SUMMARY
For successful infection, bacteriophages must overcome multiple barriers to transport the genome and proteins across the bacterial cell envelope. We use cryo-electron tomography to study infection initiation of phage P22 in Salmonella enterica sv. Typhimurium, revealing how a channel forms to allow genome translocation into the cytoplasm. Our results show free phages initially attaching obliquely to the cell through interactions between the O antigen and two of the six tailspikes; the tail needle also abuts the cell surface. The virion then orients to the perpendicular and the needle penetrates the outer membrane. The needle is released and the internal head protein gp7* is ejected and assembles into an extra-cellular channel extending from the gp10 baseplate to the cell surface. A second protein, gp20, is ejected and assembles into a structure that extends the extra-cellular channel across the outer membrane into the periplasm. Insertion of the third ejected protein gp16 into the cytoplasmic membrane likely completes the overall trans-envelope channel into the cytoplasm. Construction of a trans-envelope channel is an essential step during infection by all short-tailed phages of Gram-negative bacteria because such virions cannot directly deliver their genome into the cell cytoplasm.
Keywords: P22 phage, Cryo-electron tomography, ejection proteins, trans-envelope channel, structural intermediates, infection initiation
INTRODUCTION
Bacteriophages are the most abundant biological entity in the biosphere. About 96% of phages are tailed, implying that tails provide an evolutionary advantage 1. The phage tail is a complex macromolecular machine, responsible for host-cell recognition, attachment, cell envelope penetration, and genome ejection 2–4, although details of how the machine operates remain to be elucidated.
Phage tails are classified into three fundamental types: siphophages possess non-contractile tails of greatly varying length. The tail is usually terminated by a simple baseplate to which a tail fiber(s) or tailspike(s) is attached. The fiber/tailspike constitutes the initial adsorption organelle and, together with baseplate proteins constitute the major part(s) of the ejection machine 3. Myophages have a more complex baseplate structure with a contractile tail sheath. Irreversible adsorption accompanies sheath contraction, which drives the inner tail tube into the infected cell 5,6. Podophages have non-contractile tails that are too short to span the cell envelope. During infection, ϕ29 enzymatically digests the Gram-positive cell wall, a process that ultimately juxtaposes the body of the phage with the cytoplasmic membrane 7,8. T7 contains internal head proteins that are ejected into the cell where they assemble into a cell envelope-spanning structure 9–12. P22 also contains internal head proteins that become cell-associated after infection 13, but the mechanisms of trans-envelope channel formation remain poorly understood for most phages 14.
The short-tailed phage P22 is one of the best-studied model systems for viral assembly and infection 2,15. Both the mature P22 virion and isolated tail machine have been studied extensively by cryo-electron microscopy (cryo-EM) 16–20. The tail machine, which is inserted at a five-fold vertex of the capsid, comprises the portal protein gp1, baseplate proteins gp4 and gp10, the tail needle gp26, and the six tailspikes gp9. Together with crystal structures of gp1, gp4 21, gp9 22–24 and gp26 25, a high resolution cryo-EM structure provided a better understanding of the mature P22 virion and its tail machine 26.
Tailspikes hydrolyze the repeating units of O antigen 23, and hydrolysis is required for infection 27,28. Each tailspike is a trimer and thus has three O antigen binding sites. The crystal structure of gp9 complexed to O antigen reveals the sugar-binding pocket in atomic detail 23. The gp26 needle is a plug that prevents DNA from leaking out of the capsid 29–32, and the protein becomes cell-associated during infection 13. Gp26 shares structural similarity with coiled-coil class I membrane fusion proteins 25 but it is not known how the needle attaches to and penetrates the cell envelope. The non-contractile P22 tail is required, but is insufficient for genome ejection during infection. Three proteins (gp7*, gp16 and gp20) are ejected (E proteins) from the capsid to host cells 13. Full-length gp7 is processed by the PrlC (OpdA) protease, removing the N-terminal 20 residues to yield the packaged form gp7* 34,35. From experiments using osmotic suppression to prevent DNA ejection in vitro, protein ejection during infection was suggested to precede genome translocation 33. All three proteins are essential for infectivity but their exact functions are unknown.
Here we used Salmonella enterica sv. Typhimurium LT2 minicells as hosts for infection by both wild-type and mutant P22. High-throughput cryo-electron tomography (cryo-ET), subtomogram averaging and classification were then employed to reveal structural information that provides new insights into P22 infection initiation.
RESULTS
Visualization of P22 in different infection states
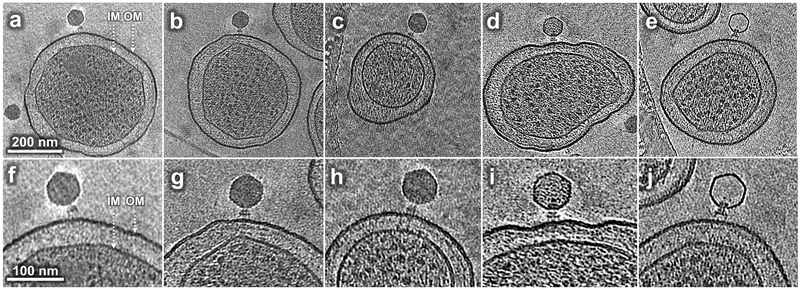
To study changes in virion structure during the initiation of infection, we infected S. Typhimurium minicells with wild-type P22 at 37°C and observed several distinct structures (Fig. 1). Adsorption to minicells was asynchronous but common structures at each sampling time can be interpreted in a pathway of phage development. However, absolute times are altered relative to an infection of normal cells (Table 1). After 3 min infection, many virions are seen bound obliquely to the minicell surface (Fig. 1a, f); this structure is less common later in infection when most particles are perpendicular (Fig. 1b, g; Supplementary Table 1). At longer infection times, electron density spanning the periplasm is visible juxtaposed to the infecting virion. Some particles appear to contain a full complement of DNA (Fig. 1c, h); in others the electron density from encapsidated DNA is less intense (Fig. 1d, i), presumably due to ongoing DNA ejection. The last intermediate observed lacks any significant electron density from DNA in the capsid (Fig. 1e, j) but this class was not common under our experimental conditions and is not further analyzed here.
