Rice qGL3 negatively regulates brassinosteroid signaling by regulating the phosphorylation level and stability of OsGSK3, which modulates OsBZR1 phosphorylation status and subcellular distribution.
Abstract
Brassinosteroids (BRs) are steroid hormones that play essential roles in plant growth and development. We previously cloned qGL3, a major quantitative trait locus regulating grain length in rice (Oryza sativa). The O. sativa japonica var N411 has extra-large grains compared with the O. sativa indica var 9311, and the recessive qgl3 allele from N411 contributes positively to grain length. qGL3 encodes a putative protein phosphatase with Kelch-like repeat domains, an ortholog of Arabidopsis (Arabidopsis thaliana) brassinosteroid-insensitive1 SUPPRESSOR1 (BSU1). BSU1 positively regulates BR signaling, while overexpression of qGL3 induced BR loss-of-function phenotypes. Both qGL3N411 and qGL39311 physically interact with the rice glycogen synthase kinase 3 (GSK3)/SHAGGY-like kinase 3 (OsGSK3), an ortholog of Arabidopsis BR INSENSITIVE2 (BIN2). qGL39311 dephosphorylates OsGSK3, but qGL3N411 lacks this activity. Knocking out OsGSK3 enhances BR signaling and induces nuclear localization of O. sativa BRASSINAZOLE RESISTANT1 (OsBZR1). Unlike the dephosphorylation of BIN2 (which leads to protein degradation) in Arabidopsis, qGL3 dephosphorylates and stabilizes OsGSK3 in rice. These results demonstrate that qGL3 suppresses BR signaling by regulating the phosphorylation and stability of OsGSK3, which modulates OsBZR1 phosphorylation and subcellular distribution. Our study clarifies the role of qGL3 in the regulation of grain length and provides insight into BR signaling, including the differences between rice and Arabidopsis.
INTRODUCTION
Brassinosteroids (BRs) are involved in diverse biological processes such as cell elongation, cell division, and cell differentiation (Clouse and Sasse, 1998; Müssig et al., 2003; Clouse, 2011; Yang et al., 2011; Wei and Li, 2016). In rice (Oryza sativa), the BR signal is perceived by two receptors and transduced to O. sativa BR SIGNALING KINASE3 (OsBSK3; Zhang et al., 2016), which phosphorylates an unidentified phosphatase and represses rice glycogen synthase kinase 3 (GSK3)/SHAGGY-like kinase 2 (OsGSK2) activity (Tong et al., 2012). OsGSK1 and OsGSK2 are homologs of Arabidopsis (Arabidopsis thaliana) BR INSENSITIVE 2 (BIN2) and repress BR signaling (Koh et al., 2007; Tong et al., 2012) by negatively regulating the levels of proteins, including O. sativa BRASSINAZOLE RESISTANT1 (OsBZR1; Bai et al., 2007) and DWARF AND LOW-TILLERING ([DLT]; Tong et al., 2012). The rice quantitative trait locus (QTL) THOUSAND GRAIN WEIGHT3 controls grain size and encodes OsGSK5 (Hu et al., 2018). OsGSK5 and OsGSK2 have different roles in BR responses, and the interaction between OsGSK5 and OsBZR1 is very weak in yeast cells (Hu et al., 2018). OsBZR1, a homolog of Arabidopsis brassinosteroid-insensitive1 (bri1)-ETHYL METHANESULFONATE SUPPRESSOR1/BZR1 (BES1/BZR1), plays a positive role in rice BR signaling, and suppression of OsBZR1 leads to semi-dwarfism and an erect-leaf phenotype (Bai et al., 2007). OsBZR1 directly regulates yield-determining genes to control multiple biological processes (Tong and Chu, 2018). OsBZR1 binds to the promoter of O. sativa INCREASE LEAF INCLINATION1 (OsILI1) and OsILI1 BINDING BASIC HELIX-LOOP-HELIX/bHLH1 to induce OsILI1 and repress OsILI1 BINDING BASIC HELIX-LOOP-HELIX/bHLH1 expression (Zhang et al., 2009a, 2009b). The rice LEAF and TILLER ANGLE INCREASED CONTROLLER interacts with and antagonizes OsBZR1 to negatively regulate BR responses (Zhang et al., 2012a). REDUCED LEAF ANGLE1/SMALL ORGAN SIZE1 interacts with OsBZR1 to modulate its transcription activity (Qiao et al., 2017). OsBZR1 also directly binds to the promoters of GA (GIBBERELLIN) 20ox (oxidase)-2, GA3ox-2, and GA2ox-3 and influences gibberellin biosynthesis in rice (Tong et al., 2014).
Recent studies have highlighted the importance of BRs in the control of grain yield (Divi and Krishna, 2009). BRs affect many agronomic traits related to yield, including plant height, leaf angle, root development, tiller number, and grain size (Zuo and Li, 2014; Wei and Li, 2018). In rice, GRAIN LENGTH2 (GL2) affects grain size and is involved in BR responses. GL2 encodes O. sativa GROWTH REGULATION FACTOR4 (OsGRF4) and GL2/OsGRF4 interacts with OsGSK2, which is capable to inhibit GL2/OsGRF4 transcription activation activity to regulate grain length (Che et al., 2015). The QTL SEED WIDTH5/GRAIN WIDTH5 strongly affects grain width and encodes a calmodulin binding protein. GRAIN WIDTH5 physically interacts with OsGSK2 and represses its kinase activity, resulting in accumulation of dephosphorylated OsBZR1 and DLT in the nucleus to mediate BR-responsive genes expression (Liu et al., 2017). Therefore, uncovering the mechanisms of BR signaling may facilitate efforts to increase grain yield in rice.
Protein phosphatases, such as Ser/Thr phosphatases closely related to protein phosphatase 1 (PP1), play critical roles in BR signaling (Di Rubbo et al., 2011). PP1 has a characteristic of C-terminal catalytic domain linked to an N-terminal Kelch-repeat domain (Kutuzov and Andreeva, 2002; Moorhead et al., 2009; Oh et al., 2009). Based on these characteristics, these PP1-related phosphatases are called protein phosphatases with Kelch-like domains (PPKLs) (Maselli et al., 2014). PPKLs are present only in plants and alveolates (Kutuzov and Andreeva, 2002). In Arabidopsis, bri1 SUPPRESSOR1 (BSU1) was the first PPKL functionally characterized as a positive effector of BR signaling (Mora-García et al., 2004). Overexpression of BSU1 suppressed the phenotype of the bri1 mutant and led to the accumulation of dephosphorylated BES1/BZR1 (Kim et al., 2009).
We previously cloned the major grain-length QTL qGL3 and showed that qGL3 encodes a putative protein phosphatase with Kelch-like repeat domains (OsPPKL1), an ortholog of BSU1. N411, an O. sativa japonica variety with extra-large grains, has a rare qGL3 allele, qgl3, that produces its long-grain phenotype. Field trials showed that rice plants carrying the qgl3 allele had significantly increased grain yield due to its effect on grain length, filling, and weight (Zhang et al., 2012b). The qGL3 protein in N411 has two amino acid changes compared with qGL3 in 9311, a high-quality elite O. sativa indica variety with relatively smaller grains: an Asp-to-Glu substitution in a conserved AVLDT motif of the second Kelch domain and a His-to-Tyr substitution (Zhang et al., 2012b). GL3.1, encoded by an allele of qGL3, modulates grain length by dephosphorylating the cell cycle protein Cyclin-T1;3 (Qi et al., 2012). Although qGL3 plays a critical role in modulating grain length and yield, the precise functions of qGL3 remain largely unknown.
RESULTS
qGL3 Functions as a Negative Regulator in the BR Signaling Pathway
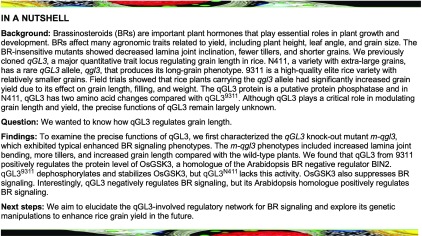
To examine the precise functions of qGL3, we first characterized the qGL3 knockout mutant m-qgl3, which exhibited typical enhanced BR signaling phenotypes (Figure 1A). These phenotypes were similar to those of the BR-sensitive mutant m107 and DLT-overexpressing plants (Tanabe et al., 2005; Tong et al., 2012). The m-qgl3 phenotypes included increased lamina joint bending, more tillers, and reduced plant height compared with the wild-type plants (Dongjin [DJ], japonica) (Figures 1A, 1B, and 1D). The angles between the leaf sheath and leaf blade were substantially increased in the m-qgl3 mutant (Figures 1A, 1C, and 1G). These results indicated that BR signaling was likely enhanced in the m-qgl3 mutant.
Figure 1.
qGL3 Is Involved in the Rice BR Signaling.
(A) Morphology of the m-qgl3 mutant and qGL3-OX1 plant at the reproductive phase.
(B), (D), and (E) Quantification of plant height (B), tiller number (D), and grain length (E) of the m-qgl3 mutant and qGL3-OX1 compared with the wild type. For quantification of plant height and tiller number, data are shown as means ± se (n = 10). For quantification of grain length, data are shown as means ± se (n = 30). The data were compared by Student’s t test. ***P < 0.001.
(C) Lamina inclination responding to 1 μM BL of the wild-type, m-qgl3 mutant, and qGL3-OX plants.
(F) Coleoptile elongation of the wild-type, m-qgl3 mutant, and qGL3-OX plants in response to 1 μM BL. The red hyphen indicates the top of coleoptiles.
(G) Statistical data of lamina inclination responding to 1 μM BL of the m-qgl3 mutant and qGL3-OX plants compared with the wild type. Data are shown as means ± se (n = 20). The data were compared by Student’s t test. ***P < 0.001.
(H) Statistical analysis of coleoptile length undertreatment with BL. Data are shown as means ± se (n = 20). The data were compared by Student’s t test. ***P < 0.001.
