Abstract
Commensal microorganisms in the mammalian gut play important roles in host health and physiology, but a central challenge remains in achieving a detailed mechanistic understanding of specific microbial contributions to host biochemistry. New function-based approaches are needed that analyze gut microbial function at the molecular level by coupling detection and measurements of in situ biochemical activity with identification of the responsible microbes and enzymes. We developed a platform employing β-glucuronidase selective activity-based probes to detect, isolate, and identify microbial subpopulations in the gut responsible for this xenobiotic metabolism. We find that metabolic activity of gut microbiota can be plastic and that between individuals and during perturbation, phylogenetically disparate populations can provide β-glucuronidase activity. Our work links biochemical activity with molecular-scale resolution without relying on genomic inference.
In terms of metabolic capacity, the gut microbiome can be considered a separate organ that expands the repertoire of biotransformation reactions possible in the host.1,2 These reactions impact the outcome of exposure or efficacy of therapeutics, and variability in gut microbiota composition may explain individual variability and susceptibility in response to drug treatment or environmental contaminant exposure.3,4 However, our understanding of host-microbiota–xenobiotic interaction is limited by a lack of tools capable of coupling microbe–enzyme-scale resolution offered by metagenome-based studies with detection and measurement of biochemical activity.5 Current bioinformatics strategies rely on accurate annotation and gene prediction. Unfortunately, approximately 50% of genes in the microbiome remain uncharacterized.6 Omics-based expression studies assume that the abundance of a gene, transcript, or protein correlates to functional activity. This assumption fails to account for regulation of expression, translation, or differing activities between similarly annotated enzymes.7–12 Approaches capable of coupling measurement of biochemical activity to identification of active microbes and enzymes are needed. 1,3,13
Activity-based probes (ABPs) are uniquely suited to address this gap.13 ABPs are small-molecule substrates that, upon activation by a catalytically active target enzyme, form a covalent bond with that enzyme.14–17 Because the probes only label an active enzyme, ABPs can demonstrate activity in lysate or live cells. Importantly, ABPs can be used to identify unannotated proteins based on a specific activity, connecting enzymes to function in poorly defined systems.13 Furthermore, the use of generalized bio-orthogonal tags allows labeling events to be enriched and measured by proteomics or fluorescently tagged for imaging, SDS-PAGE, or fluorescence-activated cell sorting (FACS). FACS-based approaches can be particularly useful for studying activity in microbial communities.18,19 By combining ABPs with FACS (ABP-FACS), subpopulations with active enzyme can be sorted in a function-dependent manner and identified by sequencing. This reveals specific subpopulations of microbes based on detected, not inferred, activity.
A key pathway modulated by the gut microbiome is glucuronidation. Glucuronidation facilitates mammalian Phase II metabolism and clearance of xenobiotics via conjugation of a glucuronic acid (GlcA) to xenobiotics and endogenous metabolites. Gut microbial β-glucuronidases can hydrolyze conjugates back to parent compounds, leading to altered pharmacodynamics, failure of therapeutics, or severe side effects.2,20,21 Recent work has identified conserved motifs to improve annotation of β-glucuronidases;12,22 however, these genes are widespread, complicating prediction of specific taxa responsible for deglucuronidation.
