Abstract
The primary female sex hormones, estrogens, are responsible for the control of functions of the female reproductive system, as well as the development of secondary sexual characteristics that appear during puberty and sexual maturity. Estrogens exert their actions by binding to specific receptors, the estrogen receptors (ERs), which in turn activate transcriptional processes and/or signaling events that result in the control of gene expression. These actions can be mediated by direct binding of estrogen receptor complexes to specific sequences in gene promoters (genomic effects), or by mechanisms that do not involve direct binding to DNA (non-genomic effects). Whether acting via direct nuclear effects, indirect non-nuclear actions, or a combination of both, the effects of estrogens on gene expression are controlled by highly regulated complex mechanisms. In this chapter, we summarize the knowledge gained in the past 60 years since the discovery of the estrogen receptors on the mechanisms governing estrogen-mediated gene expression. We provide an overview of estrogen biosynthesis, and we describe the main mechanisms by which the female sex hormone controls gene transcription in different tissues and cell types. Specifically, we address the molecular events governing regulation of gene expression via the nuclear estrogen receptors (ERα, and ERβ) and the membrane estrogen receptor (GPER1). We also describe mechanisms of cross-talk between signaling cascades activated by both nuclear and membrane estrogen receptors. Finally, we discuss natural compounds that are able to target specific estrogen receptors and their implications for human health and medical therapeutics.
Keywords: nuclear estrogen receptor, G-protein coupled estrogen receptor, Steroidogenesis, transcriptional control, gene expression
1. ESTROGENS: DEFINITION AND HISTORY
The term “estrogens” refers to a group of female hormones, including estrone, estradiol, estriol, and estretrol (Figure 1). Chemically, estrogens belong to the family of organic compounds known as steroids. As such, their core structure is composed of 17 carbon-carbon bonds arranged as four fused rings (three cyclohexane rings and a cyclopentane ring). All four estrogens contain 18 carbons (C18H24O2) and are collectively known as C18 steroids. They consist of one benzene ring, a phenolic hydroxyl group, and a ketone group (estrone), or one (17β-estradiol), two (estriol), or three (estretrol) hydroxyl groups.
Figure 1. Chemical structures of endogenous estrogens.
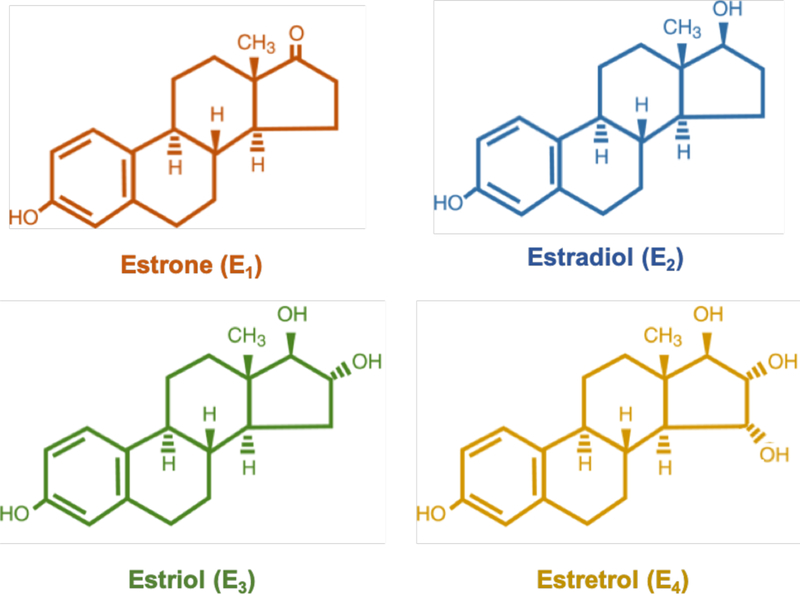
Estrone (E1; orange), estradiol (E2; blue), estriol (E3; green) and estretrol (E4; yellow).
Estrogens are primarily synthesized in the ovaries, but also in the adrenal glands and adipose tissue. They were discovered in the early 1900s, when ovarian extracts (“liquour folliculi”) from cattle and hogs were injected in rodents, and found to be effective in inducing sexual activity or “estrus” (Allen & Doisy, 1983). It was later determined that the hormone was produced by mature ovarian follicles, and that it was likely common to all female animals. The term estrogen derives from the Greek words oistros (frenzy, in heat) and gennan (to produce). As mentioned above, estrogens are a group of C18 hormones with similar chemical structures and function (Figure 1). In addition, all four estrogens are able to bind to both nuclear and membrane estrogen receptors, with different affinity and strength of the response (Watson, Jeng, & Kochukov, 2008). However, the word estrogen is commonly used to refer to estradiol (or 17β-estradiol), due to its physiological relevance and predominance during reproductive years. While females produce all estrogens throughout life, the hormones 16-hydroxyestradiol (estriol) and 15α-hydroxyestriol (estretrol) are predominantly found during pregnancy, and estrone is usually found at higher levels during menopause (Samavat & Kurzer, 2015).
Estradiol, the predominant circulating estrogen in humans, it is mainly secreted by the granulosa cells of the ovarian follicles, and the corpora lutea. On the other hand, estretrol is synthesized exclusively by the fetal liver and reaches maternal circulation through the placenta (Coelingh Bennink, Holinka, Visser, & Coelingh Bennink, 2008; Holinka, Diczfalusy, & Coelingh Bennink, 2008). Estrone, which is produced by aromatization of androstenedione in extraglandular tissues, can be reversibly transformed to estradiol by the enzyme 17β-hydroxysteroid dehydrogenase in peripheral tissues (Bulun, Zeitoun, Sasano, & Simpson, 1999; RYAN, 1959).
2. ESTROGEN BIOSYNTHESIS
The main substrate for steroid hormone biosynthesis is dietary cholesterol, specifically low-density lipoprotein (LDL)-cholesterol (Carr, MacDonald, & Simpson, 1982). Through a process called steroidogenesis, cholesterol is converted to the 21-carbon (pregnanes, progestogens), 19-carbon (androstanes), and 18-carbon (estranes) steroid hormones in gonads, adrenal cortex, and adipose tissue (Miller, 2017). The main site of estrogen synthesis is the ovaries, and specifically the granulosa cells (Figure 2).
Figure 2. Association of theca and granulosa cell in estrogen synthesis.
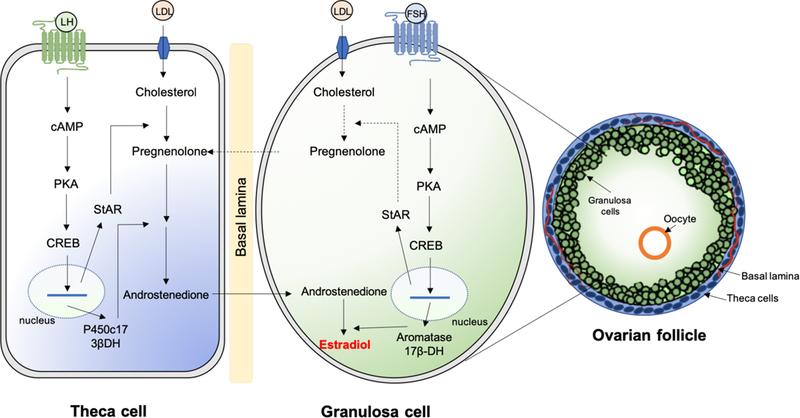
The luteinizing hormone (LH) induces the production of androgens in theca cells. The follicle-stimulating hormone (FSH) stimulates granulosa cells via aromatization of androgens to estrogens and by using cholesterol to produce pregnenolone. The process occurs in the ovarian follicle, which is composed of granulosa cells, oocyte, basal lamina and theca cells. CREB, cyclic AMP response element binding protein; PKA, protein kinase A; LDL, low density lipoproteins; cAMP, cyclic adenosine monophosphate; StAR, steroid acute regulatory protein; P450c17, 17α-hydroxylase/lyase; 17βHSD, 17β-hydroxysteroid dehydrogenase.
The first step in the biosynthesis of steroid hormones is the translocation of cholesterol into the inner mitochondrial membrane, a process regulated by the steroidogenic acute regulatory protein STARD1 (also known as StAR), which is believed to act as a shuttle enzyme (Miller & Strauss, 1999). This is the rate-limiting step of steroidogenesis in all tissues. The expression of StAR is controlled by a mechanism involving binding of luteinizing hormone (LH) to its G protein-coupled receptor in the theca cells of the ovary and stimulation of adenylate cyclase, which catalyzes the production of cyclic adenosine monophosphate (cAMP) from adenosine triphosphate (ATP). The cAMP produced activates protein kinase A, which catalyzes phosphorylation of cAMP response element binding protein (CREB) leading to activation of transcription of StAR and other factors associated with steroid hormone production (Figure 2). At the inner mitochondrial membrane, cholesterol is converted to pregnenolone by the enzyme P450scc, or cholesterol side-chain cleavage enzyme, encoded by the CYP11A1 gene (Belfiore, Hawkins, Wiltbank, & Niswender, 1994). Pregnenolone then acts as a precursor for all steroid hormones (Figure 3), and can diffuse between adjacent granulosa and theca cells of the ovary. The synthesis continues with the conversion of pregnenolone to androstenedione by the enzymes CYP17A1 (steroid 17-α-hydroxylase/17,20-lyase) and 3β-HSD (3β-hydroxysteroid dehydrogenase/Δ5−4 isomerase), via dehydroepiandrosterone (DHEA). Androstenedione can be either converted to other androgens, such as testosterone and dihydrotestosterone, or diffuse to the granulosa cells through the basal lamina (Figure 2). At the granulosa cells, androstenedione is converted to estrone by the enzyme CYP19A1 (also known as aromatase). Estrone is then converted to estradiol by the enzyme 17β-HSD (17β-hydroxysteroid dehydrogenase). In the granulosa cells, the expression of both aromatase and 17β-HSD is controlled by follicle stimulating hormone (FSH) stimulation. Interestingly, testosterone can be metabolized to estradiol and estrone by the action of aromatase in peripheral tissues, including adipose cells and bone (Simpson et al., 2002). Males also produce local estrogen by aromatization in cells of the reproductive tract, including Sertoli cells, Leydig cells, and mature spermatocytes. Overall, estrogens are normally produced by the ovaries and in smaller amounts by other tissues such as the liver, pancreas, adrenal glands, adipose tissue, and breast (Barakat, Oakley, Kim, Jin, & Ko, 2016). In specific physiological conditions, such as pregnancy, estrogen is also synthesized by the placenta. However, the biosynthesis of estrogen in non-gonadal sites follows rather unusual mechanisms, since these tissues are not able to generate C19 steroids from cholesterol. In these tissues, estrogen production is largely dependent on C19 steroids transported from other tissues and conversion by local CYP19A1 aromatase (Labrie et al., 1998; Nelson & Bulun, 2001).
Figure 3. Estrogen biosynthesis pathway.
