A RING-type E3 ligase ubiquitinates and degrades both itself and ACS2, thereby controlling the biosynthesis of ethylene, which fine-tunes cell division and fruit elongation in a dose-dependent manner.
Abstract
Plant organ growth and development are determined by a subtle balance between growth stimulation and inhibition. Fruit size and shape are important quality traits influencing yield and market value; however, the underlying mechanism regulating the balance of fruit growth to achieve final size and shape is not well understood. Here, we report a mechanistic model that governs cucumber (Cucumis sativus) fruit elongation through fine-tuning of ethylene homeostasis. We identified a cucumber mutant that bears short fruits owing to repressed cell division. SF1 (Short Fruit 1) encodes a cucurbit-specific RING-type E3 ligase, and the mutation resulted in its enhanced self-ubiquitination and degradation, but accumulation of ACS2 (1-aminocyclopropane-1-carboxylate synthase 2), a rate-limiting enzyme for ethylene biosynthesis. The overproduction of ethylene contributes to the short-fruit phenotype of sf1. Dysfunction of ACS2 resulted in reduced ethylene production, but still repressed cell division and shorter fruit, suggesting that ethylene is still required for basal fruit elongation. SF1 ubiquitinates and degrades both itself and ACS2 to control ethylene synthesis for dose-dependent effect on cell division and fruit elongation. Our findings reveal the mechanism by which ethylene dosage is regulated for the control of cell division in developing fruit.
INTRODUCTION
Fruit, corresponding to the plant ovary, protects the developing seeds, and provides humans with a source of food and nutrition. Cucurbits are a large and diverse plant family that supply many important fruits (Bisognin, 2002). Cucurbit fruits develop from inferior ovaries and are well known for their extreme diversity in fruit size and shape (Grumet and Colle, 2016; Colle et al., 2017), which are also important agronomic traits for determining crop yield and external quality. Classic fruit morphogenesis is determined by four physiological phases of early development: ovary growth, fruit set, rapid cell division, and subsequent cell expansion (Grumet and Colle, 2016). However, the genetic and molecular basis in these developmental phases is largely unknown.
Plant organ growth is determined by a subtle balance between growth stimulation and inhibition, which confer optimal plasticity in response to endogenous or environmental cues. Cucurbit fruit development initiates with the formation of a female or bisexual flower, during which ethylene plays an essential role (Chen et al., 2016). The gaseous hormone ethylene coordinates numerous aspects of plant growth and responses to stress (Wen, 2014). The inhibition of cell expansion is a well-described ethylene response (Kieber et al., 1993). The role of ethylene in fruit development has recently been investigated. In tomato (Solanum lycopersicum), ethylene plays a role in suppressing fruit set in coordination with auxin and gibberellin (Shinozaki et al., 2015). In zucchini (Cucurbita pepo), exogenous application of ethylene inhibitors promotes parthenocarpic fruit growth (Martínez et al., 2013). Although ethylene is generally considered to inhibit growth, it also stimulates growth in specific tissues and cells or at low concentrations. For instance, in Arabidopsis (Arabidopsis thaliana), ethylene modulates the cell division and quiescence of stem cells in the roots (Ortega-Martínez et al., 2007). In cucumber (Cucumis sativus) seedlings, transient treatment with physiological concentrations of ethylene stimulates cell division and alters cell division polarity (Kazama et al., 2004). Therefore, the fine tuning of ethylene production in different tissues at different developmental phases is critical for plant development, influencing both the stimulation and repression of growth (Pierik et al., 2006). However, little is known about the genetic regulation of ethylene dosage for regulating the balance of fruit growth, and the mechanistic basis for the dose-dependent ethylene response remains speculative.
Here, we report that the RING-type E3 ligase short fruit 1 (SF1) regulates fruit cell division in cucumber by modulating ethylene dosage via ubiquitination and 26S proteasome-dependent degradation of both itself and 1-aminocyclopropane-1-carboxylate synthase 2 (ACS2). We showed dosage effects of ethylene on cell division and fruit elongation in cucumber using exogenous treatments involving a series of ethylene concentrations. The findings demonstrate a genetic basis of ethylene dose-dependent control for the balance of cell division in developing fruit.
RESULTS
A Cucurbit-Specific RING-Type E3 Ligase
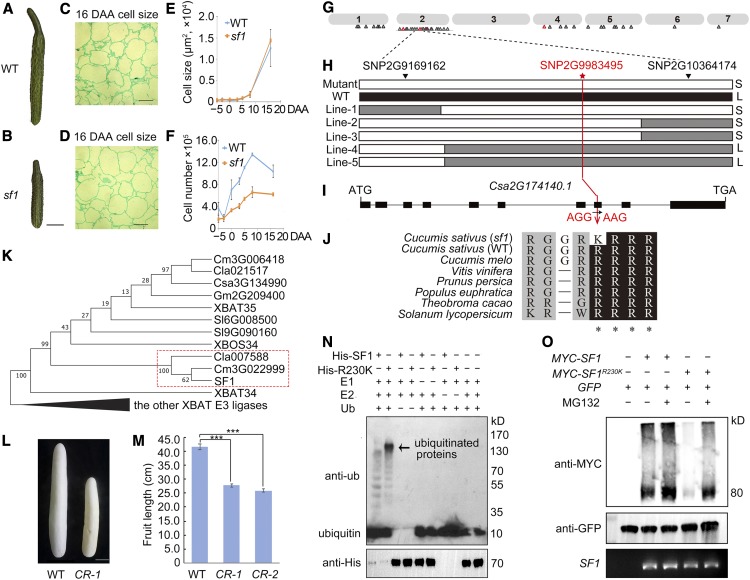
Using ethyl methanesulfonate mutagenesis, we identified a cucumber mutant bearing shortened fruits (sf1). Fruit morphogenesis was then characterized during early development stages (Figures 1A and 1B; Supplemental Figure 1A). We observed that the fruit length of the wild type (406 line) exhibits more rapid growth rate than that of the mutant (sf1) from 0 d after anthesis (DAA) to 8 DAA, whereas the fruit diameter does not significantly differ between the mutant and wild type (Supplemental Figures 1B and 1C). Considering that the average cell size of the sf1 mutant was similar to that of the wild type along the longitudinal section (Figures 1C and 1D), we inferred that early cell division is repressed in sf1. Further investigation revealed no significant difference in cell number or cell size between the wild type and sf1 along the transversal axis (Supplemental Figures 1D and 1E). However, on the longitudinal axis, although the average cell size was also identical (Figure 1E), the cell number was clearly lower in sf1 (Figure 1F). The 8 DAA fruit of the sf1 mutant contained nearly 50% fewer cells than did that of the wild-type plants (Figure 1F). These results indicate that the decreased fruit length associated with sf1 results from reduced cell numbers along the longitudinal axis but not from cell expansion.
Figure 1.
Characterization and Identification of SF1.
(A) and (B) When compared with wild type (WT; A), sf1 (B) bears short fruit. Scale bar = 5 cm.
(C) and (D) Mesocarp cells of wild type (C) and sf1 (D) at 16 DAA along a longitudinal section. Scale bar = 50 μm.
(E) The average cell size shows no significant difference between wild type and sf1. Bars = means from three independent fruits ± sem.
(F) The cell proliferation rate is substantially arrested in the sf1 fruit compared with wild type along a longitudinal axis. Bars = means from three independent fruits ± sem.
(G) Genomic distribution of homozygous SNPs. Hollow triangles indicate SNPs.
(H) Linkage analysis of the F2 population using dCAPS markers. The red star indicates the only SNP cosegregating with the short fruit phenotype.
(I) Gene structure of Csa2G174140.
(J) Alignment of Csa2G174140 homologs from diverse species.
(K) SF1 is a cucurbit-specific branch of an XBAT-related E3 ligase.
(L) and (M) Both the SF1 null-mutant plants CR-1 and CR-2 displayed significantly decreased fruit length. Bars = means of three fruits from three independent plants ± sem. Scale bar = 5 cm. *** P < 0.01 (t test, one tail).
(N) In vitro ubiquitination reactions. Those proteins were purified from E. coli extracts; (+) and (−), respectively, represent participation and absence in the reaction. Proteins were separated by standard SDS-PAGE and analyzed by immunoblotting using anti-ubi and anti-His. Arrows indicate the ubiquitinated proteins.
(O) Agroinfiltration expression in Nicotiana benthamiana was performed to confirm the self-ubiquitination ability of CsSF1 in vivo. The (+) and (−), respectively, represent participation and absence in the reaction. The SF1 mRNA expression levels and the GFP protein amount that were detected as internal controls remained consistent across all lanes.