To further investigate the functions of qGL3, we examined the transgenic plants overexpressing qGL3 under the cauliflower mosaic virus 35S promoter (qGL3-OX). The qGL3-OX plants displayed a typical BR loss-of-function phenotype with erect leaves, fewer tillers, and shorter grains compared with the wild type (Zhonghua 11 [ZH11], japonica; Figures 1A, 1D, and 1E). These phenotypes were similar to dwarf2 (d2), dwarf11 (d11), and OsBZR1-RNAi plants (Hong et al., 2003; Tanabe et al., 2005; Bai et al., 2007). We also observed leaf lamina joint bending in rice seedlings at the three-leaf stage (Figure 1C). The m-qgl3 mutant showed increased leaf inclination, and the qGL3-OX lines showed reduced leaf angles (Figures 1C and 1G).
To analyze the expression of qGL3 in rice, we constructed a fusion of the qGL3 promoter to the β-glucuronidase (GUS) reporter (ProqGL3:GUS). GUS activity was found in vascular tissues, the root-hypocotyl junction, and glumes (Supplemental Figure 1A). In situ mRNA hybridization with an antisense probe targeting the 3′-subterminal region of qGL3 showed that qGL3 was expressed in spikelet apical meristems (Supplemental Figure 1B).
Considering that the m-qgl3 mutant showed the classic phenotype of mutants with enhanced BR signaling, and the qGL3-OX line showed the classic BR loss-of-function response, we hypothesized that qGL3 is involved in modulating BR signaling. Therefore, we examined the brassinolide (BL) sensitivity of the wild type, the m-qgl3 mutant, the qGL3-OX lines, and the near-isogenic line NILqgl3 carrying the qGL3N411 allele in the 9311 background. We used the lamina inclination assay to quantify the BR sensitivity of these seedlings. BL significantly increased the lamina joint bending of m-qgl3 (Figures 1C and 1G) and NILqgl3 (Supplemental Figures 2A and 2B) seedlings compared with the wild type and had the opposite effect on qGL3-OX plants (Figures 1C and 1G). In addition, coleoptile growth experiments showed that the m-qgl3 mutant (Figures 1F and 1H) and NILqgl3 (Supplemental Figures 2C and 2D) were more sensitive to BL than the wild type. The qGL3-OX lines showed the opposite phenotypes (Figures 1F and 1H).
BR represses the expression of two BR biosynthetic genes, D2 and D11, and a BR signaling gene, DLT (Hong et al., 2003; Tanabe et al., 2005; Tong et al., 2012); we found that these genes were downregulated in the m-qgl3 mutant compared with the wild type and upregulated in the qGL3-OX lines. DLT showed a similar expression profile to that of D2 and D11 (Supplemental Figure 3). These results provide further evidence for the involvement of qGL3 as a regulator of BR responses.
qGL3 Interacts with and Dephosphorylates OsGSK3
The different responses of the qGL3-OX lines, the m-qgl3 mutant, and the NILqgl3 plants to BL treatment prompted us to further explore the functions of qGL3 in BR signaling. We performed a yeast two-hybrid (Y2H) assay to test the interaction between qGL3 and OsGSKs and found that qGL3 interacted with nine OsGSKs (Supplemental Figure 4). We obtained T-DNA insertion mutants for OsGSK1 to OsGSK5 and found that the osgsk3 mutant showed significantly increased grain length compared with the wild type (Supplemental Figure 5).
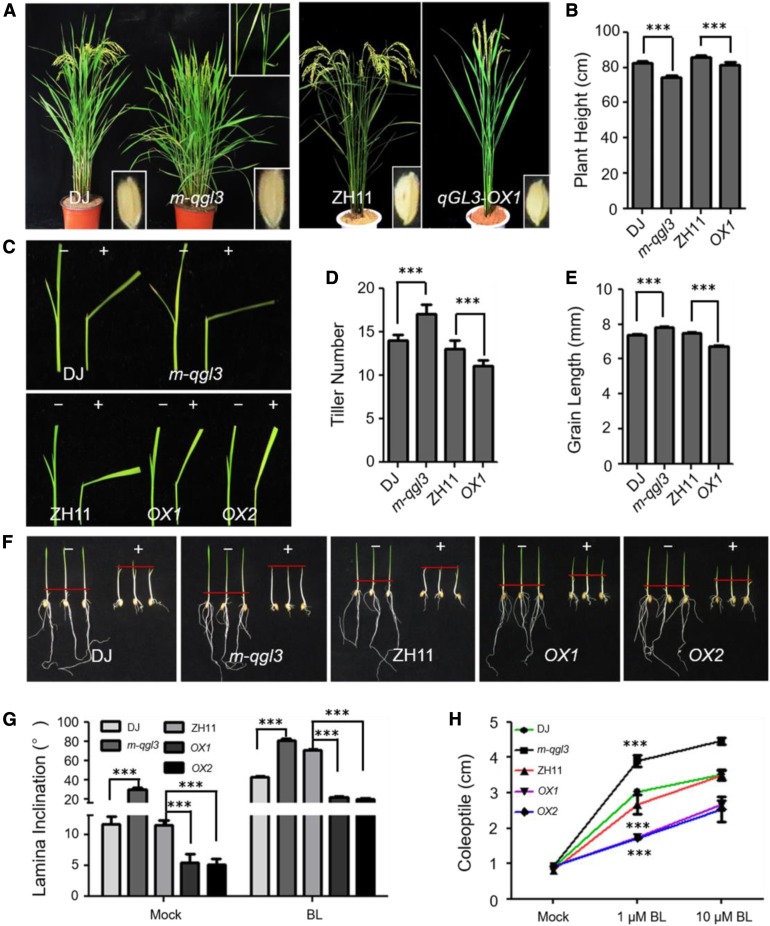
We determined the critical region of qGL3 that mediated the interaction with OsGSK3. The Y2H assay showed that the Protein Phosphatase 2A (PP2A) and regions between Kelch domain and PP2A domain (hereafter referred as regions IN) of qGL39311 (the allele from the shorter grain indica var 9311) interacted with OsGSK3, but the PP2A domain alone did not interact with OsGSK3 (Figure 2A). The two amino acid substitutions in the Kelch domain and the regions IN of qGL3N411 (the allele from the longer grain japonica var N411) did not affect the interaction between qGL3 and OsGSK3.
Figure 2.
qGL3 Interacts with OsGSK3.
(A) Y2H analysis of qGL3 and OsGSK3. "In" indicates the regions between Kelch domain and PP2A domain; -R indicates qGL39311; -P indicates qGL3N411.
(B) BiFC analysis of the qGL3 and OsGSK3 interaction. The YFP fluorescence signals and autofluorescence signals from chloroplasts are pseudo-colored as yellow and red, respectively. Bars = 20 μm. YN indicates YFP N terminal; YC indicates YFP C terminal..
(C) GST pull-down assay of the qGL3 and OsGSK3 interaction. Anti-HIS antibody was used to detect the output protein.
Bimolecular fluorescence complementation (BiFC) analysis was used to determine whether qGL3 interacts with OsGSK3 in vivo. In Nicotiana benthamiana leaf epidermal cells coexpressing the N-terminal half of yellow fluorescent protein (YFP) fused to qGL39311 or qGL3N411, and the C-terminal half of YFP fused to OsGSK3, strong YFP fluorescence was observed in the cytoplasm (Figure 2B). Moreover, glutathione S-transferase (GST) pull-down assays were used to confirm the in vitro interaction between qGL3 and OsGSK3, in which GST-OsGSK3 was used to pull down His-qGL3 proteins, which were detected by an anti-His antibody. GST alone did not pull down His-qGL3, but GST-OsGSK3 did interact with qGL39311 and qGL3N411 (Figure 2C), suggesting that the two amino acid substitutions in qGL3N411 did not affect the interaction of qGL3 and OsGSK3.
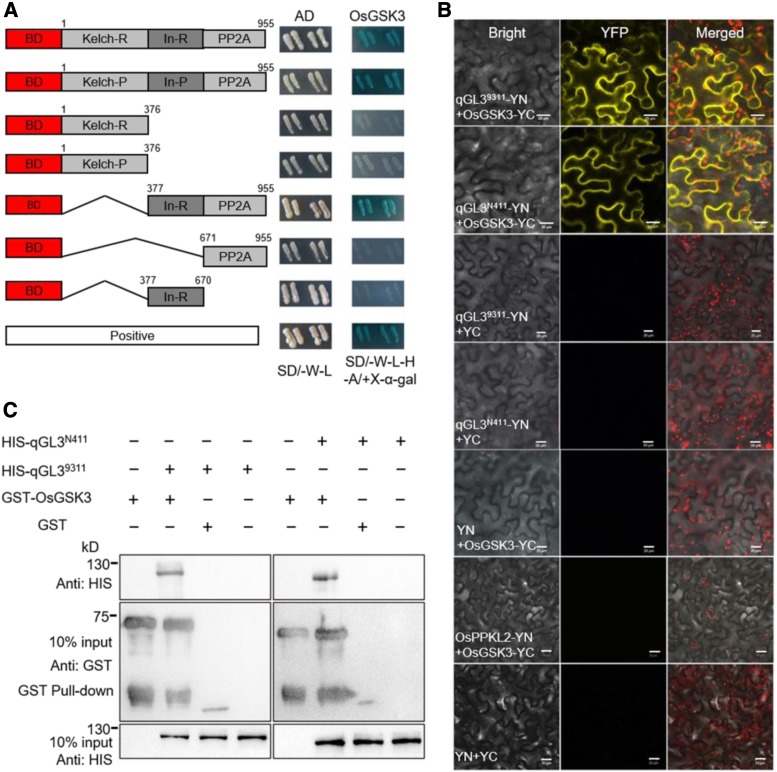
We then performed phosphorylation and dephosphorylation assays to determine whether qGL3 can dephosphorylate OsGSK3 in vitro. The purified GST-OsGSK3 was first self-phosphorylated. We used the activated GST-OsGSK3 in a dephosphorylation reaction with His-qGL39311 and HIS-qGL3N411. We detected the phosphorylation status of OsGSK3 using a Phos-tag assay in which phosphorylated proteins are visualized as bands on a nitrocellulose membrane containing the Phos-tag reagent. The GST-OsGSK3 band incubated with His-qGL39311 was fainter than the control band. However, the band of GST-OsGSK3 incubated with His-qGL3N411 showed no differences compared with that of the control incubation (Figure 3A). These results indicate that qGL39311 dephosphorylates OsGSK3 in vitro. Additionally, we performed a protein migration assay to further confirm the dephosphatase activity of qGL3 on OsGSK3. When His-qGL39311 and GST-OsGSK3 were incubated in vitro, little phosphorylated OsGSK3 was detected. These data showed that His-qGL39311, but not His-qGL3N411, dephosphorylated GST-OsGSK3, as indicated by the shifted GST-OsGSK3 band detected with anti-GST antibody (Figure 3A).