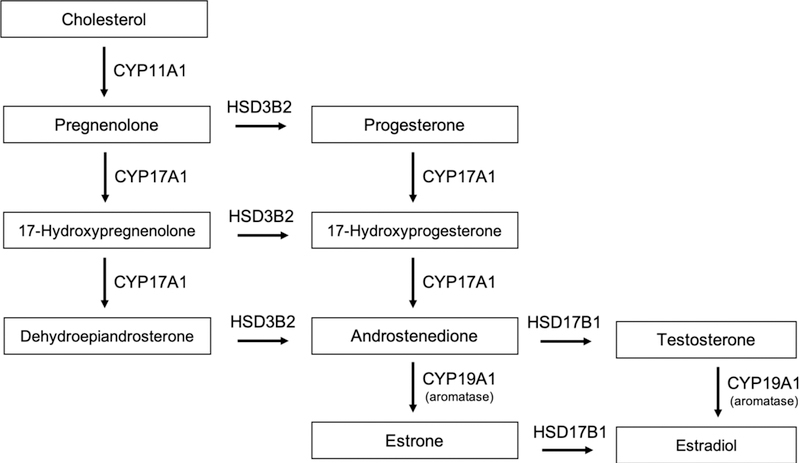
The estrogen biosynthetic pathway involves the conversion of cholesterol to progestogens, androgens and finally estrogens. The conversion of androgen to estrone (E1) and estradiol (E2) catalyzed by aromatase is the final step for synthesis of estrogen.
Estradiol, the predominant circulating estrogen in humans, it is mainly secreted by the granulosa cells of the ovarian follicles, and the corpora lutea, by the mechanisms indicated above. On the other hand, estretrol is synthesized exclusively during pregnancy by the fetal liver and reaches maternal circulation through the placenta (Coelingh Bennink et al., 2008; Holinka et al., 2008). Estriol, which is also primarily synthesized during pregnancy, is almost exclusively produced by the placenta. To produce estriol, dietary cholesterol is converted to pregnenolone and progesterone in the placenta, and these steroids are further metabolized to DHEA and DHEA-sulfate (DHEA-S) in the fetal adrenal glands. DHEA-S is later hydroxylated to 16α-OH-DHEA-S in the fetal liver by the action of the CYP3A7 enzyme, and transported back to the placenta where it is converted to 16α-OH-DHEA by the steroid sulfatase. The enzyme 3β-HSD1 converts 16α-OH-DHEA into 16α-OH-androstenedione, which is later aromatized to 16α-OH-estrone. In the final step, 16α-OH-estrone is converted to estriol by the 17β-HSD enzyme, and secreted into maternal circulation (ITTRICH & NEUMANN, 1963; WILSON, ERIKSSON, & DICZFALUSY, 1964). In non-pregnant women, estriol is produced mainly in the liver by 16α-hydroxylation of estradiol and estrone by CYP enzymes (Samavat & Kurzer, 2015; Tsuchiya, Nakajima, & Yokoi, 2005). Finally, estrone is mainly produced during menopause by aromatization of androstenedione in extra-glandular tissues, where it can act locally as a paracrine or intracrine factor (Simpson, 2003). Estrone can also be transformed to estradiol by the enzyme 17β-hydroxysteroid dehydrogenase in peripheral tissues, including adipose and breast tissue, vascular endothelium, smooth muscle cells, brain tissue, and bone cells, where it is metabolized or enters the circulation in small quantities (Bulun et al., 1999; RYAN, 1959; Simpson, 2003).
3. ESTROGEN METABOLISM
Physiologically, the metabolic conversion of estrogens allows their excretion from the body via urine, feces, and/or bile, along with the production of estrogen analogs, which have been shown to present antiproliferative effects (Tsuchiya et al., 2005). In target cells, there are different pathways capable of metabolizing estradiol and estrone. Members of the cytochrome P450 superfamily of enzymes (CYP1A1, CYP1B1, and CYP1A2) catalyze hydroxylation of estrone and estradiol at positions C2, C4 and C16. Due to the high expression of these enzymes in the liver, a large proportion of estrogen metabolism occurs in this tissue, although CYP1B1 is also expressed in target tissues such as mammary gland, uterus, kidney, brain, and pituitary gland, where estradiol and estrone can also be metabolized. Estradiol hydroxylation is followed by conversions to 2-hydroxyestrone, 4-hydroxyestrone, 2-hydroxyestradiol, 4-hydroxyestradiol, and 16α-hydroxyestrone, which are also known as catechol estrogens, due to their presence of pharmacological properties of both catecholamines and estrogens. The hydroxylation of estradiol or 16α-hydroxyestrone forms estriol. In addition, catechol estrogens can be methylated via the catechol-O-methyltransferase (COMT) enzyme to methoxy estrogens (Samavat & Kurzer, 2015). These compounds have gained significant attention due to their little estrogenic effects, antiproliferative properties, and ability to control estrogen synthesis (Purohit & Reed, 2002; Purohit et al., 2006). Moreover, catechol estrogens can also be conjugated by estrogen sulfotransferases and UDP-glucuronyltransferases (Cheng et al., 1998; Garbacz, Jiang, & Xie, 2017). In a conjugation reaction, hormones become water soluble and excreted from the body (Lakhani, Venitz, Figg, & Sparreboom, 2003).
4. PHYSIOLOGICAL FUNCTIONS OF ESTROGENS
Estrogens are sex steroid hormones, and as such display a broad spectrum of physiological functions. These include regulation of the menstrual cycle and reproduction, bone density, brain function, cholesterol mobilization, development of breast tissue and sexual organs, and control of inflammation (Liang & Shang, 2013). While estrogens play diverse roles in normal male and female physiology, in certain physiological situations they can play similar roles in both sexes (Rotstein). In females, estrogens are responsible for primary and secondary sexual characteristics. Estradiol promotes epithelial cell proliferation in the uterine endometrium and mammary glands starting in puberty (Gruber, Tschugguel, Schneeberger, & Huber, 2002; Koos, 2011; Simpson et al., 2005). During pregnancy, estrogens produced by the placenta help prepare the mammary gland for milk production (Voogt, 1978). On the other hand, lower levels of estrogens produced in men are essential for functions including sperm maturation, erectile function and maintenance of a healthy libido (Schulster, Bernie, & Ramasamy, 2016). It is important to mention here that all the estrogenic physiological functions previously described are mediated by estrogen receptors, which we describe in the next sections.
5. THE ESTROGEN RECEPTORS: HISTORY AND DISCOVERY
In 1958, Elwood Jensen discovered the estrogen receptor, the first receptor ever encountered for any hormone, by showing that reproductive female tissues were able to uptake estrogen from the circulation by binding to proteins. He later demonstrated that estrogen-bound receptors were able to migrate to the nucleus, where they could stimulate gene transcription (Jensen et al., 1967; Jensen et al., 1968). More than 20 years later, the first human estrogen receptor (known today as ERα) was cloned using RNA from the human breast cancer cell line MCF-7 (Green et al., 1986; Greene et al., 1986). Similarly, the second estrogen receptor (known today as ERβ) was described ten years later by the research team lead by Dr. Jan-Ake Gustafsson (Kuiper, Enmark, Pelto-Huikko, Nilsson, & Gustafsson, 1996). Gustafsson’s lab discovered that a newly identified protein that was mainly expressed in the secretory epithelial cells of the prostate and in the granulosa cells of the ovary, shared a high degree of homology with the ERα (DNA-binding domain, 95%; ligand-binding domain, 55%). As a result of these similarities, the team suggested for the protein be named ERβ.
More recently, a new type of estrogen binding protein was discovered in target cells: The G Protein-Coupled Estrogen Receptor GPER1, or membrane estrogen receptor. Unlike the nuclear estrogen receptors ERα and ERβ, which were isolated by traditional biochemical approaches, GPER1 was identified by molecular cloning methods (E. J. Filardo & Thomas, 2012). Almost two decades ago, several research laboratories had reported the isolation of a G Protein-Coupled Receptor homologue, which was ascribed the orphan term GPR30 (Carmeci, Thompson, Ring, Francke, & Weigel, 1997; Feng & Gregor, 1997; Kvingedal & Smeland, 1997; O’Dowd et al., 1998; Owman, Blay, Nilsson, & Lolait, 1996; Takada, Kato, Kondo, Korenaga, & Ando, 1997). It was assumed that the ligand for GPR30 was a hormone or chemotactic peptide due to its structural similarities to the receptors for angiotensin II and other peptides such as such as interleukin-8, monocyte chemotactic proteins, and complement factors (E. J. Filardo & Thomas, 2012). However, after screening of multiple chemotactic peptides and factors, no molecules with binding affinities to GPR30 were found, the receptor continued to be classified as orphan (Feng & Gregor, 1997). However, in the year 2000, a research team was able to show that fast estrogen-mediated activation of extracellular signal-regulated kinases (ERKs) was dependent on GPR30 (E. J. Filardo, Quinn, Bland, & Frackelton, 2000). Five years later, this and other groups were able to demonstrate direct binding of 17β-estradiol to GPR30 in GPR30-transfected cells and breast cancer cell lines (Revankar, Cimino, Sklar, Arterburn, & Prossnitz, 2005; Thomas, Pang, Filardo, & Dong, 2005). Finally, in 2007 GPR30 was officially named G protein-coupled estrogen receptor 1 (also known as GPER or GPER1), and its role in mediating fast responses to estrogens and overall physiological and pathological processes has been studied extensively in human and animal models (Boonyaratanakornkit & Edwards, 2007; E. Filardo et al., 2007; Molina, Figueroa, Bhoola, & Ehrenfeld, 2017; Prossnitz & Barton, 2014; Sharma & Prossnitz, 2016).
6. STRUCTURAL PROPERTIES OF ESTROGEN RECEPTORS
The full-length size of ERα is 595 amino acids and 67kDa. ERβ is 530 amino acids in length and 59kDa. The main difference between the two proteins is that ERβ has a shorter amino terminal domain than ERα (Figure 4).
Figure 4. Structural organization of estrogen receptors.
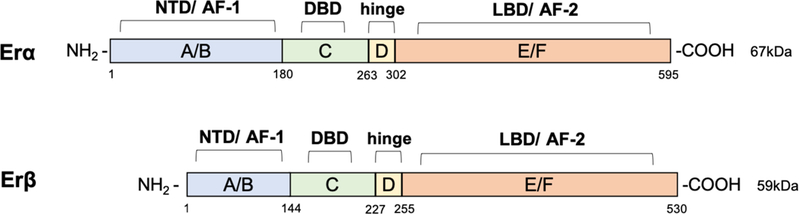
Structural domains of estrogen receptor α (ERα) (595aa) and ERβ (530aa) are labeled A-F. Both receptors have 6 different structural and functional domains: N- terminal (NTD, A/B domains, AF-1), DNA binding domain (DBD, C domain), the hinge (D domain), the C-terminal region containing the ligand binding domain (LBD, E/F domain, AF-2).
As members of the nuclear hormone receptors superfamily of transcription regulators, the structures of the estrogen receptors ERα and ERβ are composed of various functional domains and have several structural regions in common (Schwabe & Teichmann, 2004). The principal functional domains are termed A/B, C, D, and E/F, and are present in both receptor full-length structures (Figure 4). The A/B region represents the amino-terminal domain (NTD), which is involved in gene transcription transactivation, and contains a zinc-finger that mediates binding to target sequences. The C region corresponds to the DNA binding domain (DBD), which contributes to estrogen receptor dimerization and binding to specific sequences in the chromatin. These canonical sequences known collectively as estrogen response elements (ERE) (Scheidereit et al., 1986; Truss & Beato, 1993). The D domain is a hinge region that connects the C and E domains, and is able to bind to chaperone proteins. This region also contains the nuclear localization signal, that is unmasked upon estrogen binding, allowing for the receptor-ligand complexes to translocate to the nucleus. In the carboxy-terminal E/F region, also known as the ligand binding domain, contains the estrogen binding area, along with binding sites for coactivators and corepressors. Finally, two additional regulators of the estrogen receptor transcriptional activity known as activation function (AF) domains AF1 and AF2, are located within the NTD and DBD, respectively (Kumar et al., 2011). The mechanisms of transcriptional regulation mediated by these receptors appear to involve a synergistic effect of AF1 and AF2 (Tora et al., 1989). Contrarily to AF2, AF1 does not require binding to hormones or steroids to be activated (Kumar et al., 2011).