To genetically map the sf1 locus, an F2 population was constructed by crossing sf1 plants with wild-type plants. All F1 plants exhibited the same normal fruit-length phenotype as the wild-type plants. In the F2 population of 130 individuals, 23 plants displayed the short-fruit trait, whereas the remaining 107 plants were normal. The segregation pattern fit the Mendelian ratio of 3:1 (χ2= 3.70, P < 0.01, t test, one tail), suggesting that sf1 is a single recessive allele (Supplemental Table 1). To clone the candidate gene, whole-genome resequencing (> 10× coverage) of pooled genomic DNA samples from 20 short-fruit individuals (mutant pool) and 20 normal individuals (normal pool) from the F2 population was performed, and we identified 543 G-to-A (or C-to-T) single-nucleotide polymorphisms (SNPs) with a SNP index of > 0.7 between the two pools (Supplemental Data Set 1). We considered that the genomic region on chromosome 2 enriched with SNPs harbors the causal mutation for sf1 (Figure 1G). Linkage analysis within the F2 population further delimited the sf1 locus to a 1.2-Mb interval between SNP2G9169162 and SNP2G10364174, in which only SNP2G9983495 cosegregated with a short-fruit phenotype (Figure 1H). Within the interval, no other homozygous variants were discovered in the mutant pool. These results indicate that SNP2G9983495 is the causative mutation of the short-fruit phenotype.
SNP2G9983495 (G to A) is located within Csa2G174140 (Figure 1I), which is annotated as encoding a putative RING-type E3 ligase, SF1. The G-to-A mutation results in an amino acid change from Arg (R) to Lys (K) at residue 230 (R230K). Alignment of the SF1 amino acid sequence with that of homologous proteins from various plant species showed that R-230 is conserved among land plants (Figure 1J). Phylogenetic analysis revealed that SF1 and its orthologs in melon and watermelon constitute a cucurbit-specific branch of the E3 ligase that belongs to the XB3 ortholog 4 in Arabidopsis thaliana (XBAT) family (Figure 1K; Supplemental Table 2; Supplemental Data Set 2; Nodzon et al., 2004).
To confirm that SF1 is the causative gene genetically, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) was used to generate several SF1-edited plants, including two null mutants – one with a homozygous deletion of 4 bp and another with a homozygous deletion of 10 bp in the second exon of SF1 (Supplemental Figures 2A and 2B). Both the SF1 null-mutant plants CR-1 (CRISPR-line 1) and CR-2 displayed a 40% reduction in fruit length (Figures 1L and 1M), which was consistent with the short-fruit phenotype of sf1. Thus, Csa2G174140 is responsible for the observed short-fruit phenotype.
To determine the mRNA expression pattern of SF1, the number of fragments per kilobase of transcript per million mapped reads from the RNA sequencing (RNA-seq) data of different cucumber tissues was assessed. SF1 mRNA was expressed ubiquitously (Supplemental Figure 3A). To determine SF1 protein expression, rabbit polyclonal antibodies for SF1 were raised against synthetic peptides, and the antibody specificity was confirmed (Supplemental Figures 3B and 3C). Interestingly, SF1 was detected only in 0 DAA fruit and stem (Supplemental Figure 3D), suggesting a posttranslational regulation of SF1.
To investigate whether CsSF1 is a functional E3 ligase and whether the R230K mutation affects its activity, the His-tagged or glutathione S-transferase (GST)-tagged recombinant SF1, Ubiquitin conjugating enzyme 2 (E2), and Ubiquitin conjugating enzyme 8, the AtUBC8 homolog (CsUBC8) were purified from Escherichia coli extracts (Supplemental Figure 4A) and used for in vitro ubiquitination reactions. The results showed that CsSF1 can self-ubiquitinate. A polyubiquitination signal was observed by immunoblotting with an anti-ubiquitin antibody (Figure 1N). As negative controls, no polyubiquitination of CsSF1 was detected when E1, E2, ubiquitin, or His-CsSF1 was excluded from the reaction (Figure 1N). Unexpectedly, self-ubiquitination appeared to be enhanced by the R230K mutation (Figure 1N). Because E3 ubiquitin ligase is usually activated by dimerization, we also investigated the capacity of CsSF1 to self-interact, showing that both CsSF1 and the CsSF1R230K proteins could homodimerize (Supplemental Figure 4B). To confirm the self-ubiquitination ability of CsSF1 in vivo, we performed agroinfiltration expression in Nicotiana benthamiana and observed that SF1R230K was easier to degrade than was SF1 and that the degradation could be repressed by MG132, a 26S proteasome inhibitor (Figure 1O). The SF1 mRNA expression levels and the green fluorescent protein (GFP) amount that were detected as internal controls remained consistent across all lanes (Figure 1O). Together, these results demonstrated that SF1 encodes a functional cucurbit-specific RING-type E3 ligase and that the R230K mutation results in its enhanced self-ubiquitination and degradation by the 26S proteasome, indicating that sf1 is a loss-of-function mutant.
Overproduction of Ethylene in sf1
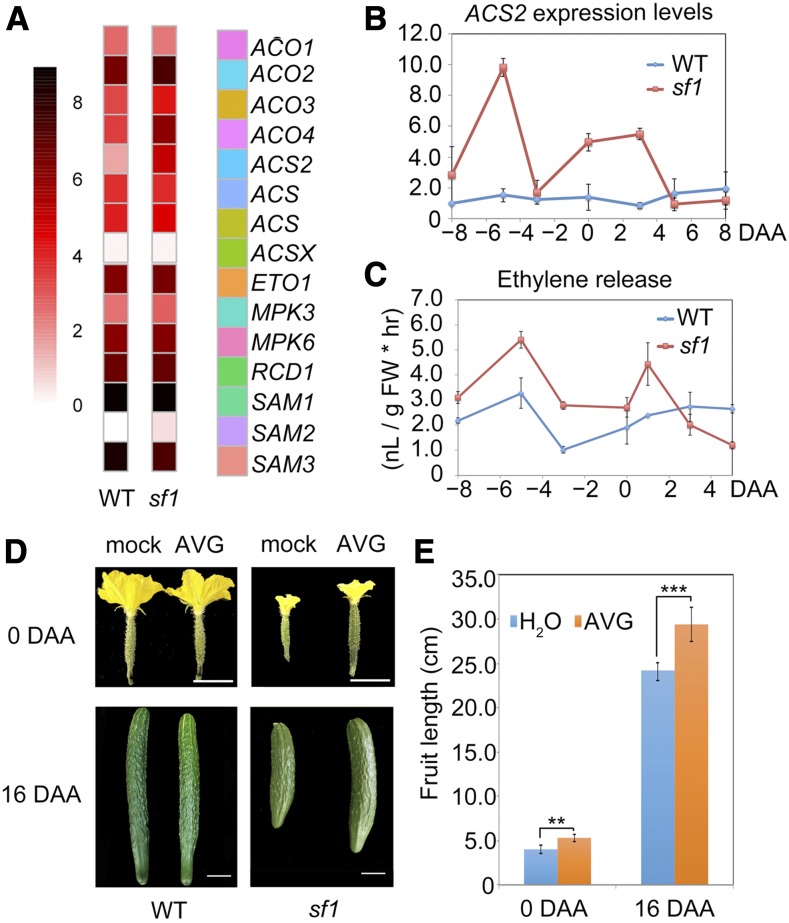
To further understand the molecular mechanism by which SF1 contributes to rapid fruit cell division, we examined the genome-wide gene expression in fruit cells at 0 DAA in sf1 and wild-type plants. A total of 904 genes were significantly differentially expressed (> 1.5-fold or < 0.67-fold; P < 0.05), including 523 and 381 genes that were up- and downregulated, respectively, in sf1 fruit (Supplemental Data Set 3). Interestingly, we found that a group of genes involved in ethylene production, such as the genes encoding aminocyclopropane-1-carboxylic acid synthase (ACC synthase, ACS), ACC oxidase, and S-adenosyl-l-methionine, are enriched in the upregulated gene set (Figure 2A). ACS2, which encodes 1-aminoacyclopropane-1-carboxylic acid synthase, is an important gene that determines bisexual flower formation in cucumber, and its expression was significantly upregulated by 7-fold in sf1 (Figure 2A; Supplemental Data Set 3). We next determined that ACS2 expression levels were higher in the sf1 plants than in the wild-type plants during early fruit development (Figure 2B). As ACS proteins catalyze the rate-limiting step in ethylene synthesis, with the exception of 3 and 5 DAA, the ethylene production of the sf1 fruits was ∼50% greater than that of the wild-type fruits during the early fruit growth stage (Figure 2C).
Figure 2.
The Short-Fruit Phenotype of sf1 Results from Increased Production of Ethylene.
(A) The genes controlling ethylene production are enriched in the upregulated gene set in 0 DAA fruits of sf1, showing ACS2 expression was significantly increased. Scaled log2 expression values are shown. SAM, S-adenosyl-l-methionine; ACO, ACC oxidase; MPK, Mitogen-activated protein kinases; RCD, Radical-induced cell death.
(B) ACS2 expression levels in fruits were upregulated in sf1 during early development. Bars = means from three independent pools of fruits ± sem.
