Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants.
Abstract
Nitrogen (N) and phosphorus (P) are key macronutrients sustaining plant growth and crop yield and ensuring food security worldwide. Understanding how plants perceive and interpret the combinatorial nature of these signals thus has important agricultural implications within the context of (1) increased food demand, (2) limited P supply, and (3) environmental pollution due to N fertilizer usage. Here, we report the discovery of an active control of P starvation response (PSR) by a combination of local and long-distance N signaling pathways in plants. We show that, in Arabidopsis (Arabidopsis thaliana), the nitrate transceptor CHLORINA1/NITRATE TRANSPORTER1.1 (CHL1/NRT1.1) is a component of this signaling crosstalk. We also demonstrate that this crosstalk is dependent on the control of the accumulation and turnover by N of the transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1), a master regulator of P sensing and signaling. We further show an important role of PHOSPHATE2 (PHO2) as an integrator of the N availability into the PSR since the effect of N on PSR is strongly affected in pho2 mutants. We finally show that PHO2 and NRT1.1 influence each other’s transcript levels. These observations are summarized in a model representing a framework with several entry points where N signal influence PSR. Finally, we demonstrate that this phenomenon is conserved in rice (Oryza sativa) and wheat (Triticum aestivum), opening biotechnological perspectives in crop plants.
INTRODUCTION
Nitrogen (N) and phosphorus (P) are key macronutrients affecting plant growth and development (Heuer et al., 2017). In natural and agricultural ecosystems, plants are faced with issues of P and N availability, and this availability is linked to mineral mobility in soil. Nitrate (NO3−), the preferred N source of plants in aerobic soils, tends to leach from the soil, while inorganic phosphate (HPO4− [Pi]) is relatively immobile. Increasing the supply of N and P through fertilizer use helps meet the growing demand for food; however, this also leads to increased N and P leaching into the biosphere, leading to pollution that threatens ecosystems. The estimated cost of inefficient chemical fertilizer use is several billion euros a year to the European community and likely more worldwide (Sutton et al., 2011). Furthermore, while N availability is considered virtually infinite, owing to the Haber–Bosch process, the global P reserves are increasingly becoming scarce; consequently, a potential Pi crisis looms over 21st century agriculture. Thus, understanding the plant response and adaptation to availability of these two key nutrients is crucial for future reduction in fertilizer use.
Research into mineral nutrition over the past 50 years has been focused mainly on investigations of plant responses to single mineral availability. This led to an in-depth understanding of how plants perceive and adapt to N or P fluctuations. More recent investigations have started to look at interactions between two or more nutrients (Kellermeier et al., 2014; Pal et al., 2017; Kisko et al., 2018). Recently, omics approaches have highlighted the strong interconnection between nutrient metabolism and signaling pathways at different integrative levels (transcriptome, proteome, metabolome, and ionome; Kellermeier et al., 2014; Li and Lan, 2015; Medici et al., 2015; Ristova et al., 2016). It is now clear that understanding the effects of crosstalk between nutritional signals, rather than single nutrient effects, is key to understanding and engineering plant adaptive responses to a fluctuating nutritional environment.
The phosphate starvation response (PSR) has been largely studied in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), where several components of the signaling pathway have been identified (Puga et al., 2017). The molecular backbone of this pathway in Arabidopsis is made of SPX DOMAIN-CONTAINING PROTEIN1 (SPX1), PHOSPHATE STARVATION RESPONSE1 ([PHR1]; and its homologous gene PHR1-LIKE 1 [PHL1]), miR399, INDUCED BY PHOSPHATE STARVATION1 (IPS1), and PHOSPHATE2 ([PHO2]; Bari et al., 2006; Ham et al., 2018). In this pathway, a P-dependent interaction between PHR1 and SPX1 likely perceives the levels of available Pi in cells (Puga et al., 2014; Wang et al., 2014; Qi et al., 2017; Jung et al., 2018). Upon Pi limitation, PHR1 activates the majority of phosphate starvation–induced (PSI) genes (i.e., miR399, IPS1, SPX1) via binding to the cis-regulatory element (Bustos et al., 2010; Puga et al., 2014). PHR1 induces miR399 that represses PHO2, an E2 ubiquitin conjugase, thereby triggering degradation of several phosphate transporters belonging to the PHOSPHATE TRANSPORTER1 (PHT1) family and PHO1 (Bari et al., 2006; Liu et al., 2012; Park et al., 2014). This allows plants to increase their capacity for Pi uptake and transfer from roots to shoots.
Like phosphate, NO3− acts as both a nutrient and a signaling molecule (Crawford and Glass, 1998; Wang et al., 2004). Interestingly, NO3− signaling is branched, as it occurs through several interconnected signaling pathways (O’Brien et al., 2016). The first pathway is named primary nitrate response ([PNR]; Medici and Krouk, 2014). It corresponds to the rapid (within minutes) and NO3−-specific activation of sentinel genes (NITRATE REDUCTASE1, NITRITE REDUCTASE1, GLUCOSE-6-PHOSPHATE DEHYDROGENASE, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING1 [HRS1]). This pathway is the best documented, and it includes regulators such as the NO3− sensor NRT1.1 (Ho et al., 2009); several kinases and phosphatases (calcineurin B-like [CBL]-interacting protein kinase8 and CBL-interacting protein kinase23 [CIPK8 and CIPK23]; Ca-dependent protein kinase10 [CPK10], CPK30, and CPK32; ABSISIC ACID INSENSITIVE2 [ABI2]; Ho et al., 2009; Hu et al., 2009; Léran et al., 2015; Liu et al., 2017); a Ca relay (Riveras et al., 2015); and several nuclear factors (NIN-likeprotein6 [NLP6], NIN-like protein7 [NLP7], NITRATE REGULATORY GENE2 [NRG2], squamosa promoter binding protein-like9 [SPL9]; Castaings et al., 2009; Wang et al., 2009; Krouk et al., 2010b; Marchive et al., 2013; Xu et al., 2016; Guan et al., 2017).
The second pathway can be qualified of N starvation response (NSR). It is characterized by a relatively slow activation of very high-affinity transporters (NRT2.4 and NRT2.5; within 24 to 48 h) following N removal from the growth medium. Several genes have been found or have been hypothesized to be involved in plant response to NSR. These include LATERAL ORGAN BOUNDARIES DOMAIN GENE36 (LBD36), LBD37, and LBD38 (Rubin et al., 2009), CBL7 (Ma et al., 2015), miR169, and NUCLEAR FACTOR Y SUBUNIT A (NFYA; Zhao et al., 2011) and HRS1 and HRS1 homologs (Kiba et al., 2018; Maeda et al., 2018; Safi et al., 2018).
Finally, N-related signaling pathways include long-distance signals named N-demand and N-supply (Ruffel et al., 2011, 2016; Li et al., 2014; Poitout et al., 2018). Cytokinin biosynthesis, C-terminally encoded peptides (CEP), and glutaredoxins have all been shown to be important in these long-distance signals (Tabata et al., 2014; Ohkubo et al., 2017). It is noteworthy that these signaling pathways are not independent from one another and their interconnectivity is still rather elusive.
The interaction between N and P signaling has already been documented in several instances. The first hints were provided by the NITROGEN LIMITATION ADAPTION (NLA) and PHO2 genes (encoding two ubiquitin conjugases) that control phosphate transporter trafficking, resulting in N-dependent Pi accumulation in shoots (Peng et al., 2007; Kant et al., 2011; Lin et al., 2013). A GOLDEN2, ARR-B, Psr1 (GARP) transcription factor NITRATE-INDUCIBLE, GARP-TYPE TRANSCRIPTIONAL REPRESSOR 1.4 (NIGT1.4; aka HRS1) was also shown to affect primary root growth according to NO3− and phosphate ion signals via dual control of transcription and protein accumulation, respectively (Medici et al., 2015). Recently, PHR1 was found to control AtNIGT1/HRS1 as a central regulator of the high-affinity transport system of NO3− (Kiba et al., 2018; Safi et al., 2018). This control is responsible for the P regulation of the NO3− transport via NRT2.1 in particular (Maeda et al., 2018). These results represent the first mechanistic insights into the potential molecular mechanisms by which N and P signaling pathways interact.