In humans, the ERα is encoded by the gene ESR1, located on chromosome 6, locus 6q25.1 (Gosden, Middleton, & Rout, 1986). In addition to the full-length ERα isoform (66kDa), several shorter isoforms (36kDa, 46kDa) have been identified as a result of the presence of alternate start codons, or as productos of alternative splicing (Figure 5). Some of these shorter isoforms do not have the NTD and thus lack the AF-1 domain. Therefore, they cannot activate transcription. Instead, they are able to form heterodimers with the full-length ERα and inhibit its ability to control transcriptional. The shorter isoform, ERα−36, lacks both AF-1 and AF-2 transcriptional activation domains, and it has been shown to exert membrane-initiated signaling events upon binding to estradiol, estriol, and estretrol (Y. Gu et al., 2014), as well as to medicate GPER1 responses (Arnal et al., 2017; Romano & Gorelick, 2018).
Figure 5. Estrogen receptor alpha (ERα) isoforms.
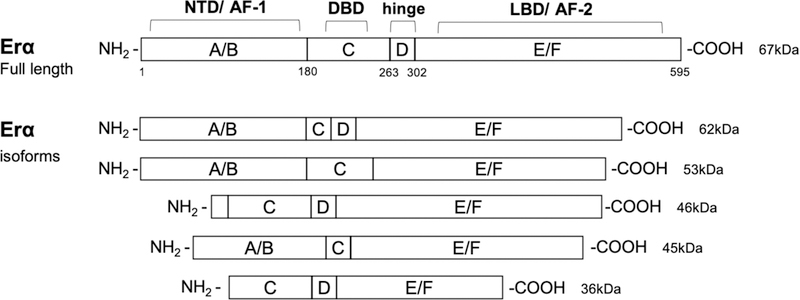
The domain organization of the full-length 595 amino acid ERα (67kDa), and truncated shorter isoforms (62kDa, 53kDa, 46kDa, 45kDa, and 36kDa) resulting from alternative splicing and/or alternate translation start sites are illustrated. Protein domains are labeled as A to F with numbering denoting amino acid sequence number based on the full-length protein (595 aa). ERα domains: N-terminal (NTD, A/B domains, AF-1), DNA binding domain (DBD, C domain), hinge (D) domain, and C-terminal region containing the ligand binding domain (LBD, E/F domain, AF-2).
On the other hand, ERβ is encoded by the ESR2 gene located in chromosome 14 (14q23–24), and has five known isoforms (Enmark et al., 1997) (Figure 6). The main difference between the full-length ERβ and the shorter ERβ isoforms is on the C-terminal LBD. Therefore, ERβ isoforms that have no transcriptional activity can also suppress ERα signaling by dimerizing with ERα (Vrtačnik, Ostanek, Mencej-Bedrač, & Marc, 2014).
Figure 6. Estrogen receptor beta (ERβ) isoforms.
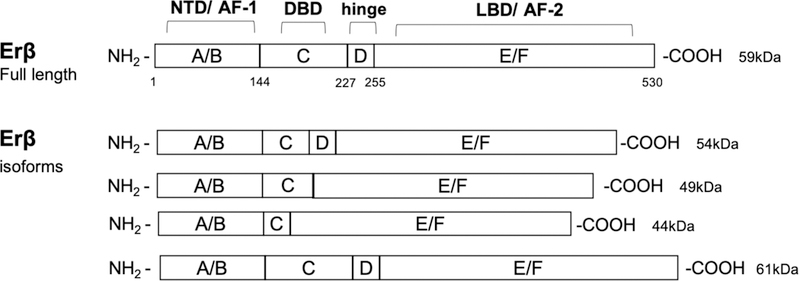
The domain organization of the full-length 530 amino acid ERβ (59kDa), truncated shorter isoforms (54 kDa, 49 kDa, and 44 kDa), and elongated isoform (61kDa), resulting from alternative splicing and/or alternate translation start sites are illustrated. Protein domains are labeled as A to F with numbering denoting amino acid sequence number based on the full-length protein (595 aa). ERβ domains: N-terminal (NTD, A/B domains, AF-1), DNA binding domain (DBD, C domain), hinge (D) domain, and C-terminal region containing the ligand binding domain (LBD, E/F domain, AF-2).
Finally, the gene coding for the membrane receptor GPER1 is located in chromosome 7 (locus 7p22.3). In terms of structure, GPER1 does not share similarities with ERα or ERβ. As a typical G protein coupled receptor, its structure consists of 7 transmembrane α-helical regions, 4 extracellular segments, and 4 cytosolic segments (Barton et al., 2018). This receptor has low binding affinity (17B-estradiol) when compared to other estrogen receptors (Prossnitz & Barton, 2014). However, this may be important as GPER1 is accountable for rapid responses to estrogen, and activation of intracellular signaling cascades mediated by second messengers (E. J. Filardo & Thomas, 2012).
7. MECHANIMS OF ESTROGEN RECEPTOR SIGNALING
As a steroid hormone, estrogen can enter the plasma membrane and interact with intracellular ERα and ERβ to exert direct effects by binding to DNA sequences. Alternatively, estrogen can activate intracellular signaling cascades via interaction with the GPER1 and/or ERα and ERβ. Due to differences in the cellular and molecular events leading to gene expression regulation in which estrogen-receptor complexes can either bind directly or indirectly to DNA, estrogen-mediated signaling events ca be divided into genomic and non-genomic. Genomic effects are those involving migration of the estrogen-receptor complexes to the cell nucleus, and direct interaction with chromatin at specific DNA sequences known as estrogen response elements (EREs). While EREs have been identified in several gene promoters and regulatory regions, it has been reported than more than one third of human genes regulated by estrogen receptors do not contain ERE sequence elements (O’Lone, Frith, Karlsson, & Hansen, 2004). On the other hand, non-genomic effects involve indirect regulation of gene expression through a variety of intracellular signaling events. The known mechanisms for genomic and non-genomic control of gene expression by estrogens are described below.
8. NUCLEAR ESTROGEN RECEPTORS: DIRECT GENOMIC SIGNALING
Direct genomic signaling is known as the classical mechanism of estrogen signaling. In this process, the nuclear estrogen receptors ERα and ERβ act as ligand-activated transcription factors (Marino, Galluzzo, & Ascenzi, 2006; O’Malley, 2005). Upon binding of estradiol to ERα or ERβ in the cytoplasm, a conformational change occurs inducing receptor dimerization (Le Dily & Beato, 2018) (Figure 7). This complex is then translocated to the nucleus, where it binds to the chromatin at ERE sequences, enhancer regions within or close to promoters, and/or 3’-untranlated regions of target genes (Klinge, 2001).
Figure 7. Genomic and non-genomic estrogen signaling pathways.
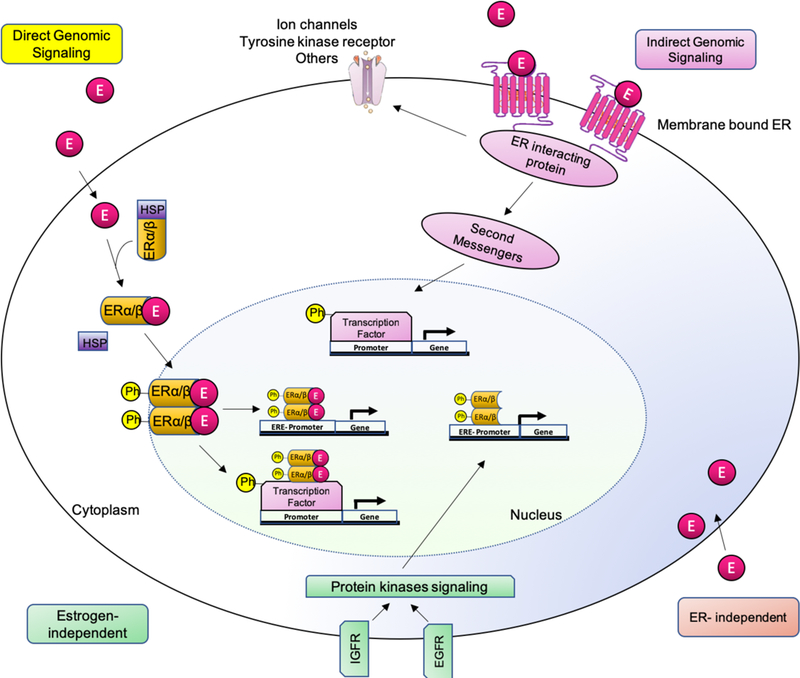
There are different estrogen-mediated signaling mechanisms. 1) Direct genomic signaling: estrogen binds to ERs. The complex dimerizes and translocate to the nucleus inducing transcriptional changes in estrogen-responsive genes with or without EREs. 2) Indirect genomic signaling: the membrane bound receptor induces cytoplasmic events such as modulation of membrane-based ion channels, second-messenger cascades and transcription factors. 3) ER-independent: estrogen exerts antioxidant effects in an ER-independent manner. 4) Estrogen independent: ligand-independent genomic events.
Recent advances in computational biology have facilitated the identification of EREs in many gene promoters, and allowed prediction of genes regulated by estrogen and other hormones in the genomes of many species (Bajic et al., 2003; Bourdeau et al., 2004). A recent genome-wide screening study identified over 70,000 EREs in the human and mouse genomes (Bourdeau et al., 2004). Interestingly, 17,000 of these EREs were located near mRNA transcriptional start sites, and only 660 were conserved sites. The efficacy of this computational approach was further supported by functional validation of estrogen receptor interaction sites (Carroll & Brown, 2006). While these elements share a high degree of sequence similarity, it is important to recognize that the intrinsic sequence composition of the EREs can alter the affinity of the receptor to bind DNA. For example, ERα has a high binding affinity for the canonical ERE sequence located within the vitellogenin A2 gene, but with less affinity for the EREs located in the oxytocin gene (Sausville, Carney, & Battey, 1985). This moderately explains why differences in ERE sequences, such as those resulting from inter-individual gene variability or mutations, can affect the activation of gene expression (Loven, Wood, & Nardulli, 2001; Yi et al., 2002). In addition, specific ERE sequences can cause allosteric changes in the receptor’s structure, and thus alter the ability of the complex to recruit coactivators and transcription factors that may contribute to ER biological activity (Hall, McDonnell, & Korach, 2002; Yaşar, Ayaz, User, Güpür, & Muyan, 2017).
9. NUCLEAR ESTROGEN RECEPTORS: INDIRECT GENOMIC SIGNALING
As mentioned earlier, the transcription of several genes that do not contain EREs in their promoter regions can also be regulated by estradiol, without direct binding of the estrogen receptors to the DNA. According to the most recent reports, an estimated 35% of genes targeted by estrogen lack ERE-like sequences (Marino et al., 2006; Vrtačnik et al., 2014). In these, the mechanisms by which estrogen affects gene expression are collectively known as “indirect genomic signaling” or “transcriptional cross-talk”, and are based on activation of gene expression by estrogen receptors not binding DNA directly. Rather, the estrogen receptor complexes act through protein-protein interactions with other transcription factors and response elements (Aranda & Pascual, 2001; Göttlicher, Heck, & Herrlich, 1998). In this way, estrogens indirect signaling influences activation or suppression of target gene expression.