Figure 3.
qGL39311, Not qGL3N411, Dephosphorylates OsGSK3.
(A) In vitro dephosphorylation analysis. Phosphorylation was detected with biotin-pendant Zn2+ Phos-tag (BTL-111). For the protein migration assay, the anti-GST antibody was used to detect the positions of dephosphorylated OsGSK3 and phosphorylated OsGSK3. The red line separates the phosphorylated and dephosphorylated OsGSK3. The corresponding bottom panels show Coomassie blue staining of the proteins used for the dephosphorylation assay.
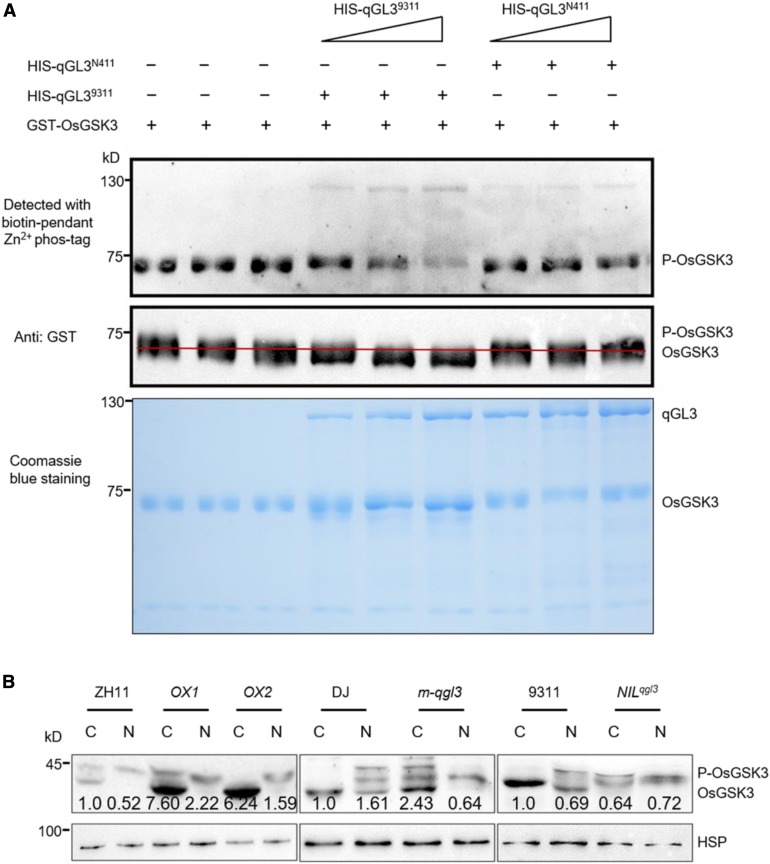
(B) Comparison of the level of OsGSK3 in the wild-type, m-qgl3 mutant, qGL3-OX lines, and NILqgl3 plants. Proteins were separately extracted from the cytoplasm and nucleus. The quantification of the immunoblot results was normalized to HSP and the relative protein levels of OsGSK3 in cytoplasm in the wild type were defined as 1. C, cytoplasm; N, nucleus.
In BiFC with qGL3 and OsGSK3, the YFP signal was observed in the cytoplasm, indicating that qGL3 may dephosphorylate OsGSK3 in the cytoplasm of rice cells. To test this, the nuclear and cytoplasmic fractions of the transgenic plants were isolated and examined by immunoblots using OsGSK3 antibody. Figure 3B shows that the phosphorylated OsGSK3 accumulated in the nuclei in the wild-type plants. In m-qgl3 plants, the phosphorylated OsGSK3 also accumulated in the cytoplasm, but dephosphorylated OsGSK3 increased in the cytoplasm in qGL3-OX lines (Figure 3B). Moreover, the dephosphorylated OsGSK3 decreased and the phosphorylated OsGSK3 increased in the cytoplasm in NILqgl3 compared with 9311 (Figure 3B). In contrast to qGL39311, qGL3N411 could not dephosphorylate OsGSK3 in rice plants.
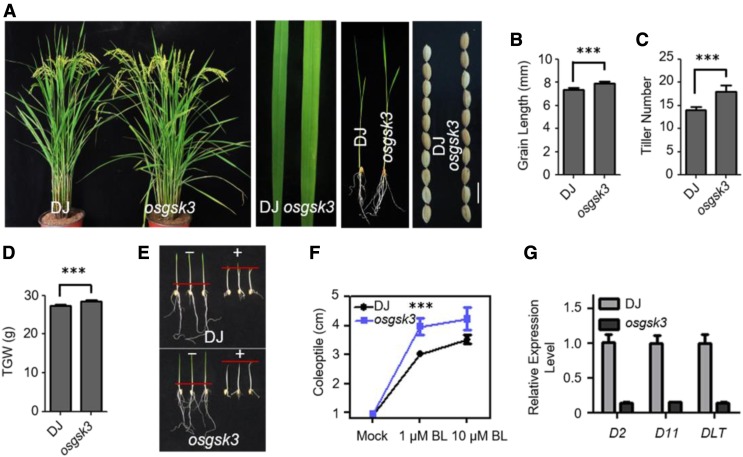
Mutation of OsGSK3 Leads to Enhanced BR Signaling Phenotypes
Loss of function of OsGSK3 (Supplemental Figures 6A and 6B) led to larger angles between the leaf sheath and leaf blade compared with the wild type (Figure 4A). In the osgsk3 mutant compared with the wild type, the tiller number significantly increased, leaves and seeds were longer, and leaves were lighter green (Figures 4A–4C). The osgsk3 mutant showed an increase in the 1000-grain weight compared with the wild type (Figure 4D). In addition, we analyzed the sensitivity of osgsk3 to BL using coleoptile growth experiments. As expected, the osgsk3 mutant was hypersensitive to BL (Figures 4E and 4F). In addition, we used the lamina inclination assay to quantify the BR sensitivity of the wild type and osgsk3 mutants. BL significantly increased the lamina joint bending of osgsk3 seedlings compared with the wild type (Supplemental Figures 6C and 6D). Similar to the m-qgl3 mutant, the expression levels of D2, D11, and DLT significantly decreased in the osgsk3 mutant, further supporting a negative role of OsGSK3 in BR signaling (Figure 4G).
Figure 4.
OsGSK3 Is Involved in BR Signaling in Rice.
(A) Morphology of the osgsk3 mutant at the reproductive phase. Leaf width and leaf color of the osgsk3 mutant. Phenotypes of the osgsk3 mutant seedling. Seed morphology of the wild type and osgsk3 mutant. Bar = 1 cm.
(B) to (D) Quantification of grain length (B), tiller number (C), and 1000-grain weight (D) of the osgsk3 mutant compared with the wild type. For quantification of grain length, data are shown as means ± se (n = 30). For quantification of tiller number and 1000-grain weight, data are shown as means ± se (n = 10). The data were compared by Student’s t test. ***P < 0.001. TGW, Thousand-Grain Weight.
(E) Coleoptile elongation of the wild type and osgsk3 mutant in response to 1 μM BL. The red hyphen indicates the top of coleoptiles.
(F) Quantification of coleoptile length of the osgsk3 mutant compared with the wild type. Data are shown as means ± se (n = 20). The data were compared by Student’s t test. ***P < 0.001.
(G) Expression patterns of BR biosynthesis and signaling-related genes in the seedlings of the wild type and osgsk3 mutant. Data are shown as means ± se (n = 3).
OsBZR1 is a downstream transcription factor in BR signaling and plays crucial roles in rice development (Bai et al., 2007). We found that OsBZR1 interacted with OsGSK3 in the Y2H analysis and GST pull-down assay (Supplemental Figures 7A and 7B). When we performed an in vitro kinase assay with GST-OsGSK3 and GST-OsBZR1, OsGSK3 indeed phosphorylated OsBZR1. Lithium chloride (LiCl) has been found to inhibit the kinase activities of BIN2 in Arabidopsis (Ye et al., 2012). We then analyzed the effect of LiCl treatment on the phosphorylation activity of OsGSK3. The result showed that Li+ can inhibit OsGSK3 phosphorylation on OsBZR1 as well as OsGSK3 autophosphorylation (Supplemental Figure 7C). Next, we analyzed the phosphorylated status of OsGSK3 in the seedlings under 10 mM LiCl treatment. The wild-type (ZH11) seedlings were grown on brassinazole (BRZ)-containing medium for 1 week and transferred then to liquid medium containing 10 mM LiCl. The results showed that the phosphorylated OsGSK3 increased when treated with BRZ, and the phosphorylated proteins then decreased after being treated with LiCl. We also found that the treatment of LiCl inhibited the accumulation of phosphorylated OsBZR1 (Supplemental Figure 7D). This result provides the further evidence of the involvement of OsGSK3 in BR signaling.
qGL3 Regulates OsGSK3 and OsBZR1 Levels
To investigate the regulation of OsGSK3 by qGL3, we performed a subcellular localization assay to determine the effect of qGL3 on the localization of OsGSK3. We produced a fusion construct in which green fluorescent protein (GFP) was fused to the C terminus of OsGSK3 and expressed the fusions in rice protoplasts. OsGSK3-GFP localized in the cytoplasm and nucleus in the wild type (DJ and ZH11; Supplemental Figures 8A and 8C). In m-qgl3, the OsGSK3-GFP signals were weaker than in DJ cells (Supplemental Figure 8B). In qGL3-OX cells, the OsGSK3-GFP signals intensified compared with ZH11, indicating that qGL3 affects the subcellular localization of OsGSK3 (Supplemental Figure 8D). These data suggest that qGL3 is a positive regulator of OsGSK3.