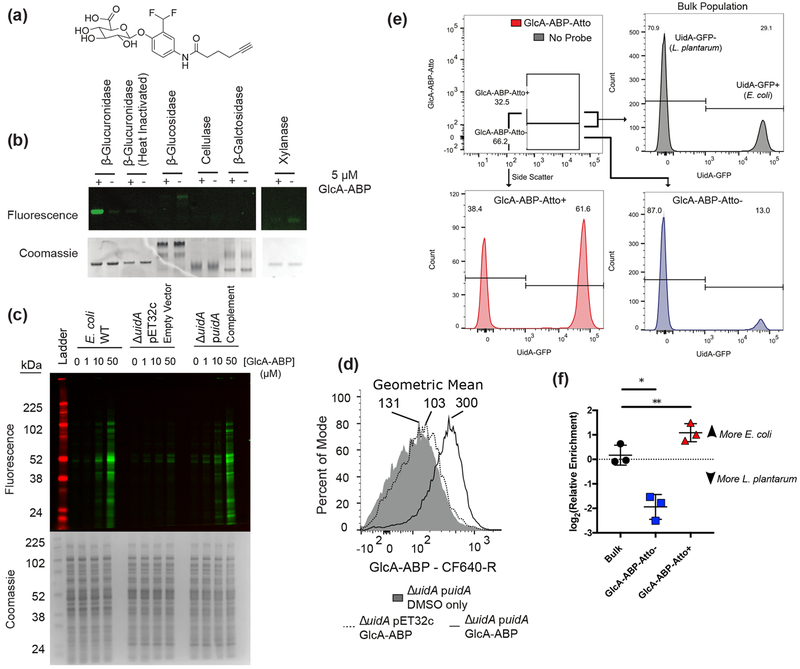
Given the ubiquity of β-glucuronidase genes, we hypothesized that activity would be spread throughout phylogenetically distinct taxa. For in situ activity detection, we synthesized GlcA-ABP which mimics a glucuronidated metabolite, and bears a reactive group attached to the anomeric position of GlcA and an alkyne for reporter group attachment (Figure 1a). When an active β-glucuronidase reacts with the probe, an electrophilic o-quinone methide forms and covalently reacts with a nearby nucleophilic residue (Figure S1).23,24 The alkyne handle of GlcA-ABP enables fluorophore attachment via copper-catalyzed azide–alkyne cycloaddition (CuAAC).25 To demonstrate probe efficacy, we performed in vitro labeling with recombinant β-glucuronidase from Escherichia coli. Enzymes were treated with GlcA-ABP, tagged with rhodamine-azide, and analyzed by SDS-PAGE. Importantly, neither heat-inactivated glucuronidases nor glycosidases as β-galactosidase, cellulase, xylanase, and β-glucosidase are labeled, confirming selective activity-dependent labeling by GlcA-ABP (Figure 1b,c).
Figure 1.
GlcA-ABP labels β-glucuronidase-active proteins and cells. (a) GlcA-ABP. (b) Selectivity of GlcA-ABP against a panel of glycosidases. (c) GlcA-ABP labeling of E. coli lysates. (d) Whole cell E. coli were labeled with GlcA-ABP and tagged with CF640-R and sorted. Geometric mean is reported above each population. (e) Mixture of L. plantarum and E. coli::pUidA-GFP labeled with GlcA-ABP-Atto. Numbers represent percent of the population in the UidA-GFP− and UidA-GFP+ gates. (f) DNA from sorted cells was used as template for quantitative PCR to determine the relative abundance of E. coli and L. plantarum in the sorted fractions. Lines and error bars represent the mean and standard error of the mean, respectively. * p = 0.0374; ** p = 0.0093 by repeated measures one-way ANOVA with Tukey’s multiple comparisons test, n = 3.
We validated in vivo labeling using an E. coli strain lacking uidA (ΔuidA) which encodes for the only β-glucuronidase in the E. coli genome. Cell lysate was GlcA-ABP-treated, tagged with rhodamine-azide, and analyzed (Figure 1c). We observed dose-dependent labeling in wild type (WT) E. coli, no labeling in ΔuidA transformed with the empty vector (ΔuidA pET32c), and restoration of labeling in ΔuidA complemented with E. coli uidA (ΔuidA puidA), both in lysate and in whole cells by FACS (Figure 1d).
