea, 100 mM NaH2PO4, and 100 mM Tris-HCl, pH 8.0). The same amounts of total proteins were subjected to SDS-PAGE. Anti-Myc (C3956, Sigma-Aldrich), anti-TCP4 (this study), and anti-REGULATORY PARTICLE NON-ATPASE 6 (RPN6; Kwok et al., 1999) antibodies with a 1000-fold dilution were used for protein gel blot analysis.
Generation of Endogenous TCP4 Antibody
A segment of TCP4 coding sequencing was amplified using primer pair TCP4F2f/TCP4F2r (Supplemental Table 1) and cloned into pET28a at the BamHI and XhoI sites, yielding pET28a-TCP4F2. The plasmid was then transformed into Escherichia coli BL21 cells to express His-TCP4F2 recombinant protein. His-tag purification was performed, and 1 mg of recombinant protein was used as antigen to repeatedly immunize the host rabbit for four times. Rabbit serum was collected and validated for its ability to detect the antigen by protein gel blot analysis.
DNA Affinity Purification Assay
The coding sequence (CDS) of TCP4 was cloned into the pT7CFE1-Myc vector using NdeI and SalI restriction enzyme sites, yielding pT7CFE1-TCP4-Myc. TCP4-Myc protein was synthesized using a Human In Vitro Translation kit (88,881; Pierce Chemical Co.). The full-length PIF3 CDS and the PIF3 CDS without the bHLH domain (PIF3 delbHLH) were cloned into the pGEX-4T-1 vector using EcoRI and XhoI restriction enzyme sites, yielding pGEX-4T-1 PIF3 and pGEX-4T-1 PIF3delbHLH. pGEX-4T-1 was transformed into E. coli BL21 cells for GST protein expression and purification, while pGEX-4T-1 PIF3 and pGEX-4T-1 PIF3delbHLH were transformed into ArcticExpress (DE3)RIL competent cells for GST-PIF3 protein expression and purification.
DAP was performed as described previously (Bartlett et al., 2017), with minor modifications. Genomic DNA (gDNA) was extracted from 7-d-old light-grown Col seedlings following the procedure of the DNeasy plant mini kit (69,104; Qiagen). The gDNA was sonicated to a fragment size of 200 to 800 bp. TCP4-Myc protein was bound to anti-Myc monoclonal antibody agarose beads (20,168; Thermo Fisher Scientific) and incubated with 200 ng of fragmented gDNA in the presence of GST, GST-PIF3, or GST-PIF3 delbHLH for 1 h at room temperature in 1 mL of 1× PBS buffer containing 0.005% Nonidet P-40. After incubation, the beads were washed and DNA was recovered following the ChIP DNA recovery protocol. Primers used are listed in Supplemental Table.
In Vitro Pull-Down Assay
The TCP4-Myc and RGA-Myc proteins were produced using a Human Coupled IVT kit-DNA (88,881; Pierce Chemical Co.). MALTOSE BINDING PROTEIN (MBP) and MBP-PIF3 proteins were expressed and purified from E. coli. MBP and MBP-PIF3 were incubated with TCP4-Myc or RGA-Myc for 1 h, and amylose resin was used to pull down MBP and MBP-PIF3. The proteins were eluted by boiling at 95°C in 1× sample buffer for 5 min and analyzed using anti-MBP (E8038, New England Biolabs) or anti-Myc (C3956, Sigma-Aldrich) antibodies with a 1000-fold dilution.
Bimolecular Fluorescence Complementation Assays
The mTCP4 coding sequence was amplified from pENTR-mTCP4 using primer pair TCP4FLDORf/TCP4FLDORr and cloned into pDonor207 using BP Clonase (Invitrogen), yielding pDonor-mTCP4. PIF3 and FBH1 coding sequence was amplified from Arabidopsis cDNA using primer pairs PIF3FLDORf/PIF3FLDORr and FBH1FLDORf/FBH1FLDORr, respectively. The PCR products were then cloned into pDonor207 using BP Clonase, yielding pDonor-PIF3 and pDonorFBH1. pEarlygate-mTCP4-YN and pEarlygate-PIF3-YN were obtained by LR reaction using pDonor-mTCP4/pEarlygate-YN and pDonor-PIF3/pEarlygate-YN, respectively. pEarlygate-FBH1-YC and pEarlygate-PIF3-YC were obtained by LR reaction using pDonor-FBH1/pEarlygate-YC and pDonor-PIF3/pEarlygate-YC, respectively. The resulting plasmids were transformed into Agrobacterium strain GV3101 and co-infiltrated into wild tobacco (Nicotiana benthamiana) leaves for the interaction test.