An important mediator of indirect genomic signaling is the stimulating protein-1 (Sp-1). Binding of this transcription factor to promoter regions at GC-rich sites is enhanced by the presence of estrogen receptors (Bajic et al., 2003; O’Lone et al., 2004). Examples of genes induced by estrogen via the Sp-1 mechanism are: low-density lipoprotein (LDL) receptor (C. Li, Briggs, Ahlborn, Kraemer, & Liu, 2001), progesterone receptor B (O’Lone et al., 2004), endothelial nitric oxide synthase (eNOS) (Chambliss & Shaul, 2002), GATA binding protein 1 (GATA1), signal transducer and activator of transcription 5 (STAT5) (Björnström & Sjöberg, 2005), and the retinoic acid receptor-1α genes (Sun, Porter, & Safe, 1998). A few studies have shown that ERα can also interact with the c-rel subunit of the nuclear factor-κB (NF-κB) complex, preventing NF-κB from binding to cytokine genes promoters (Galien & Garcia, 1997; Kalaitzidis & Gilmore, 2005). Moreover, ERα can also interact with other transcriptional modulators such as the activating transcription factor (ATF)-2, c-jun, the ATF-1/cAMP response element binding protein (ATF-1/CREB), and the nuclear transcription factor-Y (NF-Y) (O’Lone et al., 2004).
The nuclear estrogen receptors also induce the expression of genes containing the activator protein-1 (AP-1) sites though protein-protein interactions (Gaub, Bellard, Scheuer, Chambon, & Sassone-Corsi, 1990). AP-1 is a transcription factor that regulates key cellular processes such as cell differentiation, proliferation, and apoptosis. The structure of AP-1 consists of a heterodimer composed of proteins belonging to the c-Fos, c-Jun, ATF, and the Jun dimerization partners (JDP) families (Piu, Aronheim, Katz, & Karin, 2001). The ERα also interacts with c-Fos and c-Jun at these binding regions (O’Lone et al., 2004). Some examples of genes induced by ERα via the AP-1 mechanism are insulin-like growth factor-1 (IGF1), collagenase, IGF1-receptor, ovalbumin, and cyclin D1 (Fujimoto, Honda, & Kitamura, 2004; Marino, Acconcia, Bresciani, Weisz, & Trentalance, 2002). However, previous studies have shown that ERα and ERβ signal in different ways depending on the ligand and response elements present at the AP-1 sites. In fact, 17β-estradiol activates AP-1-dependent transcription via ERα, whereas ERβ inhibits this mechanism (Paech et al., 1997). Likewise, 17β-estradiol binding to ERα induces transcription when linked to Sp-1 in GC-rich regions, but not when 17β-estradiol is bound to ERβ. One example of this is the contrasting action of ERα and ERβ on the control of cyclin D1 gene expression (Liu et al., 2002), where estrogen-bound ERβ suppresses cyclin D1 expression (Marino et al., 2006) and blocks ERα-mediated production when both receptors are present (Acconcia et al., 2005; Matthews & Gustafsson, 2003). The diversity of mechanisms of transcriptional regulation in different cells by the two estrogen receptors and their interactions with local transcription factors may explain the differences observed in tissue specific biologic responses to estrogens.
10. MEMBRANE RECEPTOR: INDIRECT NON-GENOMIC SIGNALING
As mentioned above, not all estrogen responses fit the classical genomic model of steroid action. The observation of excessively fast estrogen-induced biological responses led to the development of the hypothesis that estrogen could be acting by mechanisms not involving direct target gene transcription and protein synthesis, and the subsequent discovery of the GPER1 (Prossnitz & Barton, 2011). Non-genomic actions of estrogen often involve activation of signal-transduction mechanisms with the subsequent production of intracellular second messengers, cAMP regulation and protein-kinase activation of signaling cascades that result in indirect changes in gene expression (Lösel & Wehling, 2003) (Figure 7). The protein-kinase cascades can be classified into four major ones: 1) the phospholipase C (PLC)/protein kinase C (PKCs) pathway (Marino, Pallottini, & Trentalance, 1998), 2) the Ras/Raf/MAPK cascade (Dos Santos et al., 2002; Watters, Campbell, Cunningham, Krebs, & Dorsa, 1997), 3) the phosphatidyl inositol 3 kinase (PI3K)/Akt kinase cascade (Marino, Acconcia, & Trentalance, 2003), and 4) the cAMP/protein kinase A (PKA) signaling pathway (Q. Gu & Moss, 1996; Picotto, Massheimer, & Boland, 1996). Additionally, GPER1 binding to estrogens promotes estrogen-dependent activation of adenylyl cyclase and epidermal growth factor receptor (EGFR). Subsequent phosphorylation of transcription factors by the protein kinases mentioned above can alter their function and ability to bind to genomic sequences to affect gene expression. Examples of transcription factors that are affected by these signaling mechanisms include: Elk-1, CREB, CCAAT-enhancer-binding protein beta (C/EBPβ), the NF-κB complex, and the signal transducer and activator of transcription (STAT) family (Cavalcanti, Lucas, Lazari, & Porto, 2015; Fox, Andrade, & Shupnik, 2009; Furth, 2014; Kousteni et al., 2003; Laliotis et al., 2013; Ozes et al., 1999; Romashkova & Makarov, 1999). Thus, by activating these non-genomic to genomic mechanisms, the estrogen receptors ERα and ERβ indirectly regulate gene transcription at alternative DNA response elements, in addition to the abovementioned genomic effects involving direct binding to EREs (Figure 7). Another interesting fact is that both ERα and ERβ are also targets for phosphorylation by protein kinases including MAPKs, indicating that non-genomic actions of estrogens may also involve self-regulation of receptor expression (de Leeuw, Neefjes, & Michalides, 2011; Kato et al., 1995).
Both the membrane bound estrogen receptor GPER1, and some variants of the ERα and ERβ have been associated to non-genomic estrogen signaling (Barton et al., 2018; E. J. Filardo & Thomas, 2012). It has been suggested that non-genomic actions of the ERα and ERβ could be mediated through a sub-population of receptors that located at the cell membrane and can activate intracellular signaling cascades (Razandi, Pedram, Merchenthaler, Greene, & Levin, 2004). At the cell membrane, the ERα and ERβ can interact with scaffold proteins such as caveolin-1 and MNAR/PELP-1 (modulator of non-genomic activity of estrogen receptor) (Chambliss et al., 2000; Cheskis et al., 2008; Shaul & Anderson, 1998). By proximity, the ERα and ERβ also interact with G proteins, various membrane receptors (e.g. tyrosine kinase, insulin growth factor 1, and epidermal growth factor receptors), and signaling molecules including ras, Src and PI3 kinases, ErbB2 (HER-2/neu) and Shc that are located at or near the membrane (Boonyaratanakornkit, 2011; L. Li et al., 2007; Migliaccio et al., 1996; Song et al., 2010; Song, Zhang, Chen, Bao, & Santen, 2007; Song, Zhang, & Santen, 2005). Interactions with these molecules promotes intracellular activation of mitogen activated protein kinases (MAPK) and protein kinase B (Akt) signaling pathways that can affect transcriptional regulation (Y. Li et al., 2010). While there is no clear consensus among the experts in the field about binding of ERα and ERβ to the plasma membrane, it appears that the mechanisms described above are cell-type specific and activated under certain physiological events, and by specific receptor variants (L. Li, Haynes, & Bender, 2003).
11. GENOMIC AND NON-GENOMIC SIGNALING CROSSTALK
As exemplified in the previous sections, it is evident that the mechanisms of action of estrogen in the various cell targets represent a combination of complex multifactorial processes. Besides the independent genomic and non-genomic pathways described above, many authors have proposed the existence of additional convergent pathways involving both genomic and non-genomic factors that result in regulation of gene transcription (Björnström & Sjöberg, 2005; Silva, Kabil, & Kortenkamp, 2010; Vrtačnik et al., 2014). Two mechanisms of “cross-talk” have been described, and involve protein-protein interactions of components of both pathways. In one mechanism, estrogen-bound nuclear estrogen receptor complexes are dimerized and translocated to the nucleus, where they bind to phosphorylated transcription factors resulting from GPER1-mediated signaling. The complexes then bind to either ERE sequences via the nuclear estrogen receptors, or to AP-1, STATs, ATF-2/c-Jun, Sp1, and/or NF-κB cognate DNA binding sites (Björnström & Sjöberg, 2005). In the second mechanism, interaction of GPER1 and ERα and ERβ located at the plasma membrane activate protein kinase cascades that result in phosphorylation of AP-1, STATs, Elk-1, CREB, and NF-κB, and other transcription factors, as well as estrogen receptors themselves, that can then interact with DNA sequences to regulate transcription (Björnström & Sjöberg, 2005). Thus, convergence of the two classical estrogen receptor regulation pathways can result in enhanced transcriptional activity in specific tissues and physiological processes.
12. ESTROGEN RECEPTOR LIGAND INDEPENDENT SIGNALING
An interesting phenomenon observed in many cells is that estrogen receptors can actually be activated in the absence of estrogens or other receptor agonists (Bennesch & Picard, 2015; Maggi, 2011; Vrtačnik et al., 2014). This ligand-independent estrogen receptor activation is mainly triggered by phosphorylation on specific residues (e.g. serine and tyrosine) in the receptors themselves, or their association with coregulators (described below). This independent mechanism requires the action of regulatory molecules necessary for phosphorylation, such as protein kinase A (PKA), protein kinase C (PKC), MAPK phosphorylation cascade components, as well as inflammatory cytokines (e.g. interleukin-2), cell adhesion molecules (e.g. heregulin), cell cycle regulators (e.g. RAS p21 protein activator cyclins A and D1), and peptide growth factors including EGF, insulin, IGF1, and transforming growth factor beta (TGFβ) (Nilsson et al., 2001).
13. ESTROGEN RECEPTOR COREGULATORS AND TRANSCRIPTIONAL CONTROL
In addition to the regulatory pathways described above, the cell also expresses a battery of coregulators that can either enhance or decrease transcriptional activity of steroid hormone receptors. These are called estrogen receptor coactivators and corepressors, respectively. Coregulators are involved in many steps of the gene expression process, including chromatin modification and remodeling, transcription initiation, elongation of RNA chains, mRNA splicing, mRNA translation, miRNA processing, and degradation of the activated NR-coregulator complexes (Lonard & O’malley, 2007). Currently, there are hundreds of coregulators of nuclear receptors described that play a key role in promoting gene expression and transcriptional activity. Coregulators are a dynamic group of proteins able to act as integrators of signals from steroid hormones, and have been linked to many diseases affected by sex hormones, such as cancer (Lonard & O’Malley, 2006). One of the first coregulators of ERα, known as steroid receptor coactivator (SRC-1), was identified in 1995 (Oñate, Tsai, Tsai, & O’Malley, 1995). Since then, many additional coregulators have been discovered for ERα, although very few are known for ERβ (Lonard & O’Malley, 2006). Coregulators for ERα comprise members of the steroid receptor coactivator (SRC)/p160 group, the histone acetyltransferase cAMP responsive element binding protein (CREB)-binding protein (CBP)/p300, ATP-dependent chromatin remodeling complexes like SWI/SNF, E3 ubiquitin-protein ligases, and steroid RNA activator (SRA) (Lonard & O’Malley, 2006; Manavathi, Samanthapudi, & Gajulapalli, 2014). Therefore, as indicated above, even though both nuclear estrogen receptors are able to use estradiol as their physiological ligand, they exert multiple effects and functions in different cells and tissues that are mediated by several intermediaries and differential utilization of coregulators (Manavathi et al., 2014).