Quinone methides, while highly electrophilic, have been shown to be able to diffuse from the enzyme active site resulting in labeling of nearby proteins in addition to the activating enzyme.26,27 This can be utilized as a signal amplification strategy.26 We initially observed diffusion in vitro with E. coli lysate (Figure 1c), and between cells during in vivo labeling (Figure S2). Thus, we sought to leverage the quinone methide for signal amplification while ensuring the reactive intermediate does not leave the activating cell. To achieve this, we tagged GlcA-ABP with the charged small-molecule fluorophore Atto633 (GlcA-ABP-Atto) and confirmed this had no impact on labeling specificity (Figure S1). We then constructed and transformed E. coli with a plasmid encoding a glucuronidase-GFP fusion (E. coli::pUidA-GFP) to identify glucuronidase-positive cells within a heterogeneous population. Samples of E. coli::pUidA-GFP only, the glucuronidase-negative Lactobacillus plantarum only, or a mixture were labeled with GlcA-ABP-Atto or treated with Atto633 only (“No Probe”) (Figure 1e). Cells were analyzed by flow cytometry to detect GlcA-ABP-Atto, and the GFP signal was used to identify glucuronidase expressing cells. The glucuronidase-active E. coli::pUidA-GFP was enriched in the GlcA-ABP-Atto+ population and depleted in the GlcA-ABP-Atto− population (Figure 1e). To confirm that GlcA-ABP-Atto enabled accurate sorting, we performed quantitative PCR on sorted cells using species-specific primers. The GlcA-ABP-Atto+ population was significantly enriched for E. coli while the GlcA-ABP-Atto− population was significantly enriched for L. plantarum, validating the approach (Figure 1f).
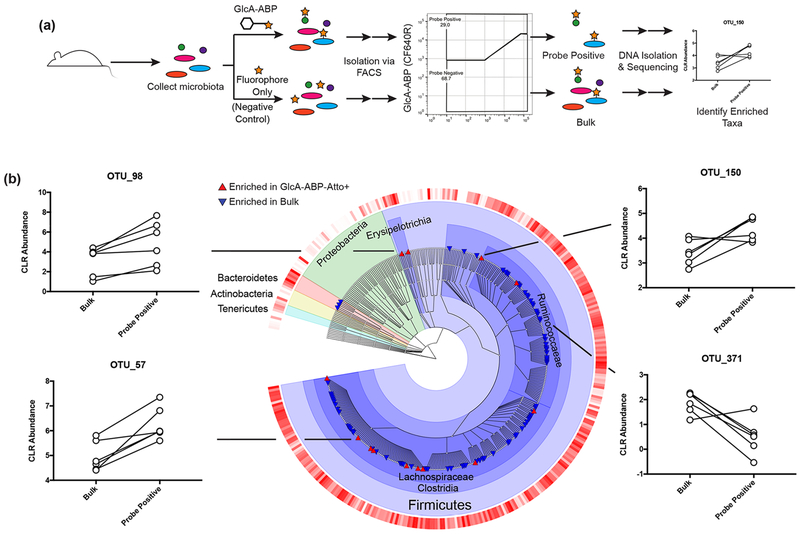
We then sought to identify glucuronidase-active members of the gut microbiota. Microbes isolated from the mouse gastrointestinal tract were incubated with GlcA-ABP-Atto or Atto633 only (“No Probe”) under anaerobic conditions. Cells were fixed and sorted into GlcA-ABP-Atto+, GlcA-ABP-Atto−, and Bulk cell populations (Figure 2a; Figure S3). Community composition was determined for each population by 16S rRNA gene amplicon sequencing, and differentially abundant operational taxonomic units (OTUs) were identified (Figure S4).28 OTUs with statistically enriched abundance in the GlcA-ABP-Atto+ population compared to the bulk population were considered glucuronidase-active. This identified 13 glucuronidase-active OTUs, all Firmicutes in the classes Erysipelotrichia (2 OTUs) or Clostridia (11 OTUs; Figure 2b) of distinct genera such as Ruminiclostridium, Tyzzerella, and Roseburia. Additionally, OTUs with significantly decreased abundance in the GlcA-ABP-Atto+ compared to the bulk population were considered glucuronidase-inactive. This group was also taxonomically diverse, with representative sequences from Bacteroidetes and Firmicutes. Interestingly, some groups at the level of family or even genus contained both glucuronidase-active and glucuronidase-inactive OTUs. This finding supports the notion that in vivo metabolic activity cannot be ascribed based solely on phylogeny.