Yeast Two-Hybrid Assays
Two different systems were used to test the interaction of TCP4 with PIF3 in yeast. One system uses pDEST32 (PQ1000101, Invitrogen) and pDEST22 (PQ1000101, Invitrogen) as backbones. The TCP4 coding sequencing was amplified from Arabidopsis cDNA and truncated into TCP4NT132, TCP4NT321, TCP4bHLH, and TCP4CT942 using primer pairs TCP4-1-F/TCP4-132-R, TCP4-1-F/TCP4-bHLH-R, TCP4-bHLH-F/TCP4-bHLH-R, and TCP4-322-F/TCP4-1263-R, respectively. The products were cloned into pENTR/D-TOPO (K240020, Invitrogen). All the TCP4-truncated entry plasmids were cloned into pDEST32 by LR reaction as baits. PIF3 was cloned into pDEST22 by LR reaction as prey. Bait and prey plasmids were cotransformed into yeast strain AH109. Empty vectors were used as a negative control. Synthetic defined medium lacking leucine, tryptophan, and histidine (SD−Leu−Trp−His) plates containing 5 mM 3-aminotriazole (3-AT) were used for DBD-TCP4CT942 selection. SD−Leu−Trp−His plates without 3-AT were used to select other bait plasmids.
The other system uses pGADT7-GW and pGBKT7-GW as backbones (Lu et al., 2010). The PIF3W96A mutation was introduced into pDonor-PIF3 using PIF3m1/PIF3m2, yielding pDonor-PIF3W96A. PIF3CT was amplified from Arabidopsis cDNA using primer pair PIF3-NDORf/PIF3FLDORr and cloned into pDonor207, yielding pDonor-PIF3CT. pGBKT7-FBH1, pGBKT7-PIF3W96A, and pGBKT7-PIF3CT bait plasmids were obtained by LR reaction using pGBKT7 with pDonor-FBH1, pDonor-PIF3W96A, and pDonor-PIF3CT, respectively. pGADT7-mTCP4 and pGADT7-PIF3 prey plasmids were obtained by LR reaction using pGADT7-GW with pDonor-mTCP4 and pDonor-PIF3, respectively. Bait and prey plasmids were cotransformed into yeast strain Y2HGold. SD−Leu−Trp−His plates containing 5 mM 3-AT were used for selection. The primers are listed in Supplemental Table 1.
Statistical Analysis
For all phenotypic quantification, data are shown as the mean ± se. Statistical analysis was performed by two-tailed Student’s t test, with P-values > 0.05 being considered not significant and P-values < 0.05 being considered significant for the analyzed data: *, P < 0.05; **, P < 0.01; ***, P < 0.001. For cotyledon opening percentages, n = 4, which represents four independent areas. For cotyledon opening angles, the angles were measured using ImageJ software, and n represents the total number of cotyledons used. In Figure 1G, n = 29 for all genotypes. In Figure 3C, n represents the following: Col, 18; mTCP4, 20; saur50/mTCP4#1, 23; saur50/mTCP4#2, 20; saur16 saur50/mTCP4#1, 23; saur16 saur50/mTCP4#2, 22; and saur16 saur50 saur77 saur78/mTCP4 and saur16 saur50 saur62 saur77 saur78/mTCP4, 23. For ChIP-qPCR, RT-qPCR, and DAP assays, n = 3, indicating that three independent experiments were performed and defined as biological replicates. Each biological replicate was measured using at least two repeats of qPCR.
