Abstract
West Nile virus (WNV) is an emerging mosquito-borne flavivirus, related to dengue virus and Zika virus. To gain insight into host pathways involved in WNV infection, we performed a systematic affinity-tag purification mass spectrometry (AP-MS) study to identify 259 WNV-interacting human proteins. RNAi screening revealed 26 genes that both interact with WNV proteins and influence WNV infection. We found that WNV, dengue and Zika virus capsids interact with a conserved subset of proteins that impact infection. These include the exon-junction complex (EJC) recycling factor, PYM1, which is antiviral against all three viruses. The EJC has roles in nonsense-mediated decay (NMD), and we found that both the EJC and NMD are antiviral and the EJC protein RBM8A directly binds WNV RNA. To counteract this, flavivirus infection inhibits NMD and the capsid-PYM1 interaction interferes with EJC protein function and localization. Depletion of PYM1 attenuates RBM8A binding to viral RNA, suggesting that WNV sequesters PYM1 to protect viral RNA from decay. Together, these data suggest a complex interplay between the virus and host in regulating NMD and the EJC.
Introduction
West Nile virus (WNV) is a member of the flavivirus genus, comprised of globally important emerging and re-emerging pathogens, including dengue virus (DENV), Zika virus (ZIKV), Japanese encephalitis virus (JEV) and Yellow Fever virus (YFV) 1. Flavivirus are small, positive-sense RNA viruses that are translated as a single polyprotein and processed into structural (capsid, prM, Env) and nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) proteins. During infection, flaviviruses utilize host machinery to carry out replication and must subvert antiviral Type I interferon and cell-intrinsic pathways. Screening strategies have provided a wealth of information regarding host restriction and susceptibility factors in WNV infection 2–9. However, it is unclear if these factors interface with viral proteins to impact infection, and the plethora of factors identified is likely incomplete.
Here, we combine a mass spectrometry-based approach for mapping protein-protein interactions with genetic screening to identify host factors that physically interact with WNV proteins and influence infection. This approach is a powerful strategy to uncover mechanisms of viral infection and subversion of cell-intrinsic restriction pathways 10–14. In total, we identified 259 WNV-interacting host proteins by co-immunoprecipitation of WNV proteins coupled with in-solution mass spectrometry 12,15. As WNV is one of a larger genus of flaviviruses, we compared our WNV-interactome with DENV and ZIKV and discovered a statistically significant overlap between flavivirus capsid proteins (p<0.01).
We selected 122 host factors, including conserved capsid interactors, for siRNA screening to determine their role in WNV, DENV and ZIKV infection. We identified 26 genes that impact WNV infection; 13 were specific to WNV, while the remaining 13 impact WNV and DENV or ZIKV. In total, we identified 40 genes with a phenotype in at least one flaviviruses. Notably, eight WNV-interacting proteins impacted infection of all three viruses. We focused on PYM1, which interacts with flavivirus capsids. PYM1 is an exon-junction complex (EJC)-associated protein with a role in nonsense-mediated decay (NMD), a cellular RNA degradation pathway 16–21. We show that flaviviruses inhibit nonsense-mediated decay and components of both the EJC and NMD pathway are antiviral against WNV, DENV and ZIKV. EJC association with mRNA elicits NMD and we demonstrate that the EJC protein RBM8A binds to WNV RNA, suggesting that NMD targets viral RNA. Moreover, WNV antagonizes this process through PYM1, relocalizing the EJC and inhibiting interactions with viral RNA. Collectively, the results of our proteomics/genomics approach identifies new facets of the virus-host arms race.
RESULTS
Constructing an WNV-Host Protein-Protein Map
To identify human proteins that physically interact with WNV proteins, we cloned each of the 10 WNV proteins expressing a C-terminal 2X-Strep affinity tag (Fig. 1a). Using immunoblotting, we verified expression of the affinity-tagged WNV proteins, as well as expression of DENV and ZIKV capsid, by transient transfection into HEK293 cells (Supplementary Fig. 1). Proteins not robustly detected by western blot or mass spectrometry (NS2A, NS2B) were excluded from further analyses (Supplementary Fig. 2). WNV proteins were affinity-purified and eluted to collect WNV bait and associated host prey proteins (Fig. 1b). Eluates were analyzed by western blotting and silver staining (Fig. 1c), followed by in-solution mass spectrometry. In total, we identified ~7000 co-purifying host proteins from HEK293 cells from quadruplicate replicates for all WNV baits (Supplementary Table 1).