However, the actual gene regulation following N and P combinatorial deprivation has not been reported. Therefore, it is still unclear whether PSR, PNR, and NSR interact in a reciprocal manner, as well as what the potential molecular hubs of these interactions may be. Here, we show that PSR is strongly and actively controlled by N provision. We report several observations leading to a working model describing convergent points of N signals into the PSR signaling pathway.
RESULTS
The PSR Strongly Depends on N Provision
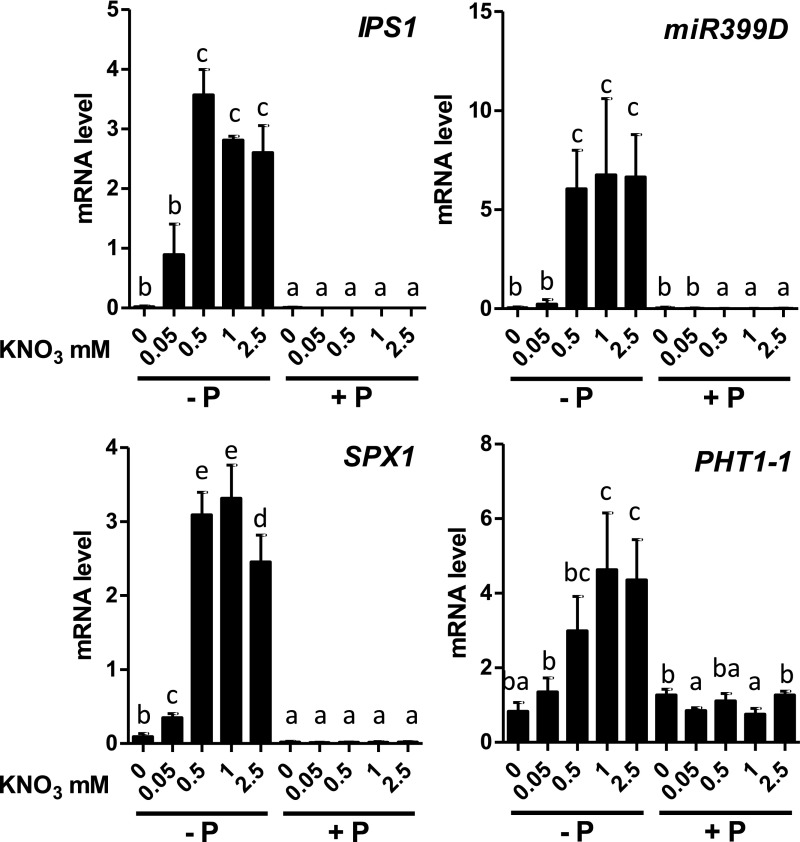
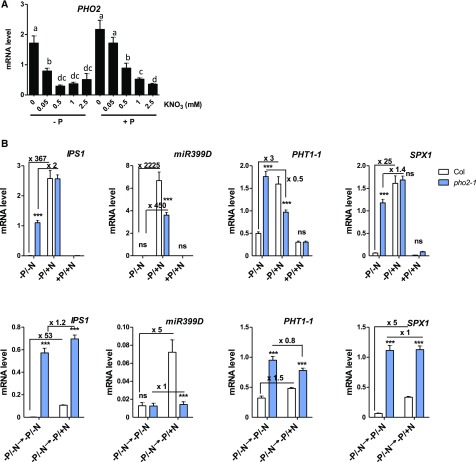
In the course of our investigations into the N and P crosstalk mediated by HRS1, we treated Arabidopsis plants with an array of NO3− and Pi under various conditions to observe any changes in root development and gene expression (Medici et al., 2015). During these investigations, we noticed in the wild-type plants that PSR marker gene (IPS1, SPX1, miR399D, PHT1-1) responses were dependent on N provision (Figure 1). Indeed, transcripts of PSI genes accumulated in P-depleted conditions only in the presence of at least 0.05 mM NO3−, and these transcripts displayed an extremely low abundance at 0 mM NO3− (Figure 1).
Figure 1.
PSR Is Repressed by a Lack of N.
Plants were grown on combinations of P (0 or 0.5 mM, KH2PO4) and N (0, 0.05, 0.5, 1, or 2.5 mM KNO3) for 14 d. Roots were harvested for measurements of PSI genes by RT-qPCR. Values are mean ± se (n = 3). Results are from three independent experiments. Different letters indicate significant differences as determined by ANOVA followed by Tukey test (P < 0.05). Plant phenotypes are reported in Supplemental Figure 1.
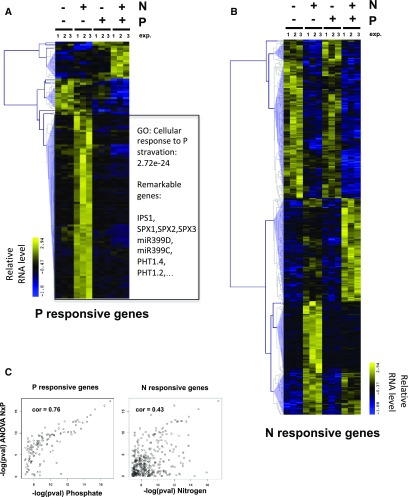
We performed a transcriptomic analysis to evaluate the genomic impact of such crosstalk. Three independent experiments were analyzed on Affymetrix whole genome arrays (Supplemental Data Set 1). Data were modeled using analysis of variance ([ANOVA]; see “Methods” for details; Supplemental Data Set 2). We retrieved 125 nonambiguous P-regulated genes (Figure 2A; gene lists are provided in Supplemental Data Set 3) and 350 nonambiguous N-regulated genes (Figure 2B; Supplemental Data Sets 1 and 4) using a very stringent P-value cutoff (<0.001; false discovery rate < 5%). Interestingly, we observed that the vast majority (∼85%) of the P-regulated genes are also significantly influenced by N (Figure 2A). The reciprocal (P effect of N response) is also true, but less dramatic (∼45%; Figure 2B). This effect is statistically observable when P-values for each factor (N, P, and NxP, Supplemental Data Sets 2 to5) effect are plotted against each other (Figure 2C). Thus, the crosstalk observed on maker genes (Figure 1) is a general genome-wide phenomenon (Figure 2). We then further investigated the molecular mechanisms that may be at the core of this significant crosstalk.
Figure 2.
PSR throughout the Genome Is Controlled by N.
Plants were grown on combinations of P and N for 14 d. Roots were harvested for transcriptomic analysis using Affymetrix chips. Results are from three independent experiments (exp. 1, 2, 3).
(A) Cluster of genes controlled by P (ANOVA P-value < 0.001).
(B) Cluster of genes controlled by N (ANOVA P-value < 0.001). Heatmaps report high (yellow) and low expression (blue).
(C) Correlation between −log(pval) between P and NxP (left panel) and N and NxP (right panel). pval, P-value.