The mechanisms by which coregulators control the actions of estrogen receptors are still a topic of ongoing research. From studies in cancer cells, we have learned that a large group of coregulators have specific structural motifs that than affect their contact with ER ligand-binding domains (Heery, Kalkhoven, Hoare, & Parker, 1997). The specific motifs are called NR boxes or LXXLL motifs (X, any amino acid; L, leucine). On the other hand, we know that corepressors block ER-mediated gene transcription via 1) direct interaction with unbound estrogen receptors; 2) using their corepressor nuclear receptor box; 3) competing with coactivators (X. Hu & Lazar, 1999). It has also been reported that the concentration of several coregulators depends on estrogen induced-transcriptional regulation via the estrogen receptors (Mishra, Balasenthil, Nguyen, & Vadlamudi, 2004). Additionally, several post-translational modifications such as phosphorylation, methylation, ubiquitination, SUMOylation, and acetylation can impact the action of coregulators targeting gene expression (Han, Lonard, & O’Malley, 2009; Lonard & O’malley, 2007; O’Malley & McKenna, 2008).
14. ENDOGENOUS AND EXOGENOUS ESTROGEN RECEPTORS LIGANDS
Apart from the estrogens that are naturally produced by gonadal and other tissues in the body, there is a diverse variety of organic and inorganic molecules that are able to recognize the estrogen receptors ligand-binding domains in a precise manner (Table 1). Most of these ligands display higher selectivity toward ERα, however, several selective compounds for ERβ have recently been described (Farooq, 2015). There are five main classes of ER ligands: endoestrogens, phytoestrogens, xenoestrogens, selective estrogen receptor modulators (SERMs) and metalloestrogens.
Table 1.
Types of Estrogen Receptor Ligands
| Endoestrogens | Phytoestrogens | Xenoestrogens | SERMs | Metalloestrogens |
|---|---|---|---|---|
| estrone 17β-estradiol estriol estretrol |
Isoflavones: genistein, daidzein, formononetin, glycitein Coumestans: coumestrol, repensol, trifoliol Lignans: lariciresinol, matairesinol, pinoresinol, secoisolariciresinol, podophyllotoxin, steganacin |
Medicinal drugs: diethylstilbestrol, ethinyl estradiol Food additives: butylated hydroxyanisole, erythrosine Body cosmetics: 4-methylbenzylidene camphor, methylparaben, ethylparaben, propylparaben Environmental pesticides: atrazine, dichlorodiphenyldichloroethylene, dichlorodiphenyltrichloroethane, methoxychlor, dieldrin, endosulfan, heptachlor, lindane Industrial chemicals: bisphenol A, nonylphenol, monochlorobiphenyl and dichlorobiphenyl, di-2-ethylhexyl phthalate, diisodecyl phthalate, diisononyl phthalate |
Tamoxifen Clomifene Toremifene Raloxifene Ormeloxifene |
Cations: aluminum (Al3+), antimony (Sb3+), barium (Ba2+), cadmium (Cd2+), chromium (Cr2+), cobalt (Co2+), copper (Cu2+), lead (Pb2+), mercury (Hg2+), nickel (Ni2+) Anions: arsenite (AsO33−), selenite (SeO32−), vanadate (VO43−) |
Endoestrogens are physiological estrogens that are endogenously produced by the body. Most endoestrogens (i.e. estradiol, estriol, estretrol, and estrone) were previously discussed in the chapter. Briefly, endoestrogens are steroidal compounds produced from cholesterol in the male and female gonads and other organs (Farooq, 2015). In contrast, phytoestrogens are non-steroidal compounds produced by plants. There are three known groups of phytoestrogens: isoflavones, coumestans, and lignans (Basu & Maier, 2018). Because phytoestrogens are chemically and structurally similar to estradiol, they can participate in both estrogenic and antiestrogenic effects through activation or blocking of the estrogen receptor ligand-binding domains (Turner, Agatonovic-Kustrin, & Glass, 2007). Interestingly, the phytoestrogens genistein, coumestrol, and liquiritigenin have been reported to display more affinity towards ERβ than to ERα, but the implications of these differences remain unknown (Kuiper et al., 1998; Manas, Xu, Unwalla, & Somers, 2004; Mersereau et al., 2008; Nilsson, Kuiper, & Gustafsson, 1998).
Xenoestrogens are another group of ligands that comprise an extensive variety of non-natural synthetic chemical compounds with estrogenic effects. The family of xenoestrogens can be divided into five major types: medicinal drugs, food additives, body cosmetics, environmental pesticides, and industrial chemicals (Farooq, 2015). Drugs such as diethylstilbestrol (DES) and ethinyl estradiol were specifically synthesized to mimic the action of endoestrogens, and have been extensively to treat many conditions in women (Gennari, Merlotti, Valleggi, Martini, & Nuti, 2007; Maximov, Lee, & Jordan, 2013). However, it has been found that these compounds can affect cellular and molecular processes leading to severe effects on health, and their use in medical therapeutics remains controversial (Aravindakshan, Gregory, Marcogliese, Fournier, & Cyr, 2004; Aravindakshan, Paquet, et al., 2004; Arukwe, Celius, Walther, & Goksøyr, 2000; Christin et al., 2004; Golden et al., 1998; Iorga et al., 2017; Vajda et al., 2008; Williams, Lech, & Buhler, 1998). In the past few years, a wealth of evidence has been accumulated demonstrating that estrogens regulate many facets of the inflammatory response and the immune system via complex molecular mechanisms that are also sex dependent (Khan & Ansar Ahmed, 2015). It is now plausible that any immune cell that expresses estrogen receptors can potentially respond to ligand binding in a context-dependent manner, which will affect the outcome of the overall immune response. Thus, given the known spatial and temporal expression of the estrogen receptors, it is important to consider this aspect when designing potential therapeutic therapies targeting the estrogen receptor signaling pathways (Arnal et al., 2017). Additionally, precise timing of treatment initiation and duration may be required to determine the true efficacy of estrogen treatment (Burns & Korach, 2012; Hamilton, Hewitt, Arao, & Korach, 2017).
The selective estrogen receptor modulators (SERMs) are another type of estrogen receptor ligands. The main difference between SERMs and xenoestrogens relies on the fact that SERMs present functional duality and are able to act both as agonists and antagonists of the estrogen receptors in different tissues (Martinkovich, Shah, Planey, & Arnott, 2014; Shang & Brown, 2002; Smith & O’Malley, 2004). At the molecular level, SERMs employ their antagonistic actions by competing with estradiol for binding to an inner hydrophobic pocket within the ligand-binding domain of ERα (Bourguet, Germain, & Gronemeyer, 2000; Shiau et al., 1998; Wärnmark et al., 2002). Binding of this estradiol agonist induces a conformational change in the LBD that results in sealing the ligand binding pocket. Some of the most important SERMs include tamoxifen, raloxifene, clomifene, ormeloxifene, and toremifene (Farooq, 2015). One of the most used SERMs in the treatment of breast cancer, tamoxifen, acts as an antagonist in breast tissue, but as an agonist in the uterus. Therefore, while tamoxifen is often the selected treatment for ER-positive breast cancer, it can also stimulate endometrial cell growth leading to uterine cancer (R. Hu, Hilakivi-Clarke, & Clarke, 2015). While most SERMs are mainly selective for ERα, there are a few synthetic steroidal analogs that can regulate the actions of ERβ, or both receptors (Blizzard, Gude, Chan, et al., 2007; Blizzard, Gude, Morgan, et al., 2007; Blizzard et al., 2006; Papapetropoulos, 2007).
Finally, in addition to the organic ligands mentioned above, there are also inorganic compounds in the form of heavy metal ions that present estrogenic activity. These are collectivelly known as metalloestrogens. Examples of these include: aluminum (Al3+), antimony (Sb3+), barium (Ba2+), cadmium (Cd2+), chromium (Cr2+), cobalt (Co2+), copper (Cu2+), lead (Pb2+), mercury (Hg2+), nickel (Ni2+), arsenite (AsO3 3-), selenite (SeO3 2-) and vanadate (VO4 3-) (Farooq, 2015). Studies have have shown that these metalloestrogens are able to coordinate to specific amino acid residues within the ligand-binding domain of the nuclear estrogen receptors, thus blocking binding of estradiol in a non-competitive manner (Stoica, Katzenellenbogen, & Martin, 2000; Stoica, Pentecost, & Martin, 2000a, 2000b).
15. DISCUSSION
Estrogen receptors regulate a multitude of biological and physiological processes. These are tightly controlled by complex mechanisms involving either genomic nuclear direct binding to specific DNA sequences, or activation of intracellular cascades resulting in non-genomic control of transcription. Over the past 60 years since the discovery of the first nuclear estrogen receptors, and the almost 20 years since the discovery of the membrane receptor, multiple mechanisms of action have been discovered and characterized. These involve a multitude of intracellular kinases, transcription and growth factors, membrane receptors, coregulators, and natural and synthetic ligands. The information obtained in these studies has helped in the design of therapeutic strategies for diseases involving the estrogen receptors such as many cancers, as well as in the treatment of endocrine conditions affecting fertility and resulting from menopause. While there are still many diseases for which estrogens have been implicated but the role of their receptors has not been elucidated, the knowledge gained in the past six decades together with new advances in precision medicine and molecular diagnostic techniques will allow for the development of more personalized strategies to prevent and treat conditions that are affected by estrogens and other steroid hormones.
Acknowledgments
This work was supported by funding from NIH K01HL133520 (PS), NIH R03HL141618 (PS), Center for Research on Women and Newborn Health (PS), and the American Physiological Society Porter Physiology Development Program (NF).