Figure 1. Tomograms reveal P22 intermediates at different stages of infection.
(a, f) A phage obliquely attached to the minicell envelope after 3 min infection; (b, g) A phage attached to the minicell surface perpendicularly after 15 min infection; (c, h) A phage attached to the minicell surface showing extended density across the cell envelope after 15 min infection; (d, i) a phage with less than a complete genome in its capsid after 30 min infection; (e, j) a phage with an apparently empty capsid after 60 min infection. IM: inner (cytoplasmic) membrane; OM: outer membrane, including O antigen. Comparable tomograms have been obtained in at least three experiments using independent preparations of minicells and phage.
Table 1.
Cryo-ET data and parameters used in this study
| Genotype | Time of Incubation (min) |
Number of Tomograms | Number of extracted sub- tomograms |
Number of particles for class average with trans-envelope channel |
|---|---|---|---|---|
| Wild type | Unadsorbed | 35 | 450 | - |
| 3 | 30 | 235 | - | |
| 15 | 161 | 1,353 | - | |
| 30 | 229 | 3,432 | - | |
| 30-60 | 1,199 | 43,195 | 6,201 | |
| 30-60 | 78 (data collected on a Titan Krios with K2 detector and energy filter; used for the reconstruction shown in Fig. 4b) | 7,803 | 2,250 | |
| Gp16-defective | 30-60 | 247 | 19,115 | 2,886 |
| Gp20-defective | 30-60 | 224 | 10,802 | -- |
| Gp7-defective | 30-60 | 149 | 6,636 | -- |
| Sum | -- | 2,352 | 93,021 | 11,337 |
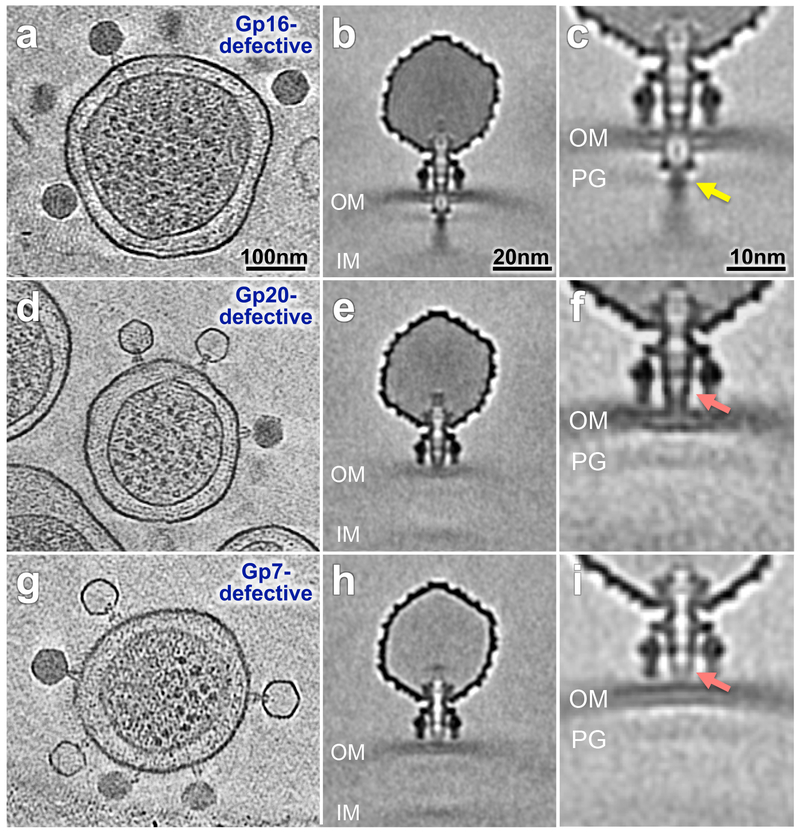
Figure 5. Channel formed by E protein-defective particles.
Tomograms of minicells infected for 60 min by gp16-defective (a-c), gp20-defective (d-f), and gp7-defective particles (g-i). Comparable tomograms have been obtained in five experiments using independent preparations of minicells and mutant phage particles. Class averages with both inner and outer membrane visible (b) gp16-defective, (e) gp20-defective and (h) gp7-defective. Classification reveals details of the interaction between the extracellular channel and the outer membrane (c) gp16-defective, (f) gp20-defective and (i) gp7-defective particles. The arrows in panels c, f and i highlight differences between the three mutants.