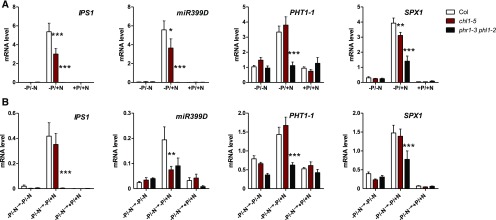
To further investigate N/P signaling crosstalk, we first wanted to ensure that the lack of PSR under N depletion conditions was not simply due to the harsh plant growth conditions (plants were grown for 14 d on N- and P-varying media; under the −N/−P conditions plants were stunted; Supplemental Figure 1), but rather due to an active loss of PSR under −N conditions. We therefore decided to conduct transfer experiments (Figures 3A and 3B). We grew plants under −N/−P conditions (Figure 3A) for 11 d and then transferred them for 3 d to replenishing media combining N and P provisions (Figure 3B, see wild-type bars). In agreement with a previous observation, we noted that N provision to plants starved for P was able to reactivate the PSR, as reported by an increase in steady state transcript levels of PSI genes (Figure 3B). However, when this experiment was performed in N and P signaling mutants for NRT1.1 (chl1-5) and PHR1/PHL1 (phr1 phl1), respectively (Ho et al., 2009; Puga et al., 2014), we noticed that the phr1 single and phr1 phl1 double mutant have a strong effect on PSR activation (Supplemental Figure 2A and Figure 3A, respectively) and upon N provision (Figure 3B). We also observed a moderate effect of chl1-5 mutation on the regulation of PSR under constant nutrient conditions experiments (Figure 3A), and only a limited effect upon plant transfer to N-containing media (Figure 3B). This moderate effect of chl1 mutation can easily be explained as it has been previously shown that the effect of the chl1 mutation can be bypassed if N starvation precedes N provision (Wang et al., 2009). As the plants have been N starved for 11 d, it is likely that most NO3− sensing mechanisms on N transfer conditions still rely on an unidentified sensing protein that bypasses NRT1.1 activity (fully discussed in Wang et al., 2009; Medici and Krouk, 2014).
Figure 3.
PSR Is Reactivated upon NO3− Provision and NRT1.1 and PHR1/PHL1 Affect PSR in N Varying Conditions.
(A) The wild-type, chl1-5, and phr1-phl1 genotypes were grown on combinations of P (0 or 0.5 mM, KH2PO4) and N (0, 2.5 mM KNO3) for 11 d. Col, Columbia (wild type).
(B) Eleven-day-old plants from −P/−N conditions are transferred toward P and/or N replenished media. Values are mean ± se (n = 3). Asterisks indicate significant differences from the wild-type plants (*P < 0.05; ***P < 0.001; Student’s t test).
Taken together, these results strongly support the idea that phosphate and NO3− signaling pathways are tightly linked and that the P starvation signaling pathway downstream of PHR1/PHL1 activity is partly dependent on NO3− uptake and/or signaling mediated by the NO3− sensor NRT1.1.
phr1 phl1 Double Mutant Still Perceives NO3−
Because the phr1 phl1 double mutant is insensitive to N-controlled reactivation of PSR (Figure 3), we wanted to rule out the possibility that the missing activation was due to a lack of NO3− sensing in the phr1 phl1 double mutant. To do so, we examined whether the phr1 phl1 double mutant was able to perceive NO3− as the PNR is conserved in this genetic background (Supplemental Figure 2B). We confirmed that NO3− provision was able to induce NIR and NRT1.1, and we therefore conclude that it is not possible to explain the default of PSR activation by default NO3− sensing in the phr1 phl1 background. This prompted us to propose that the effect of N on the PSR is likely upstream of the PHR1 sensing activity.
N Controls PSR via Local and Systemic Signals
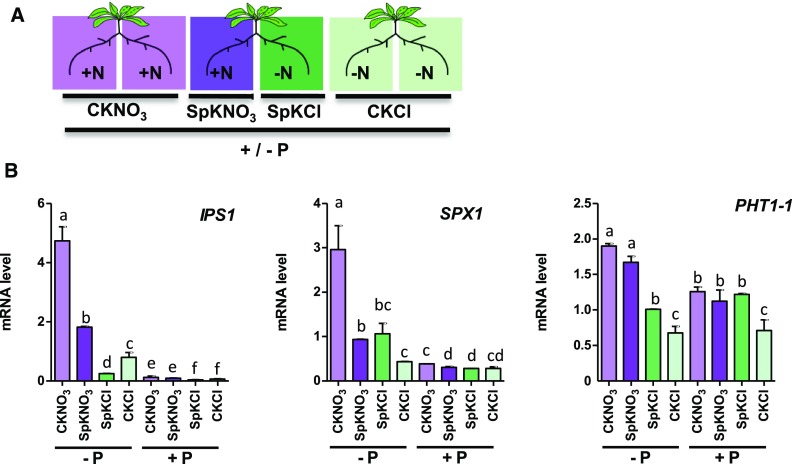
In plants, the responses to NO3− provision depend not only on local but also on long-distance signals, which allow their adaptation to nonhomogeneous soils (Gansel et al., 2001; Ruffel et al., 2011, 2016; Li et al., 2014; Poitout et al., 2018). Two kinds of long-distance signals can be distinguished. The N-demand signal informs roots replete with N to compensate for roots growing in NO3−-deprived regions of the soil. The N-supply signal informs the N-deprived roots to adapt their physiology according to the fact that other roots are foraging for N. To determine whether PSR depends on these N-related long-distance signals, we set up a split-root experiment in which we tested the response to phosphate deficiency in heterogeneous N conditions (Figure 4A). The plants were cultivated for 11 d on N- and P-containing media and then subjected to double deficiency (−N and −P) for 3 d. Finally, a first batch of plants was transferred for three more days to media without phosphate and +N or −N in homogenous (control) or heterogeneous (split) conditions. Another batch of plants was transferred to the same media plates but containing 1 mM Pi. The expression of PSI genes (Figure 4B) in the roots grown in the absence of phosphate confirmed that in homogeneous conditions the presence of N switches the PSR on, and the prolonged N starvation switches the PSR off (marked by PSI genes high expression in −P control potassium nitrate [CKNO3] and low expression in −P control potassium chloride [CKCl] in Figure 4B). In heterogeneous conditions, the N-supply and N-demand long-distance signals (Ruffel et al., 2011) also influence PSR. Indeed, IPS1 and SPX1 were inhibited by the N-demand signal coming from the compartment without N (compareCNO3 and SpNO3 in Figure 4B). N-supply controls PSR genes in different ways: for IPS1, the N-supply signal acts as an IPS1 repressor, since RNA accumulation is stronger in CKCl than in split potassium chloride (SpKCl). On the other hand, SPX1 and PHT1-1 show induction in roots grown under SpKCl compared with CKCl (Figure 4B). This demonstrates that the N-supply long-distance signal indeed controls the PSR genes, but that this regulation is dependent on the gene identity.
Figure 4.
Local N Provision and N-Demand Long-Distance Signaling Controls PSR.
(A) Split-root conditions on N (schematic) were applied on P varying contexts. Roots in the same local environment are compared to evaluate the influence of distant treatments. CKNO3 and CKCl indicate control conditions (homogeneous KNO3 or KCl supply). SpKNO3 and SpKCl indicate heterogeneous conditions with KNO3 supply for one-half of the root system and KCl supply for the other part. Differences recorded between CKNO3 and SpKNO3 or between SpKCl and CKCl are due to treatments applied and sensed by the other side of the split root.
(B) Measurements of PSI genes by RT-qPCR in the N split-root context on two P conditions (see text for details of the experimental procedure). Values are mean ± se (n = 3). Results are from three independent experiments. Different letters indicate significant ANOVA followed by Tukey test (P < 0.05).
Taken together, these results strongly suggest that the PSR is under the control of N-related long-distance systemic signals (Ruffel et al., 2011). This constitutes an independent demonstration that PSR is actively shut down when N is lacking. This active repression of PSR relies at least partly on already defined local (Figures 1 to 3) and long-distance (Figure 4) signaling pathways. This opens perspectives for further studies on the molecular actors involved, since several genes have already been identified in these N-related signaling pathways (Ruffel et al., 2011, 2016; Li et al., 2014; Tabata et al., 2014; Ohkubo et al., 2017; Poitout et al., 2018).
PHO2 Is Transcriptionally Controlled by N Starvation
PHO2 is an E2 conjugase involved in the phosphate response as it is targeted by miR399 under P limiting conditions (Bari et al., 2006). PHO2 triggers the degradation of PHT1 transporter family members and PHO1 (Liu et al., 2012; Huang et al., 2013) under +P conditions. To investigate the existence of a link between PHO2 and N-related signals, we studied the steady state expression of PHO2 in roots of plants grown in the presence of different NO3− concentrations. P provision does not strongly impact PHO2 expression level, as previously described (Bari et al., 2006). Only a small but significant decrease in PHO2 transcripts was recorded in −P at 0.05 mM and 0.5 mM KNO3 (Figure 5A). Conversely, and more importantly, PHO2 transcript accumulation is regulated by N provision. Indeed, PHO2 shows mRNA accumulation in N-depleted conditions and, conversely, strong mRNA depletion by increasing concentration of NO3− in the media (Figure 5A). This N regulation is dependent on NRT1.1 activity, since it is partly lost in the chl1-5 mutant (Supplemental Figure 3). Thus, PHO2 is (i) repressed posttranscriptionally by P starvation (as shown in Bari et al., 2006) and (ii) downregulated by NO3− provision.