References
- Acconcia F, Totta P, Ogawa S, Cardillo I, Inoue S, Leone S, … Marino M (2005). Survival versus apoptotic 17beta-estradiol effect: role of ER alpha and ER beta activated non-genomic signaling. J Cell Physiol, 203(1), 193–201. [DOI] [PubMed] [Google Scholar]
- Allen E, & Doisy EA (1983). Landmark article Sept 8, 1923. An ovarian hormone. Preliminary report on its localization, extraction and partial purification, and action in test animals. By Edgar Allen and Edward A. Doisy. JAMA, 250(19), 2681–2683. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/6355545. [DOI] [PubMed] [Google Scholar]
- Aranda A, & Pascual A (2001). Nuclear hormone receptors and gene expression. Physiol Rev, 81(3), 1269–1304. [DOI] [PubMed] [Google Scholar]
- Aravindakshan J, Gregory M, Marcogliese DJ, Fournier M, & Cyr DG (2004). Consumption of xenoestrogen-contaminated fish during lactation alters adult male reproductive function. Toxicol Sci, 81(1), 179–189. [DOI] [PubMed] [Google Scholar]
- Aravindakshan J, Paquet V, Gregory M, Dufresne J, Fournier M, Marcogliese DJ, & Cyr DG (2004). Consequences of xenoestrogen exposure on male reproductive function in spottail shiners (Notropis hudsonius). Toxicol Sci, 78(1), 156–165. [DOI] [PubMed] [Google Scholar]
- Arnal JF, Lenfant F, Metivier R, Flouriot G, Henrion D, Adlanmerini M, … Katzenellenbogen J (2017). Membrane and Nuclear Estrogen Receptor Alpha Actions: From Tissue Specificity to Medical Implications. Physiol Rev, 97(3), 1045–1087. [DOI] [PubMed] [Google Scholar]
- Arukwe A, Celius T, Walther BT, & Goksøyr A (2000). Effects of xenoestrogen treatment on zona radiata protein and vitellogenin expression in Atlantic salmon (Salmo salar). Aquat Toxicol, 49(3), 159–170. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/10856602. [DOI] [PubMed] [Google Scholar]
- Bajic VB, Tan SL, Chong A, Tang S, Ström A, Gustafsson JA, … Liu ET (2003). Dragon ERE Finder version 2: A tool for accurate detection and analysis of estrogen response elements in vertebrate genomes. Nucleic Acids Res, 31(13), 3605–3607. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12824376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat R, Oakley O, Kim H, Jin J, & Ko CJ (2016). Extra-gonadal sites of estrogen biosynthesis and function. BMB Rep, 49(9), 488–496. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/27530684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M, Filardo EJ, Lolait SJ, Thomas P, Maggiolini M, & Prossnitz ER (2018). Twenty years of the G protein-coupled estrogen receptor GPER: Historical and personal perspectives. J Steroid Biochem Mol Biol, 176, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu P, & Maier C (2018). Phytoestrogens and breast cancer: In vitro anticancer activities of isoflavones, lignans, coumestans, stilbenes and their analogs and derivatives. Biomed Pharmacother, 107, 1648–1666. [DOI] [PubMed] [Google Scholar]
- Belfiore CJ, Hawkins DE, Wiltbank MC, & Niswender GD (1994). Regulation of cytochrome P450scc synthesis and activity in the ovine corpus luteum. J Steroid Biochem Mol Biol, 51(5–6), 283–290. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7826890. [DOI] [PubMed] [Google Scholar]
- Bennesch MA, & Picard D (2015). Minireview: Tipping the balance: ligand-independent activation of steroid receptors. Mol Endocrinol, 29(3), 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnström L, & Sjöberg M (2005). Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol, 19(4), 833–842. [DOI] [PubMed] [Google Scholar]
- Blizzard TA, Gude C, Chan W, Birzin ET, Mojena M, Tudela C, … Hammond ML (2007). Bridged androstenediol analogs as ER-beta selective SERMs. Bioorg Med Chem Lett, 17(10), 2944–2948. [DOI] [PubMed] [Google Scholar]
- Blizzard TA, Gude C, Morgan JD, Chan W, Birzin ET, Mojena M, … Hammond ML (2007). Androstene-3,5-dienes as ER-beta selective SERMs. Bioorg Med Chem Lett, 17(22), 6295–6298. [DOI] [PubMed] [Google Scholar]
- Blizzard TA, Gude C, Morgan JD, Chan W, Birzin ET, Mojena M, … Hammond ML (2006). Androstenediol analogs as ER-beta-selective SERMs. Bioorg Med Chem Lett, 16(4), 834–838. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V (2011). Scaffolding proteins mediating membrane-initiated extra-nuclear actions of estrogen receptor. Steroids, 76(9), 877–884. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, & Edwards DP (2007). Receptor mechanisms mediating non-genomic actions of sex steroids. Semin Reprod Med, 25(3), 139–153. [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Deschênes J, Métivier R, Nagai Y, Nguyen D, Bretschneider N, … Mader S (2004). Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol, 18(6), 1411–1427. [DOI] [PubMed] [Google Scholar]
- Bourguet W, Germain P, & Gronemeyer H (2000). Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci, 21(10), 381–388. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11050318. [DOI] [PubMed] [Google Scholar]
- Bulun SE, Zeitoun K, Sasano H, & Simpson ER (1999). Aromatase in aging women. Semin Reprod Endocrinol, 17(4), 349–358. [DOI] [PubMed] [Google Scholar]
- Burns KA, & Korach KS (2012). Estrogen receptors and human disease: an update. Arch Toxicol, 86(10), 1491–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeci C, Thompson DA, Ring HZ, Francke U, & Weigel RJ (1997). Identification of a gene (GPR30) with homology to the G-protein-coupled receptor superfamily associated with estrogen receptor expression in breast cancer. Genomics, 45(3), 607–617. [DOI] [PubMed] [Google Scholar]
- Carr BR, MacDonald PC, & Simpson ER (1982). The role of lipoproteins in the regulation of progesterone secretion by the human corpus luteum. Fertil Steril, 38(3), 303–311. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7117556. [DOI] [PubMed] [Google Scholar]
- Carroll JS, & Brown M (2006). Estrogen receptor target gene: an evolving concept. Mol Endocrinol, 20(8), 1707–1714. [DOI] [PubMed] [Google Scholar]
- Cavalcanti FN, Lucas TF, Lazari MF, & Porto CS (2015). Estrogen receptor ESR1 mediates activation of ERK½, CREB, and ELK1 in the corpus of the epididymis. J Mol Endocrinol, 54(3), 339–349. Retrieved from [DOI] [PubMed] [Google Scholar]
- Chambliss KL, & Shaul PW (2002). Rapid activation of endothelial NO synthase by estrogen: evidence for a steroid receptor fast-action complex (SRFC) in caveolae. Steroids, 67(6), 413–419. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11960616. [DOI] [PubMed] [Google Scholar]
- Chambliss KL, Yuhanna IS, Mineo C, Liu P, German Z, Sherman TS, … Shaul PW (2000). Estrogen receptor alpha and endothelial nitric oxide synthase are organized into a functional signaling module in caveolae. Circ Res, 87(11), E44–52. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11090554. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Rios GR, King CD, Coffman BL, Green MD, Mojarrabi B, … Tephly TR (1998). Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7. Toxicol Sci, 45(1), 52–57. [DOI] [PubMed] [Google Scholar]
- Cheskis BJ, Greger J, Cooch N, McNally C, Mclarney S, Lam HS, … Freedman LP (2008). MNAR plays an important role in ERa activation of Src/MAPK and PI3K/Akt signaling pathways. Steroids, 73(9–10), 901–905. [DOI] [PubMed] [Google Scholar]
- Christin MS, Ménard L, Gendron AD, Ruby S, Cyr D, Marcogliese DJ, … Fournier M (2004). Effects of agricultural pesticides on the immune system of Xenopus laevis and Rana pipiens. Aquat Toxicol, 67(1), 33–43. [DOI] [PubMed] [Google Scholar]
- Coelingh Bennink F, Holinka CF, Visser M, & Coelingh Bennink HJ (2008). Maternal and fetal estetrol levels during pregnancy. Climacteric, 11 Suppl 1, 69–72. [DOI] [PubMed] [Google Scholar]
- de Leeuw R, Neefjes J, & Michalides R (2011). A role for estrogen receptor phosphorylation in the resistance to tamoxifen. Int J Breast Cancer, 2011, 232435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos EG, Dieudonne MN, Pecquery R, Le Moal V, Giudicelli Y, & Lacasa D (2002). Rapid nongenomic E2 effects on p42/p44 MAPK, activator protein-1, and cAMP response element binding protein in rat white adipocytes. Endocrinology, 143(3), 930–940. [DOI] [PubMed] [Google Scholar]
- Enmark E, Pelto-Huikko M, Grandien K, Lagercrantz S, Lagercrantz J, Fried G, … Gustafsson JA (1997). Human estrogen receptor beta-gene structure, chromosomal localization, and expression pattern. J Clin Endocrinol Metab, 82(12), 4258–4265. [DOI] [PubMed] [Google Scholar]
- Farooq A (2015). Structural and Functional Diversity of Estrogen Receptor Ligands. Curr Top Med Chem, 15(14), 1372–1384. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/25866274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, & Gregor P (1997). Cloning of a novel member of the G protein-coupled receptor family related to peptide receptors. Biochem Biophys Res Commun, 231(3), 651–654. [DOI] [PubMed] [Google Scholar]
- Filardo E, Quinn J, Pang Y, Graeber C, Shaw S, Dong J, & Thomas P (2007). Activation of the novel estrogen receptor G protein-coupled receptor 30 (GPR30) at the plasma membrane. Endocrinology, 148(7), 3236–3245. [DOI] [PubMed] [Google Scholar]
- Filardo EJ, Quinn JA, Bland KI, & Frackelton AR (2000). Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Mol Endocrinol, 14(10), 1649–1660. [DOI] [PubMed] [Google Scholar]
- Filardo EJ, & Thomas P (2012). Minireview: G protein-coupled estrogen receptor-1, GPER-1: its mechanism of action and role in female reproductive cancer, renal and vascular physiology. Endocrinology, 153(7), 2953–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EM, Andrade J, & Shupnik MA (2009). Novel actions of estrogen to promote proliferation: integration of cytoplasmic and nuclear pathways. Steroids, 74(7), 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N, Honda H, & Kitamura S (2004). Effects of environmental estrogenic chemicals on AP1 mediated transcription with estrogen receptors alpha and beta. J Steroid Biochem Mol Biol, 88(1), 53–59. [DOI] [PubMed] [Google Scholar]
- Furth PA (2014). STAT signaling in different breast cancer sub-types. Mol Cell Endocrinol, 382(1), 612–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galien R, & Garcia T (1997). Estrogen receptor impairs interleukin-6 expression by preventing protein binding on the NF-kappaB site. Nucleic Acids Res, 25(12), 2424–2429. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9171095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbacz WG, Jiang M, & Xie W (2017). Sex-Dependent Role of Estrogen Sulfotransferase and Steroid Sulfatase in Metabolic Homeostasis. Adv Exp Med Biol, 1043, 455–469. [DOI] [PubMed] [Google Scholar]
- Gaub MP, Bellard M, Scheuer I, Chambon P, & Sassone-Corsi P (1990). Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell, 63(6), 1267–1276. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/2124518. [DOI] [PubMed] [Google Scholar]
- Gennari L, Merlotti D, Valleggi F, Martini G, & Nuti R (2007). Selective estrogen receptor modulators for postmenopausal osteoporosis: current state of development. Drugs Aging, 24(5), 361–379. [DOI] [PubMed] [Google Scholar]
- Golden RJ, Noller KL, Titus-Ernstoff L, Kaufman RH, Mittendorf R, Stillman R, & Reese EA (1998). Environmental endocrine modulators and human health: an assessment of the biological evidence. Crit Rev Toxicol, 28(2), 109–227. [DOI] [PubMed] [Google Scholar]
- Gosden JR, Middleton PG, & Rout D (1986). Localization of the human oestrogen receptor gene to chromosome 6q24----q27 by in situ hybridization. Cytogenet Cell Genet, 43(3–4), 218–220. [DOI] [PubMed] [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, & Chambon P (1986). Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature, 320(6058), 134–139. [DOI] [PubMed] [Google Scholar]
- Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, & Shine J (1986). Sequence and expression of human estrogen receptor complementary DNA. Science, 231(4742), 1150–1154. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/3753802. [DOI] [PubMed] [Google Scholar]
- Gruber CJ, Tschugguel W, Schneeberger C, & Huber JC (2002). Production and actions of estrogens. N Engl J Med, 346(5), 340–352. Retrieved from [DOI] [PubMed] [Google Scholar]