Phylogenetic Analysis
The protein sequences of TCP transcription factors were downloaded from TAIR. The sequences were aligned using an online MUSCLE program (https://www.ebi.ac.uk/Tools/msa/muscle/) provided by The European Bioinformatics Institute (EMBL), and the neighbor-joining phylogenetic tree was built using MEGA-X version 10.0.5 using default parameters: bootstrap method, 1000 of bootstrap replications, Poisson model, uniform rates, and pairwise deletion for gap/missing data treatment. A text file of the alignment is provided in Supplemental File.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: TCP3 (AT1G53230), TCP4 (AT3G15030), TCP10 (AT2G31070), TCP2 (AT4G18390), TCP5 (At5g60970), TCP13 (AT3G02150), TCP17 (AT5G08070), TCP24 (AT1G30210), SAUR14 (AT4G38840), SAUR16 (AT4G38860), SAUR50 (AT4G34760), SAUR62 (AT1G29430), SAUR77 (AT1G17345), SAUR78 (AT1G72430), PIF3 (AT1G09530), PIF1 (AT2G20180), PIF4 (AT2G43010), PIF5 (AT3G59060), HY5 (AT5G11260), HYH (AT3G17609), COP1 (AT2G32950), DET1 (AT4G10180), Arabidopsis RESPONSE REGULATOR16 (AT2G40670), LIPOXYGENASE2 (AT3G45140), ACT2 (AT3G18780) and TA3 (AT1G37110). ChIP-seq and RNA-seq data have been deposited to National Center for Biotechnology Information’s Gene Expression Omnibus under ID code GSE115589.
Supplemental Data
Supplemental Figure 1. CIN-like TCPs are predominantly expressed in cotyledons.
Supplemental Figure 2. Kinetics of changes in cotyledon opening angle and cotyledon surface area during de-etiolation.
Supplemental Figure 3. ChIP-seq of mTCP4.
Supplemental Figure 4. RNA-seq analyses of mTCP4 lines.
Supplemental Figure 5. The saur16 saur50 mutant lines exhibited a delay in light-induced cotyledon opening.
Supplemental Figure 6. Confirmation of SAUR mutations and SAUR50 overexpression in plants used for phenotypic observation.
Supplemental Figure 7. Regulation of TCP by light.
Supplemental Figure 8. Verification of endogenous TCP4 antibody.
Supplemental Figure 9. PIFs regulate PIL1 expression, but not TCP4 expression.
Supplemental Figure 10. HY5/HYH might not assist TCP4 with binding to the SAUR promoters.
Supplemental Figure 11. Characterization of PIF3 overexpression in mTCP4.
Supplemental Figure 12. Results of interaction testing between PIF3 and TCP4.
Supplemental Figure 13. The binding motifs of TCP4 and PIF3 are close to each other in the SAUR promoters.
Supplemental Table. Primers used in this study.
Supplemental Data Set 1. List of TCP4 binding sites in the genome.
Supplemental Data Set 2. List of TCP4-regulated genes in cotyledons.
Supplemental Data Set 3. List of genes bound and regulated by TCP4.
Supplemental File. Protein sequence alignment used to construct the TCP family phylogenetic tree shown in
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Tomotsugu Koyama for kindly providing the tcp mutant materials, Liumin Fan for hy5 hyh materials, and Jian-kang Zhu and Qi-Jun Chen for CRISPR/Cas9 vectors. We also thank Fei Zhang for critically reading the articlet and providing valuable suggestions, and Jie Liu, Jiajun Wang and Zihao Lin for technical assistance. This work was supported in part by the National Institutes of Health (grant R01GM047850), the National Key R&D Program of China (grant 2017YFA0503800),the National Natural Science Foundation of China (grants 31621001 and 31330048), and open funds of the State Key Laboratory of Plant Physiology and Biochemistry (grant SKLPPBKF1806).
AUTHOR CONTRIBUTIONS
J.D., N.S., H.C., and N.W. designed the experiments; J.D. and N.S. performed the biological experiments; J.Y., Z.D., and H.H. analyzed the ChIP-seq and RNA-seq data; G.Q. and J.L. generated the mTCP4 overexpression lines and performed the yeast two-hybrid assay; V.F.I. and X.W.D provided the resources; J.D., H.C., and N.W. wrote the article; and V.F.I., X.W.D., J.D., H.C., and N.W. edited the article.
Footnotes
Articles can be viewed without a subscription.