Figure 5.
PHO2 Is Regulated by N and Integrates the N Signal into the PSR Pathway.
(A) Plants were grown on combinations of P (0 or 0.5 mM, KH2PO4) and N (0, 0.05, 0.5, 1, 2.5 mM KNO3) for 14 d. Roots were harvested for measuring PHO2 by RT-qPCR measurements. Values are mean ± se (n = 3). Results are from three independent experiments.
(B) The wild-type and pho2-1 genotypes were grown on combinations of P (0 or 0.5 mM, KH2PO4) and N (0, 2.5 mM KNO3) for 11 d (top panel). Eleven-day-old plants from −P/−N conditions were transferred to −P and +/− N replenished media (bottom panels). Roots were harvested for measuring PSI gene transcription by RT-qPCR. Values are mean ± se (n = 3). Asterisks indicate significant differences from the wild-type plants (*P < 0.05; ***P < 0.001; Student’s t test). Nitrogen effect (ratio between +N/−N conditions) is reported in the panels. Col, Colombia (wild type).
PHO2 Is Responsible for the N-Dependent Control of PSR
Given the strong N-dependent transcriptional regulation of PHO2 (Figure 5A), we tested the hypothesis that PHO2 could integrate the N signal during the P starvation plant response. To this end, we studied the N control of PSR in pho2 mutants. We performed the same steady state and transfer experiments on the wild-type and pho2 genotypes. We observed that, indeed, in pho2 mutants the PSR of the vast majority of PSI genes is no longer N controlled (Figure 5B). In other words, the repressive effect of N depletion on the PSR response is strongly affected by pho2 mutation (Figure 5B). These results further demonstrate that PHO2 functions as an integrator of the N signal into the PSR. This observation was valid for IPS1, PHT1-1, and SPX1, as these genes appeared to be strongly derepressed and no longer N regulated in the pho2 mutant (Figure 5B). Interestingly, miR399D remained under N control in the pho2 background, revealing that the N effect on PSR contains PHO2-dependent and -independent branches (Figure 5B).
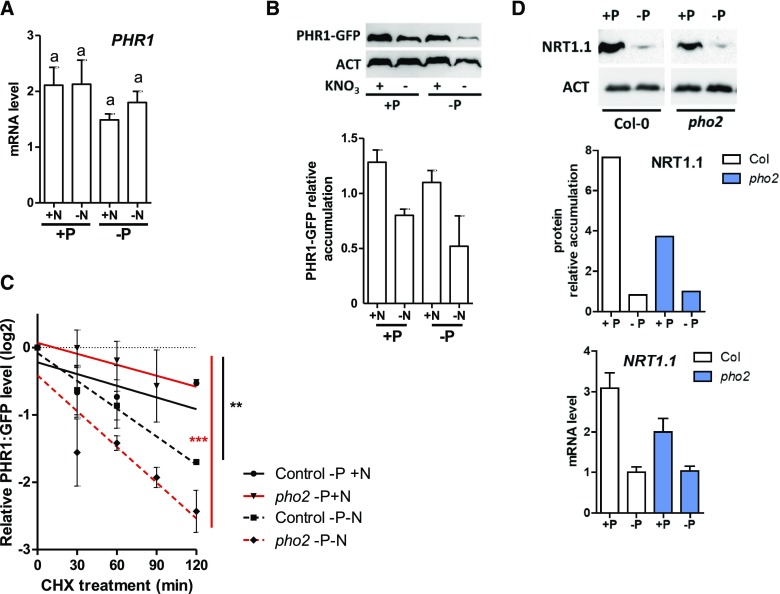
N Controls PHR1 Protein Accumulation and Half-Life
It is widely accepted that PSR is controlled by the PHR1 transcription factor (Bari et al., 2006; Franco-Zorrilla et al., 2007; Liu et al., 2012; Puga et al., 2014; Figure 2). We also hypothesized that the N/P crosstalk likely takes place upstream of the P signaling pathway since the phr1 phl1 double mutant retains NO3− sensing capabilities (Supplemental Figure 2B). We therefore tested whether PSR could be repressed in −N conditions through the regulation of PHR1 (being central to PSR; Puga et al., 2014). First, we studied the transcriptional regulation of PHR1 under varying N and P conditions. The results of this experiment did not yield a clear conclusion concerning the transcriptional regulation of this gene either by P or by N signals (Figure 6A), as previously reported by others (Rubio et al., 2001). We, therefore, investigated whether P/N signals affect PHR1 accumulation. To this end, a functional pPHR1:PHR1:GFP transgenic line was studied using antibodies directed against the GFP tag (Supplemental Figure 4). We detected a clear and reproducible decrease in PHR1 accumulation in N-deprived conditions (Figure 6B). Furthermore, we evaluated PHR1 turnover in response to N starvation (Figure 6C). We observed that the protein half-life (determined using cycloheximide [CHX] treatments) is decreased under −N conditions, explaining the potential protein accumulation as shown in Figure 6B.
Figure 6.
N Influences PHR1 Accumulation and Turnover; Phosphate via PHO2 Affects NRT1.1 Protein Accumulation.
(A) PHR1 mRNA accumulation in N and P varying conditions. Experimental conditions correspond to those described in Figure 3A. None of the responses display a significant difference.
(B) PHR1-GFP immunoblot and quantification relative to actin in three independent experiments. ACT, actin.
(C) PHR1-GFP turnover (following CHX treatment) is measured in the wild-type and pho2 genetic backgrounds in +N or −N conditions. This is a compilation of three independent experiments. Comparison of linear models to zero were performed using analysis of covariance (Prism, GraphPad [**P < 0.01; ***P < 0.001]).
(D) NRT1.1 immunoblot in response to P starvation in the wild-type and pho2-1 genetic backgrounds. NRT1.1 protein quantification and mRNA level in the same conditions. Uncropped version of the blot showing the bands on the same membrane are provided Supplemental Figure 6. Col-0, Columbia-0 (wild type).
These results demonstrate that N deprivation modifies both PHR1 half-life and accumulation, which likely explains the repression of PSR activity by the N depletion signal (Figure 1).
PHR1 Accumulation in Response to N Is Independent of PHO2
Having shown that (i) PHR1 is destabilized in response to −N and that (ii) PHO2 (a ubiquitin conjugase) is key for the downregulation of PSR by −N signal, we hypothesized that PHO2 might be responsible for the stability of PHR1 in response to −N. To this end, we also monitored the PHR1 GFP-tagged protein in the pho2 background. Interestingly, we did not observe a clear and reproducible misregulation of the protein accumulation or stability in the pho2 background compared with the wild type (Figure 6C). This observation led us to conclude that PHR1 stability is indeed controlled by −N (Figure 6), and PHO2 is a central point of crosstalk between the N response and the PSR (Figure 5); however, this does not occur through any direct regulation of PHR1 by PHO2.