- Gu Q, & Moss RL (1996). 17 beta-Estradiol potentiates kainate-induced currents via activation of the cAMP cascade. J Neurosci, 16(11), 3620–3629. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/8642406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Chen T, López E, Wu W, Wang X, Cao J, & Teng L (2014). The therapeutic target of estrogen receptor-alpha36 in estrogen-dependent tumors. J Transl Med, 12, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlicher M, Heck S, & Herrlich P (1998). Transcriptional cross-talk, the second mode of steroid hormone receptor action. J Mol Med (Berl), 76(7), 480–489. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9660166. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP, & Korach KS (2002). Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol, 16(3), 469–486. [DOI] [PubMed] [Google Scholar]
- Hamilton KJ, Hewitt SC, Arao Y, & Korach KS (2017). Estrogen Hormone Biology. Curr Top Dev Biol, 125, 109–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Lonard DM, & O’Malley BW (2009). Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab, 20(1), 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, & Parker MG (1997). A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387(6634), 733–736. [DOI] [PubMed] [Google Scholar]
- Holinka CF, Diczfalusy E, & Coelingh Bennink HJ (2008). Estetrol: a unique steroid in human pregnancy. J Steroid Biochem Mol Biol, 110(1–2), 138–143. [DOI] [PubMed] [Google Scholar]
- Hu R, Hilakivi-Clarke L, & Clarke R (2015). Molecular mechanisms of tamoxifen-associated endometrial cancer (Review). Oncol Lett, 9(4), 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, & Lazar MA (1999). The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature, 402(6757), 93–96. [DOI] [PubMed] [Google Scholar]
- Iorga A, Cunningham CM, Moazeni S, Ruffenach G, Umar S, & Eghbali M (2017). The protective role of estrogen and estrogen receptors in cardiovascular disease and the controversial use of estrogen therapy. Biol Sex Differ, 8(1), 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITTRICH G, & NEUMANN H (1963). [PARTICIPATION OF THE FETUS IN ESTROGEN METABOLISM OF THE MOTHER]. Cesk Gynekol, 28, 454–456. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/14091393. [PubMed] [Google Scholar]
- Jensen EV, Desombre ER, Kawashima T, Suzuki T, Kyser K, & Jungblut PW (1967). Estrogen-binding substances of target tissues. Science, 158(3800), 529–530. [DOI] [PubMed] [Google Scholar]
- Jensen EV, Suzuki T, Kawashima T, Stumpf WE, Jungblut PW, & DeSombre ER (1968). A two-step mechanism for the interaction of estradiol with rat uterus. Proc Natl Acad Sci U S A, 59(2), 632–638. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/5238991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzidis D, & Gilmore TD (2005). Transcription factor cross-talk: the estrogen receptor and NF-kappaB. Trends Endocrinol Metab, 16(2), 46–52. [DOI] [PubMed] [Google Scholar]
- Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, … Chambon P (1995). Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science, 270(5241), 1491–1494. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7491495. [DOI] [PubMed] [Google Scholar]
- Khan D, & Ansar Ahmed S (2015). The Immune System Is a Natural Target for Estrogen Action: Opposing Effects of Estrogen in Two Prototypical Autoimmune Diseases. Front Immunol, 6, 635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM (2001). Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res, 29(14), 2905–2919. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11452016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koos RD (2011). Minireview: Putting physiology back into estrogens’ mechanism of action. Endocrinology, 152(12), 4481–4488. [DOI] [PubMed] [Google Scholar]
- Kousteni S, Han L, Chen JR, Almeida M, Plotkin LI, Bellido T, & Manolagas SC (2003). Kinase-mediated regulation of common transcription factors accounts for the bone-protective effects of sex steroids. J Clin Invest, 111(11), 1651–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, & Gustafsson JA (1996). Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A, 93(12), 5925–5930. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/8650195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, … Gustafsson JA (1998). Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology, 139(10), 4252–4263. [DOI] [PubMed] [Google Scholar]
- Kumar R, Zakharov MN, Khan SH, Miki R, Jang H, Toraldo G, … Jasuja R (2011). The dynamic structure of the estrogen receptor. J Amino Acids, 2011, 812540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvingedal AM, & Smeland EB (1997). A novel putative G-protein-coupled receptor expressed in lung, heart and lymphoid tissue. FEBS Lett, 407(1), 59–62. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9141481. [DOI] [PubMed] [Google Scholar]
- Labrie F, Bélanger A, Luu-The V, Labrie C, Simard J, Cusan L, … Candas B (1998). DHEA and the intracrine formation of androgens and estrogens in peripheral target tissues: its role during aging. Steroids, 63(5–6), 322–328. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9618795. [DOI] [PubMed] [Google Scholar]
- Lakhani NJ, Venitz J, Figg WD, & Sparreboom A (2003). Pharmacogenetics of estrogen metabolism and transport in relation to cancer. Curr Drug Metab, 4(6), 505–513. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/14683478. [DOI] [PubMed] [Google Scholar]
- Laliotis A, Vrekoussis T, Kafousi M, Sanidas E, Askoxilakis J, Melissas J, … Stathopoulos EN (2013). Immunohistochemical study of pElk-1 expression in human breast cancer: association with breast cancer biologic profile and clinicopathologic features. Breast, 22(1), 89–95. [DOI] [PubMed] [Google Scholar]
- Le Dily F, & Beato M (2018). Signaling by Steroid Hormones in the 3D Nuclear Space. Int J Mol Sci, 19(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Briggs MR, Ahlborn TE, Kraemer FB, & Liu J (2001). Requirement of Sp1 and estrogen receptor alpha interaction in 17beta-estradiol-mediated transcriptional activation of the low density lipoprotein receptor gene expression. Endocrinology, 142(4), 1546–1553. [DOI] [PubMed] [Google Scholar]
- Li L, Haynes MP, & Bender JR (2003). Plasma membrane localization and function of the estrogen receptor alpha variant (ER46) in human endothelial cells. Proc Natl Acad Sci U S A, 100(8), 4807–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hisamoto K, Kim KH, Haynes MP, Bauer PM, Sanjay A, … Bender JR (2007). Variant estrogen receptor-c-Src molecular interdependence and c-Src structural requirements for endothelial NO synthase activation. Proc Natl Acad Sci U S A, 104(42), 16468–16473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang JP, Santen RJ, Kim TH, Park H, Fan P, & Yue W (2010). Estrogen stimulation of cell migration involves multiple signaling pathway interactions. Endocrinology, 151(11), 5146–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, & Shang Y (2013). Estrogen and cancer. Annu Rev Physiol, 75, 225–240. [DOI] [PubMed] [Google Scholar]
- Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, … Kushner PJ (2002). Opposing action of estrogen receptors alpha and beta on cyclin D1 gene expression. J Biol Chem, 277(27), 24353–24360. [DOI] [PubMed] [Google Scholar]
- Lonard DM, & O’Malley BW (2006). The expanding cosmos of nuclear receptor coactivators. Cell, 125(3), 411–414. [DOI] [PubMed] [Google Scholar]
- Lonard DM, & O’malley BW (2007). Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell, 27(5), 691–700. [DOI] [PubMed] [Google Scholar]
- Loven MA, Wood JR, & Nardulli AM (2001). Interaction of estrogen receptors alpha and beta with estrogen response elements. Mol Cell Endocrinol, 181(1–2), 151–163. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11476949. [DOI] [PubMed] [Google Scholar]
- Lösel R, & Wehling M (2003). Nongenomic actions of steroid hormones. Nat Rev Mol Cell Biol, 4(1), 46–56. [DOI] [PubMed] [Google Scholar]
- Maggi A (2011). Liganded and unliganded activation of estrogen receptor and hormone replacement therapies. Biochim Biophys Acta, 1812(8), 1054–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manas ES, Xu ZB, Unwalla RJ, & Somers WS (2004). Understanding the selectivity of genistein for human estrogen receptor-beta using X-ray crystallography and computational methods. Structure, 12(12), 2197–2207. [DOI] [PubMed] [Google Scholar]
- Manavathi B, Samanthapudi VS, & Gajulapalli VN (2014). Estrogen receptor coregulators and pioneer factors: the orchestrators of mammary gland cell fate and development. Front Cell Dev Biol, 2, 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino M, Acconcia F, Bresciani F, Weisz A, & Trentalance A (2002). Distinct nongenomic signal transduction pathways controlled by 17beta-estradiol regulate DNA synthesis and cyclin D(1) gene transcription in HepG2 cells. Mol Biol Cell, 13(10), 3720–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino M, Acconcia F, & Trentalance A (2003). Biphasic estradiol-induced AKT phosphorylation is modulated by PTEN via MAP kinase in HepG2 cells. Mol Biol Cell, 14(6), 2583–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino M, Galluzzo P, & Ascenzi P (2006). Estrogen signaling multiple pathways to impact gene transcription. Curr Genomics, 7(8), 497–508. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/18369406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino M, Pallottini V, & Trentalance A (1998). Estrogens cause rapid activation of IP3-PKC-alpha signal transduction pathway in HEPG2 cells. Biochem Biophys Res Commun, 245(1), 254–258. [DOI] [PubMed] [Google Scholar]
- Martinkovich S, Shah D, Planey SL, & Arnott JA (2014). Selective estrogen receptor modulators: tissue specificity and clinical utility. Clin Interv Aging, 9, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J, & Gustafsson JA (2003). Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol Interv, 3(5), 281–292. [DOI] [PubMed] [Google Scholar]
- Maximov PY, Lee TM, & Jordan VC (2013). The discovery and development of selective estrogen receptor modulators (SERMs) for clinical practice. Curr Clin Pharmacol, 8(2), 135–155. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/23062036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersereau JE, Levy N, Staub RE, Baggett S, Zogovic T, Zogric T, … Leitman DC (2008). Liquiritigenin is a plant-derived highly selective estrogen receptor beta agonist. Mol Cell Endocrinol, 283(1–2), 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio A, Di Domenico M, Castoria G, de Falco A, Bontempo P, Nola E, & Auricchio F (1996). Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. EMBO J, 15(6), 1292–1300. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/8635462. [PMC free article] [PubMed] [Google Scholar]
- Miller WL (2017). Steroidogenesis: Unanswered Questions. Trends Endocrinol Metab, 28(11), 771–793. [DOI] [PubMed] [Google Scholar]
- Miller WL, & Strauss JF (1999). Molecular pathology and mechanism of action of the steroidogenic acute regulatory protein, StAR. J Steroid Biochem Mol Biol, 69(1–6), 131–141. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/10418987. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Balasenthil S, Nguyen D, & Vadlamudi RK (2004). Cloning and functional characterization of PELP1/MNAR promoter. Gene, 330, 115–122. [DOI] [PubMed] [Google Scholar]
- Molina L, Figueroa CD, Bhoola KD, & Ehrenfeld P (2017). GPER-1/GPR30 a novel estrogen receptor sited in the cell membrane: therapeutic coupling to breast cancer. Expert Opin Ther Targets, 21(8), 755–766. [DOI] [PubMed] [Google Scholar]
- Nelson LR, & Bulun SE (2001). Estrogen production and action. J Am Acad Dermatol, 45(3 Suppl), S116–124. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11511861. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Kuiper G, & Gustafsson JA (1998). ERbeta: a novel estrogen receptor offers the potential for new drug development. Trends Endocrinol Metab, 9(10), 387–395. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/18406312. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Mäkelä S, Treuter E, Tujague M, Thomsen J, Andersson G, … Gustafsson JA (2001). Mechanisms of estrogen action. Physiol Rev, 81(4), 1535–1565. [DOI] [PubMed] [Google Scholar]