Figure 2.
ABP-FACS identifies β-glucuronidase-active taxa in the gut microbiota. (a) Experimental design. (b) Phylogenetic distribution of taxa enriched or depleted in the GlcA-ABP-Atto+ and bulk populations. The outer ring indicates centered log-ratio abundance, where darker red indicates higher abundance. Taxa with significantly increased or decreased abundance in the ABP+ population are shown by triangles. Centered-log-ratio (CLR) normalized abundances of three GlcA-ABP-Atto enriched taxa (left and top right) and one GlcA-ABP-Atto depleted taxon (bottom right) are shown. Lines connect populations from the same sample. Taxa are differentially abundant where Benjamini–Hochberg adjusted p < 0.05 using a generalized linear model (n = 6).
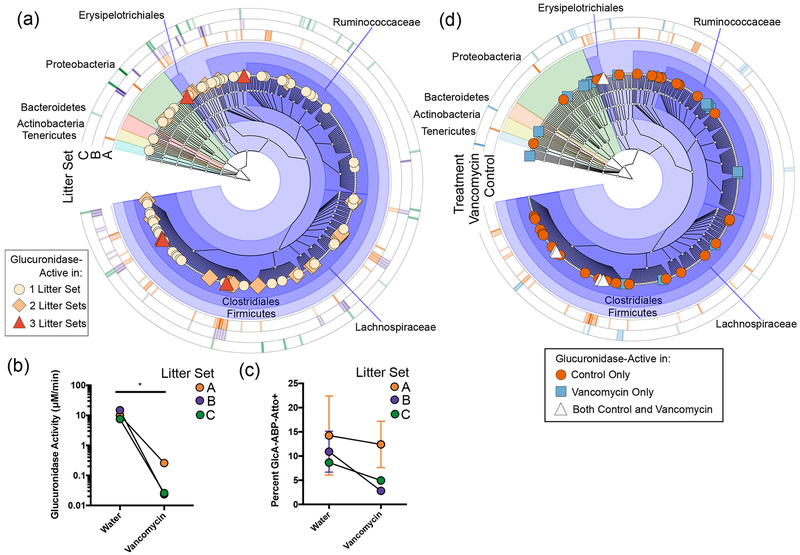
To investigate populations responsible for a given activity across different individual hosts, functional approaches are needed. To determine if ABP-FACS can provide such a tool, we compared glucuronidase-active taxa across different mouse litters. Because of limitations on litter size, the number of mice was too small to apply the statistical approach used in Figure 2. Instead, we considered any taxa that had a 2-fold increase in relative abundance in the GlcA-ABP-Atto+ population compared to the Bulk population to be glucuronidase-active (Figure 3a; see also the SI). We identified 5 taxa as enriched in all three Litter Sets, including three Lachnospiraceae OTUs we identified as glucuronidase-active in our initial, more stringent search. We also identified 24 OTUs enriched in two of three Litter Sets (light orange diamonds), and 55 OTUs enriched in only one Litter Set (tan circles). Interestingly, we observed clades where distinct OTUs are active in each Litter Set. This supports the idea that distinct but related taxa may possess a similar function (i.e., hydrolysis of glucuronides) among different hosts and demonstrates the utility of tools capable of linking functional activity to taxa in situ. While all litters had similar community composition, the active populations differ. Approaches like ABP-FACS will be necessary to more directly connect genetic information to function in a manner that accounts for interindividual variability.
Figure 3.