Oblique attachment
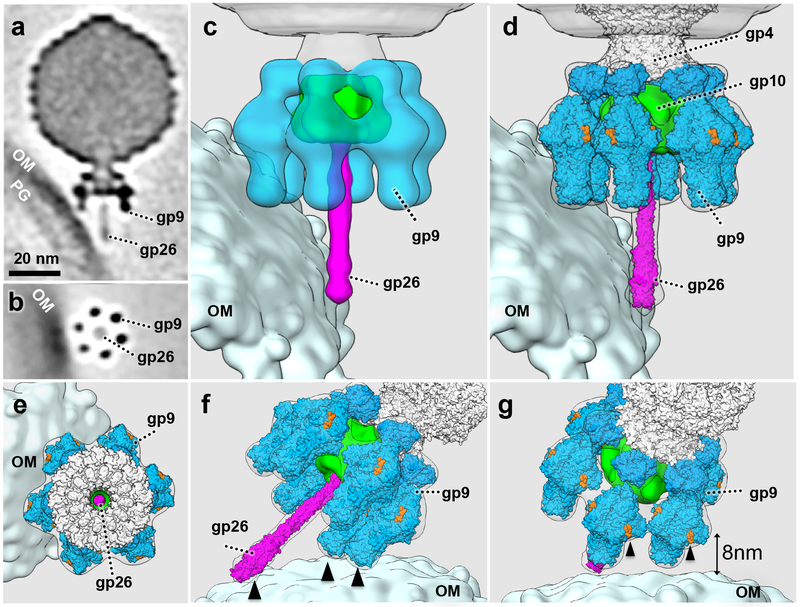
Early in infection, many phages are bound obliquely to the minicell through two adjacent tailspikes, with the tail needle abutting the cell surface (Fig. 1a, f; Fig. 2). To understand this interaction in more detail, we used sub-tomogram classification and averaging of 361 particles to obtain an asymmetric reconstruction of this phage-host complex. 3-D classification reveals that virions are bound to the outer membrane in six distinct orientations (Supplementary Fig. 1), suggesting that any two of the six tailspikes, plus the needle tip are in contact with the cell surface. Compared to free virions (Supplementary Fig. 2), the tail machine of adsorbed phage has not undergone obvious structural changes because the X-ray structures of the P22 tailspike and needle are readily fitted into the averaged map (Fig. 2d-g). A top view from the phage capsid highlights the most exposed O antigen-binding cleft on each gp9 trimer (Fig. 2e). Rotating the view of the adsorbed phage-cell complex clearly shows three points of contact with the cell surface: the tail needle and the tips of two tailspikes (Fig. 2f). The distance between the closest O antigen-binding sites on the two bound tailspikes and the cell surface core lipopolysaccharide (LPS) is ~8 nm (Fig. 2g), about the length of five O antigen repeat units (the limit product for tailspike hydrolysis of O antigen attached to LPS 27). Importantly, this angled orientation of the adsorbed phage is much less common after longer times of infection (Supplementary Table 1), allowing the inferences that it is the earliest structure sufficiently stable for analysis by cryo-ET and that it represents an early infection intermediate.
Figure 2. P22 binds obliquely to the cell surface.
(a) A central section of an averaged structure of P22 (361 particles used, obtained from at least five experiments using independent preparations of minicells and phage) bound obliquely to the minicell surface; (b) a cross-section shows two of the six tailspikes attached to the outer membrane; (c) A 3-D surface view with two bound tailspikes and the needle abutting the outer membrane; (d) Atomic structures of the tailspikes (gp9, PDB ID: 1TYX and 1LKT) and the needle (gp26, PDB ID: 2POH) are docked into the cryo-ET density, and (e) a top view of the complex; (f) and (g) two additional surface views highlight the multiple host-contact sites with the P22 tail machine (black arrows). The O antigen binding sites on gp9 are colored in orange. The closest distance between the O antigen binding sites and the outer membrane is 8nm. Only those sites associated with black arrows can be associated with five repeat units of O antigen, the limit product of tailspike enzyme activity on O antigen attached to LPS 27. OM: outer membrane; PG: peptidoglycan cell wall.
Conformational changes of the tail machine during adsorption
At longer times of infection essentially all particles are oriented perpendicularly (Supplementary Table 1). Considering only those particles that appear to contain a full complement of DNA, applying a local classification around the cell envelope revealed four distinct conformations (Fig. 3). Each structure corresponds to only 1–8% of all the particles analyzed but they likely represent distinct intermediate states during infection. The structures are presented in what we consider to be a logical sequence of events during phage infection although we emphasize that no evidence is presented that the structures actually lie on a direct kinetic pathway.
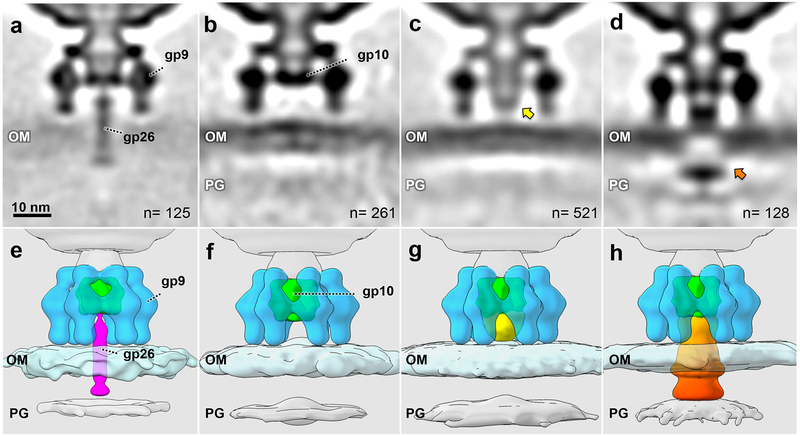
Figure 3. Intermediate structures during commitment to infection.
(a, e) An averaged structure of 125 perpendicularly adsorbed virions with the tail needle penetrating the outer membrane; (b, f) an averaged structure from 261 particles without the needle; (c, g) an averaged structure from 521 particles with an extracellular channel (yellow arrow) between the distal end of the baseplate gp10 and the outer membrane; (d, h) an averaged structure from 128 particles with channel extension and membrane cavity (orange arrow). OM: outer membrane, including O antigen; PG: peptidoglycan cell wall. All intermediates were observed in at least three experiments using independent preparations of minicells and phage.