References
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Bartlett A., O’Malley R.C., Huang S.C., Galli M., Nery J.R., Gallavotti A., Ecker J.R. (2017). Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 12: 1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat C., Barbez E., Feraru M.I., Lucyshyn D., Kleine-Vehn J. (2017). Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat. Plants 3: 17105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum D.E., Neff M.M., Van Volkenburgh E. (1994). Light-stimulated cotyledon expansion in the blu3 and hy4 mutants of Arabidopsis thaliana. Plant Physiol. 105: 1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K., Isaacs C.G., Reeves P.H., Maloney G.S., Muday G.K., Nagpal P., Reed J.W. (2012). Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 71: 684–697. [DOI] [PubMed] [Google Scholar]
- Challa K.R., Aggarwal P., and Nath U. (2016). Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28: 2117–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Dong J., Ni W., Yu R., Deng X.W., Chen H., Wei N. (2017). Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr. Biol. 27: 2420–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Han S.K., Kim H.J., Wu M.F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liu T., Qin B., Zhang Y., Liu X.S. (2012). Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D.L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., Zhu J.K. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Zhao X., Kong F., Zuo Z., Liu X. (2016). TCP2 positively regulates HY5/HYH and photomorphogenesis in Arabidopsis. J. Exp. Bot. 67: 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Irish V.F. (2015). Temporal control of plant organ growth by TCP transcription factors. Curr. Biol. 25: 1765–1770. [DOI] [PubMed] [Google Scholar]
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91: 29–66. [DOI] [PubMed] [Google Scholar]
- Kim J., Kang H., Park J., Kim W., Yoo J., Lee N., Kim J., Yoon T.Y., Choi G. (2016). PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 28: 1388–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen M.V., Schmid-Siegert E., Trevisan M., Petrolati L.A., Sénéchal F., Müller-Moulé P., Maloof J., Xenarios I., Fankhauser C. (2016). Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Zhu Y., Gao C., She W., Lin W., Chen Y., Han N., Bian H., Zhu M., Wang J. (2013). Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 54: 609–621. [DOI] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A., et al. (2017). TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 13: e1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S.F., Staub J.M., Deng X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285: 85–95. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.F., He J.X. (2016). BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 9: 113–125. [DOI] [PubMed] [Google Scholar]
- Liu J., Cheng X., Liu P., Li D., Chen T., Gu X., Sun J. (2017). MicroRNA319-regulated TCPs interact with FBHs and PFT1 to activate CO transcription and control flowering time in Arabidopsis. PLoS Genet. 13: e1006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen C.Y., Wang K.C., Luo M., Tai R., Yuan L., Zhao M., Yang S., Tian G., Cui Y., Hsieh H.L., Wu K. (2013). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25: 1258–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W., Harada J.J., Rothstein S.J., Cui Y. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61: 259–270. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39. [DOI] [PubMed] [Google Scholar]
- McClure B.A., Guilfoyle T. (1987). Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 9: 611–623. [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.L., González-Grandío E., Chalkley R.J., Huhmer A.F.R., Burlingame A.L., Wang Z.Y., Quail P.H. (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8: 15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Ren H., Gray W.M. (2015). SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentandreu M., Martín G., González-Schain N., Leivar P., Soy J., Tepperman J.M., Quail P.H., Monte E. (2011). Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23: 3974–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhong S., Mo X., Liu N., Nezames C.D., Deng X.W. (2013). HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Lyu M., Luo Y., Liu S., Li Y., He H., Wei N., Deng X.W., Zhong S. (2018). Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. USA 115: 6482–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Lee S.H., Wenger J.P., Gonzalez N., Itoh H., Inzé D., Peer W.A., Murphy A.S., Overvoorde P.J., Gray W.M. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Wang J., Gao Z., Dong J., He H., Terzaghi W., Wei N., Deng X.W., Chen H. (2016). Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. USA 113: 6071–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., Zhang J., Wei T., Gu H., Qu L.J., Qin G. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A., Deng X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 215–243. [DOI] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Kwok S.F., von Arnim A.G., Lee A., McNellis T.W., Piekos B., Deng X.W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6: 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Song Z., Li C., Jiang J., Zhou Y., Wang R., Wang Q., Ni C., Liang Q., Chen H., Fan L.M. (2018). RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 14: e1007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Cui X., Su L., Fang S., Chu J., Gong Q., Yang J., Zhu Z. (2017). Jasmonate inhibits COP1 activity to suppress hypocotyl elongation and promote cotyledon opening in etiolated Arabidopsis seedlings. Plant J. 90: 1144–1155. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Hare P.D., Yang S.W., Zeidler M., Huang L.F., Chua N.H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43: 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang D., An J., Yin H., Fang S., Chu J., Zhao Y., Li J. (2018). TCP transcription factors regulate shade avoidance via directly mediating the expression of both PHYTOCHROME INTERACTING FACTORs and auxin biosynthetic genes. Plant Physiol. 176: 1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]