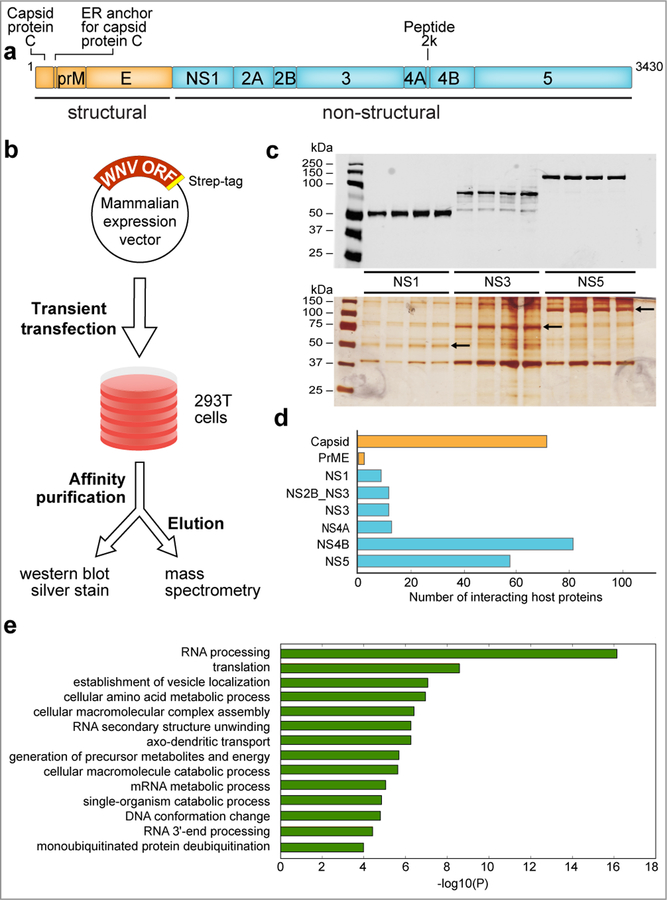
Fig. 1. Generation of the WNV-host protein interaction map.
a, Schematic of WNV genome and the 10 proteins derived from the viral polyprotein. b, Flowchart outlining the experimental strategy used to generate the WNV-host protein-protein interactome. c, Anti-strep western blot (upper panel) and silver staining (lower panel) of representative WNV (NS1, NS3, NS5) proteins after transient transfection and affinity purification from HEK293 cells. Silver-stained bands corresponding to WNV bait proteins are indicated by arrows. Experiments for western blot and silver stain images were repeated four times with similar results. d, The number of interacting host proteins present in the final WNV-host protein-protein interaction map is indicated for each bait protein. e, Bar graph representing enriched terms using the following resources: KEGG Pathway, GO Biological Processes, GO Molecular Function, Reactome Gene Sets, Canonical Pathways and CORUM biological processes. Further details of enrichment analysis are outlined in the Methods section. The magnitude of the bars in the graph represent statistical significance (-Log(P) value). For all enrichment analyses, n = four biologically independent experiments. This data was generated with all host interacting proteins for all WNV bait proteins that were above our selected cut-off (Supplementary Table 2) using Metascape. P-values were calculated based on accumulative hypergeometric distribution and the most statistically significant term within a cluster was chosen as the representative category for each cluster.
High-Confidence WNV-Interacting Host Proteins
To prioritize reproducible, bait-specific PPIs, we analyzed four replicates for each bait protein with two different AP-MS scoring algorithms: mass spectrometry interaction statistics (MIST) 12 and COMPPASS 15. The WNV proteins with the highest number of interacting host proteins were capsid, NS4B and NS5 (Fig. 1d). To identify the pathways targeted by WNV, we performed gene ontology (GO) enrichment analysis on host proteins, revealing an enrichment of interactors associated with RNA processing, vesicle localization, and ER protein processing, consistent with the intricate relationship between flaviviruses and the ER (Fig. 1e; Supplementary Table 3)22. Following our analysis, we identified 259 high-confidence interacting host proteins for WNV capsid, prME, NS1, NS2B-NS3, NS3, NS4A, NS4B and NS5 (Fig. 2; Supplementary Table 2). Our high-confidence interactome revealed a total of 89 (~35%) of the WNV-interacting proteins identified in this study also interacted with the analogous viral proteins in previous flavivirus AP-MS studies, and 31 (~12%) affected flavivirus infection in previous genetic screens (Supplementary Table 4) 2–9.