ABP-FACS can detect interindividual variability. (a) Taxa that were 2-fold enriched in the GlcA-ABP-Atto+ population from each litter were identified (n = 2 mice per Litter Set). Outer rings indicate relative fold enrichment (darker = more enriched) for Litter Sets. Glucuronidase activity (b) and percent GlcA-ABP-Atto+ population (c) in gut microbiota of control (Water) or vancomycin-treated mice. Paired groups of two littermates (n = 3 groups of 2) are connected by lines. * p = 0.0386 by two-sided paired Student’s t test. Data are the average of the two littermates per condition. Error bars represent standard error of the mean. (d) Taxa that were 2-fold enriched in the GlcA-ABP-Atto+ population from either control or vancomycin-treated mice from Litter Set A are shown (n = 2 littermates per condition). Outer rings indicate relative fold enrichment (darker = more enriched) for either control or vancomycin-treated mice.
One major hurdle for microbiome research is difficulty determining how perturbation-induced changes in community composition relate to changes in activity. ABP-FACS is well-suited to address this problem. To demonstrate this, we exposed pairs of littermates to water with or without vancomycin, an antibiotic that differentially impacts taxa in the gut microbiota (n = 2 littermates per condition, 3 litters total).29 Vancomycin treatment significantly reduced glucuronidase activity within the gut microbiota to below the limit of quantification for 4 of 6 samples (Figure 3b). The percent GlcA-ABP-Atto+ population decreased as well (Figure 3c). Using the approach in Figure 2, we sought to identify taxa significantly enriched or depleted in the GlcA-ABP-Atto+ population compared to the bulk population in vancomycin-treated mice. In accordance with the reduced activity, no taxa were identified as significantly enriched (seven taxa were significantly depleted).
We then investigated the impact of vancomycin on the individual Litter Sets. Interestingly, two Litter Sets (B and C) exhibited a strong population shift in response to vancomycin exposure, while one Litter Set (A) did not (Figure S5). The vancomycin-treated samples in Litter Set A also had higher glucuronidase activity and percent GlcA-ABP-Atto+ population than the other vancomycin-treated samples, but lower than the littermate controls (Figure 3b,c). Disparate responses in microbiota to perturbation have been described,30,31 as have cage effects on littermates (Figure S6).32,33 We utilized ABP-FACS to provide more insight into the active subpopulations in these samples (Figure 3d). While similar community composition based on 16S rRNA gene sequencing suggests that the glucuronidase-active populations should be similar, we found the active populations in these samples were largely distinct. While 25 OTUs were enriched only in the vancomycin-treated samples, 40 OTUs were enriched only in the controls; 5 OTUs were enriched in both, including four Lachnospiraceae and one Erysipelotrichaceae OTU. Regarding vancomycin resistance, we observe dominant probe enrichment in Litter Set B of a Lactococcus sp., while the dominant taxon in Litter Set C is a Paenibacillus sp. Vancomycin resistant Lactococcus have not been reported; however their taxonomic similarity to Enterococcus and known horizontal gene transfer in Lactobacilli in general suggests a real possibility for resistance. Paenibacillus strains have shown vancomycin resistance.34–36
While the sample size limits the statistical power, our findings highlight the capacity of function-based approaches to provide a detailed description of active populations compared to solely 16S rRNA gene sequencing. Revealing the functional change perturbations cause to microbiota will enable improved prediction on host health impacts. While genetic and in vitro analyses of gut microbiota isolates demonstrated that genes encoding glucuronidases are taxonomically widespread,37,38 ABP-FACS, for the first time, links molecular-scale detection of in situ activity with identification of the responsible, functionally active taxa. The use of ABPs targeting other key functions such as proteases, other glycosidases, or bile acid metabolism will someday serve as important tools in functional gut microbiome measurements.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Redinbo (University of North Carolina) for purified β-glucuronidase. Research was supported by the Microbiomes in Transition Lab Directed Research and Development Program at PNNL, and the NIEHS (ES029319). This research employed capabilities supported by the NIGMS (GM103493). PNNL is operated by Battelle for the DOE under Contract DE-AC06-76RL01830.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b09668.