- O’Dowd BF, Nguyen T, Marchese A, Cheng R, Lynch KR, Heng HH, … George SR (1998). Discovery of three novel G-protein-coupled receptor genes. Genomics, 47(2), 310–313. [DOI] [PubMed] [Google Scholar]
- O’Lone R, Frith MC, Karlsson EK, & Hansen U (2004). Genomic targets of nuclear estrogen receptors. Mol Endocrinol, 18(8), 1859–1875. [DOI] [PubMed] [Google Scholar]
- O’Malley BW (2005). A life-long search for the molecular pathways of steroid hormone action. Mol Endocrinol, 19(6), 1402–1411. [DOI] [PubMed] [Google Scholar]
- O’Malley BW, & McKenna NJ (2008). Coactivators and corepressors: what’s in a name? Mol Endocrinol, 22(10), 2213–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owman C, Blay P, Nilsson C, & Lolait SJ (1996). Cloning of human cDNA encoding a novel heptahelix receptor expressed in Burkitt’s lymphoma and widely distributed in brain and peripheral tissues. Biochem Biophys Res Commun, 228(2), 285–292. [DOI] [PubMed] [Google Scholar]
- Ozes ON, Mayo LD, Gustin JA, Pfeffer SR, Pfeffer LM, & Donner DB (1999). NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature, 401(6748), 82–85. [DOI] [PubMed] [Google Scholar]
- Oñate SA, Tsai SY, Tsai MJ, & O’Malley BW (1995). Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science, 270(5240), 1354–1357. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7481822. [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, & Scanlan TS (1997). Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science, 277(5331), 1508–1510. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9278514. [DOI] [PubMed] [Google Scholar]
- Papapetropoulos A (2007). A ginseng-derived oestrogen receptor beta (ERbeta) agonist, Rb1 ginsenoside, attenuates capillary morphogenesis. Br J Pharmacol, 152(2), 172–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotto G, Massheimer V, & Boland R (1996). Acute stimulation of intestinal cell calcium influx induced by 17 beta-estradiol via the cAMP messenger system. Mol Cell Endocrinol, 119(2), 129–134. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/8807632. [DOI] [PubMed] [Google Scholar]
- Piu F, Aronheim A, Katz S, & Karin M (2001). AP-1 repressor protein JDP-2: inhibition of UV-mediated apoptosis through p53 down-regulation. Mol Cell Biol, 21(9), 3012–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz ER, & Barton M (2011). The G-protein-coupled estrogen receptor GPER in health and disease. Nat Rev Endocrinol, 7(12), 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz ER, & Barton M (2014). Estrogen biology: new insights into GPER function and clinical opportunities. Mol Cell Endocrinol, 389(1–2), 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit A, & Reed MJ (2002). Regulation of estrogen synthesis in postmenopausal women. Steroids, 67(12), 979–983. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12398994. [DOI] [PubMed] [Google Scholar]
- Purohit A, Tutill HJ, Day JM, Chander SK, Lawrence HR, Allan GM, … Reed MJ (2006). The regulation and inhibition of 17beta-hydroxysteroid dehydrogenase in breast cancer. Mol Cell Endocrinol, 248(1–2), 199–203. [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Merchenthaler I, Greene GL, & Levin ER (2004). Plasma membrane estrogen receptors exist and functions as dimers. Mol Endocrinol, 18(12), 2854–2865. [DOI] [PubMed] [Google Scholar]
- Revankar CM, Cimino DF, Sklar LA, Arterburn JB, & Prossnitz ER (2005). A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science, 307(5715), 1625–1630. [DOI] [PubMed] [Google Scholar]
- Romano SN, & Gorelick DA (2018). Crosstalk between nuclear and G protein-coupled estrogen receptors. Gen Comp Endocrinol, 261, 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romashkova JA, & Makarov SS (1999). NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature, 401(6748), 86–90. [DOI] [PubMed] [Google Scholar]
- Rotstein A. Sex hormone synthesis, regulation, and function. Retrieved from http://www.pathophys.org/sexhormones/
- RYAN KJ (1959). Biological aromatization of steroids. J Biol Chem, 234(2), 268–272. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/13630892. [PubMed] [Google Scholar]
- Samavat H, & Kurzer MS (2015). Estrogen metabolism and breast cancer. Cancer Lett, 356(2 Pt A), 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausville E, Carney D, & Battey J (1985). The human vasopressin gene is linked to the oxytocin gene and is selectively expressed in a cultured lung cancer cell line. J Biol Chem, 260(18), 10236–10241. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/2991279. [PubMed] [Google Scholar]
- Scheidereit C, Krauter P, von der Ahe D, Janich S, Rabenau O, Cato AC, … Beato M (1986). Mechanism of gene regulation by steroid hormones. J Steroid Biochem, 24(1), 19–24. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/3009974. [DOI] [PubMed] [Google Scholar]
- Schulster M, Bernie AM, & Ramasamy R (2016). The role of estradiol in male reproductive function. Asian J Androl, 18(3), 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe JW, & Teichmann SA (2004). Nuclear receptors: the evolution of diversity. Sci STKE, 2004(217), pe4 [DOI] [PubMed] [Google Scholar]
- Shang Y, & Brown M (2002). Molecular determinants for the tissue specificity of SERMs. Science, 295(5564), 2465–2468. [DOI] [PubMed] [Google Scholar]
- Sharma G, & Prossnitz ER (2016). GPER/GPR30 Knockout Mice: Effects of GPER on Metabolism. Methods Mol Biol, 1366, 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul PW, & Anderson RG (1998). Role of plasmalemmal caveolae in signal transduction. Am J Physiol, 275(5 Pt 1), L843–851. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9815100. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, & Greene GL (1998). The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell, 95(7), 927–937. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9875847. [DOI] [PubMed] [Google Scholar]
- Silva E, Kabil A, & Kortenkamp A (2010). Cross-talk between non-genomic and genomic signalling pathways--distinct effect profiles of environmental estrogens. Toxicol Appl Pharmacol, 245(2), 160–170. [DOI] [PubMed] [Google Scholar]
- Simpson ER (2003). Sources of estrogen and their importance. J Steroid Biochem Mol Biol, 86(3–5), 225–230. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/14623515. [DOI] [PubMed] [Google Scholar]
- Simpson ER, Clyne C, Rubin G, Boon WC, Robertson K, Britt K, … Jones M (2002). Aromatase--a brief overview. Annu Rev Physiol, 64, 93–127. [DOI] [PubMed] [Google Scholar]
- Simpson ER, Misso M, Hewitt KN, Hill RA, Boon WC, Jones ME, … Clyne CD (2005). Estrogen--the good, the bad, and the unexpected. Endocr Rev, 26(3), 322–330. [DOI] [PubMed] [Google Scholar]
- Smith CL, & O’Malley BW (2004). Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev, 25(1), 45–71. [DOI] [PubMed] [Google Scholar]
- Song RX, Chen Y, Zhang Z, Bao Y, Yue W, Wang JP, … Santen RJ (2010). Estrogen utilization of IGF-1-R and EGF-R to signal in breast cancer cells. J Steroid Biochem Mol Biol, 118(4–5), 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RX, Zhang Z, Chen Y, Bao Y, & Santen RJ (2007). Estrogen signaling via a linear pathway involving insulin-like growth factor I receptor, matrix metalloproteinases, and epidermal growth factor receptor to activate mitogen-activated protein kinase in MCF-7 breast cancer cells. Endocrinology, 148(8), 4091–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RX, Zhang Z, & Santen RJ (2005). Estrogen rapid action via protein complex formation involving ERalpha and Src. Trends Endocrinol Metab, 16(8), 347–353. [DOI] [PubMed] [Google Scholar]
- Stoica A, Katzenellenbogen BS, & Martin MB (2000). Activation of estrogen receptor-alpha by the heavy metal cadmium. Mol Endocrinol, 14(4), 545–553. [DOI] [PubMed] [Google Scholar]
- Stoica A, Pentecost E, & Martin MB (2000a). Effects of arsenite on estrogen receptor-alpha expression and activity in MCF-7 breast cancer cells. Endocrinology, 141(10), 3595–3602. [DOI] [PubMed] [Google Scholar]
- Stoica A, Pentecost E, & Martin MB (2000b). Effects of selenite on estrogen receptor-alpha expression and activity in MCF-7 breast cancer cells. J Cell Biochem, 79(2), 282–292. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/10967555. [DOI] [PubMed] [Google Scholar]
- Sun G, Porter W, & Safe S (1998). Estrogen-induced retinoic acid receptor alpha 1 gene expression: role of estrogen receptor-Sp1 complex. Mol Endocrinol, 12(6), 882–890. [DOI] [PubMed] [Google Scholar]
- Takada Y, Kato C, Kondo S, Korenaga R, & Ando J (1997). Cloning of cDNAs encoding G protein-coupled receptor expressed in human endothelial cells exposed to fluid shear stress. Biochem Biophys Res Commun, 240(3), 737–741. [DOI] [PubMed] [Google Scholar]
- Thomas P, Pang Y, Filardo EJ, & Dong J (2005). Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology, 146(2), 624–632. [DOI] [PubMed] [Google Scholar]
- Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, & Chambon P (1989). The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell, 59(3), 477–487. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/2805068. [DOI] [PubMed] [Google Scholar]
- Truss M, & Beato M (1993). Steroid hormone receptors: interaction with deoxyribonucleic acid and transcription factors. Endocr Rev, 14(4), 459–479. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, & Yokoi T (2005). Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett, 227(2), 115–124. [DOI] [PubMed] [Google Scholar]
- Turner JV, Agatonovic-Kustrin S, & Glass BD (2007). Molecular aspects of phytoestrogen selective binding at estrogen receptors. J Pharm Sci, 96(8), 1879–1885. [DOI] [PubMed] [Google Scholar]
- Vajda AM, Barber LB, Gray JL, Lopez EM, Woodling JD, & Norris DO (2008). Reproductive disruption in fish downstream from an estrogenic wastewater effluent. Environ Sci Technol, 42(9), 3407–3414. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/18522126. [DOI] [PubMed] [Google Scholar]
- Voogt JL (1978). Control of hormone release during lactation. Clin Obstet Gynaecol, 5(2), 435–455. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/361330. [PubMed] [Google Scholar]
- Vrtačnik P, Ostanek B, Mencej-Bedrač S, & Marc J (2014). The many faces of estrogen signaling. Biochem Med (Zagreb), 24(3), 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CS, Jeng YJ, & Kochukov MY (2008). Nongenomic actions of estradiol compared with estrone and estriol in pituitary tumor cell signaling and proliferation. FASEB J, 22(9), 3328–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters J, Campbell J, Cunningham M, Krebs E, & Dorsa D (1997). Rapid membrane effects of steroids in neuroblastoma cells: effects of estrogen on mitogen activated protein kinase signalling cascade and c-fos immediate early gene transcription. Endocrinology, 138(9), 4030–4033. [DOI] [PubMed] [Google Scholar]
- Williams DE, Lech JJ, & Buhler DR (1998). Xenobiotics and xenoestrogens in fish: modulation of cytochrome P450 and carcinogenesis. Mutat Res, 399(2), 179–192. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9672659. [DOI] [PubMed] [Google Scholar]
- WILSON R, ERIKSSON G, & DICZFALUSY E (1964). OESTRIOL METABOLISM IN PREGNANT WOMEN. Acta Endocrinol (Copenh), 46, 525–543. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/14202469. [DOI] [PubMed] [Google Scholar]
- Wärnmark A, Treuter E, Gustafsson JA, Hubbard RE, Brzozowski AM, & Pike AC (2002). Interaction of transcriptional intermediary factor 2 nuclear receptor box peptides with the coactivator binding site of estrogen receptor alpha. J Biol Chem, 277(24), 21862–21868. [DOI] [PubMed] [Google Scholar]
- Yaşar P, Ayaz G, User SD, Güpür G, & Muyan M (2017). Molecular mechanism of estrogen-estrogen receptor signaling. Reprod Med Biol, 16(1), 4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Driscoll MD, Huang J, Bhagat S, Hilf R, Bambara RA, & Muyan M (2002). The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ER alpha and ER beta. Mol Endocrinol, 16(4), 674–693. [DOI] [PubMed] [Google Scholar]