One structure reveals the tail needle penetrating the outer membrane (Fig. 3a, e). The tail needle appears intact as the crystal structure of gp26 fits well into the density (Supplementary Fig. 3). This likely represents the initial conformational state following virion reorientation and the definitive commitment to infection; particles lacking gp26 are unstable and spontaneously release their packaged genome 29–32. We cannot determine whether the needle has diffused in the outer membrane or whether it has been released into the periplasm. In all other structures the needle has disappeared, initially leaving a region with no electron density between the P22 baseplate gp10 and the cell surface (Fig. 3b, f).
In the third intermediate, electron density is present between the baseplate gp10 and the cell surface, forming what we call the extracellular channel (Fig. 3c, g). This density does not appear to breach the outer membrane. The fourth structure shows that the extracellular channel has extended and now disrupts the outer membrane (Fig. 3d, h). A central slice through this structure reveals a significant cavity in the outer membrane that occupies the center of the infecting tail machine; additional electron density is visible just below the inner surface of the outer membrane. This density does not extend far into the periplasm but may reach the cell wall. The structures of he portal-baseplate complex and tailspikes do not appear to have undergone major conformational changes and we show below that the additional density is due to ejection of E proteins into the cell. The tailspikes have also been drawn closer to the outer membrane (Fig. 3d, h), perhaps suggesting that a secondary binding surface on the tailspikes is interacting more tightly with the cell surface or that the affinity of the E proteins for both the outer membrane and the body of the virion is sufficiently high to pull the latter closer to the cell.
Visualization of the trans-envelope channel
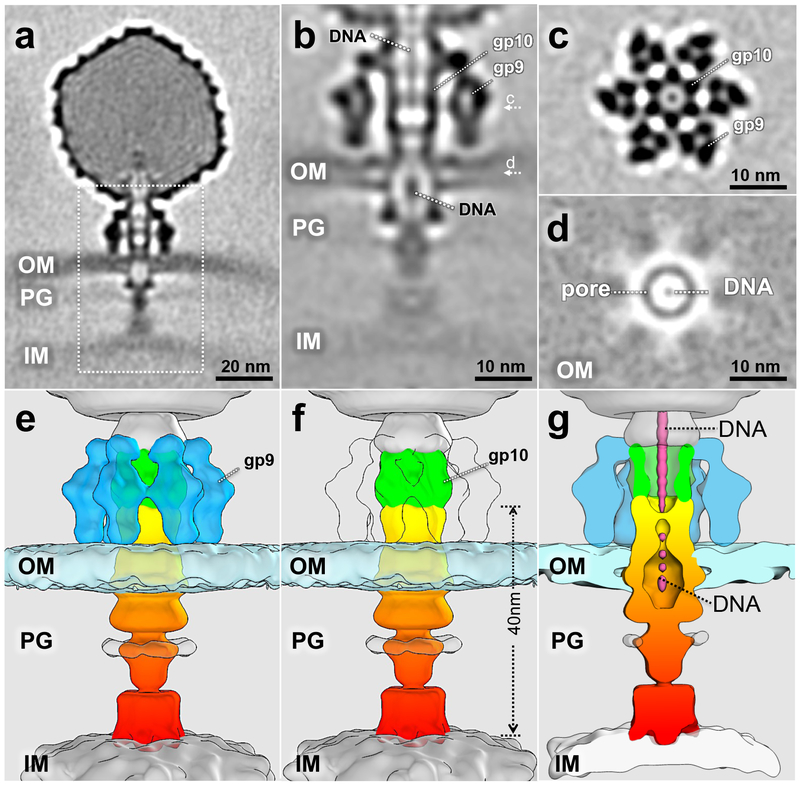
The majority of adsorbed particles observed exhibit a trans-envelope channel bridging the outer and inner membranes. However, this structure exhibited substantial heterogeneity in both orientation and length. To obtain a high-resolution structure of the intact trans-envelope channel, 6,201 particles selected by classification were used to generate an averaged structure (Fig. 4a). Electron density due to the phage genome suggests that most or all DNA is still inside the capsid. After additional local refinement and classification, we obtained a structure of the trans-envelope channel with an overall resolution of 2.7 nm, although the bottom part of the channel remains poorly defined (Fig. 4b-g). A central slice through the structure (Fig. 4b) and a cross-section (Fig. 4c) show that the orientation of the tailspikes and the gp10 baseplate are indistinguishable from those in free or adsorbed virions (cf. Fig. 3 and Supplementary Fig. 2). The channel extends from the distal end of the gp10 baseplate to the outer face of the cytoplasmic membrane, a distance of ~40 nm. The width of the complete channel structure is irregular with an average diameter of ~8 nm, varying from ~5 – 10 nm (Fig. 4f). The outer membrane completely surrounds the cavity or pore of the genome ejection channel (Fig. 4b, d, g). The cavity appears ellipsoid with dimensions of ~4 – 8 nm, the smaller value being co-planar with the membrane. A slice through the center plane of the outer membrane highlights this P22-mediated cavity, which contains electron density consistent with dsDNA in its center (Fig. 4d). In cross-sectional slices, density consistent with dsDNA is also seen extending from the phage capsid into the extracellular channel (Fig. 4b, d, g).
Figure 4. 3-D structure of the trans-envelope channel.
(a) A class averaged structure from 6201 particles, obtained from ten experiments using independent preparations of minicells and phage, showing a complete trans-envelope channel; (b) the trans-envelope channel structure after focused refinement. Density that may correspond to the ejecting genome is highlighted. c and d refer to the plane of the cross-section in the corresponding panels; (c) a cross-section shows the interaction between gp9 and gp10; (d) a cross-section shows the pore structure formed by the E proteins in the outer membrane; (e) and (f) surface views of the tail machine trans-envelope channel; (g) a central section showing DNA in the channel.