Detailed synthetic and experimental procedures, and additional figures including SDS-PAGE, mass spectra, HPLC trace, side-scattering, principle coordinates analysis, population shifts, and NMR results (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Patterson AD; Turnbaugh PJ Microbial determinants of biochemical individuality and their impact on toxicology and pharmacology. Cell Metab. 2014, 20 (5), 761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Claus SP; Guillou H; Ellero-Simatos S The gut microbiota: a major player in the toxicity of environmental pollutants? Npj Biofilms And Microbiomes 2016, 2, 16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Koppel N; Maini Rekdal V; Balskus EP Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356 (6344), eaag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Haiser HJ; Gootenberg DB; Chatman K; Sirasani G; Balskus EP; Turnbaugh PJ Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 2013, 341 (6143), 295–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sousa T; Paterson R; Moore V; Carlsson A; Abrahamsson B; Basit AW The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm 2008, 363 (1–2), 1–25. [DOI] [PubMed] [Google Scholar]
- (6).Joice R; Yasuda K; Shafquat A; Morgan XC; Huttenhower C Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab. 2014, 20 (5), 731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Vogel C; Marcotte EM Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet 2012, 13 (4), 227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stitt M; Gibon Y Why measure enzyme activities in the era of systems biology? Trends Plant Sci. 2014, 19 (4), 256–65. [DOI] [PubMed] [Google Scholar]
- (9).Rocca JD; Hall EK; Lennon JT; Evans SE; Waldrop MP; Cotner JB; Nemergut DR; Graham EB; Wallenstein MD Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J. 2015, 9 (8), 1693–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Franzosa EA; Hsu T; Sirota-Madi A; Shafquat A; Abu-Ali G; Morgan XC; Huttenhower C Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat. Rev. Microbiol 2015, 13 (6), 360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Xiao M; Yang J; Feng Y; Zhu Y; Chai X; Wang Y Metaproteomic strategies and applications for gut microbial research. Appl. Microbiol. Biotechnol 2017, 101 (8), 3077–3088. [DOI] [PubMed] [Google Scholar]
- (12).Wallace BD; Roberts AB; Pollet RM; Ingle JD; Biernat KA; Pellock SJ; Venkatesh MK; Guthrie L; O’Neal SK; Robinson SJ; Dollinger M; Figueroa E; McShane SR; Cohen RD; Jin J; Frye SV; Zamboni WC; Pepe-Ranney C; Mani S; Kelly L; Redinbo MR Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem. Biol 2015, 22 (9), 1238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Biteen JS; Blainey PC; Cardon ZG; Chun M; Church GM; Dorrestein PC; Fraser SE; Gilbert JA; Jansson JK; Knight R; Miller JF; Ozcan A; Prather KA; Quake SR; Ruby EG; Silver PA; Taha S; van den Engh G; Weiss PS; Wong GC; Wright AT; Young TD Tools for the Microbiome: Nano and Beyond. ACS Nano 2016, 10 (1), 6–37. [DOI] [PubMed] [Google Scholar]
- (14).Sadler NC; Wright AT Activity-based protein profiling of microbes. Curr. Opin. Chem. Biol 2015, 24, 139–44. [DOI] [PubMed] [Google Scholar]
- (15).Niphakis MJ; Cravatt BF Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem 2014, 83, 341–77. [DOI] [PubMed] [Google Scholar]
- (16).Phillips CI; Bogyo M Proteomics meets microbiology: technical advances in the global mapping of protein expression and function. Cell. Microbiol 2005, 7 (8), 1061–76. [DOI] [PubMed] [Google Scholar]
- (17).Cravatt BF; Wright AT; Kozarich JW Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem 2008, 77, 383–414. [DOI] [PubMed] [Google Scholar]