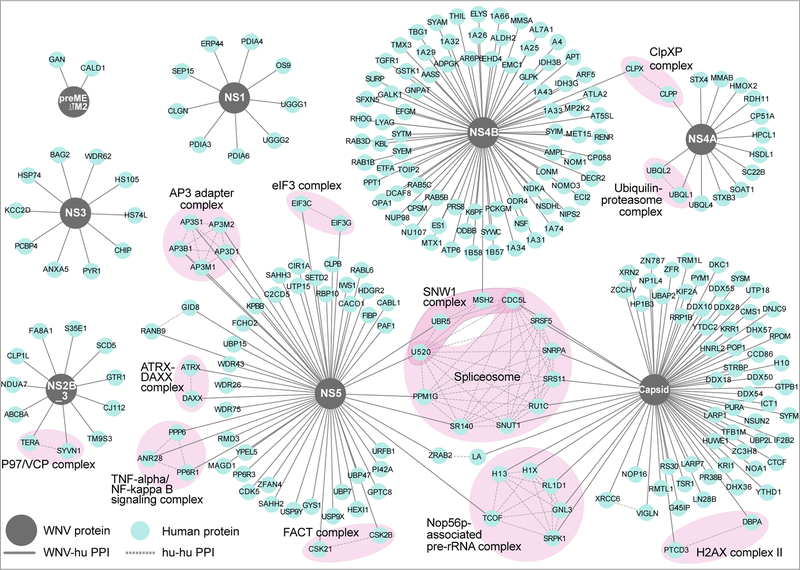
Fig. 2. The WNV-human protein-protein interaction network.
259 high confidence WNV-human protein-protein interactions are displayed. Viral baits (grey circles), human prey (blue circles), virus-host interactions (solid grey lines), host-host interactions (dashed grey lines). The virus-host interaction map was derived from four biologically independent experiments.
Host Processes and Complexes interact with WNV proteins
We used a database of known human-human protein interactions to overlay additional connections between host proteins to identify multiprotein complexes (Fig. 2). We identified several complexes that interact with WNV, including interactions between NS5 and the AP3 adapter complex, previously shown to be required for flaviviral infection 8,23. We also find components of the spliceosome interacting with WNV NS5 and capsid; a recent protein-protein interaction study found connections between DENV NS5 and core components of the host splicing machinery 24. Interestingly, we identify three members of the TNF-alpha/NF-kappa B signaling complex (ANR28, PPP6, PP6R1) as interaction partners of WNV NS5.
Capsid interactions are conserved between flaviviruses
To identify shared flavivirus-host protein interactions, we compared the WNV host interactors with similar datasets from DENV (serotype 2 strain 16681) and ZIKV (French Polynesia 2013 H/FP/2013). This revealed that the most significant overlap between all three viruses was shared between capsid-interacting host proteins (p<0.01, Fig. 3a,b; Supplementary Table 5). An examination of the protein sequence similarity between the capsid proteins of these flaviviruses revealed ~ 40% similarity by pairwise analysis (Fig. 3c). In contrast, NS5 is the most conserved across these flaviviruses (~70%) (Supplementary Fig. 3). Although the conservation between capsid proteins is not high, the significance of overlap between interactors suggests significant structural and functional conservation between these proteins.
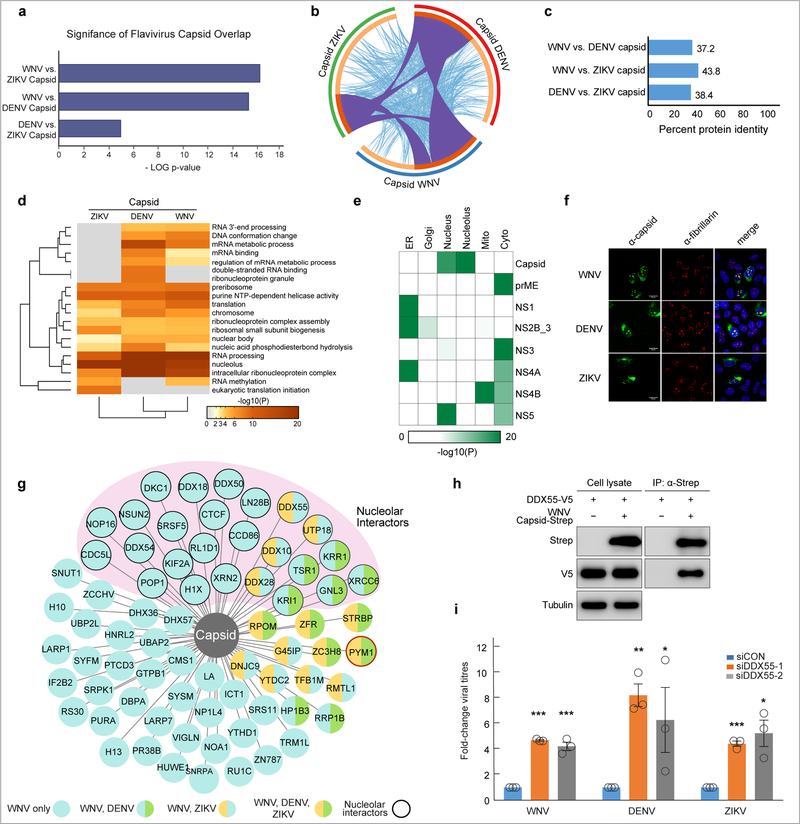
Fig. 3. Flaviviral capsid proteins interact with overlapping host factors.