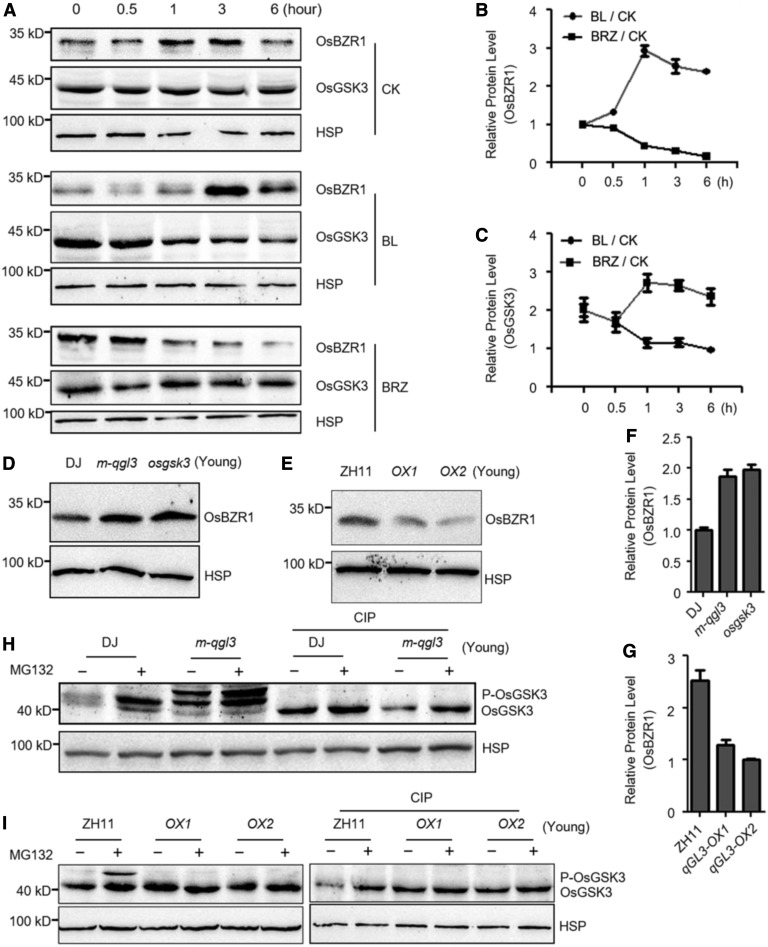
Since qGL3 interacts with OsGSK3, the regulation of OsGSK3 by BR likely occurs through the qGL3 phosphatase activity. To further investigate this, we determined the level of OsGSK3 in the m-qgl3 mutant and the qGL3-OX lines. First, we examined the effects of BL and BRZ treatment on the levels of OsGSK3 and OsBZR1. OsBZR1 substantially accumulated and OsGSK3 decreased in the wild-type plants treated with 1 μM BL (Figures 5A–5C). By contrast, BRZ treatment decreased OsBZR1 accumulation and increased OsGSK3 expression level (Figures 5A–5C). We then examined the effect of N-(benzyloxycarbonyl)-Leu-Leu-Leu-al ([MG132], a proteasome inhibitor) on the levels of OsGSK3. The phosphorylated form of OsGSK3 increased after treatment with MG132 in the wild-type seedlings, suggesting that phosphorylated OsGSK3 was targeted for proteasomal degradation (Figures 5H and 5I).
Figure 5.
Protein Level of OsGSK3 And OsBZR1 Are Modulated By qGL3.
(A) Accumulation of OsGSK3 and OsBZR1 in rice seedlings under BL and BRZ treatment. OsGSK3 and OsBZR1 were detected by immunoblot assays with anti-OsGSK3 and anti-OsBZR1 antibodies. Rice HSP (∼90 kD) was used as the internal control.
(B) and (C) Quantification analysis for (A). The relative levels of the protein level of OsBZR1 (B) and OsGSK3 (C) in response to mock were defined as 1. Data are shown as means ± se (n = 3).
(D) and (E) Comparison of the level of OsBZR1 in the seedlings of the wild type, m-qgl3 and osgsk3 mutants (D), and qGL3-OX plants (E).
(F) Quantification analysis for (D). The relative levels of the DJ were defined as 1. Data are shown as means ± se (n = 3).
(G) Quantification analysis for (E). The relative levels of the qGL3-OX2 were defined as 1. Data are shown as means ± se (n = 3).
(H) Effect of MG132 treatment on the level and phosphorylation status of OsGSK3 in the wild type and m-qgl3 mutant.
(I) Effect of MG132 treatment on the level and phosphorylation status of OsGSK3 in the wild-type and qGL3-OX plants.
We analyzed the levels of OsGSK3 in the m-qgl3 mutant and the qGL3-OX plants and found that the dephosphorylated form of OsGSK3 accumulated in the qGL3-OX plants, while the phosphorylated form of OsGSK3 increased in the m-qgl3 mutant compared with the wild type (Figures 5H and 5I). The slower migrating bands disappeared when the total protein samples were treated with calf intestine alkaline phosphatase (CIP), indicating that the slower migrating bands represent phosphorylated OsGSK3. OsGSK2 is an ortholog of BIN2, and overexpression of mutated OsGSK2 led to the typical BR loss-of-function phenotypes (Tong et al., 2012). The protein level and phosphorylated status of OsGSK2 in the m-qgl3 mutant and the qGL3-OX plants with OsGSK2 antibody were analyzed, and we found no significant variation of OsGSK2 in the qGL3-OX plants but higher phosphorylated OsGSK2 levels in the m-qgl3 mutant compared with the wild type (Supplemental Figure 9).
We also analyzed the level of OsBZR1 in the osgsk3 and m-qgl3 mutants and the qGL3-OX plants and found that OsBZR1 increased in the osgsk3 and m-qgl3 mutant seedlings compared with the wild type (Figures 5D and 5F) and decreased in qGL3-OX seedlings (Figures 5E and 5G). Moreover, the m-qgl3 plants slightly accumulated more dephosphorylated DLT compared with the wild type, and the total protein level of DLT decreased in qGL3-OX lines (Supplemental Figure 9). Interestingly, the osgsk3 mutant plants accumulated both the phosphorylated and dephosphorylated DLT (Supplemental Figure 9).
qGL3 and OsGSK3 Regulate the Subcellular Localization of OsBZR1
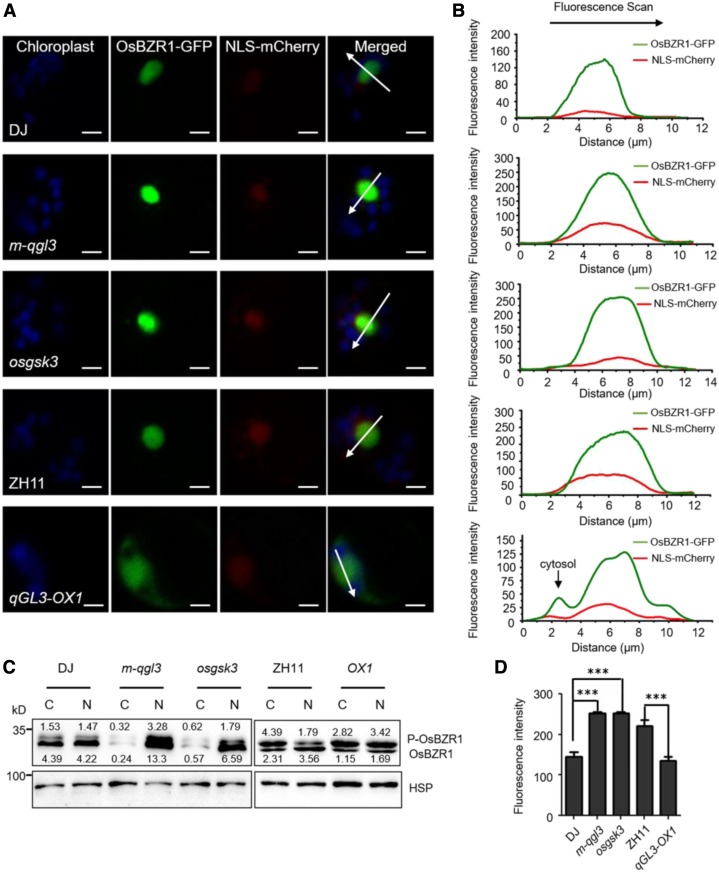
In Arabidopsis, BIN2 and BSU1 are nucleocytoplasmic regulators that modulate the subcellular localization of BZR1/BES1 (Ryu et al., 2010). In this study, we analyzed the subcellular localization of OsBZR1 in rice protoplasts. The merged image of OsBZR1-GFP and nuclear localization signal (NLS)-mCherry signals showed that OsBZR1 localized in the nucleus (Figure 6A).
Figure 6.
OsBZR1 Subcellular Distribution Is Modulated By OsGSK3 And qGL3.
(A) Subcellular localization of OsBZR1 in the protoplasts of the wild-type, m-qgl3 and osgsk3 mutants, and qGL3-OX1 plants. NLS-mCherry was used as a nuclear marker; the OsBZR1-GFP and NLS-mCherry signals match well in the nucleus. The GFP fluorescence signals, mCherry fluorescence signals, and autofluorescence signals from chloroplasts are pseudo-colored as green, red, and blue, respectively. Bars = 5 μm.
(B) Fluorescent signal was analyzed using Image Browser software. Different signal peaks of OsBZR1-GFP were detected in the nucleus and cytoplasm. The fluorescence intensities of the regions indicated by arrows are shown.
(C) Localization of phosphorylated OsBZR1 was affected by OsGSK3 and qGL3. Proteins of the wild-type, m-qgl3, osgsk3, and qGL3-OX1 plants were separately extracted from the cytoplasm and nucleus. C, cytoplasm; N, nucleus.
(D) Statistical analysis of the fluorescence intensities. For quantification of fluorescence intensities, data are shown as means ± se (n = 10). The data were compared by Student’s t test. ***P < 0.001.
To investigate the effects of qGL3 and OsGSK3 on the subcellular localization of OsBZR1, we transfected OsBZR1-GFP into the m-qgl3 and osgsk3 mutants and the qGL3-OX plants. We detected strong OsBZR1-GFP signals in the nuclei of protoplasts from the m-qgl3 and osgsk3 mutants (Figure 6A). The peak of OsBZR1-GFP signal overlapped with that of NLS-mCherry (Figure 6B). In qGL3-OX protoplasts, OsBZR1 showed nuclear and cytoplasmic localization (Figures 6A and 6B). Analysis of the OsBZR1-GFP intensities in the nucleus revealed that GFP signals significantly increased in m-qgl3 and osgsk3 mutants and decreased in qGL3-OX plants (Figure 6D).