(C) The ethylene production was elevated in sf1 fruits during early fruit growth. FW, Fresh Weight. Bars = means of three fruits from three independent plants ± sem.
(D) and (E) A 1-mM AVG treatment partially complemented the short fruit phenotype of sf1, but had no effect on wild type (WT). Scale bar = 4 cm. **P < 0.05; ***P < 0.01 (t test, one tail). Bars = means of three fruits from three independent plants ± sem.
To determine whether the short-fruit phenotype of sf1 results from increased production of ethylene, we attempted to test whether the phenotype could be rescued by the ethylene synthetic inhibitor 1-aminoethoxyvinyl glycine (AVG). A concentration of 1 mM AVG partially rescued the short-fruit phenotype, but had no significant effect on wild type, indicating that the increased ethylene is essential for the short-fruit phenotype of the sf1 mutant (Figures 2D and 2E).
To confirm the role of ethylene signaling in fruit elongation, we used 1-methylcyclopropene to block ethylene perception in sf1. Treatment with 1-methylcyclopropene at different concentrations (0, 1.0, 2.0, and 3.0 ppm) produced dose-dependent effects on the length of sf1 fruits (Supplemental Figure 5). The 1.0-ppm treatment partially complemented the short-fruit phenotype (Supplemental Figures 6A and 6B). These results further indicate that the short-fruit phenotype of the sf1 mutant is due to the increased ethylene perception.
The Short-Fruit Mutant acs2
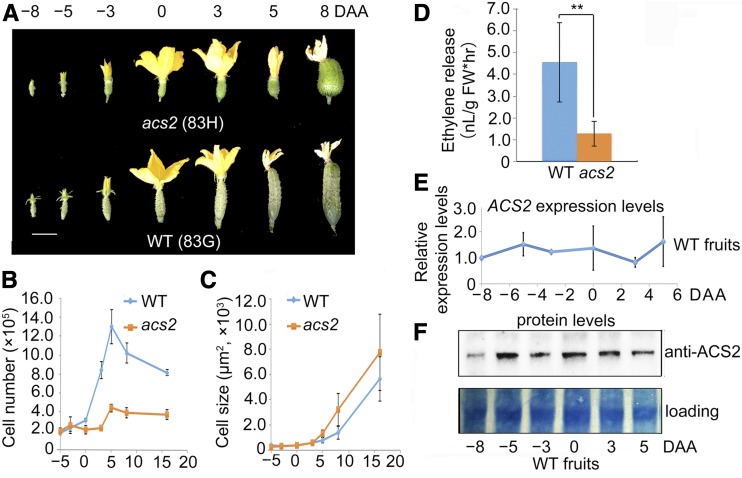
We previously cloned ACS2 (Csa1G580750) that controls bisexual expression (Li et al., 2009). To further dissect the relationship between SF1 and ACS2, we investigated the cucumber mutant acs2 (line 83H), in which ACS2 harbors a single-point mutation at the 33rd amino acid residue (Gly [G] to Cys [C]; G33C), partially abolishing the ACS enzyme activity (Li et al., 2009; Supplemental Figure 7A). In addition to the floral sex phenotype, asc2 plants also bear shorter fruits than do wild-type plants (83G, the near-isogenic line of 83H; Figure 3A; Supplemental Figures 7B and 7C). We also determined that the repressed fruit elongation resulted from a reduced cell number along the longitudinal axis rather than from cell expansion (Figures 3B and 3C); moreover, the ethylene release in acs2 fruit was significantly reduced (P < 0.05; Figure 3D). To confirm the role of ACS2 in fruit development, CRISPR-Cas9 was used to generate several ACS2-edited plants, including two null mutants – one with a homozygous deletion of 1 bp and another with a homozygous deletion of 76 bp (17 bp plus 59 bp; Supplemental Figure 7D). Both cucumber ACS2 knock-out mutants displayed androecy in plants that bore only male flowers (Supplemental Figure 7E), indicating the essential role of ACS2 in cucumber fruit formation.
Figure 3.
The Short-Fruit Mutant acs2 Produces Less Ethylene Than Does Wild Type (WT).
(A) Wild-type (83G) and acs2 (83H) fruits during early fruit development. Scale bar = 3 cm.
(B) Decrease of cell number in acs2 fruit compared with that in wild type along the longitudinal axis. Bars = means from three independent fruits ± sem.
(C) The average cell size shows no significant difference between wild type and acs2. Bars = means from three independent fruits ± sem.
(D) The ethylene release was significantly reduced in 0 DAA fruits of acs2. FW, Fresh Weight. Bars = means of three fruits from three independent plants ± sem. **P < 0.05 (t test, one tail).
(E) The mRNA expression levels during early fruit development in wild-type fruits. Bars = means of three independent pools of fruits from three independent plants ± sem.
(F) The protein expression of ACS2 in wild-type fruits increased significantly from −5 to 3 DAA. Proteins were analyzed by immunoblotting using anti-ACS2. Coomassie brilliant blue (Coomassie) staining of the bottom part of each blot is shown as a loading control.
To determine the ACS2 protein expression pattern, rabbit polyclonal antibodies of ACS2 were raised against specific synthetic peptides (Supplemental Figure 8A), and the antibody specificity was confirmed (Supplemental Figure 8B). We observed that the G33C mutation in acs2 significantly affects the accumulation of ACS2 (Supplemental Figure 8B), indicating that the decreased enzyme activity of ACS2G33C downregulates its expression in a positive feedback manner, in agreement with a previous report (Li et al., 2012). The results also demonstrated that acs2 is a loss-of-function mutant. We further observed that, in wild-type fruits, in contrast with the stable mRNA expression patterns (Figure 3E), ACS2 protein was preferentially accumulated in fruit tissues (Supplemental Figure 8C); moreover, its protein levels increased from −8 to −5 DAA, and then decreased after 3 DAA (Figure 3F). These results suggest a posttranslational regulation of ACS2, which supports ACS2 being a potential substrate of SF1 for fruit length regulation.
Interaction between SF1 and ACS2
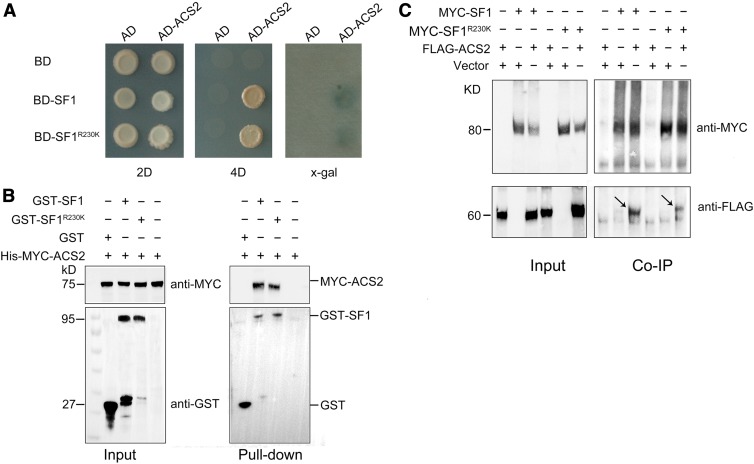
To confirm that ACS2 is a ubiquitinated substrate of SF1 for proteasomal degradation, we first determined their interaction in vitro and in vivo. For yeast two-hybrid assays, the RING domain of SF1 and SF1R230K was made nonfunctional by mutating two essential zinc ligand residues, Cys-336 and His-338, to Ala (SF1-AA and SF1R230K-AA) to prevent the ubiquitination and degradation of itself (Nodzon et al., 2004). Both SF1-AA and SF1R230K-AA could interact with ACS2 to induce Trp1, Leu2, His3, Ade2, and LacZ reporter genes in AH109 yeast lines (Figure 4A).
Figure 4.
SF1 Interacts With ACS2.
(A) Interaction between SF1 and ACS2 verified by yeast two-hybrid assays. Interaction between SF1 and ACS2 was identified, showing growth on medium lacking Leu, Try, His, and Ade (4D), and showing blue color in an X-Gal assay. BD, Binding domain; AD, Activation domain; 2D, SD-Leu-Trp; 4D, SD-Ade-Leu-Trp-His; X-gal, bromochloroindoxyl galactoside.
(B) Pull-down assays confirming the interaction between GST-SF1 or GST-SF1R230K and His-MYC-ACS2 in vitro. GST protein was used as the negative control.
(C) Coimmunoprecipitation assays indicating the interaction between SF1 and ACS2 in vivo. Arrows indicate the immunoprecipitated FLAG-ACS2 by MYC-SF1 and MYC-SF1R230K. The (+) and (−) represent participation and absence, respectively, in the reaction.