PHO2 Controls the Nitrate Transceptor NRT1.1 and Vice Versa
Since PHO2 showed no influence on PHR1 accumulation, we tested the hypothesis that PHO2 could interfere with the NO3− signaling pathway instead (and affect PSR that way). To do so, we studied NRT1.1 accumulation (using specific antibodies; Medici et al., 2015) in the wild type and pho2 mutant grown on +P/+N media and then transferred (for 3 d) on +P or −P and +N media (Figure 6D). We confirmed that NRT1.1 protein accumulation is strongly repressed by P starvation conditions, as previously reported (Medici et al., 2015). Furthermore, we demonstrated that PHO2 acts a positive regulator of NRT1.1 since in the pho2 mutant, NRT1.1 levels are lower than in the wild type. This observation is consistent with previous proteomic analysis that determined that NRT1.1 is indeed strongly downregulated in the pho2 background (Huang et al., 2013). This finding demonstrates once more the complexity and connectivity between P and N signals. As NRT1.1 is at the forefront of the NO3− sensing controlling PNR in plants, we tested the pho2 mutant response to PNR and NSR (as a control, since NRT1.1 is not known to be involved in NSR). We found that pho2 mutants are affected in the late phase of PNR (Supplemental Figure 5A) and maintain a normal NSR response (Supplemental Figure 5B). This validates that (i) pho2 influences the NO3− response and (ii) pho2 might affect the PSR partly through its control of NRT1.1.
The Dependency of the PSR on Nitrogen Is Conserved in Crop Species
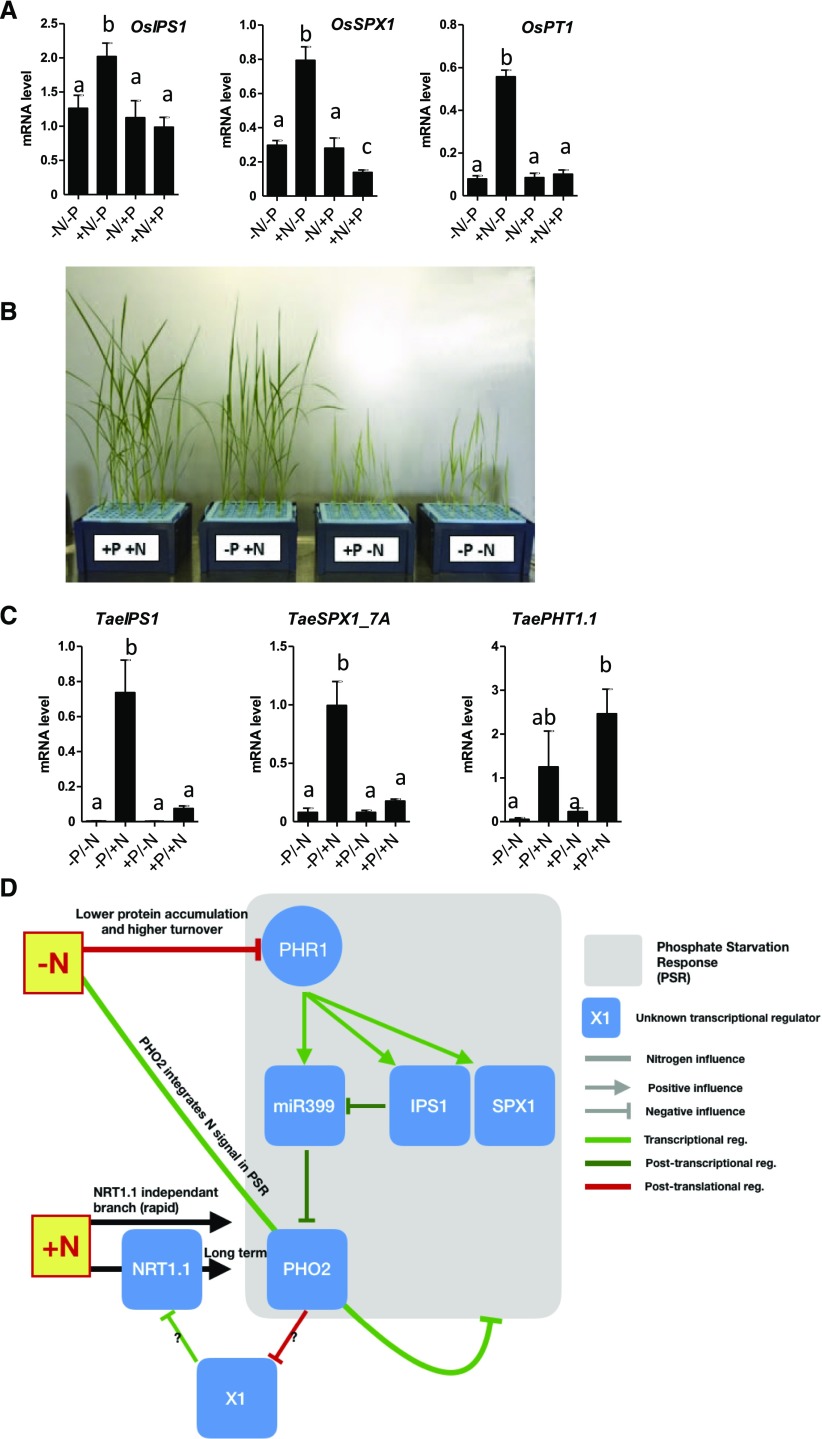
Given the strong similarity between the PSR pathways in Arabidopsis and rice, we examined whether the dependence of PSR on N was conserved in monocots such as rice and wheat (Triticum aestivum). First, we grew rice plantlets in media composed of four combinations of N and P, and we tested the response of several PSI genes (Figure 7A). As shown for Arabidopsis, rice PSI genes (OsIPS1, OsSPX1, OsPT1 [Os for O. sativa]) were upregulated by P starvation only in the presence of N. Thus, in rice, N deprivation also prevents activation of PSR (Figure 7A). Interestingly, during these investigations we noted that N deprivation strongly affected shoot growth in rice. However, the effect of N deprivation seems less important for plants deprived of P (compare −N/+P versus −N/−P conditions; Figure 7B). This highlights another important interaction between N and P at the level of rice shoot growth, which will be the topic of future investigations.
Figure 7.
N Control of PSR Operates in Rice and Wheat; Proposed Model of N Effect on PSR (Based on Arabidopsis Results).
(A) PSI gene response to N/P combinations in rice (see “Methods” for experimental conditions). Roots were harvested for measurements of PSI genes by RT-qPCR. Values are mean ± se (n = 3). Different letters indicate significant differences as determined by ANOVA followed by Tukey test (P < 0.05).
(B) Shoot phenotypes of rice grown on N/P combinations display a growth enhancement in −N/−P conditions compared with +P/−N.
(C) PSI gene response to N/P combinations in wheat (see “Methods” for experimental conditions). Roots were harvested for measurements of PSI genes by RT-qPCR. Values are mean ± se (n = 3). Different letters indicate significant differences as determined by ANOVA followed by Tukey test (P < 0.05).
(D) Model summarizing the different molecular entry points of N signals in the control of PSR. PHO2/NRT1.1 co-control is hypothesized to be central in this crosstalk. reg, regulation.
We also evaluated N control of PSR genes in wheat. To do so, we grew wheat (cv Chinese Spring) plants in −P/−N media for 11 d and then studied the PSR in roots of plants transferred for 24h onto media containing different combinations of P and NO3−. The response of IPS1 and SPX1 wheat homologs (TaeIPS1, TaeSPX1_7A [Tae for T. aestivum]) confirmed that the PSR is repressed in −P/−N (Figure 7C). However, TaePHT1.1 does not display the prototypical N/P crosstalk response since it seems to be controlled solely by N provision.
Taken together, these investigations demonstrate that the control of PSR by N is a general mechanism that is likely conserved across a wide range of plant species. The potential conservation of the molecular mechanisms that we uncovered in Arabidopsis is discussed below.
DISCUSSION
Here, we report a discovery of a conserved mechanism of PSR control by N availability in plants. Previous studies have reported such evidence for N and P interaction in the control of ACID PHOSPHATASE5 (ACP5), which encodes an acid phosphatase involved in P recycling (Cerutti and Delatorre, 2013); however, an advanced mechanistic explanation for these observations was lacking. Using Arabidopsis, we provide insights into the molecular actors involved in N and P signaling crosstalk, leading to a model presented in Figure 7D. Briefly, N control of PSR is dependent on the NO3− sensing– and Pi sensing–related proteins NRT1.1 and PHR1/PHL1, respectively. This demonstrates that this crosstalk is rooted in the key components of both signaling pathways, the nutrient sensors. Moreover, we show that the PHO2 gene is an essential element of the N signal transduction into the PSR as (i) the PHO2 gene is transcriptionally activated upon N deprivation and (ii) pho2 mutant displays an important derepression of the PSI genes that includes a desensitization of the PSI genes to N (Figure 5). In other words, N effect on PSR is strongly affected in the pho2 genetic background, consistent with the proposed role for PHO2 as the integrator of the two signals (Figure 5).