Ejection proteins have distinct roles in trans-envelope channel formation
The three intra-capsid E proteins are essential for infectivity. Mutant particles containing a defective E protein in the capsid adsorb normally but do not allow the P22 genome to enter the cell cytoplasm 15. We hypothesized that these proteins are the building elements of the 40 nm trans-envelope channel. To test this idea, we prepared P22 gp7-defective, gp20-defective, and gp16-defective particles, and incubated them with minicells prior to cryo-ET imaging. Representative tomograms are shown for each mutant (Fig. 5a, d, g). Defective gp16-particles form the 40 nm ejection channel (Fig. 5b, c); at our level of resolution no significant difference from the averaged structure of wild-type phage could be detected (cf. Fig. 4a, b). However, unlike in a productive infection by wild-type phage, essentially all mutant virions contain DNA (Fig. 5a).
Infections by gp20-defective or gp7-defective particles do not show electron density spanning the periplasm (Fig. 5e, f, h, i). In contrast to gp16-defective infections, most gp7- and some gp20-defective capsids appear empty and have therefore ejected their DNA (Fig. 5d, g). There is a subtle difference between the two mutant virions: electron density associated with the extracellular channel is less intense and less complete with the gp7-defective particle (cf. Fig. 5h, i with Fig. 5e, f). Additional classification and subtomogram averaging of the gp7-defective and gp20-defective particles revealed that the extracellular channel of the gp7-defective particle does not fully interact with the outer membrane (Fig. 5i), whereas density associated with the gp20-defective particle is clearly contiguous. We tentatively ascribe the extracellular channel to the ejected gp7* (the mature form of gp7) and propose that gp20 is responsible for the majority, perhaps all, of the structure in the periplasm.
DISCUSSION
Most bacteriophages utilize elaborate tail machines for host-cell recognition, attachment, cell envelope penetration, and DNA ejection. Despite strikingly different tail morphologies and different strategies for genome translocation, all phages of Gram-negative bacteria face similar barriers: two membranes and a cell wall. P22 is one of the best-studied model systems for viral assembly and infection 2,15. Here we used Salmonella minicells as a host to study P22 adsorption and DNA translocation. Our structures provide the first evidence that phage P22 virion undergoes coordinated conformational changes in assembling a 40 nm trans-envelope channel to allow genome ejection into the cell cytoplasm. A simplified overview of the infection process revealed by this study is shown in Supplementary Figure 4, although we stress that the pathway depicted has not been rigorously established.
Initial adsorption
P22 infection starts with the specific interaction between the tailspike and the repeating units of O antigen on the distal end of the LPS. At least five repeats are necessary for hydrolysis on the Salmonella cell surface 27,36, and hydrolysis is essential for a productive infection 27,28. The majority of O antigen chains contain 19–34 repeats 37,38 that could extend 27–48 nm away from the cell surface. However, the rate of tailspike endorhamnosidase activity on oligosaccharide substrates is only ~two per min 39, which is too slow to facilitate efficient P22 infection. As adsorption of P22 to O antigen on bacteria also appears irreversible 40,41, it was proposed that adsorbed phages approach the host cell surface by rapid release and rebinding 39. Alternatively, it is possible that enzymatic activity is faster on longer, native O antigen chains than on model substrates, or that coordination between the six tailspikes on a virion leads to an increased rate of hydrolysis.
The homotrimeric tailspike contains two domains with a hinge between the enzymatic C-terminal domain and the N-terminal domain that binds to the virion 22,24. In the mature virion the hinge is bent so that the elongated C-terminal domain points approximately orthogonally away from the head. Each tailspike has three enzymatic active sites 22–24; receptor O antigen bound in these sites would orient the virion so that the tail is closest to the bacterial surface if the flexible polysaccharide is stretched outward from the cell surface.
P22 particles initially bind to the cell surface at an oblique angle. This seems obvious in hindsight, even obligatory. The tail needle extends 13.7 nm below the distal plane of the tailspikes and the common cartoon of phage infection cannot reflect the initial specific interaction of P22 with the cell surface. Interestingly, ϕ29, whose tail knob extends below the plane of its appendages, also initially binds to a Bacillus subtilis cell at an oblique angle 7. In contrast, SP6, whose tailspikes are homologous to P22 but which do not extend below the plane of the tail knob, was not observed to bind obliquely 42. There are almost certainly earlier interactions between phage and cell that are non-specific, or less so than the obliquely bound state; for example, the capsid portion of the phage may make the initial collision. However, any such structures may be too unstable to be captured in our experiments.
The central needle and two adjacent tailspikes form an asymmetric tripod that interacts with O antigen and the outer membrane (Fig. 2). Our data show that tailspikes likely interact with both O antigen and core LPS and they provide the first structural evidence that the P22 needle tip interacts directly with the outer membrane. It is not known whether the tip is bound to a specific moiety. The P22 needle tip does not confer host specificity but alterations in its sequence can cause a delay in the infection process 43. The crystal structure of tailspike bound to O antigen 23 fits well into the electron density of an obliquely attached P22 virion and reveals that in this orientation only one of the three active sites directly faces the outer membrane. At this stage of infection only two adjacent spikes are seen closely interacting with the outer membrane, suggesting that two enzymatic active sites (one on each spike) are making contacts with O antigen. Assuming that O antigen chains are fully extended away from the LPS, obliquely adsorbed phages must either have hydrolyzed several repeat units or have undergone rapid cycles of bind and release for two tailspikes on the infecting P22 to approach ~8 nm from the outer core LPS surface (Fig. 2g). Bind and release 39 seems likely, particularly if at any given instant during this early stage of specific adsorption only two of the 18 active sites on the tailspikes are actually bound to O antigen.