- (18).Maurice CF; Haiser HJ; Turnbaugh PJ Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013, 152 (1–2), 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hatzenpichler R; Connon SA; Goudeau D; Malmstrom RR; Woyke T; Orphan VJ Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (28), E4069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wallace BD; Wang H; Lane KT; Scott JE; Orans J; Koo JS; Venkatesh M; Jobin C; Yeh LA; Mani S; Redinbo MR Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330 (6005), 831–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Carmody RN; Turnbaugh PJ Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Invest 2014, 124 (10), 4173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pollet RM; D’Agostino EH; Walton WG; Xu Y; Little MS; Biernat KA; Pellock SJ; Patterson LM; Creekmore BC; Isenberg HN; Bahethi RR; Bhatt AP; Liu J; Gharaibeh RZ; Redinbo MR An Atlas of beta-Glucuronidases in the Human Intestinal Microbiome. Structure 2017, 25 (7), 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chauvigne-Hines LM; Anderson LN; Weaver HM; Brown JN; Koech PK; Nicora CD; Hofstad BA; Smith RD; Wilkins MJ; Callister SJ; Wright AT Suite of activity-based probes for cellulose-degrading enzymes. J. Am. Chem. Soc 2012, 134 (50), 20521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tsai CS; Li YK; Lo LC Design and synthesis of activity probes for glycosidases. Org. Lett 2002, 4 (21), 3607–10. [DOI] [PubMed] [Google Scholar]
- (25).Kolb HC; Finn MG; Sharpless KB Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed 2001, 40 (11), 2004–2021. [DOI] [PubMed] [Google Scholar]
- (26).Polaske NW; Kelly BD; Ashworth-Sharpe J; Bieniarz C Quinone Methide Signal Amplification: Covalent Reporter Labeling of Cancer Epitopes using Alkaline Phosphatase Substrates. Bioconjugate Chem. 2016, 27 (3), 660–6. [DOI] [PubMed] [Google Scholar]
- (27).Modica E; Zanaletti R; Freccero M; Mella M Alkylation of amino acids and glutathione in water by o-quinone methide. Reactivity and selectivity. J. Org. Chem 2001, 66 (1), 41–52. [DOI] [PubMed] [Google Scholar]
- (28).Fernandes AD; Reid JN; Macklaim JM; McMurrough TA; Edgell DR; Gloor GB Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Schubert AM; Sinani H; Schloss PD Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. mBio 2015, 6 (4), No. e00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Smits SA; Marcobal A; Higginbottom S; Sonnenburg JL; Kashyap PC Individualized Responses of Gut Microbiota to Dietary Intervention Modeled in Humanized Mice. mSystems 2016, 1 (5), e00098–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Mamantopoulos M; Ronchi F; McCoy KD; Wullaert A Inflammasomes make the case for littermate-controlled experimental design in studying host-microbiota interactions. Gut Microbes 2018, 9 (4), 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Franklin CL; Ericsson AC Microbiota and reproducibility of rodent models. Lab Anim (NY) 2017, 46 (4), 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Laukens D; Brinkman BM; Raes J; De Vos M; Vandenabeele P Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol Rev. 2016, 40 (1), 117–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Devirgiliis C; Zinno P; Perozzi G Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front. Microbiol 2013, 4, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Patel R Enterococcal-type glycopeptide resistance genes in non-enterococcal organisms. FEMS Microbiol. Lett. 2000, 185 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- (36).Guardabassi L; Perichon B; van Heijenoort J; Blanot D; Courvalin P Glycopeptide resistance vanA operons in Paenibacillus strains isolated from soil. Antimicrob. Agents Chemother. 2005, 49 (10), 4227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Dabek M; McCrae SI; Stevens VJ; Duncan SH; Louis P Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 2008, 66 (3), 487–95. [DOI] [PubMed] [Google Scholar]
- (38).Gloux K; Berteau O; El Oumami H; Beguet F; Leclerc M; Dore J A metagenomic beta-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (S1), 4539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.