The specific interaction between SF1 and ACS2 in vitro was verified via GST pull-down assays. We observed that Myelocytomatosis (MYC)-ACS2 interacted with GST-SF1 and GST-SF1R230K, whereas no interaction signals were observed when GST protein was used as a control (Figure 4B). To obtain evidence of the SF1 and ACS2 interaction in vivo, we performed coimmunoprecipitation. The pairwise FLAG-ACS2/MYC-SF1 or FLAG-ACS2/MYC-SF1R230K agrobacteria were transiently expressed in N. benthamiana leaves supplemented with MG132. FLAG-ACS2 formed a protein complex with MYC-SF1 and MYC-SF1R230K (Figure 4C). All of these results demonstrate that SF1 specifically interacts with ACS2.
Ubiquitination and Degradation of ACS2 by SF1
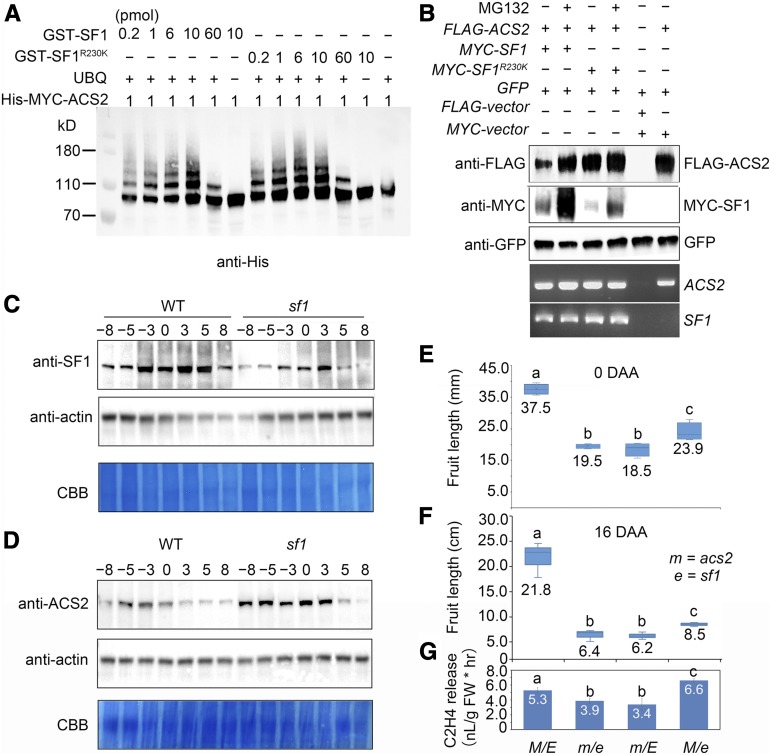
To examine the possibility that SF1 mediates the direct ubiquitination of ACS2, we performed in vitro ubiquitination assays using recombinant proteins. ACS2 was found to be effectively ubiquitinated by SF1 and SF1R230K in the presence of ubiquitin (Figure 5A). There was no perceivable difference between the ubiquitination ability of SF1 or SF1R230K. Moreover, as the ratio of GST-SF1:ACS2 or GST-SF1R230K:ACS2 increased from 1 to 10, the intensity of the ubiquitinated bands gradually increased; however, when the ratio reached 60, the ubiquitination of ACS2 was repressed, indicating that SF1 catalyzes the ubiquitination of ACS2 in a dose-dependent manner (Figure 5A).
Figure 5.
ACS2 is a Substrate of SF1.
(A) Ubiquitination of ACS2 by SF1 in vitro. The above two lines of numbers represent the concentration of GST-SF1 and GST-SF1R230K. Reaction were analyzed by immunoblotting using anti-His.
(B) SF1 promotes ACS2 degradation via 26S proteasomes. When ACS2 and SF1R230K were coexpressed, the dramatic degradation of SF1R230K proteins resulted in a significant accumulation of ACS2. SF1 and ACS2 were expressed in N. benthamiana leaves treated with (+) or without (−) MG132. GFP was used as the control for the infiltration event. Bottom panels, RNA levels of ACS2 and SF1.
(C) and (D) Expression profiles of SF1 (C) and ACS2 proteins (D) in wild-type (WT) and sf1 fruit during early fruit development, showing decreased SF1 protein levels while increased ACS2 protein levels in sf1. Actin was used as an internal control. Coomassie brilliant blue (CBB) staining of the bottom part of each blot is shown as a loading control.
(E) and (F) Boxplots show the epistasis of acs2 (m) to sf1 (e) genetically. Twenty fruits of each genotype at 0 DAA and 16 DAA are shown: wild type (M E), m e, m E, and M e fruit from 20 independent plants were identified to measure fruit length. Bars = means ± sem (n = 20). Different lowercase letters indicate significant difference, P < 0.05, Tukey’s test.
(G) The ethylene release in wild type (M E), m e, m E, and M e fruits at 0 DAA, showing that m E and m e fruits produced relatively less ethylene, and M e fruits produced more ethylene than in wild type. FW: Fresh Weight. Bars = means from three independent fruits ± sem. Different lowercase letter indicates significant difference, P < 0.01, Tukey’s test.
We then used a cell-free degradation assay to monitor the turnover of the enzyme in the presence and absence of MG132 in vitro. In the absence of MG132, His-MYC-ACS2 gradually disappeared when incubated with protein extracts from wild-type fruits, and the degradation rate of His-MYC-ACS2 protein decreased when incubated with protein extracts from sf1 fruits. This degradation can be blocked by MG132 (Supplemental Figures 9A and 9B). Thus, ACS2 can be degraded by SF1 via 26S proteasomes.
To further confirm that the SF1-mediated ubiquitination of ACS2 promotes protein degradation in vivo, we transiently expressed FLAG-ACS2, MYC-SF1, MYC-SF1R230K, and a GFP internal control protein by agroinfiltration in N. benthamiana (Figure 5B). We assessed the ACS2 and SF1 mRNA expression levels and the GFP internal control protein levels to determine whether they remained consistent in related lanes. When compared with the separate infiltration of FLAG-ACS2, coexpression of FLAG-ACS2 and MYC-SF1 in the same leaf region resulted in significant degradation of FLAG-ACS2. When MG132 was infiltrated into the same area, the degradation of both FLAG-ACS2 and MYC-SF1 was repressed (Figure 5B), indicating that SF1 targets ACS2 for ubiquitin-dependent degradation by the 26S proteasome. Intriguingly, when FLAG-ACS2 and MYC-SF1R230K were coexpressed, the SF1R230K proteins were substantially self-degraded, resulting in a significant accumulation of FLAG-ACS2 (Figure 5B). To confirm the above results in planta, total protein extracts from fruits were prepared for immunoblotting. The overall amount of SF1R230K protein was decreased during fruit development in sf1 (Figure 5C), which is consistent with the transient expression results from N. benthamiana (Figures 1O and 5B). It was further demonstrated ACS2 indeed accumulated to a greater extent in the sf1 fruits than in the wild-type fruits (Figure 5D), which explained our previous observation that ethylene was overproduced in sf1 fruits (Figure 2C).
The above results demonstrated that SF1 and ACS2 interact biochemically. Because the genetic backgrounds of sf1 (East Asian background) and acs2 (Eurasian background) are distinct, to confirm that SF1 and ACS2 interact genetically, we developed a series of BC2S1 lines using 1983H (acs2) as the donor and line 406 as the recurrent parent (sf1 and wild type, each for backcrossing once). Plants with a genotype of M E (wild type, ACS2/SF1), m E (acs2), m e (acs2/sf1), or M e (sf1) were identified as the near-isogenic lines for measuring the fruit length and ethylene release (Supplemental Figure 10). The phenotypic analysis revealed that, when compared with the other plants, the wild-type (M E) plants produced longer fruits, and the fruits of M e plants were significantly shorter. The length of the m E fruits was reduced the most, whereas the fruit length of the m e double mutant was similar to that of m E but not similar to that of the synthetic m plus e (Figures 5E and 5F; Supplemental Figure 10). These results indicate that acs2 (m) is genetically epistatic to sf1 (e). The ethylene release in 0 DAA fruits of M E (wild type), m E, m e, and M e was consistent with the observation on acs2 and sf1 mutants. Thus, m E and m e produced relatively less ethylene, and M e produced more ethylene than the wild type. By contrast, the difference in ethylene release between m E and m e is not significant (Figure 5G). As in the double mutant m e, the mutation of E3 ligase activity in sf1 that results in an increase in ACS2 protein cannot overcome the reduction in ethylene biosynthetic activity in the acs2 mutant, further demonstrating the epistasis of acs2.
Together, the above results demonstrate that SF1 targets both itself and ACS2 for ubiquitin-dependent degradation to regulate both the ethylene dosage and balance of cell division in cucumber fruit.
Ethylene Dose-Dependent Fruit Elongation
When compared with wild-type plants, sf1 plants bear shorter fruits due to the overproduction of ethylene, whereas acs2 plants produce less ethylene but also bear shorter fruits. These lines of evidence suggest that the tight control of ethylene production is responsible for cucumber fruit elongation.