An infection intermediate where P22 is bound obliquely to the cell surface provides an opportunity for exposed O antigen-binding sites on other tailspikes to be recognized by the flexible chains of their substrate. It has long been established that at least three tailspikes must be bound to heads for a productive infection 40,44, and thus at least three tailspikes must bind the cell. Binding of additional tailspikes to O antigen, beyond the pair closest to the cell surface when the phage is obliquely bound, likely requires reorientation of the virion. Reorientation also renders more accessible all three binding sites on each tailspike and thus may be accompanied by an increase from two to as many as 18 tailspike-O antigen complexes. Reorientation of the virion must also force the tail needle through the outer membrane. The energetics of virion reorientation and needle penetration are unknown but will likely include not only O antigen binding but also its hydrolysis.
Ejection of the needle and formation of the 40nm transmembrane channel
Tailspikes are attached to the cylindrical tail hub formed by the dodecameric portal gp1, dodecameric gp4, hexameric gp10 and the trimeric needle gp26. The needle plugs the central channel of the tail hub 25, and particles lacking gp26 spontaneously release encapsidated DNA 29–32. It has been suggested that the P22 tailspikes might spread out like the petals of a flower in bloom 18,24 so that the bottom of the baseplate could abut the host cell outer membrane like the tail knob of podophages T7 and SP6 12,42. However, the structures and orientations of gp10, gp4, gp9 and gp1 do not detectably change during infection from those in the mature virion, and needle release therefore results in a gap between the bottom of the baseplate and the cell surface. This suggests either that subtle changes, undetectable with our current cryo-ET approach, occur in the tail to cause needle ejection or that as a consequence of the needle tip penetrating the outer membrane a signal is transmitted along the entire length of the needle to its connection with the baseplate.
The gap between the gp10 baseplate and the cell surface resulting from needle release is subsequently filled by what we have called the extracellular channel, one that may be necessary to protect the phage genome from attack by environmental nucleases during its translocation into the cell interior. This extracellular channel is subsequently extended and assembled into a 40 nm structure that spans the cell envelope extending from the tail hub to the inner membrane. The proteins that constitute this structure are the E proteins that, in mature virions, are inside the phage capsid.
Electron density spanning the cell envelope appears the same following infection by wild-type or gp16-defective particles, suggesting that gp16 is embedded in the cytoplasmic membrane. This conclusion is in agreement with partitioning of gp16 into the S. Typhimurium membrane fraction and forming a dye-permeable channel in liposomes 45. No electron density was found in the periplasm following infection by gp20- or gp7-defective particles. However, the gp7-defective extracellular channel is less complete and there is no clear fusion with the outer membrane. This suggests that the extracellular channel contains gp7*. Gp20-defective particles appear to form a complete extracellular channel but the cavity in the outer membrane is not formed. Ejected gp20 therefore likely creates this cavity in addition to forming the structure that spans the periplasm. This last conclusion is bolstered by the structure of the purified and in vitro refolded Sf6 gp12 46, the homologous protein coded by a close relative of P22. However, further studies are necessary to evaluate more fully the roles of the E proteins when inside the cell.
MATERIAL AND METHODS
Bacterial and phage strains
P22 strains were grown on IJ612 (= MS1868 [S. Typhimurium LT2 leuA414(Am) hsdSB (R–M+) (Fels2–)]) or its isogenic supE (Su2+) derivative IJ613 (= MS1883). Both strains were originally from M. M. Susskind. The lysis-defective phage P22 13amH101 was used to prepare wild-type virions. P22 7amH1363 (codon 78), 7amH1375 (codon 155), 16amN121 (codon 172), and 20amN20 (codon 79) have been described 47,48, and were obtained, some as double mutants with 13amH101, from M. M Susskind, S. R. Casjens, and A. R. Poteete. Gp7-, gp20- and gp16-defective particles were made in MS1868 and purified by a discontinuous, followed by an equilibrium, CsCl-density gradient. Phages were stored in CsCl and dialyzed immediately prior to use for infection and cryo-ET sample preparation.
Minicells and preparation of frozen infected samples
To obtain high-resolution structures of phage-infected cells by cryo-ET it is necessary to reduce sample thickness. We achieved this by preparing minicells, whose diameter is 30–50% that of short axis of the mother cell. Freshly prepared minicells are metabolically active 49, and have been shown to support phage growth 50,51.
IJ2299 (= TH16943), a wild-type LT2 strain except that the araBAD coding sequences had been replaced with ftsZ+ has been described 42. Growth in the presence of arabinose results in the production of minicells. IJ2299 was grown at 37˚C in 50 ml LB medium containing 0.2% arabinose and minicells isolated as reported 42. Briefly, a 50ml overnight culture was centrifuged at 10,000 × g for 10min. The supernatant was centrifuged again at 40,000 x g for 20min and the minicell pellet was resuspended in 30 μl LB medium containing 400 μg/ml rifampicin and incubated at 37°C for 5min. Minicells were then infected at a multiplicity of ~5–10. Rifampicin serves to inhibit gene expression that potentially could affect minicell structure during the infection. Samples were removed at various times, mixed with 10 nm colloidal gold and deposited onto freshly glow-discharged, holey carbon grids for 1 min. Grids were blotted with filter paper and rapidly frozen in liquid ethane, cooled with liquid nitrogen, using a homemade gravity-driven plunger apparatus. Grids were stored under liquid nitrogen.