We also demonstrate that N affects PHR1 protein accumulation and half-life, which likely explains the effect of N on PSR (Figure 6). Interestingly, we did not find a robust and reproducible effect of PHO2 on PHR1 protein turnover and accumulation. This led us to hypothesize that the control of the PSR by N is likely multipronged (Figure 7D), with an important branch of the pathway consisting of control of both PHO2 and NRT1.1 via feedback control of each other (Figures 6D and 7D; Supplemental Figure 3).
From a PSR-only perspective, our results also indirectly imply that PHO2, being downstream of the PSR (Figure 7D; Briat et al., 2015), is also likely upstream of it. Indeed, in the pho2 mutant, we not only recorded a loss of N effect but also a very strong upregulation of the PSI genes (Figure 5). Thus, in the light of these results, the apparent linearity of the PSR (Briat et al., 2015; Puga et al., 2017) should be revised. A similar feedback mechanism proposed by Liu et al. (2010) in rice, where OsPHO2 negatively control OsSPX1 and OsIPS1, supports our model. We believe that PHO2 can be at the same time downstream and upstream of the PSR, defining a potential circular signaling pathway.
A Suggested Priority of N Signal on P
Our work provides evidence for an apparent prioritization of the plant for N. Indeed, in our experiments we demonstrated a very strong influence of N starvation on PSR, when the reciprocal (P effect of NSR) is of more modest effect in Arabidopsis (Figure 2B). Thus, plants seem to bypass PSR in order to wait for more N favorable conditions. Indeed, in the same set of experiments (Figure 2), we observe that the core N starvation is still active regardless of P provision (Figure 2B). Explanations for such an apparent priority may reside in the fact that P foraging and retrieval are likely very dependent on plant capacity to grow, since P is very immobile in the soil. Thus, plant may invest in growth and P retrieval only if N is available potentially provided by rain, leaching, and bacterial burst activity in the soil. Since P and N responses are linked to hormonal signals on many aspects, it will be interesting to investigate whether this N/P crosstalk is dependent on hormones such as auxin (Krouk et al., 2010a; Krouk, 2016; Ristova et al., 2016), cytokinins (Franco-Zorrilla et al., 2002, 2005; Ruffel et al., 2011; Poitout et al., 2018; Ruffel, 2018), or strigoloactones (Mayzlish-Gati et al., 2012), for instance.
From a molecular systems perspective, the phenomenon that we describe here resembles an electronic logic gate. Indeed, PSR genes get activated only when N, and not P, signals are combined. This echoes recent studies (Kellermeier et al., 2014; Ristova et al., 2016) that revealed that plants indeed perceive external and internal signals in combinations and that it is likely to be a rule rather that an exception.
In conclusion, we believe that the identification of the molecular actors (PHR1, PHO2, NRT1.1) that integrate N and P signals brings us closer to the understanding of an evolved logic gate in plant cells. This may have also important consequences in agricultural practices, or biotechnology, by opening research avenues toward the uncoupling of these signaling pathways to adapt genotypes to particular agricultural conditions.
METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana; Columbia-0) background mutants were previously described: chl1-5 (Tsay et al., 1993), phr1-3 phl1-2 (Bournier et al., 2013), and pho2-1 (Bari et al., 2006). The pPHR1:PHR1-GFP construct was prepared by PCR amplification from the wild-type plants of a genomic fragment comprising the PHR1 gene promoter region (up to 2063 bp upstream of the start site), and all exons and introns, using the following primers: pPHR1HindIII.BF, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATAAGCTTTCAGCAACAGAGGAAGAGGTG-3′ and pPHR1HindIII.BR, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAACGAAGCTTCATTGTTGTCCTGCAAGAG-3′. The PCR product was cloned using Gateway technology (Invitrogen) into the pDONR207 (Invitrogen) plasmid by the BP reaction and subsequently transferred by the LR reaction to the pGWB4 plasmid, for fusion to GFP. The resulting construct was used to transform Arabidopsis phr1 mutant plants by the floral dip method. After T1 selection based on hygromycin resistance, the T2 segregating progenies were selected based on the complementation of phr1 mutant phenotypes to isolate homozygous plants for the construct. pPHR1:PHR1:GFP × pho2-1 plants were obtained by crossing the two homozygous parental lines. The generated F2 plants were verified for the presence of the GFP and the homozygous mutation of the pho2-1 locus by sequencing the specific region (using primers listed in Supplemental Table 1). Expression of PSI genes in rice (Oryza sativa) and wheat (Triticum aestivum) was measured respectively in rice plants (cv Nipponbare) and in wheat plants (cv Chinese Spring).
Each experiment was performed at least twice (most of them three times). Representative results are reported in the figures. Figures 1, 2, 5, and 6C represent a compilation of three independent experiments with several hundreds of plants pooled per sample.
Growth Conditions and Treatments
For Arabidopsis root transcript accumulation analysis, plants were grown on vertical plates. The wild-type and mutant seeds were sterilized and sown on the surface of solid media consisting of N- and P-free Murashige and Skoog basal salt medium supplemented with KNO3 at different concentrations (0.05, 0.5, 1, and 2.5 mM), 3 mM Suc, MES (0.5 g/L), and 0.8% (w/v) agarose. For the P-sufficient condition, 0.5 mM KH2PO4 was added. Different volumes of 1 M KCl solution were added to the media to keep the K+ concentration constant across the different conditions. Plants were grown vertically for 9 d in day/night cycles (at 16/8 h; 90 μmol photons m−2 s−1) at 22°C. Light is provided by a mix of sodium-vapor and metal halide 400-W lamps (in growth chambers used for hydroponic culture) and Osram 18-W 840 Lumilux neon tubes (for in vitro plant growth).
For the transfer experiment, 9-d-old seedlings were transferred to new plates and grown for three more days under the same conditions. The conditions for the P or N starvation experiments in a sterile hydroponic system were the same as described in Medici et al. (2015), with 2.5 mM KNO3. P starvation treatment was applied for 72 h. Every sample contains at least 30 plant roots from different plants. Experiments were repeated at least twice
For rice experiments, the wild-type rice (cv Nipponbare) plants were grown in a hydroponic nonsterile system. After overnight soaking in deionized water in the dark, the seeds were transferred to 1/4 full-strength Yoshida media for 10 d. For treatment conditions, plants were transferred to a Yoshida modified nutrient solution [1.43 mM NH4NO3, 1.64 mM MgSO4, 0.75 mM CaCl2, 0.51 mM K2SO4, 0.33 mM NaH2PO4, 0.02 mM H3BO3, 0.01 mM MnCl2, 0.04 mM Fe-NaEDTA, 2.5 µM ZnSO4, 0.16 µM CuSO4, 0.08 µM (NH4)6Mo7O24, and 2.5 µM MES buffer, pH 5.5], with P- or N-modified contents (see legends of the figures) that was renewed every 5 d. Plants were cultivated for 10 d in these N/P-varying media under a light/dark cycle (14/10 h) at 28/25°C and 80% RH. Light is provided by a mix of sodium-vapor and metal halide 400-W lamps.