We further analyzed the protein level and status of OsBZR1 in the nucleus and cytoplasm. Nuclear and cytoplasmic fractions were prepared from the mature leaves of the qGL3-OX, m-qgl3, osgsk3, and wild-type plants. Immunoblotting showed that the protein level of OsBZR1 in the cytoplasm was largely reduced in m-qgl3 and osgsk3 mutants compared with DJ. In qGL3-OX plants, the phosphorylated OsBZR1 accumulated in the cytoplasm and dephosphorylated OsBZR1 decreased in the nucleus (Figure 6C). These results indicated that qGL3 and OsGSK3 influenced the nuclear localization of OsBZR1.
Genetic Analysis of qGL3, OsGSK3, and OsBZR1
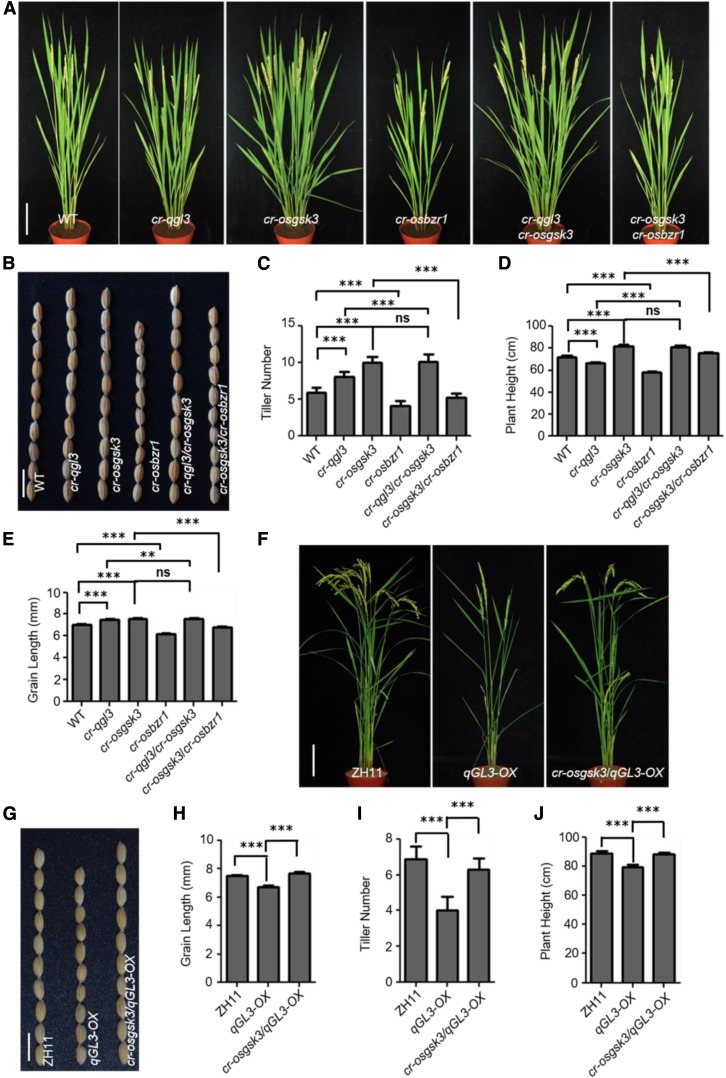
To analyze the genetic interactions, we used the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR associated protein 9 (Cas9) system to create multiple mutants of qGL3, OsGSK3, and OsBZR1 (specific target sites on each gene are shown in Supplemental Table). Moreover, we used CRISPR/Cas9 to knock out OsGSK3 in qGL3-OX plants and obtained the homozygous cr-osgsk3/qGL3-OX line. To ascertain the genotype and heritability of each line, we sequenced the target genes to confirm the genotypes (Supplemental Figure 10). Compared with the wild type, cr-qgl3, qGL3-OX, and cr-osbzr1 displayed a dwarf phenotype, while cr-osgsk3 showed increased plant height compared with the wild type. The cr-qgl3/cr-osgsk3 plants were similar in height to cr-osgsk3 plants (Figures 7A and 7D). The cr-osgsk3/qGL3-OX plants showed a wild-type–like phenotype (Figures 7F and 7J). In the cr-osgsk3/cr-osbzr1 double mutants, knocking out OsBZR1 suppressed the plant height of cr-osgsk3 (Figures 7A and 7D).
Figure 7.
Genetic Analysis of qGL3, OsGSK3, and OsBZR1.
(A) Morphological features of the wild-type, cr-qgl3, cr-osgsk3, cr-osbzr1, cr-qgl3/cr-osgsk3, and cr-osgsk3/cr-osbzr1 plants at the mature stage. Bar = 10 cm. WT, wild type.
(B) Grain phenotypes of the wild-type, cr-qgl3, cr-osgsk3, cr-osbzr1, cr-qgl3/cr-osgsk3, and cr-osgsk3/cr-osbzr1 plants. Bar = 1 cm. WT, wild type.
(C) to (E) Quantification of tiller number (C), plant height (D), and grain length (E) of the CRISPR lines. For quantification of tiller number and plant height, data are shown as means ± se (n = 10). For quantification of grain length, data are shown as means ± se (n = 30). Data were compared by Student’s t test. ***P < 0.001; **P < 0.01; ns, not significant. WT, wild type.
(F) Morphological features of the wild-type, qGL3-OX2, and cr-osgsk3/qGL3-OX2 plants at the mature stage. Bar = 10 cm.
(G) Grain phenotype of the wild-type, qGL3-OX2, and cr-osgsk3/qGL3-OX2 plants. Bar = 1 cm.
(H) to (J) Quantification of grain length (H), tiller number (I), and plant height (J) of the wild-type, qGL3-OX2, and cr-osgsk3/qGL3-OX2 plants. For quantification of grain length, data are shown as means ± se (n = 30). For quantification of tiller number and plant height, data are shown as means ± se (n = 10). Data were compared by Student’s t test. ***P < 0.001.
In terms of tiller number, cr-qgl3 and cr-osgsk3 showed increased tiller numbers compared with the wild type, while compared with cr-osgsk3, the cr-qgl3 plants showed decreased tiller number (Figures 7A and 7C). The cr-qgl3/cr-osgsk3 plants formed tillers similar to cr-osgsk3 (Figures 7A and 7C) and cr-osgsk3 could also suppress the phenotype of qGL3-OX (Figures 7F and 7I). The cr-osbzr1 plants displayed decreased tiller number and suppressed the phenotype of cr-osgsk3 (Figures 7A and 7C).
For grain length, cr-qgl3/cr-osgsk3 formed grains similar to cr-osgsk3 and longer than cr-qgl3 (Figures 7B and 7E). Moreover, knocking out OsGSK3 in qGL3-OX plants rescued the shorter grains of qGL3-OX (Figures 7G and 7H). The cr-osgsk3/cr-osbzr1 plants formed shorter grains than cr-osgsk3 plants (Figures 7B and 7E). The cr-qgl3/cr-osgsk3 double mutant plants displayed phenotypes similar to cr-osgsk3 plants, indicating that OsGSK3 may act downstream of qGL3, and that OsBZR1 likely acts downstream of OsGSK3.
Next, we tested the BL sensitivity of these CRISPR/Cas9 mutants. We used the lamina inclination assay to quantify the BR sensitivity of the wild type and mutants. BL significantly increased the lamina joint bending of cr-qgl3, cr-osgsk3, and cr-qgl3/cr-osgsk3 seedlings compared with the wild type and had the opposite effect on cr-osbzr1 and cr-osgsk3/cr-osbzr1 plants (Supplemental Figures 11A and 11C). cr-osgsk3 and cr-qgl3/cr-osgsk3 displayed similar sensitivity and showed larger lamina inclination compared with cr-qgl3 (Supplemental Figure 11A and 11C). Knocking out OsGSK3 in qGL3-OX plants increased lamina inclination under BL treatment (Supplemental Figures 12A and 12B). In addition, coleoptile growth experiments showed that the cr-qgl3, cr-osgsk3, and cr-qgl3/cr-osgsk3 mutants were more sensitive to BL than the wild type. The cr-osbzr1 and cr-osgsk3/cr-osbzr1 lines displayed an insensitive phenotype under BL treatment (Supplemental Figures 11B and 11D), but cr-osgsk3/qGL3-OX plants showed a BL-sensitive phenotype (Supplemental Figures 12C and 12D).
To further test whether OsGSK3 acts downstream of qGL3, we examined the protein level of OsBZR1 and DLT and found that OsBZR1 increased in the cr-qgl3, cr-osgsk3, and cr-qgl3/cr-osgsk3 mutant seedlings compared with the wild type (Supplemental Figure 13A). Knocking out OsGSK3 in qGL3-OX plants rescued the decreasing protein level of OsBZR1 (Supplemental Figure 13B). Moreover, the cr-osgsk3 and cr-qgl3/cr-osgsk3 plants accumulated more dephosphorylated DLT compared with the wild type (Supplemental Figure 13A). DLT decreased in qGL3-OX plants, but total protein level of DLT increased in cr-osgsk3/qGL3-OX (Supplemental Figure 13B). Together with the biochemical evidence, this genetic evidence strengthened the conclusion that the pathway for rice BR signaling involves the actions of qGL3, OsGSK3, and OsBZR1, in that order.