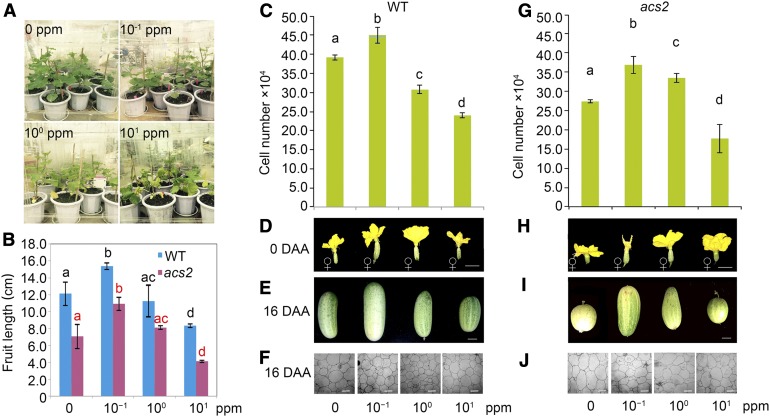
To assess this possibility, 1-month-old wild-type and acs2 cucumber plants were transferred into four acrylic chambers, with five plants of wild type and five plants of acs2 in each chamber (Wen, 2014). The four boxes were treated with different concentrations of ethylene (0, 10−1, 10°, and 101 ppm) for 1 week (Figure 6A). The ethylene treatment exerted a dose-dependent effect on fruit elongation for the wild-type and acs2 plants. Under 10−1 ppm ethylene, the acs2 fruit length significantly recovered, but the effect was diminished when 10° ppm ethylene was applied; however, under 101 ppm ethylene, the acs2 fruit length was significantly lower (Figure 6B). The same dose-dependent effect of ethylene on fruit elongation was also observed for the wild-type fruits (Figure 6B).
Figure 6.
Ethylene Dose-Dependent Effects on Cucumber Fruit Elongation.
(A) Exogenous ethylene treatment for wild type (WT) and acs2 at different concentrations of ethylene for 1 week in four acrylic boxes. Four different concentrations of ethylene in four boxes are shown.
(B) The 16 DAA fruit length of wild type and acs2 treated with different concentrations of ethylene. Bars = means of three fruits from three independent plants ± sem. Different lowercase letters indicate significant difference, P < 0.01, Tukey’s test.
(C) Cell number of 0 DAA fruits of wild type treated with different concentrations of ethylene. Bars are means of three fruits from three independent plants ± sem. Different lowercase letter indicate significant difference, P < 0.01, Tukey’s test.
(D) to (F) Phenotypes of 0 DAA (D) and 16 DAA (E) fruits of wild-type plants treated with different concentrations of ethylene. Scale bar = 2 cm. (F) Mesocarp cells of wild type at 16 DAA. Scale bar = 50 μm.
(G) Cell number of 0 DAA acs2 fruits treated with different concentrations of ethylene. Bars = means of three fruits from three independent plants ± sem. Different lowercase letters indicate significant difference, P < 0.01, Tukey’s test.
(H) to (J) Phenotypes of 0 DAA (H) and 16 DAA (I) acs2 fruits treated with different concentrations of ethylene. Scale bar = 2 cm. (J) Mesocarp cells of acs2 at 16 DAA. Scale bar = 50 μm.
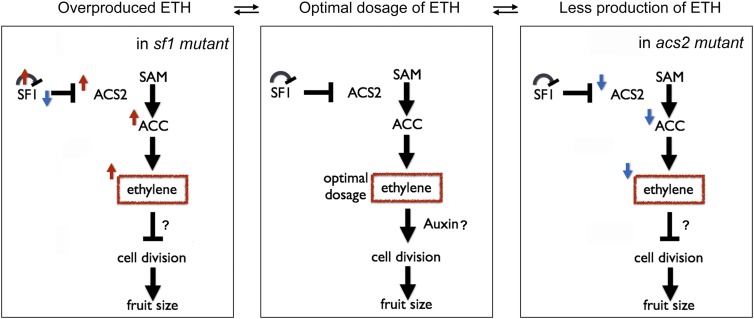
To confirm that the ethylene dose-dependent effect on fruit length is due to changes in cell division, we compared the mesocarp cell number between wild type (Figures 6C to 6F) and acs2 (Figures 6G to 6J) along the longitudinal axis using 0 DAA fruits under different ethylene concentrations. Although cell size was reduced to a certain degree in response to ethylene treatment, the number of cells increased under the relatively low ethylene doses (10−1 ppm for wild type and 10−1 and 10° ppm for acs2) but decreased under higher ethylene concentrations. We also observed that sex of the flowers produced of the acs2 mutant plants changed from bisexual to female flowers when treated with continuously increasing concentrations of ethylene; however, the effect of ethylene on sex expression is not dependent on the dose of ethylene (Figure 6H). Together, our findings provide genetic and molecular evidence that SF1 ubiquitinates and degrades both itself and ACS2 to tightly control ethylene synthesis for dose-dependent cell division and fruit elongation in cucumber (Figure 7).
Figure 7.
Regulatory Mechanism of Ethylene Dosage for Cell Division in Developing Cucumber Fruit.
SF1 regulates the ethylene dosage by targeting both itself and ACS2 for ubiquitin-dependent degradation. The ethylene biphasic response regulates the balance of cell division and fruit growth. Ethylene is highlighted in red rectangles. Black arrows and bar-ended lines represent positive and negative interactions, respectively. Red and blue arrows represent activation and repression, respectively. ETH, ethylene; SAM, S-adenosyl-l-methionine.
DISCUSSION
Plant hormones are key factors required for plant growth, and among them, ethylene plays essential roles in various plant developmental processes, both by stimulating and inhibiting growth (Vandenbussche et al., 2012). In this study, cucumber fruit constituted an attractive system for understanding the dose-dependent control of the subtle balance of cell division by ethylene.
Ethylene contributes to the control of the flower sex type of cucurbit plants (Rudich et al., 1969). ACS catalyzes the rate-limiting step in ethylene biosynthesis, producing ACC from S-adenosylmethionine (Adams and Yang, 1979; Yang and Hoffman, 1984). Dysfunction of CsACS2 and CmACS7 in cucumber and melon (Cucumis melo) leads to the development of hermaphroditic flowers (Boualem et al., 2015), whereas bisexual flowers are associated with round-fruit phenotypes (Liu et al., 2008; Monforte et al., 2014), suggesting that ethylene promotes cucurbit fruit elongation.
Ethylene has long been recognized as a growth inhibitor; however, evidence is accumulating that ethylene can also promote growth depending on endogenous and environmental conditions and plant developmental stage (Dugardeyn and Van Der Straeten, 2008). The application of ethylene to dark-grown seedlings inhibits hypocotyl elongation (Guzmán and Ecker, 1990); this growth inhibition is caused by a decrease in cell elongation (Le et al., 2005). Under low nutrient conditions, low concentrations of ethylene treatment increase hypocotyl growth (Smalle et al., 1997) by stimulating additional cortex cell division but not cell elongation (Saibo et al., 2003). Studies suggest that auxin transport (Vandenbussche et al., 2003) or brassinosteroid signaling (De Grauwe et al., 2005) is involved in ethylene-mediated hypocotyl elongation. Recent studies on tomato and zucchini fruits showed that ethylene suppresses fruit set; however, those studies used ACC or ethephon for treatment. ACC and ethephon treatments may not accurately reproduce natural ethylene responses in some physiological processes (Zhang and Wen, 2010).
Although numerous quantitative trait loci for fruit length and shape in cucurbit plants have been identified (Qi et al., 2013; Wei et al., 2014; Bo et al., 2015; Weng et al., 2015; Pan et al., 2017), the genetic and molecular basis of these traits remain largely unknown. In the current study, using two shortened fruit mutants, sf1, which exhibits reduced cell division and overproduces ethylene, and acs2, which exhibits reduced cell division but produces relatively less ethylene, we revealed the genetic and molecular control of ethylene dosage for cucumber fruit elongation. We demonstrated the effects of different ethylene dosage on fruit elongation using ethylene gas as a direct ethylene treatment, providing major insight into the molecular basis of cucurbit fruit morphogenesis. Cell size was increased in acs2 fruits along the transversal axis (Supplemental Figure 11), and the suppression effect of ethylene on diameter appears to be different from the dose-dependent effect on length, suggesting the complex and pleiotropic effects of ethylene on cell division rate, cell division polarity, and cell expansion. Our findings may also provide a potentially useful method to generate a series of fruits with a gradient of lengths via physiological treatment or genome editing methods. Because fruit size preferences vary among consumer populations, this research could guide cucumber breeding practices.