For wheat experiments, seeds of wheat (cv Chinese Spring) were surface sterilized using sodium hypochlorite (4% [w/v]), preimbibed for 3 d, and transferred to a hydroponic setup where the seeds were germinated and grown for 11 d in deionized water in growth conditions of 24°C and 16 h (light)/8 h (dark). The seedlings were then transferred to four treatment levels: +P+N, −P+N, +P−N, and −P−N. Modified Hoagland solution [2 mM Ca(NO3)2, 0.75 mM MgSO4, 0.7 mM K2SO4, 0.5 mM KH2PO4, 0.1 mM KCl, 100 μM FeEDTA, 5 μM ZnSO4, 1 μM MnSO4, 1 μM H3BO3, 0.2 μM CuSO4, and 0.01 μM (NH4)6Mo7O24, pH 5.8] was used for +P+N condition. For −N treatment, Ca(NO3)2 was replaced with CaSO4 at 0.5 mM; for −P treatment, KH2PO4 was replaced with KCl at 0.5 mM; and for −P−N treatment, Ca(NO3)2 and KH2PO4 were replaced with 0.5 mM CaSO4 and 0.5 mM KCl, respectively. The plants were allowed to grow in the four treatment levels for 24 h. Next, total roots were harvested, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction. For each treatment level, three biological replicates were performed, with each sample containing root samples from four to six plants. RT quantitative PCR (qPCR) was performed on cDNA samples using a CFX 384 Real-Time System machine (C1000Touch, Bio-Rad) and whenever possible homeologue-specific primers were used. EF1a was used as the housekeeping gene.
Split-Root Growth Conditions
The split-root experiment was performed in square Petri dishes (12 × 12 cm) based on a previously published protocol (Ruffel et al., 2011). Arabidopsis Columbia-0 seeds were surface sterilized and sown on a basal salt medium, with 3 mM Suc, MES (0.5 g/L), 1% (w/v) agarose, 1 mM KH2PO4, 0.5 mM NH4-succinate, and 0.1 mM KNO3. After 12 d, the primary root was cut below the second emerging lateral root. Three days later, the two newly formed secondary roots were separated on the same media to allow proper development. Plants were the transferred on media with four different combinations of N and P: −P/−N (KCl, 3 mM), +P/+N (KNO3, 2.5 mM; KH2PO4, 0.5 mM), −P/+N (KCl, 0.5 mM; KNO3, 2.5 mM), and +P/−N (KH2PO4, 0.5 mM; KCl, 2.5 mM). KCl, KNO3, and KH2PO4 solutions were spread independently on each side of the solidified media. All the root samples were collected after 3 d.
Real-Time qPCR Analysis
Total RNA was isolated from Arabidopsis, rice, and wheat root tissues, previously harvested and stored in liquid N. DNase treatment, RT, and qPCR conditions were the same as described in Medici et al. (2015). Specific primer pairs used for RT-qPCR analysis in Arabidopsis and rice are listed in Supplemental Table 2.
Soluble Protein Isolation and Immunoblot Analysis
Protein isolation and immunoblot with GFP antibody were performed and quantified as described previously (Medici et al., 2015). For protein loading normalization, anti-actin antibody (1:5000, AS132640, Agrisera) and goat anti-rabbit IgG (1:10,000, AS09602, Agrisera) were used. CHX was applied at 100 µM in DMSO solution (used as mock control).
Transcriptomic Analyses
Total RNA was extracted from frozen and ground root tissues using TRIzol reagent (Thermo Fisher Scientific). RNA integrity and concentration were determined using a 2100 Bioanalyzer Instrument (Agilent) and RNA 6000 Nano kit (5067-1511, Agilent). DNA was removed by digestion with DNase I (AMPD1, Sigma). Gene expression measurements were performed using Arabidopsis Affymetrix Gene1.1 ST array strips. Biotin-labeled and fragmented cRNAs were obtained using GeneChip WT PLUS Reagent kit (902280, Thermo Fisher Scientific) following the manufacturer’s instructions. Hybridization on array strips was performed for 16 h at 48°C. Arrays are washed, stained, and scanned using a GeneAtlas HWS kit (901667, Thermo Fisher Scientific) on the GeneAtlas Fluidics and Imaging Station.
All data manipulations were performed on R (http://www.r-project.org/). The ANOVA was performed using the R aov() function on GC Robust Multi-Array Average data (logged data). A probe signal has been modeled as follows: Yi = α1.N + α2.P + α3.N*P + ε, where α1 to α3 represent the coefficient quantifying the effect of each of the factors (N, P) and their interactions (N*P), and ε represents the nonexplained variance. The false discovery rate was determined to be <5%. We determined a gene as regulated by N (Figure 2B) if the P-value associated with α1 was <0.001. We determined a gene as regulated by P (Figure 2A) if the P-value associated with α2 was <0.001. Raw data and whole genome ANOVA results are provided in Supplemental Data Set 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CHL1/NRT1.1/NPF6.3 (At1g12110); PHO2 (At2g33770), PHR1 (At4g28610), and PHL1 (AT5G29000).
Supplemental Data
Supplemental Figure 1. Plant phenotypes following 11 days of growth on P/N varying conditions.
Supplemental Figure 2. phr1 and phr1 phl1 mutations affect N/P crosstalk but primary nitrate response (PNR) is still active in the phr1 phl1 double mutant.
Supplemental Figure 3. PHO2 mRNA level response to N is affected by the chl1-5 mutation.
Supplemental Figure 4. Characterization of pPHR1:PHR1:GFP transgenic plants.
Supplemental Figure 5. pho2 mutation affects PNR and not NSR consistently with its effect of NRT1.1 protein level.
Supplemental Figure 6. Un-cropped version of the immunoblot presented in Figure 6D.
Supplemental Table 1. Primer pair for genotyping pPHR1:PHR1:GFP (pho2-1) plants.
Supplemental Table 2. qRT-PCR primer pairs for A. thaliana and rice.
Supplemental Data Set 1. Transcriptomic data.
Supplemental Data Set 2. ANOVA Tukey test results.
Supplemental Data Set 3. List of P regulated genes.
Supplemental Data Set 4. List of N regulated genes.
Supplemental Data Set 5. List of NxP regulated genes.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported in the Honude group (Biochemistry & Plant Molecular Physiology) by Agence Nationale de la Recherche (IMANA ANR-14-CE19-0008 with a doctoral fellowship to A.S.), by the Centre National de la Recherche Scientifique (CNRS LIA-CoopNet to G.K.), and by the National Science Foundation (NSF IOS 1339362-NutriNet). Research in V.R.’s laboratory was funded by the Ministry of Economy and Competitiveness and AEI/FEDER/European (grants BIO2013-46539-R and BIO2016-80551-R).
Author Contributions
G.K., A.M., and W.S. designed the project. A.M., W.S., S.R., P.D., A.S., I.M.D., C.S., A.E., V.R., H.R., and G.K. performed the experiments and analyzed the data. A.M., W.S., B.L., S.R., M.T., H.R., and G.K. contributed to the study design during the project course. G.K., A.M., M.T., and H.R. wrote the article.