DISCUSSION
BRs have been shown to regulate various biological processes in plants including cell elongation, cell division, and cell differentiation (Clouse and Sasse, 1998; Müssig et al., 2003; Clouse, 2011; Yang et al., 2011; Wei and Li, 2016). To analyze how qGL3 regulates grain length, we examined the epidermal cells of mature grains and found that the cell density of the outer surface of glumes was not significantly different between DJ and m-qgl3 (Supplemental Figure 14). It is very likely that the long glumes of m-qgl3 result from an increase in cell numbers longitudinally. The cell density analysis in 9311 and NILqgl3 also revealed that qGL3 regulated grain length via increasing cell numbers (Zhang et al., 2012b). Furthermore, our study provides evidence that qGL3 negatively regulates BR signaling in rice, in contrast to the positive effect of BSU1 on BR signaling in Arabidopsis. First, the m-qgl3 mutant displayed enhanced BR signaling phenotypes: the angles between the leaf sheath and leaf blade were greatly increased and the tiller number was substantially increased compared with the wild type (Figures 1A and 1D). These characteristics were similar to the phenotypes of BR-sensitive mutants such as m107 and Do (Tanabe et al., 2005; Tong et al., 2012). Second, the m-qgl3 mutant was sensitive to BL in coleoptile growth (Figures 1F and 1H) and in BR-responsive gene expression (Supplemental Figure 3). Third, subcellular localization analysis demonstrated that qGL3 was required for OsBZR1-mediated BR signaling in rice (Figure 6). The accumulated qGL3 interacted with OsGSK3 to regulate the level of OsBZR1, thereby decreasing BR-responsive gene expression, supporting the key role of qGL3 in mediating BR signaling in rice. Furthermore, genetic analysis demonstrated that OsGSK3 is required for qGL3-mediated BR signaling, and OsBZR1 is required for OsGSK3-mediated BR signaling. The rice GSK3/SHAGGY-like kinases interacted with multiple proteins including O. sativa AUXIN RESPONSE FACTOR4 (Hu et al., 2018) and an APATELA2 transcription factor (Qiao et al., 2017). We hypothesize that the other proteins (such as O. sativa AUXIN RESPONSE FACTOR4 and APATELA2 transcription factor) negatively modulated by OsGSK3 play parallel roles with OsBZR1. Moreover, qGL3 and OsGSK3 repressed the protein level of DLT in mature leaves (Supplemental Figure 9); DLT might be regulated directly by OsGSK3 and indirectly by qGL3 at the protein level. Therefore, knocking out OsBZR1 in cr-osgsk3 plants could not fully repress the plant height and grain length phenotypes compared with cr-osbzr1. As expected, it was observed that, compared with cr-osgsk3, knocking out OsBZR1 in cr-osgsk3 reduced plant height and grain length.
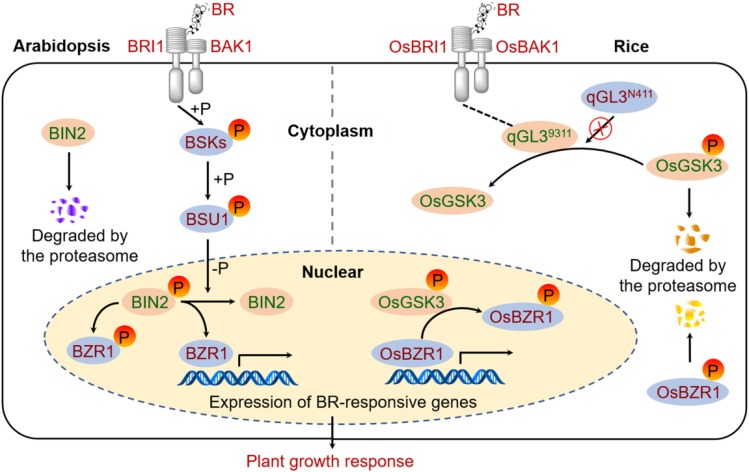
Based on our findings, we propose a model to illustrate how qGL3 acts in BR signaling in rice (Figure 8). This model illustrates a tight regulation on OsGSK3 by BR signaling through a combination of qGL3-mediated dephosphorylation and proteasome-mediated degradation. In the presence of qGL39311, it inhibits the degradation of OsGSK3 by dephosphorylating OsGSK3 and causes the OsGSK3 accumulation in the cytoplasm. Moreover, the inhibition of the OsGSK3 degradation by overexpression of qGL3 also increases the level of phosphorylated OsGSK3 in the nucleus (Figure 3B), leading to the decreased protein level of OsBZR1 and reduced BR signaling. In the absence of qGL3, the phosphorylated OsGSK3 accumulates in the cytoplasm for degradation but decreases in the nucleus, resulting in the accumulation of dephosphorylated OsBZR1 in the nucleus and enhanced BR signaling. Phosphorylated OsBZR1 is exported from the nucleus and targeted for proteasome-mediated degradation in the cytoplasm (Bai et al., 2007). In Arabidopsis, the nuclear BIN2 induced phosphorylation and nuclear export of BZR1/BES1 more efficiently than cytosolic BIN2 (Ryu et al., 2010). BIN2 is localized in the nucleus and cell periphery, but its negative effect on BR signaling occurs mainly in the nucleus (Vert and Chory, 2006). We propose that such dynamic translocations of key regulators and transcription factors enable the cell to rapidly respond to the BR signal. Plants may benefit from the regulatory mechanism by rapidly responding to environmental or developmental stimulating factors.
Figure 8.
A proposed model of BR signaling in rice as compared with that in Arabidopsis. (Left) BR signal transduction pathway in Arabidopsis. BR directly interacts with BRI1 and BAK1 to form the BRI1-BAK1 complex. The active BSKs phosphorylates BSU1. BSU1 dephosphorylates BIN2 and triggers its degradation. BZR1 is phosphorylated and inhibited by BIN2. (Right) BR signaling pathway in rice. BR binding to OsBRI1 promotes association with OsBAK1. qGL39311 dephosphorylates and stabilizes OsGSK3, but qGL3N411 lacks this activity. OsGSK3 phosphorylates OsBZR1 and inhibits its activity. Protein names in red text indicate positive roles in BR signaling, whereas those in green text denote negative roles. BAK1, BRI1-ASSOCIATED RECEPTOR KINASE1; BRI1, BRASSINOSTEROID-INSENSITIVE1; the “P” in orange circles indicate phosphorylation.
In rice, bioinformatics analysis revealed that the rice genome has nine genes (including OsGSK1 to OsGSK5) encoding BIN2-like kinases (Youn and Kim, 2015). OsGSK1 is a BIN2 homolog and is involved in BR signaling and stress responses (Koh et al., 2007). When OsGSK2 containing point mutations was overexpressed, the transgenic plants showed dark-green leaves, dwarfism, and fewer tillers (Tong et al., 2012). Hu et al. (2018) found that OsGSK5 regulated grain length and width, but OsGSK5 and OsGSK2 have different roles in BR responses, in which OsGSK5 interacts weakly with OsBZR1 in yeast cells. Here, we established the role of OsGSK3 in BR signaling and illustrated the relationships among qGL3, OsGSK3, and OsBZR1 in rice. Loss of function of OsGSK3 led to obvious phenotypes in rice and the osgsk3 mutant was hypersensitive to BL, indicating that OsGSK3 is a negative regulator in the BR signaling pathway. In this study, OsBZR1 increased in the nucleus of the m-qgl3 and osgsk3 mutants, suggesting the requirement for qGL3 and OsGSK3 in the nuclear export of OsBZR1. These data confirmed that qGL3 and OsGSK3 are negative regulators of BR signaling, indicating that qGL3 behaves differently from BSU1 in Arabidopsis.
Since OsGSK3 is a negative regulator of BR signaling, the tight regulation of OsGSK3 level is essential. We propose, if phosphorylated OsGSK3 accumulates, the ubiquitin-proteasome system may degrade the redundant phosphorylated OsGSK3. Meanwhile, the dephosphorylation of OsGSK3 by qGL3 in the cytoplasm is to maintain the OsGSK3 stability. The accumulation and activation of OsGSK3 should be tightly regulated in plants. qGL3 with the activity of dephosphorylation on OsGSK3 plays an important role in the stability of OsGSK3 and the BR signaling in rice. In Arabidopsis, BR activates the receptor kinases, triggering sequential phosphorylation of the BSK1 and BSU1; BSU1 dephosphorylates BIN2 to reduce its kinase activity, which leads to the binding to BIN2 by KINK SUPPRESSED in bzr1-1D 1 (KIB1). The binding by KIB1 excludes BIN2 association with BZR1 and facilitates the degradation of BIN2 (Zhu et al., 2017). BR-induced dephosphorylation of BZR1 likely involves at least two mechanisms in Arabidopsis: degradation of BIN2 and activation of a phosphatase (Peng et al., 2008). The different mechanisms underlying the OsGSK3 and BIN2 degradation may lead to the different models of BR signaling in rice and Arabidopsis.
To identify the potential sites where OsGSK3 was phosphorylated, we conducted mass spectroscopy analysis on autophosphorylated OsGSK3. This revealed several potential phosphorylation sites in OsGSK3 (Supplemental Figure 15). Phosphorylation of different amino acids by different kinases can have opposing effects on protein stability. For instance, OPEN STOMATA1-mediated phosphorylation at Ser278 enhances INDUCER OF CBF EXPRESSION1 stability and positively regulates freezing tolerance, but mitogen-activated protein kinase 3/mitogen-activated protein kinase 6–mediated phosphorylation at different sites decreases INDUCER OF CBF EXPRESSION1 stability and negatively regulates freezing tolerance (Li et al., 2017). In Arabidopsis, BSU1 dephosphorylates BIN2 at a conserved Tyr residue (Tyr200; Kim et al., 2009). However, through mass spectroscopic analysis of phosphorylated OsGSK3, we have not found the phosphorylated site (Tyr200) in OsGSK3 in rice (Supplemental Figure 15). The OsGSK3 kinase in rice and BIN2 kinase in Arabidopsis have different phosphorylation sites, which may indicate the differences between rice and Arabidopsis BR signaling pathways. Interestingly, the phosphorylation status of OsGSK3 differs in DJ and ZH11. DJ displayed increased tiller number and reduced plant height compared with ZH11. We therefore propose that OsGSK3 regulates plant phenotype through different phosphorylation status and sites.
To clarify the phylogenetic relationships of PPKLs in Arabidopsis and rice, we performed a search and curation of related sequences. Supplemental Figure 16 shows that BSU1 stands out as a divergent homolog of qGL3/OsPPKL1. We found that qGL3 localized in the cytoplasm and nucleus (Supplemental Figure 17), but BSU1-GFP was observed only in the nucleus and BSU1-YFP was detected predominantly in the nucleus and weakly in the cytoplasm (Maselli et al., 2014). The different subcellular localizations of qGL3 and BSU1 suggest that the two proteins may function in different cellular compartments and have different roles. Furthermore, the dephosphorylation of a particular Tyr in BIN2 (Kim et al., 2009), possibly by the action of BL-activated BSU1, triggers its degradation (Peng et al., 2008), while phosphorylated OsGSK3 is subject to proteasome-mediated degradation in rice. We hypothesize that the different phosphorylated sites of BIN2 and OsGSK3 may cause the opposite degradation patterns. The dephosphorylation effects of BSU1 on BIN2 and that of qGL3 on OsGSK3 may lead to the different phosphorylated sites.