ACS catalyzes the rate-limiting step in ethylene biosynthesis, and the amount of ACS2 protein is positively associated with ethylene production (Wen, 2014). The stability of individual ACS proteins is tightly regulated by phosphorylation, dephosphorylation, and ubiquitination-mediated proteasomal degradation (Wen, 2014), which is critical for fine tuning ethylene biosynthesis in response to various endogenous and environmental signals (Pierik et al., 2006). In Arabidopsis, the ethylene-overproducer with constituent of ubiquitin ligase 3 E3 Ligase complex targets AtACS5 and AtACS9 for degradation by the 26S proteasome (Wang et al., 2004); the RING-type E3 ligase AtXBAT32 targets AtACS7 for proteasomal degradation to regulate lateral root production (Prasad et al., 2010; Lyzenga et al., 2012). In this study, the control of ethylene doses for cucumber fruit elongation was achieved through the posttranslational degradation of ACS2 by the RING-type E3 ligase SF1 via the 26S proteasome pathway. In melon, the mutation of CmACS7 is reported to be associated with round fruit (Monforte et al., 2014; Boualem et al., 2015), suggesting that a conserved regulatory mechanism underlying ethylene dose-dependent fruit elongation exists in cucurbit plants.
The ethylene dose-dependent response has been reported for some species and tissues (Pierik et al., 2006). For example, in N. benthamiana, petiole growth and stem elongation were stimulated at low ethylene concentrations, and repressed at higher ethylene concentrations (Pierik et al., 2003, 2004). How do dose-dependent ethylene responses exert multiple roles in fruit cell division? Although the mechanistic basis for this diversification remains speculative, the biphasic ethylene response model was proposed. It is inferred that the interactions between ethylene and other signal transduction components might dependent on specific ethylene concentration, and the signal transduction pathways that stimulate growth are probably different to those that inhibit growth (Pierik et al., 2006). Cucumber fruit constitutes a potential important system for unraveling the signal transduction components involved in the different ethylene responses.
METHODS
Plant Material
The seeds of cucumber (Cucumis sativus) inbred line 406 (North China type) were treated with 1% (v/v) ethyl methanesulfonate (Sigma Aldrich) in mutagenesis experiments. The M1 plants were self-pollinated, and the short fruit line sf1 was identified in the M2 population. Then, sf1 was crossed with the wild-type 406, and an F2 population was derived from self-crossed F1 plants. The near-isogenic lines 1983G and 1983H (Liu et al., 2008) were used in this study. The cucumber inbred line CU2 was used in cucumber transformation. All cucumber plants were cultivated in the greenhouse at day/night temperatures of 24/18°C with a light regime of 16-h natural light/8-h dark in the Chinese Academy of Agricultural Sciences. Standard management was performed during cultivation.
The Nicotiana benthamiana plants were cultivated in pots in a growth chamber with a light regime of 16-h light/8-h dark at 22°C.
Derived Cleaved-Amplified Polymorphic Sequence Markers
For fine mapping of sf1, we developed derived cleaved-amplified polymorphic sequence (dCAPS) markers (Neff et al., 1998) on chromosome 2. The primers for dCAPS markers used to verify the SNP were designed by the web-based software dCAPS FINDER 2.0. PCR products, which were digested with appropriate restriction enzymes (Supplemental Table 3), were subsequently separated by electrophoresis on 8% (w/v) polyacrylamide gels.
Measurement of the Cell Number and Size
To measure the cell size and number of fruits of 406, sf1, 1983G, and 1983H, sliced samples from different periods at −5, −3, 0, 3, 5, 8, and 16 DAA were fixed in the solution of 70% (v/v) ethanol, acetic acid, and formaldehyde (90:5:5 by volume). Five-millimeter-thick slices cut from different parts of the cucumber fruit (outer, middle, and inner pericarp) were embedded in paraffin. Then, 8-μm-thick sections were prepared from both cross-sections and longitudinal direction with a microtome and stained with hematoxylin-eosin. Subsequently, these sections were imaged using a microscope (OLYMPUS BX51). The cell number and cell size in each given section were calculated using Infinity capture 6.0 and Image Proplus 5.1 software according to the method reported previously (Yang et al., 2013). All measurements were made on three sites of each tissue, for three sections from each fruit.
Phylogenetic Analysis
Multiple sequence alignment of the C3HC4 domain and typical Ankyrin repeat domain sequences, which were obtained by hidden Markov models corresponding to PF13920 and PF12796 (Gene ID shown in Supplemental Table 2), was performed using the Muscle program with default parameters (Edgar, 2004). The sequence alignments were used for the subsequent phylogenetic analysis. Phylogenetic trees were generated with MEGA7.0 software (https://megasoftware.net/) using the neighbor join algorithms, and the reliability of the obtained trees was assessed with a bootstrap value of 1000.
The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Jones-Taylor-Thornton matrix-based method and are in the units of the number of amino acid substitutions per site. The analysis involved 58 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 202 positions in the final data set. Evolutionary analyses were conducted in MEGA7.
Plasmid Construction and Plant Transformation
The binary vector pBSE402, which contained a CRISPR cassette with a functional Cas9 under a constitutive promoter (Cauliflower mosaicvirus 35S) plus a 35S-GFP expression cassette, was modified from pBSE401 (a gift from Qi-Jun Chen). To gain CRISPR/Cas9 engineering mutation in SF1 gene and ACS2 gene, single guide RNA (sgRNA) target sites from the N termini of SF1 and ACS2 were selected. The sgRNA were assembled into pBSE402 as previously described (Xing et al., 2014). Equal volumes of 100 μM forward and reverse primers were incubated at 95°C for 5 min, and slowly cooled to room temperature to form a double stranded DNA fragment. This short DNA fragment was then assembled into pBSE402 with BsaI and T4 Ligase (New England Biolabs). Primers are shown in Supplemental Table 3. The final binary vector pBSE402-sgRNA-SF1 and pBSE402-sgRNA-ACS2 were then transformed into Agrobacterium (Agrobacterium tumefaciens) strain EHA105.
Agrobacterium-mediated method was used to transform cotyledonary nodes of cucumber as previously described (Hu et al., 2017). CU2 seeds were soaked in distilled water at 50°C for 30 min. Seed coats were removed with forceps and sterilized in 75% (v/v) ethanol for 15 s and 0.6% (v/v) sodium hypochlorite solution for 15 min. The sterilized seeds were germinated in the dark overnight at 28°C in a plastic Petri dish containing 1×Murashige and Skoog medium (Phytotech) supplemented with 2 mg/L 6-benzylaminopurine (Sigma Aldrich) and 1 mg/L abscisic acid (Phytotech). Cotyledons were excised from germination seedlings and infected with Agrobacterium. Subsequently, after shoot regeneration, elongation, and rooting processes, the rooted plants were transplanted to greenhouse.
Genomic DNA was extracted with the DNeasy Plant Mini Kit (Qiagen). The target gene was respectively amplified with specific primers (Supplemental Table 3). PCR products were cloned into pEASY-Blunt Zero (TRANSGEN BIOTECH) and sequenced.
Construction of Near-Isogenic Lines
A series of BC2S1 lines were developed using 1983H (acs2) as donor and line 406 as recurrent parent (sf1 and wild type, each for once). The BC2 population were screened by whole cucumber genome chip containing 181 loci for SNPs test. We chose a BC2 line whose genotype was heterozygous for both acs2 and sf1; backgrounds were most similar to line 406 and then self-crossed to identify M E (wild type), m E, m e, and M e genotypes in a BC2S1 population with 500 plants. These plants were used for phenotypic analyses.
Recombinant Protein Expression and Purification
Recombinant proteins GST-SF1, GST-SF1R230K, GST-E2, His-E2, His-SF1, His-SF1R230K, and His-MYC-ACS2 were expressed in BL21 (DE3; TransGen). After induction with 1 mM isopropyl b-D-1-thiogalactopyranoside, cells were harvested and subsequently sonicated in the lysis buffer. The soluble proteins were affinity-purified using Ni-nitrilotriacetic acid Resin (Thermo Fisher Scientific) and Glutathione-Sepharose resin (GE) according to the manufacturer’s instructions. Purified supernatant was analyzed by 10% (w/v) SDS-PAGE.
Immunoblotting
SF1 and ACS2 antibodies were produced in rabbit (Agrisera) with synthetic peptides, which matched amino acids 344 to 360 in the SF1 sequence, and 11 to 27 in the ACS2 sequence. The antibody specificity was identified in vitro and in vivo.
Protein extracts of plant tissues were prepared using protein extraction buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 4 M Urea, 10% (v/v) glycerol, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail), followed by centrifugation at 12,000g for 30 min at 4°C. Then, 50-100 μg of supernatant was used for 10% (w/v) SDS-PAGE and subsequently transferred to a polyvinylidene fluoride membrane. Immunoblotting signal was visualized using a SuperSignal West Femto kit (Thermo Fisher Scientific).