References
- Bari R., Datt Pant B., Stitt M., Scheible W.-R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournier M., Tissot N., Mari S., Boucherez J., Lacombe E., Briat J.-F., Gaymard F. (2013). Arabidopsis Ferritin 1 (AtFer1) gene regulation by the Phosphate Starvation Response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 288: 22670–22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J.F., Rouached H., Tissot N., Gaymard F., Dubos C. (2015). Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: Potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front. Plant Sci. 6: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez-Pérez J., Solano R., Leyva A., Paz-Ares J. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., Taconnat L., Renou J.P., Daniel-Vedele F., Fernandez E., Meyer C., Krapp A. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57: 426–435. [DOI] [PubMed] [Google Scholar]
- Cerutti T., Delatorre C.A. (2013). Nitrogen and phosphorus interaction and cytokinin: responses of the primary root of Arabidopsis thaliana and the pdr1 mutant. Plant Sci. 198: 91–97. [DOI] [PubMed] [Google Scholar]
- Crawford N.M., Glass A.D.M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3: 389–395. [Google Scholar]
- Franco-Zorrilla J.M., Martin A.C., Solano R., Rubio V., Leyva A., Paz-Ares J. (2002). Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 32: 353–360. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Martín A.C., Leyva A., Paz-Ares J. (2005). Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 138: 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gansel X., Muños S., Tillard P., Gojon A. (2001). Differential regulation of the NO3− and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: Relation with long-distance and local controls by N status of the plant. Plant J. 26: 143–155. [DOI] [PubMed] [Google Scholar]
- Guan P., Ripoll J.J., Wang R., Vuong L., Bailey-Steinitz L.J., Ye D., Crawford N.M. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 114: 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham B.K., Chen J., Yan Y., Lucas W.J. (2018). Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 49: 1–9. [DOI] [PubMed] [Google Scholar]
- Heuer S., Gaxiola R., Schilling R., Herrera-Estrella L., López-Arredondo D., Wissuwa M., Delhaize E., Rouached H. (2017). Improving phosphorus use efficiency: A complex trait with emerging opportunities. Plant J. 90: 868–885. [DOI] [PubMed] [Google Scholar]
- Ho C.H., Lin S.H., Hu H.C., Tsay Y.F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hu H.C., Wang Y.Y., Tsay Y.F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57: 264–278. [DOI] [PubMed] [Google Scholar]
- Huang T.K., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.-Y., Ried M.K., Hothorn M., Poirier Y. (2018). Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr. Opin. Biotechnol. 49: 156–162. [DOI] [PubMed] [Google Scholar]
- Kant S., Peng M., Rothstein S.J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genet. 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F., Armengaud P., Seditas T.J., Danku J., Salt D.E., Amtmann A. (2014). Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Inaba J., Kudo T., Ueda N., Konishi M., Mitsuda N., Takiguchi Y., Kondou Y., Yoshizumi T., Ohme-Takagi M., Matsui M., Yano K., et al. (2018). Repression of nitrogen-starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. Plant Cell 30: 925–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisko M., Bouain N., Safi A., Medici A., Akkers R.C., Secco D., Fouret G., Krouk G., Aarts M.G., Busch W., Rouached H. (2018). LPCAT1 controls phosphate homeostasis in a zinc-dependent manner. eLife 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., et al. (2010a). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937. [DOI] [PubMed] [Google Scholar]
- Krouk G. (2016). Hormones and nitrate: a two-way connection. Plant Mol. Biol. 91: 599–606. [DOI] [PubMed] [Google Scholar]
- Krouk G., Mirowski P., LeCun Y., Shasha D.E., Coruzzi G.M. (2010b). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S., Edel K.H., Pervent M., Hashimoto K., Corratgé-Faillie C., Offenborn J.N., Tillard P., Gojon A., Kudla J., Lacombe B. (2015). Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 8: ra43. [DOI] [PubMed] [Google Scholar]
- Li W., Lan P. (2015). Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopsis roots. BMC Res. Notes 8: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Krouk G., Coruzzi G.M., Ruffel S. (2014). Finding a nitrogen niche: A systems integration of local and systemic nitrogen signalling in plants. J. Exp. Bot. 65: 5601–5610. [DOI] [PubMed] [Google Scholar]
- Lin W.Y., Huang T.K., Chiou T.J. (2013). NITROGEN LIMITATION ADAPTATION, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Wang Z., Ren H., Shen C., Li Y., Ling H.-Q., Wu C., Lian X., Wu P. (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62: 508–517. [DOI] [PubMed] [Google Scholar]
- Liu T.-Y., Huang T.-K., Tseng C.-Y., Lai Y.-S., Lin S.-I., Lin W.-Y., Chen J.-W., Chiou T.-J. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Tang R.J., Zheng X.J., Wang S.M., Luan S. (2015). The calcium sensor CBL7 modulates plant responses to low nitrate in Arabidopsis. Biochem. Biophys. Res. Commun. 468: 59–65. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Konishi M., Kiba T., Sakuraba Y., Sawaki N., Kurai T., Ueda Y., Sakakibara H., Yanagisawa S. (2018). A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 9: 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C., Roudier F., Castaings L., Bréhaut V., Blondet E., Colot V., Meyer C., Krapp A. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4: 1713. [DOI] [PubMed] [Google Scholar]
- Mayzlish-Gati E., et al. (2012). Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol. 160: 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A., Krouk G. (2014). The primary nitrate response: A multifaceted signalling pathway. J. Exp. Bot. 65: 5567–5576. [DOI] [PubMed] [Google Scholar]
- Medici A., Marshall-Colon A., Ronzier E., Szponarski W., Wang R., Gojon A., Crawford N.M., Ruffel S., Coruzzi G.M., Krouk G. (2015). AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6: 6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J.A., Vega A., Bouguyon E., Krouk G., Gojon A., Coruzzi G., Gutiérrez R.A. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9: 837–856. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y., Tanaka M., Tabata R., Ogawa-Ohnishi M., Matsubayashi Y. (2017). Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Pal S., Kisko M., Dubos C., Lacombe B., Berthomieu P., Krouk G., Rouached H. (2017). TransDetect identifies a new regulatory module controlling phosphate accumulation. Plant Physiol. 175: 916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.S., Seo J.S., Chua N.-H. (2014). NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Hannam C., Gu H., Bi Y.M., Rothstein S.J. (2007). A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 50: 320–337. [DOI] [PubMed] [Google Scholar]
- Poitout A., Crabos A., Petřík I., Novák O., Krouk G., Lacombe B., Ruffel S. (2018). Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-zeatin in shoots. Plant Cell 30: 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga M.I., et al. (2014). SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga M.I., Rojas-Triana M., de Lorenzo L., Leyva A., Rubio V., Paz-Ares J. (2017). Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Curr. Opin. Plant Biol. 39: 40–49. [DOI] [PubMed] [Google Scholar]
- Qi W., Manfield I.W., Muench S.P., Baker A. (2017). AtSPX1 affects the AtPHR1-DNA-binding equilibrium by binding monomeric AtPHR1 in solution. Biochem. J. 474: 3675–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristova D., Carré C., Pervent M., Medici A., Kim G.J., Scalia D., Ruffel S., Birnbaum K.D., Lacombe B., Busch W., Coruzzi G.M., Krouk G. (2016). Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci. Signal. 9: rs13. [DOI] [PubMed] [Google Scholar]
- Riveras E., Alvarez J.M., Vidal E.A., Oses C., Vega A., Gutiérrez R.A. (2015). The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 169: 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martín A.C., Iglesias J., Leyva A., Paz-Ares J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S. (2018). Nutrient-related long-distance signals: Common players and possible crosstalk. Plant Cell Physiol. 59: 1723–1732. [DOI] [PubMed] [Google Scholar]
- Ruffel S., Krouk G., Ristova D., Shasha D., Birnbaum K.D., Coruzzi G.M. (2011). Nitrogen economics of root foraging: Transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. USA 108: 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S., Poitout A., Krouk G., Coruzzi G.M., Lacombe B. (2016). Long-distance nitrate signaling displays cytokinin dependent and independent branches. J. Integr. Plant Biol. 58: 226–229 [DOI] [PubMed] [Google Scholar]
- Safi A., Medici A., Szponarski W., Marshall-Colon A., Ruffel S., Gaymard F., Coruzzi G., Lacombe B., Krouk G. (2018). HRS1/HHOs GARP transcription factors and reactive oxygen species are regulators of Arabidopsis nitrogen starvation response. bioRxiv 10.1101/164277. [DOI] [Google Scholar]
- Sutton M.A., Oenema O., Erisman J.W., Leip A., van Grinsven H., Winiwarter W. (2011). Too much of a good thing. Nature 472: 159–161. [DOI] [PubMed] [Google Scholar]
- Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346. [DOI] [PubMed] [Google Scholar]
- Tsay Y.-F., Schroeder J.I., Feldmann K.A., Crawford N.M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713. [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 111: 14953–14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Tischner R., Gutiérrez R.A., Hoffman M., Xing X., Chen M., Coruzzi G., Crawford N.M. (2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136: 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xing X., Wang Y., Tran A., Crawford N.M. (2009). A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 151: 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Wang R., Zhao L., Zhang C., Li Z., Lei Z., Liu F., Guan P., Chu Z., Crawford N.M., Wang Y. (2016). The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28: 485–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Ding H., Zhu J.K., Zhang F., Li W.X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190: 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]