Sequence comparisons of qGL3 between the 9311 and N411 accessions revealed two amino acid residue changes, Asp364Glu and His499Tyr in N411. These two amino acid substitutions influenced the dephosphorylation activity of qGL3, as the activity of qGL3N411 was lower than that of qGL39311. We hypothesize that two mutations may influence the protein structure of qGL3 and thus affect the dephosphorylation activity of qGL3. Alternatively, qGL39311 can be activated while qGL3N411 cannot be fully activated due to the two amino acid substitutions. More studies will be essential to clarify the roles of these two amino acid residues in qGL3 activity. In this study, NILqgl3, which contains the qgl3 allele from N411 in the 9311 background, showed hypersensitivity to BL compared with 9311 (Supplemental Figure 2), implying that the negative role of qGL3 in BR signaling is abolished in N411 and the grain length is therefore increased.
Rice has two qGL3/OsPPKL1 homologs, OsPPKL2 and OsPPKL3. OsPPKL2 is more closely related to BSU1, compared with OsPPKL1 and OsPPKL3 (Supplemental Figure 16). OsPPKL1/3 plays a negative role and OsPPKL2 plays a positive role in the regulation of grain length (Zhang et al., 2012b). Based on the phenotype analysis, OsPPKL2 seems to be OsBSU1 in rice BR signaling; however, OsPPKL2 could not interact with OsGSKs in yeast cells (Supplemental Figure 4). Interestingly, qGL3/OsPPKL1 plays an opposite role in BR signaling in rice compared with BSU1 in Arabidopsis, although it interacts with OsGSKs. Collectively, our study provides a novel mechanism in the regulation of BR signaling in rice, which differs from that in Arabidopsis. Our data support the existence of the negative qGL3-OsGSK3 regulatory module in modulating BR signaling and grain length, which may help in improving grain yield by enabling precise manipulations on the BR signaling pathway in rice.
METHODS
Plant Materials
Four T-DNA insertion mutants (m-qgl3, osgsk1, osgsk2, and osgsk3) in the rice (Oryza sativa) japonica DJ background were obtained from Kyung Hee University, Korea; osgsk1, osgsk4, and osgsk5 in the japonica ZH11 background were obtained from Huazhong Agricultural University, China (http://cbi.khu.ac.kr/RISD_DB.html). The NILqgl3 line was developed in the genetic background of indica var 9311 with the qgl3 allele from japonica N411 (Zhang et al., 2012b). The agronomic traits were determined in the field in Nanjing, China, under natural growth conditions. The gene-editing constructs for qGL3, OsGSK3, and OsBZR1 via CRISPR/Cas9 were designed as described previously in Ma et al. (2015). All constructs were confirmed by sequencing. The edited targets are given in Supplemental Table. These CRISPR mutants in the japonica Nangeng 9108 background were grown in the fields of Nanjing. To evaluate tiller number and plant height, 10 plants were used to measure these phenotypes before harvesting. For each line, the lengths of 30 filled grains were measured with an electronic digital caliper (WSEEN).
GUS Activity Assay
Plants were submerged in 90% (v/v) acetone for 30 min at 4°C, washed twice with PBS buffer, and incubated in GUS solution [1 mM X-Gluc, 0.5% dimethylformamide, 0.5% Triton X-100, 1 mM EDTA, 0.5 mM K3Fe(CN)6, and 0.5 mM K4Fe(CN)6 in PBS buffer] at 37°C overnight. The samples were washed in PBS and submerged in 70% (v/v) ethanol for chlorophyll destaining. Images were taken with a binocular microscope (Leica) or Nikon camera.
BL and BRZ Treatments
The T-DNA insertion mutants; the wild-type lines 9311, DJ, and ZH11; the qGL3-OX transgenic plants; the CRISPR mutants; and the NILqgl3 plants were grown in the field or in the greenhouse at 30°C/25°C day/night cycles. BL, BRZ, and MG132 (Sigma-Aldrich) were dissolved in DMSO to suitable concentrations as stock solutions. For lamina inclination assays, the lamina joint inclination assay and excised leaf segment assay were performed as described previously (Qiao et al., 2017). Twenty plants were used for each test. The leaf angles were measured with ImageJ. For coleoptile growth experiments, 20 rice seeds were grown on 0.3% agar medium with or without BL at 30°C under continuous dark for 5 d. For protein level tests, the BL and BRZ treatments were performed using wild-type plants (ZH11). The leaves of the 2-week-old wild-type seedlings were cut into 0.5-cm segments and immersed in 1 μM BL and 1 μM BRZ containing 0.1% Triton X-100 at different times. An identical volume of the solvent (DMSO) was used as mock treatment.
RNA Extraction and RT-qPCR
Total RNA was isolated with TRIzol reagent (Takara) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 5 μg of total RNA using oligo(dT)18 as primers. The cDNA was used as a template in a 20-μL PCR amplification. For RT-quantitative (q)PCR, SYBR Green I was added to the reaction and amplified on a real-time PCR detection system (Roche) according to the manufacturer’s instructions. The melting curve was acquired at the end. The transcript data were calculated by Roche’s software and were normalized using 18S ribosomal RNA as an internal control; the relative expression level was calculated by 2−ΔΔCt. Each experiment was performed with three replicates. The primers for RT-qPCR are listed in Supplemental Table.
Y2H Assays
For the Y2H analysis, qGL3, OsPPKL2, and OsGSK3 were cloned into the pGBKT7 vector and OsGSKs and OsBZR1 were cloned into the pGADT7 vector, resulting in qGL3-BD (Binding Domain), OsPPKL2-BD, OsGSK3-BD, OsBZR1-AD (Activation Domain), and OsGSKs-AD, respectively. The reporter gene assay (Clontech) was performed following the manufacturer’s instructions. The yeast cells were cultured on synthetic defined medium (SD)/−Trp−Leu and SD/−Trp−Leu−His−Ade containing 5-Bromo-4-Chloro-3-Indolyl-α-D-Galactoside (X-α-gal) at 30°C for 3 d in the dark. SD/−Trp−Leu is yeast culture medium without Trp and Leu. SD/−Trp−Leu−His−Ade is culture medium without Trp, Leu, His, and Ade. The PCR primers used for Y2H assays are listed in Supplemental Table.
Bimolecular Fluorescence Complementation
For the BiFC analysis, qGL3 and OsPPKL2 were cloned into the pSPYNE173 vector and OsGSK3 was cloned into the pSPYCE(M) vector, resulting in qGL3-YFPn, OsPPKL2-YFPn, and OsGSK3-YFPc, respectively. The plasmids were transformed into Agrobacterium tumefaciens strain EHA105. A. tumefaciens cells containing each construct were prepared and mixed to an OD600 of 0.5:0.5. YFP fluorescence was visualized with a confocal scanning microscope 40 to 48 h after infiltration. The PCR primers used for BiFC assays are listed in Supplemental Table.
GST Pull-Down Assay
To test the interaction between qGL3 and OsGSK3, and the interaction between OsGSK3 and OsBZR1, the full-length coding sequence of qGL3 and OsBZR1 was cloned into the pET-30a vector and transformed into the Escherichia coli strain BL21 to produce the His-qGL3 and His-OsBZR1 fusion proteins. The full-length coding sequence of OsGSK3 was cloned into the pGEX-2T vector and transformed into BL21 to produce the OsGSK3-GST fusion proteins. For pull-down, 0.5 μg of GST-OsGSK3 or GST was incubated with GST Bind Resin (70541, Novagen) at 4°C for 2 h, and then 0.5 μg of His-qGL3 and 0.5 μg of His-OsBZR1 were added. The incubation continued for 2 h, and the beads were washed three times. The beads were boiled in 1× SDS loading buffer and separated by 10% SDS-PAGE. The His antibody (9991S, Cell signaling Technology) and GST antibody (2624S, Cell Signaling Technology) were used to detect the proteins.
In Vitro Dephosphatase and Kinase Assays
His-qGL3, GST-OsGSK3, and GST-OsBZR1 proteins were expressed and purified from E. coli strain BL21 to produce fusion proteins, and the imidazole or GSH was removed from the proteins by ultrafiltration using Amicon Ultra-15 centrifugal filter unit (Millipore). The dephosphatase assays were performed as described previously (Guo et al., 2018). To prepare fully phosphorylated OsGSK3, GST-OsGSK3 was incubated in kinase buffer (20 mM Tris, pH 7.5, 1 mM MgCl2, 100 mM NaCl, and 1 mM DTT) containing ATP (100 μM) at 30°C for 1 h, and the reaction mixture was then desalted to remove free ATP. To dephosphorylate the fusion protein GST-OsGSK3, the recombinant His-qGL3 was mixed with the phosphorylated GST-OsGSK3 protein and incubated at 30°C for 3 h. The in vitro kinase assay was performed as described previously (Ye et al., 2012). GST-OsGSK3 and GST-OsBZR1 were incubated in kinase buffer containing ATP (100 μM) and LiCl (0, 10, and 50 mM, respectively) at 30°C for 1 h. Samples were separated by 8% (v/v) SDS-PAGE followed by immunoblotting with biotin-pendant Zn2+-Phos-tag (BTL-111) according to the manufacturer’s instructions (Western Blot Analysis of Phosphorylated Proteins-Chemiluminescent Detection using Biotinylated Phos-tag, Wako, Nard Institute), and immunoblotting with GST antibody (2624S, Cell Signaling Technology).