Cryo-ET data collection
Frozen grids were imaged at −170 °C on a cryo-electron microscope (Polara, FEI Company) equipped with a K2 direct electron detector (Gatan) and operated at 300kV with a magnification of 15,500x, resulting in an effective pixel size of 2.6 Å. Images were collected automatically using SerialEM 52 in dose fraction mode. The cumulative electron dose for each single-axis tilt series was ~50e/Å2, distributed over 35 images and covering an angular range of −51° to +51° with increments of 3°. Defocus ranged between −2 and −6 μm. For refinement of the trans-envelope structure, ~70 tilt series were collected on a Titan Krios, equipped with an energy filter (Gatan) with −3 μm defocus; the effective pixel size was 2.7 Å.
Tomogram reconstruction
Raw images were processed using MotionCor2 53. The tilt series were aligned automatically using IMOD 54. Defocus determination and CTF correction were performed using IMOD 54,55. CTF-corrected tomograms were generated by using TOMO3D 56.
Subtomogram analysis
Free phage particles were manually picked initially to serve as a template for automatic particle picking using Pytom 57. In total, 93,021 particles were extracted from 2,352 tomographic reconstructions (Table 1). After translational and rotational alignment based on the global average, hierarchical ascendant classification was used to analyze the structures of trans-envelope channel as described previously 5,12,42.
Three-Dimensional Visualization and Modeling
IMOD was used to take snapshots of 2-D slices from 3-D tomograms and UCSF Chimera 58 was used for surface rendering of 3-D averaged structures. The cryo-EM structure of the isolated tail machine (EMDB-5051) 17 was first docked into the cryo-ET density, then the crystal structures of gp1 and gp4 21 (PDB ID: 3LJ5 and 4V4K), gp26 25 (PDB ID: 2POH), gp9 with the sugar-binding pocket 23,24 (PDB ID: 1TYX and 1LKT) were fitted into the tailspike density map using UCSF Chimera.
Data availability
The structures derived from cryo-ET and sub-tomogram averaging were deposited to the Electron Microscopy Data Bank (EMDB). The accession codes are EMDB-9006, EMDB-9007, EMDB-9008, EMDB-9009 and EMDB-9010.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to S. Casjens for providing phage strains and amber mutant sequence data. We also thank M. M. Susskind and A. R. Poteete for phage and bacterial strains. This work was supported by GM124378 and GM110243 to IJM and JL. CW, JT and JL were also supported in part by grants AI087946 from the NIAID and AU-1714 from the Welch Foundation.
Footnotes
Competing Interests
The authors declare no competing interests.
REFERENCES
- 1.Ackermann HW Bacteriophage observations and evolution. Res Microbiol 154, 245–251, doi: 10.1016/S0923-2508(03)00067-6 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Casjens SR & Molineux IJ Short noncontractile tail machines: adsorption and DNA delivery by podoviruses. Adv Exp Med Biol 726, 143–179, doi: 10.1007/978-1-4614-0980-9_7 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Davidson AR, Cardarelli L, Pell LG, Radford DR & Maxwell KL Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol 726, 115–142, doi: 10.1007/978-1-4614-0980-9_6 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Leiman PG & Shneider MM Contractile tail machines of bacteriophages. Adv Exp Med Biol 726, 93–114, doi: 10.1007/978-1-4614-0980-9_5 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Hu B, Margolin W, Molineux IJ & Liu J Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proceedings of the National Academy of Sciences of the United States of America 112, E4919–4928, doi: 10.1073/pnas.1501064112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor NMI et al. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533, 346–+ (2016). [DOI] [PubMed] [Google Scholar]
- 7.Farley MM, Tu J, Kearns DB, Molineux IJ & Liu J Ultrastructural analysis of bacteriophage Phi29 during infection of Bacillus subtilis. J Struct Biol 197, 163–171, doi: 10.1016/j.jsb.2016.07.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Gui M, Wang D & Xiang Y The bacteriophage varphi29 tail possesses a pore-forming loop for cell membrane penetration. Nature 534, 544–547, doi: 10.1038/nature18017 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Kemp P, Garcia LR & Molineux IJ Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 340, 307–317 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Kemp P, Gupta M & Molineux IJ Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Molecular microbiology 53, 1251–1265 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Chang CY, Kemp P & Molineux IJ Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology 398, 176–186, doi: 10.1016/j.virol.2009.12.002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu B, Margolin W, Molineux IJ & Liu J The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 339, 576–579, doi: 10.1126/science.1231887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israel V E proteins of bacteriophage P22. I. Identification and ejection from wild-type and defective particles. J Virol 23, 91–97 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molineux IJ & Panja D Popping the cork: mechanisms of phage genome ejection. Nature reviews. Microbiology 11, 194–204, doi: 10.1038/nrmicro2988 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Susskind MM & Botstein D Molecular genetics of bacteriophage P22. Microbiol Rev 42, 385–413 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hryc CF et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proceedings of the National Academy of Sciences of the United States of America 114, 3103–3108, doi: 10.1073/pnas.1621152114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander GC et al. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure 17, 789–799, doi: 10.1016/j.str.2009.04.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang J, Weigele P, King J, Chiu W & Jiang W Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14, 1073–1082, doi: 10.1016/j.str.2006.05.007 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Lander GC et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312, 1791–1795, doi: 10.1126/science.1127981 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Tang J et al. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure 19, 496–502, doi: 10.1016/j.str.2011.02.010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olia AS, Prevelige PE Jr., Johnson JE & Cingolani G Three-dimensional structure of a viral genome-delivery portal vertex. Nature structural & molecular biology 18, 597–603, doi: 10.1038/nsmb.2023 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinbacher S et al. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265, 383–386 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Steinbacher S et al. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proceedings of the National Academy of Sciences of the United States of America 93, 10584–10588 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinbacher S et al. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. Journal of molecular biology 267, 865–880, doi: 10.1006/jmbi.1997.0922 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olia AS, Casjens S & Cingolani G Structure of phage P22 cell envelope-penetrating needle. Nature structural & molecular biology 14, 1221–1226, doi: 10.1038/nsmb1317 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Pintilie G, Chen DH, Haase-Pettingell CA, King JA & Chiu W Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage. Biophys J 110, 827–839, doi: 10.1016/j.bpj.2015.11.3522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andres D et al. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem 285, 36768–36775, doi: 10.1074/jbc.M110.169003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berget PB & Poteete AR Structure and functions of the bacteriophage P22 tail protein. J Virol 34, 234–243 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King J, Lenk EV & Botstein D Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. Journal of molecular biology 80, 697–731 (1973). [DOI] [PubMed] [Google Scholar]
- 30.Lenk E, Casjens S, Weeks J & King J Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology 68, 182–199 (1975). [DOI] [PubMed] [Google Scholar]
- 31.Botstein D, Waddell CH & King J Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. Journal of molecular biology 80, 669–695 (1973). [DOI] [PubMed] [Google Scholar]
- 32.Strauss H & King J Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. Journal of molecular biology 172, 523–543 (1984). [DOI] [PubMed] [Google Scholar]
- 33.Jin Y et al. Bacteriophage P22 ejects all of its internal proteins before its genome. Virology 485, 128–134, doi: 10.1016/j.virol.2015.07.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conlin CA, Trun NJ, Silhavy TJ & Miller CG Escherichia coli prlC encodes an endopeptidase and is homologous to the Salmonella typhimurium opdA gene. J Bacteriol 174, 5881–5887 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conlin CA, Vimr ER & Miller CG Oligopeptidase A is required for normal phage P22 development. J Bacteriol 174, 5869–5880 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kastowsky M, Gutberlet T & Bradaczek H Molecular modelling of the three-dimensional structure and conformational flexibility of bacterial lipopolysaccharide. J Bacteriol 174, 4798–4806 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldman RC & Leive L Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli 0111 and Salmonella typhimurium LT2. Eur J Biochem 107, 145–153 (1980). [DOI] [PubMed] [Google Scholar]
- 38.Palva ET & Makela PH Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur J Biochem 107, 137–143 (1980). [DOI] [PubMed] [Google Scholar]
- 39.Baxa U et al. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys J 71, 2040–2048, doi: 10.1016/S0006-3495(96)79402-X (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Israel V A model for the adsorption of phage P22 to Salmonella typhimurium. J Gen Virol 40, 669–673, doi: 10.1099/0022-1317-40-3-669 (1978). [DOI] [PubMed] [Google Scholar]
- 41.Israel V, Rosen H & Levine M Binding of bacteriophage P22 tail parts to cells. J Virol 10, 1152–1158 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu JG et al. Dual host specificity of phage SP6 is facilitated by tailspike rotation. Virology 507, 206–215, doi: 10.1016/j.virol.2017.04.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leavitt JC et al. The tip of the tail needle affects the rate of DNA delivery by bacteriophage P22. PloS one 8, e70936, doi: 10.1371/journal.pone.0070936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Israel V Role of the bacteriophage P22 tail in the early stages of infection. J Virol 18, 361–364 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez GL, Huynh B, Slater M & Maloy S Transport of phage P22 DNA across the cytoplasmic membrane. J Bacteriol 191, 135–140, doi: 10.1128/JB.00778-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao H et al. Structure of a Bacterial Virus DNA-Injection Protein Complex Reveals a Decameric Assembly with a Constricted Molecular Channel. PloS one 11, e0149337, doi: 10.1371/journal.pone.0149337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Botstein D, Chan RK & Waddell CH Genetics of bacteriophage P22. II. Gene order and gene function. Virology 49, 268–282 (1972). [DOI] [PubMed] [Google Scholar]
- 48.Poteete AR & King J Functions of two new genes in Salmonella phage P22 assembly. Virology 76, 725–739 (1977). [DOI] [PubMed] [Google Scholar]
- 49.Farley MM, Hu B, Margolin W & Liu J Minicells, Back in Fashion. J Bacteriol 198, 1186–1195, doi: 10.1128/JB.00901-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reeve JN Bacteriophage infection of minicells: a general method for identification of “in vivo” bacteriophage directed polypeptide biosynthesis. Mol Gen Genet 158, 73–79 (1977). [DOI] [PubMed] [Google Scholar]
- 51.Roozen KJ, Fenwick RG Jr. & Curtiss R 3rd. Synthesis of ribonucleic acid and protein in plasmid-containing minicells of Escherichia coli K-12. J Bacteriol 107, 21–33 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51, doi: 10.1016/j.jsb.2005.07.007 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastronarde DN & Held SR Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol 197, 102–113, doi: 10.1016/j.jsb.2016.07.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76, doi: 10.1006/jsbi.1996.0013 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Agulleiro JI & Fernandez JJ Tomo3D 2.0--exploitation of advanced vector extensions (AVX) for 3D reconstruction. J Struct Biol 189, 147–152, doi: 10.1016/j.jsb.2014.11.009 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Hrabe T et al. PyTom: a python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J Struct Biol 178, 177–188, doi: 10.1016/j.jsb.2011.12.003 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures derived from cryo-ET and sub-tomogram averaging were deposited to the Electron Microscopy Data Bank (EMDB). The accession codes are EMDB-9006, EMDB-9007, EMDB-9008, EMDB-9009 and EMDB-9010.