In Vitro Ubiquitination Assay
Ubiquitin was from BostonBiochem (5 mg, reconstitute at 10 mg/mL in a solution). Yeast GST-E1 was also from BostonBiochem (0.71 mg/mL). Recombinat His-E2 (CsUBC8, the AtUBC8 homologs), His- or GST-SF1, His- or GST-SF1R230K, and His-MYC-ACS2 proteins were purified from Escherichia coli, and dialyzed into buffer containing 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 5% (v/v) glycerol, 1 mM DTT, and 1 mM EDTA. In vitro ubiquitination assays were performed as follows. Each reaction (15 μL volume) contained 5 µg of ubiquitin, 50 ng of recombinant E1, 110 ng E2, and 300 ng purified His-SF1 in ubiquitination buffer (50 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 50 mM KCl, 2 mM ATP, and 1 mM DTT). After incubation at 30°C for 2 h, the reaction was stopped by adding 5 × SDS sample buffer, and analyzed by electrophoresis on 12% (w/v) SDS gels. Ubiquitinated SF1 or ACS2 was detected using anti-ubiquitin antibody or anti-His antibody.
In Vivo Ubiquitination Assay
In vivo ubiquitination assay was performed according to the protocol described by Liu et al. (Liu et al., 2010). We coinfiltrated the agrobacteria carrying the FLAG-ACS2 and MYC-SF1 plasmids into tobacco leaves, and the corresponding empty vectors were used as the controls. To keep the consistence in all loading sample, the agrobacteria carrying 35S-GFP was used as an internal control. For inhibition of 26S-proteasome, 50 mM MG132 (Sigma-Aldrich; in 10 mM MgCl2 solution) was infiltrated 12 h before leave collection. Samples were collected for protein and RNA extraction after 3 d. For protein analysis, the extracts were analyzed using anti-FLAG, anti-MYC and anti-GFP antibodies (Supplemental Table 4). For RNA level expression analysis, RT-PCR was performed.
GST-Pull Down
The pull-down assay was conducted as previously described with some modifications (Oh et al., 2012). In brief, 2 μg of GST-SF1, GST-SF1R230K, or GST bound glutathione agarose beads were incubated with 2 μg of His-MYC-ACS2 in 1× PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH7.4) at 4°C on a platform that swings up and down for 1 h. After being washed three times with 1× PBS buffer, the beads were directly added with an equal volume of 1× SDS sample buffer and w detected by immunoblotting using anti-MYC antibody.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed using the GAL4-based Two-Hybrid System (Clontech), according to the manufacturer’s instructions. The full-length cDNA of SF1, SF1R230K, and ACS2 were subcloned into the pGADT7 and pGBKT7 vectors, respectively. Yeast (Saccharomyces cerevisiae) strain AH109 was used for transformation, and then was grown in 2D (SD-Leu-Trp) medium. The transformed colonies were plated onto a 4D (SD-Ade-Leu-Trp-His) medium plates to verify the interaction. The primers used for yeast two-hybrid assays are listed in Supplemental Table 3.
Coimmunoprecipitation
Cultured Agrobacteria harboring pCAMBIA1300-FLAG-ACS2, pCAMBIA1300-MYC-SF1, or pCAMBIA1300-MYC-SF1R230K and P19 were resuspended in 10 mM MES (pH 5.8) buffer containing 10 mM MgCl2 and 200 µM acetosyringone at a ratio of 1:1:2. After 2 h incubation at room temperature in darkness, the strains were infiltrated into healthy and fully expanded leaves of 4-week-old N. benthamiana plants using a needleless syringe. The infiltrated leaves were harvested after 2 d, and total protein was extracted in IP buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% (v/v) Nonidet P-40, 5% (v/v) glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail). The extracted protein was incubated with anti-MYC agarose conjugates (Sigma Aldrich) for 2 h at 4°C. The beads were then washed three times with washing buffer (150 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl [pH 7.5]), and beads were sampled for coimmunoprecipitation analysis. Anti-MYC antibody and anti-FLAG antibody (Sigma Aldrich) were used for immunoblots (Supplemental Table 4).
RNA Seq Analysis
Total RNA was extracted from 0 DAA fruits of wild type (M E), m e, m E, and M e plants with two biological replications using a TRIzol kit (Invitrogen) according to the protocol. Sequencing cDNA libraries with an average insert size of 250-300 bp were prepared according to the manufacturer’s instructions (Illumina), and 150-bp paired-end reads were generated on an Illumina Hiseq 2000 Analyzer. The fragments per kilobase of transcript per million mapped reads expression value of each gene was calculated using StringTie (Pertea et al., 2016). Genes with a fold change >1.5 or <0.67, and a p value of < 0.05 in the t test (one-tailed) were defined as differentially expressed genes.
RT-quantitative PCR Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using 1 μg total RNA with the Moloney Murine Leukemia VirusReverse Transcriptase (Promega), and then a quantitative PCR assay was performed on an ABI 7900 (Applied Biosystems) machine using SYBR Premix (Roche) according to the manufacturer’s instructions. Three independent biological replicates were performed. Relative gene expression was performed using the 2−∆∆Ct method. The UBIQUITIN gene, Csa3G778350, was used as an internal reference gene. Primers are listed in Supplemental Table 3.
Exogenous Ethylene Treatment
One-month-old wild-type (1983G) and acs2 (1983H) cucumber plants were placed in four acrylic chambers, with five plants of wild type and five plants of acs2 in a chamber. The side of each chamber was sealed with a rubber stopper that formed a gas inlet/outlet to facilitate the injection of the ethylene gas. The ethylene source was a pressurized gas tank containing a known concentration of ethylene gas. Plants were provided with normal water and fertilizer. The concentrations of ethylene in chambers were determined using the known concentration of ethylene stock and the chamber volume (1 m3). The four chambers are individually treated with different concentrations of ethylene at 0, 10−1, 10°, and 101 ppm for 1 week. Ethylene gas was changed with fresh air twice a day, for 30 min each time. The fruits from 11 and 12 nodes were used for phenotype observation. Half of the fruits were used for data collection at 0 DAA, and half were allowed to grow for data collection at 16 DAA.
Ethylene Measurement
The 0 DAA fruits of wild type (1983G) and acs2 (1983H), wild type (406) and sf1, and wild type (M E), m e, m E, and M e plants were excised to measure the ethylene release. At least three biological replicates were measured. Each sample was placed in a 65-mL container with a piece of wet cotton at the bottom and sealed with a rubber stopper. After incubation at 23°C for 1 h, the containers were opened to volatilize the ethylene induced by cutting. Samples were incubated in a sealed container at 23°C for another 10 h in the dark, and 10 μL of gas was removed with a syringe and injected into a gas chromatograph (Agilent 7890B-5977A) equipped with a flame-ionization detector and a capillary column for ethylene measurements. All determinations were made in triplicate. The ethylene release rate (nanoliliters per gram fresh weight per hour) was calculated on the basis of fresh weight of fruits.
Statistical Analyses
In the case of comparisons between two sample groups, the Student’s t test was applied. For multiple comparison, significance analysis was performed as pairwise comparison based on ANOVA, which was followed by Tukey’s honestly significant difference test and was performed in R for statistical evaluation (Supplemental Data Set 4).
Data Availability
All data that support the findings within this article are available from the corresponding author upon reasonable request.
Accession Numbers
Sequence data from this article can be found in the Cucurbit Genomics Database (www.icugi.org) under accession numbers SF1(Csa2G174140); ACS2(Csa1G580750); and CsUBC8 (Csa6G133760).
Supplemental Data
Supplemental Figure 1. Morphological and physiological changes of WT and sf1 cucumber fruit during early development.
Supplemental Figure 2. Identification and phenotypic analysis of CRISPR-SF1 plants.
Supplemental Figure 3. mRNA and protein expression profiles of SF1 in various cucumber tissues.
Supplemental Figure 4. Purification of recombinant proteins for biochemical assays.
Supplemental Figure 5. Treatment with 1-MCP showed dose effects on sf1 fruit elongation.
Supplemental Figure 6. 1-MCP treatment partially complemented the short fruit phenotype of sf1.
Supplemental Figure 7. ACS2 knock-out cucumber plants displayed androecy.
Supplemental Figure 8. mRNA and protein expression profiles of ACS2 in various cucumber tissues.
Supplemental Figure 9. Proteasome-dependent degradation of ACS2.
Supplemental Figure 10. Fruit ontology of WT (M E), m e, m E, and M e during early development stages.
Supplemental Figure 11. Cell size difference in fruits between WT and acs2 along transversal axis.
Supplemental Table 1. Phenotypic statistics of selfed F2.
Supplemental Table 2. The gene ID for phylogenetic tree analysis.
Supplemental Table 3. Primers used in this study.
Supplemental Table 4. Antibodies used in this study.
Supplemental Data Set 1. The SNP-index from MutMap analysis.
Supplemental Data Set 2. Alignment of genes used to generate the phylogeny shown in Figure 1K.
Supplemental Data Set 3. Differential expression of genes in fruit cells at 0 DAA in sf1 mutants and WT (P < 0.05).