Protoplast Transient Expression Assay and Fluorescence Microscopy
For the transient expression assays, the protoplasts were isolated from shoots of 4-week-old rice seedlings. For transient expression analysis of OsGSK3-GFP and OsBZR1-GFP, the protoplasts were transfected with 10 μg of plasmid DNA. To test the qGL3- and OsGSK3-mediated cytosolic translocation of OsBZR1, the protoplasts from wild type (DJ and ZH11), m-qgl3, qGL3-OX1, and osgsk3 were transformed with plasmid DNA containing OsBZR1-GFP. To test the qGL3-mediated subcellular localization of OsGSK3, the protoplasts from wild type (DJ and ZH11), m-qgl3, and qGL3-OX1 were transformed with plasmid DNA containing OsGSK3-GFP. All transient expression experiments were repeated at least three times. GFP fluorescence was observed with a confocal laser scanning microscope (LSM780, Carl Zeiss). The GFP fluorescence was excited with a 488 nm laser. Emission fluorescence was captured in the frame-scanning mode with alternating GFP fluorescence via a band-pass filter at 507 nm. The mCherry fluorescence was excited with a 587 nm laser. Emission fluorescence was captured in the frame-scanning mode with alternating mCherry fluorescence via a band-pass filter at 610 nm. The fluorescent signal intensities of OsGSK3-GFP, OsBZR1-GFP, and NLS-mCherry were determined along the line drawn on the confocal images using Image Browser software (Carl Zeiss). The OsBZR1-GFP fluorescence signal intensities in the nucleus of ten cells were analyzed.
Immunoblotting
For detection of OsGSK3, the specific N-terminal sequence of OsGSK3 was cloned into the pGEX-2T vector, and polyclonal antibodies were prepared by immunizing rabbits with purified GST-OsGSK3 fusion protein (Abclonal). Commercial anti-OsBZR1 (AbP80051-A-SE), anti-OsGSK2 (AbP80050-A-SE), anti-DLT (AbP80269-A-SE), and anti–heat shock protein (HSP; AbM51099-31-PU) polyclonal antibodies (Beijing Protein Innovation; http://www.proteomics.org.cn) were used to detect OsBZR1, OsGSK2, DLT , and HSP, respectively. Detection of HSP was used as a control for equal loading. For CIP treatment, the extracts were incubated with CIP at 37°C for 30 min. Plant materials from young leaves were ground into a powder in liquid nitrogen and suspended in lysis buffer (100 mM Tris-·HCl pH 7.5, 300 mM NaCl, 2 mM EDTA, pH 8.0, 1% Triton X-100, 10% [v/v] glycerol, 1× phosphatase inhibitor cocktail, and 1× protease inhibitor cocktail). The total extracts were then centrifuged at 15,000g for 30 min at 4°C; the supernatant, which contained total protein, was analyzed by immunoblotting with appropriate antibodies. For the protein localization experiments, nuclei were separated from cytoplasm with CELLYTPN1 CelLytic PN isolation/extraction kit (Sigma-Aldrich) using the samples from mature leaves of the stage before harvesting. For data analysis, levels of protein were calculated by Tanon Image software. The quantification of the BL and BRZ treatment immunoblot results was normalized to HSP and then normalized to the mock control.
Determination of Phosphorylation Sites of OsGSK3
The phosphorylated OsGSK3-GST was recovered from the SDS-PAGE gel and subjected to in-solution alkylation/tryptic digestion followed by liquid chromatography-tandem mass spectrometry (Applied Protein Technology) as described previously (Cai et al., 2014).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: qGL3 (Os03g44500); OsGSK3 (Os02g14130); OsBZR1 (Os07g39220); OsGSK2 (Os05g11730); DLT (Os06g03710); D2 (Os01g10040); D11 (Os04g39430).
Supplemental Data
Supplemental Figure 1. The expression patterns of qGL3.
Supplemental Figure 2. Altered BR sensitivity in the 9311 and NILqgl3 plants.
Supplemental Figure 3. Expression patterns of BR biosynthesis and signaling-related genes in the seedlings of the WT, m-qgl3 mutant, and qGL3-OX plants.
Supplemental Figure 4. qGL3, not OsPPKL2, interacted with OsGSKs in yeast two-hybrid assay.
Supplemental Figure 5. Seed morphology of the WT and osgsk mutants.
Supplemental Figure 6. Expression analysis of OsGSK3 and altered BR sensitivity in the osgsk3 plants.
Supplemental Figure 7. OsGSK3 interacts with and phosphorylates OsBZR1.
Supplemental Figure 8. qGL3 affects the subcellular localization of OsGSK3.
Supplemental Figure 9. The protein level and phosphorylation status of OsGSK2 and DLT in the WT, m-qgl3, osgsk3, and qGL3-OX plants.
Supplemental Figure 10. The sequences of the target genes.
Supplemental Figure 11. Responses to BR treatment of the WT, cr-qgl3, cr-osgsk3, cr-osbzr1, cr-qgl3/cr-osgsk3, and cr-osgsk3/cr-osbzr1 plants.
Supplemental Figure 12. Responses to BR treatment of the WT, qGL3-OX, and cr-osgsk3/qGL3-OX plants.
Supplemental Figure 13. The protein level analyses of OsBZR1 and DLT in different genetic lines.
Supplemental Figure 14. Cell density analysis of glume outer surfaces of the WT and m-qgl3.
Supplemental Figure 15. Identification of OsGSK3 phosphorylation sites using LC-MS/MS.
Supplemental Figure 16. Phylogenetic analysis and sequence alignment of plant protein PPKLs (phosphatases with Kelch-like repeat domains).
Supplemental Figure 17. Subcellular localization of qGL3.
Supplemental Table. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by National Key Research and Development Program (grant 2016YFD0100400), the Fundamental Research Funds for the Central Universities (grant KYZ201804), Natural National Science Foundation of China (grants 91335106 and 31571627), and the Jiangsu Collaborative Innovation Center for Modern Crop Production and Cyrus Tang Seed Innovation Center, Nanjing Agricultural University.
AUTHOR CONTRIBUTIONS
J.H., X.G., and H.Z. designed the research; X.G. and J.-Q.Z. performed most of the experiments; X.Z., J.Z., Z.J., P.H., Z.T., Y.B., J.C., and W.Z. provided technical assistance and some experiments; X.Z. and H.T. generated the transgenic plants; and J.H., X.G., and H.Z. wrote the manuscript.
References
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Liu J., Wang H., Yang C., Chen Y., Li Y., Pan S., Dong R., Tang G., Barajas-Lopez Jde.D., Fujii H., Wang X. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., Xiao Y., Hu B., Liu L., Wang H., Zhao M., Chu C. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2: 15195. [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (2011). Brassinosteroids. The Arabidopsis Book 9: e0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Sasse J.M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451. [DOI] [PubMed] [Google Scholar]
- Di Rubbo S., Irani N.G., Russinova E. (2011). PP2A phosphatases: the “on-off” regulatory switches of brassinosteroid signaling. Sci. Signal. 4: pe25. [DOI] [PubMed] [Google Scholar]
- Divi U.K., Krishna P. (2009). Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. N. Biotechnol. 26: 131–136. [DOI] [PubMed] [Google Scholar]
- Guo T., Chen K., Dong N.Q., Shi C.L., Ye W.W., Gao J.P., Shan J.X., Lin H.X. (2018). GRAIN SIZE AND NUMBER1 Negatively Regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30: 871–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., Yoshida S., Ashikari M., Kitano H., Matsuoka M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11: 736–749. [DOI] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S., Lee S.C., Kim M.K., Koh J.H., Lee S., An G., Choe S., Kim S.R. (2007). T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 65: 453–466. [DOI] [PubMed] [Google Scholar]
- Kutuzov M.A., Andreeva A.V. (2002). Protein Ser/Thr phosphatases with kelch-like repeat domains. Cell. Signal. 14: 745–750. [DOI] [PubMed] [Google Scholar]
- Li H., Ding Y., Shi Y., Zhang X., Zhang S., Gong Z., Yang S. (2017). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 43: 630–642. [DOI] [PubMed] [Google Scholar]
- Liu J., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3: 17043. [DOI] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., Xie Y., Shen R., et al. (2015). A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Maselli G.A., Slamovits C.H., Bianchi J.I., Vilarrasa-Blasi J., Caño-Delgado A.I., Mora-García S. (2014). Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol. 164: 1527–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G.B., De Wever V., Templeton G., Kerk D. (2009). Evolution of protein phosphatases in plants and animals. Biochem. J. 417: 401–409. [DOI] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C., Shin G.H., Altmann T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.H., Wang X., Kota U., Goshe M.B., Clouse S.D., Huber S.C. (2009). Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P., Yan Z., Zhu Y., Li J. (2008). Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 1: 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P., Lin Y.S., Song X.J., Shen J.B., Huang W., Shan J.X., Zhu M.Z., Jiang L., Gao J.P., Lin H.X. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22: 1666–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S., Sun S., Wang L., Wu Z., Li C., Li X., Wang T., Leng L., Tian W., Lu T., Wang X. (2017). The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29: 292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Hwang I. (2010). Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol. Cells 29: 291–296. [DOI] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., Yoshimura A., Kitano H., Matsuoka M., Fujisawa Y., Kato H., Iwasaki Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Chu C. (2018). Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 23: 1016–1028. [DOI] [PubMed] [Google Scholar]
- Tong H., Liu L., Jin Y., Du L., Yin Y., Qian Q., Zhu L., Chu C. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Xiao Y., Liu D., Gao S., Liu L., Yin Y., Jin Y., Qian Q., Chu C. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100. [DOI] [PubMed] [Google Scholar]
- Wei Z., Li J. (2016). Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 9: 86–100. [DOI] [PubMed] [Google Scholar]
- Wei Z., Li J. (2018). Receptor-like protein kinases: Key regulators controlling root hair development in Arabidopsis thaliana. J. Integr. Plant Biol. 60: 841–850. [DOI] [PubMed] [Google Scholar]
- Yang C.J., Zhang C., Lu Y.N., Jin J.Q., Wang X.L. (2011). The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol. Plant 4: 588–600. [DOI] [PubMed] [Google Scholar]
- Ye H., Li L., Guo H., Yin Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20142–20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.H., Kim T.W. (2015). Functional insights of plant GSK3-like kinases: Multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8: 552–565. [DOI] [PubMed] [Google Scholar]
- Zhang B., et al. (2016). OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 170: 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009a). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu Y., Guo S., Zhu J., Huan Q., Liu H., Wang L., Luo G., Wang X., Chong K. (2012a). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Cai Z., Wang X. (2009b). The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang J., Huang J., Lan H., Wang C., Yin C., Wu Y., Tang H., Qian Q., Li J., Zhang H. (2012b). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 109: 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Li Y., Cao D.M., Yang H., Oh E., Bi Y., Zhu S., Wang Z.Y. (2017). The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell 66: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Li J. (2014). Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48: 99–118. [DOI] [PubMed] [Google Scholar]