Supplemental Data Set 4. ANOVA and t test.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Qi Xie from Chinese Academy of Sciences and Juan Xu from Zhejiang University for comments on the article. We also thank Jingjin Sun and Yuanchao Xu for experimental assistance from Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences. This work was supported by grants from the National Natural Science Foundation of China (NSFC) (grants 31530066 to S.H., 313220419 to Z.H.Z., and 31572117 to X.Y.) and the National key R&D Program of China (2016YFD0101007 and 2016YFD0100500). Additional support was provided by ASTIP-CAAS (CAAS-XTCX2016001), the Leading Talents of Guangdong Province Program (grant 00201515 to S.H.), and the Shenzhen Municipal, The Peacock Plan (KQTD2016113010482651) and the Dapeng district government.
AUTHOR CONTRIBUTIONS
T.X., Z.Z., and X.Y. made major contributions to biochemical assays and interpreted the data and wrote the article; T.X. and X.Y. led genetic transformation of plants; Z.Z., S.L., S.Z., and X.Y. performed the mutant screen, genetic studies, and phenotype observations; Q.L., Z.H.Z., and X.Y. led bioinformatic analyses; S.H. and X.Y. designed the research.
Footnotes
Articles can be viewed without a subscription.
References
- Adams D.O., Yang S.F. (1979). Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 76: 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisognin D.A. (2002). Origin and evolution of cultivated cucurbits. Cienc. Rural 32: 715–723. [Google Scholar]
- Bo K., Ma Z., Chen J., Weng Y. (2015). Molecular mapping reveals structural rearrangements and quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor. Appl. Genet. 128: 25–39. [DOI] [PubMed] [Google Scholar]
- Boualem A., Troadec C., Camps C., Lemhemdi A., Morin H., Sari M.-A., Fraenkel-Zagouri R., Kovalski I., Dogimont C., Perl-Treves R., Bendahmane A. (2015). A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350: 688–691. [DOI] [PubMed] [Google Scholar]
- Chen H., et al. (2016). An ACC oxidase gene essential for cucumber carpel development. Mol. Plant 9: 1315–1327. [DOI] [PubMed] [Google Scholar]
- Colle M., Weng Y., Kang Y., Ophir R., Sherman A., Grumet R. (2017). Variation in cucumber (Cucumis sativus L.) fruit size and shape results from multiple components acting pre-anthesis and post-pollination. Planta 246: 641–658. [DOI] [PubMed] [Google Scholar]
- De Grauwe L., Vandenbussche F., Tietz O., Palme K., Van Der Straeten D. (2005). Auxin, ethylene and brassinosteroids: Tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 46: 827–836. [DOI] [PubMed] [Google Scholar]
- Dugardeyn J., Van Der Straeten D. (2008). Ethylene: Fine-tuning plant growth and development by stimulation and inhibition of elongation. Plant Sci. 175: 59–70. [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumet R., Colle M. (2016). Genomic analysis of cucurbit fruit growth. In Grumet R, Katzir N, Garcia-Mas J, eds, Genetics and Genomics of Cucurbitaceae. Plant Genetics and Genomics: Crops and Models. Springer, Cham, pp 321–344. [Google Scholar]
- Guzmán P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Li D., Liu X., Qi J., Gao D., Zhao S., Huang S., Sun J., Yang L. (2017). Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant 10: 1575–1578. [DOI] [PubMed] [Google Scholar]
- Kazama H., Dan H., Imaseki H., Wasteneys G.O. (2004). Transient exposure to ethylene stimulates cell division and alters the fate and polarity of hypocotyl epidermal cells. Plant Physiol. 134: 1614–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., De Cnodder T., Van Der Straeten D., Verbelen J.-P. (2005). Cell elongation and microtubule behavior in the Arabidopsis hypocotyl: Responses to ethylene and auxin. J. Plant Growth Regul. 24: 166–178. [Google Scholar]
- Li Z., et al. (2009). Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 182: 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang S., Tao Q., Pan J., Si L., Gong Z., Cai R. (2012). A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J. Exp. Bot. 63: 4475–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Liu S., Xu L., Jia Z., Xu Y., Yang Q., Fei Z., Lu X., Chen H., Huang S. (2008). Genetic association of ETHYLENE-INSENSITIVE3-like sequence with the sex-determining M locus in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 117: 927–933. [DOI] [PubMed] [Google Scholar]
- Lyzenga W.J., Booth J.K., Stone S.L. (2012). The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 71: 23–34. [DOI] [PubMed] [Google Scholar]
- Martínez C., Manzano S., Megías Z., Garrido D., Picó B., Jamilena M. (2013). Involvement of ethylene biosynthesis and signalling in fruit set and early fruit development in zucchini squash (Cucurbita pepo L.). BMC Plant Biol. 13: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monforte A.J., Diaz A., Caño-Delgado A., van der Knaap E. (2014). The genetic basis of fruit morphology in horticultural crops: Lessons from tomato and melon. J. Exp. Bot. 65: 4625–4637. [DOI] [PubMed] [Google Scholar]
- Neff M.M., Neff J.D., Chory J., Pepper A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392. [DOI] [PubMed] [Google Scholar]
- Nodzon L.A., Xu W.H., Wang Y., Pi L.Y., Chakrabarty P.K., Song W.Y. (2004). The ubiquitin ligase XBAT32 regulates lateral root development in Arabidopsis. Plant J. 40: 996–1006. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y., Wang Z.-Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Martínez O., Pernas M., Carol R.J., Dolan L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317: 507–510. [DOI] [PubMed] [Google Scholar]
- Pan Y., Liang X., Gao M., Liu H., Meng H., Weng Y., Cheng Z. (2017). Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 130: 573–586. [DOI] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Visser E.J., de Kroon H., Voesenek L.A. (2003). Ethylene is required in tobacco to successfully compete with proximate neighbours. Plant Cell Environ. 26: 1229–1234. [Google Scholar]
- Pierik R., Cuppens M.L., Voesenek L.A., Visser E.J. (2004). Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol. 136: 2928–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Tholen D., Poorter H., Visser E.J.W., Voesenek L.A.C.J. (2006). The Janus face of ethylene: Growth inhibition and stimulation. Trends Plant Sci. 11: 176–183. [DOI] [PubMed] [Google Scholar]
- Prasad M.E., Schofield A., Lyzenga W., Liu H., Stone S.L. (2010). Arabidopsis RING E3 ligase XBAT32 regulates lateral root production through its role in ethylene biosynthesis. Plant Physiol. 153: 1587–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., et al. (2013). A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45: 1510–1515. [DOI] [PubMed] [Google Scholar]
- Rudich J., Halevy A.H., Kedar N. (1969). Increase in femaleness of three cucurbits by treatment with Ethrel, an ethylene releasing compound. Planta 86: 69–76. [DOI] [PubMed] [Google Scholar]
- Saibo N.J.M., Vriezen W.H., Beemster G.T.S., Van Der Straeten D. (2003). Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 33: 989–1000. [DOI] [PubMed] [Google Scholar]
- Shinozaki Y., et al. (2015). Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83: 237–251. [DOI] [PubMed] [Google Scholar]
- Smalle J., Haegman M., Kurepa J., Van Montagu M., Straeten D.V. (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. USA 94: 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., et al. (2003). The Arabidopsis mutant alh1 illustrates a cross talk between ethylene and auxin. Plant Physiol. 131: 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Vaseva I., Vissenberg K., Van Der Straeten D. (2012). Ethylene in vegetative development: A tale with a riddle. New Phytol. 194: 895–909. [DOI] [PubMed] [Google Scholar]
- Wang K.L.C., Yoshida H., Lurin C., Ecker J.R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950. [DOI] [PubMed] [Google Scholar]
- Wei Q., Wang Y., Qin X., Zhang Y., Zhang Z., Wang J., Li J., Lou Q., Chen J. (2014). An SNP-based saturated genetic map and QTL analysis of fruit-related traits in cucumber using specific-length amplified fragment (SLAF) sequencing. BMC Genomics 15: 1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C.-K. (2014). Ethylene in Plants. Springer, Netherlands. [Google Scholar]
- Weng Y., Colle M., Wang Y., Yang L., Rubinstein M., Sherman A., Ophir R., Grumet R. (2015). QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor. Appl. Genet. 128: 1747–1763. [DOI] [PubMed] [Google Scholar]
- Xing H.-L., Dong L., Wang Z.-P., Zhang H.-Y., Han C.-Y., Liu B., Wang X.-C., Chen Q.-J. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.F., Hoffman N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35: 155–189. [Google Scholar]
- Yang X.Y., Wang Y., Jiang W.J., Liu X.L., Zhang X.M., Yu H.J., Huang S.W., Liu G.Q. (2013). Characterization and expression profiling of cucumber kinesin genes during early fruit development: Revealing the roles of kinesins in exponential cell production and enlargement in cucumber fruit. J. Exp. Bot. 64: 4541–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wen C.-K. (2010). Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiol. Biochem. 48: 45–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that support the findings within this article are available from the corresponding author upon reasonable request.