a, An analysis of flavivirus bait-interacting proteins reveals a highly significant overlap between capsid-interacting host proteins for WNV, DENV and ZIKV. Results were derived from four biologically independent experiments and all p-values were calculated using the hypergeometric test. b, A circos plot to visualize flavivirus capsid-interacting host protein overlap. The outermost circles are proportional to the number of capsid-interacting host proteins for each virus (WNV (blue), DENV (red), ZIKV (green). Inner circles indicate proportion of capsid interacting proteins unique to each virus (light orange) and shared between flaviviruses (dark orange). Interconnecting purple lines indicate shared interacting host proteins, while interconnecting light blue lines indicate shared Gene Ontology terms between interacting host proteins. c, Pairwise comparison showing the percentage of amino acid conservation between flavivirus capsid proteins. d, Heat map representing enriched KEGG Pathway, GO Biological Processes, GO Molecular Function, Reactome Gene Sets, Canonical Pathways and CORUM biological processes of the host factors interacting with indicated flavivirus capsid proteins. The colors represent statistical significance (-Log(P) value) as indicated by the accompanying scale. e, Heat map visualizing the enrichment analysis of the GO Cellular Compartment localization of WNV-interacting host proteins for each bait. The colors represent statistical significance (-Log(P) value) as indicated by the accompanying scale. For d and e, p-values were calculated based on accumulative hypergeometric distribution and the most statistically significant term within a cluster was chosen as the representative category for each cluster. For all analyses, data are derived from four biologically independent experiments. f, Localization of flavivirus capsid proteins in infected cells. WNV, DENV and ZIKV capsid are shown in green (left panels) using virus-specific capsid antibodies at 24h post-infection. The nucleolus is shown in red (center panel) using an antibody recognizing the nucleolar protein, fibrillarin. Merged images demonstrating the overlap between flavivirus capsid and the nucleolus are shown in the right panels, with nuclear staining (Hoescht) in blue. Images are representative of three independent biological experiments repeated with similar results. g, WNV capsid-host interacting map indicates those host proteins that are WNV-specific (light blue circles), shared between WNV and DENV (light blue and green circles), shared between WNV and ZIKV (light blue and orange circles), shared between all three viruses (green and orange circles). Capsid interacting proteins that localize to the nucleolus are indicated with a thick outer line and grouped together in light pink. The capsid-interacting protein PYM1 is indicated in red. h, Immunoprecipitations in HEK293 cells expressing the indicated constructs (WNV capsid-2X strep, DDX55-V5) were performed with Strep-Tactin resin (IBA) and blotted with the indicated antibodies. Results are representative of two biologically independent experiments. i, Infectious WNV, DENV and ZIKV viral particles from DDX55-depleted and control U2OS cells were measured by TCID50 in BHK cells 24h post-infection. Shown is the mean±SE; n = three biologically independent experiments repeated with similar results. Statistical significance was calculated using an unpaired, one-tailed student’s t-test; * p < 0.05, ** p < 0.005, *** p < 0.0005.
Capsids interact with nucleolar proteins in the nucleolus
GO enrichment analysis for flavivirus capsid interactors revealed significant overlap in categories including RNA processing, ER protein processing and nucleolar proteins (Fig. 3d; Supplementary Table 6). We performed subcellular localization and GO enrichment analysis on the interactors for each WNV bait protein (Fig. 3e; Supplementary Fig. 5, Supplementary Table 7). This revealed that the nonstructural proteins, NS2B-NS3, NS4A and NS1 are enriched for ER interactions. Notably, NS4B is highly enriched for mitochondrial interactions, consistent with studies suggesting that flavivirus NS4B can localize to the mitochondria 25. NS5 has significant interactions with nuclear and cytoplasmic proteins suggesting localization in both compartments; although the NS5 proteins of DENV, JEV, YFV are thought to localize to the nucleus, nuclear localization of WNV NS5 has not been observed 26.
Flavivirus (DENV, JEV, WNV) capsid proteins can localize to the nucleus, and nucleolar localization has been reported for DENV, JEV and WNV capsid proteins 26–28. We confirmed that WNV, DENV and ZIKV capsids are in the nucleolus during infection, as they colocalize with a known nucleolar protein, fibrillarin (Fig. 3f). Ectopic expression of WNV capsid also localizes to the nucleolus (Supplementary Fig. 4a). As expected, capsid also resides in the cytoplasm, consistent with the roles for capsid in viral replication. Of the 72 WNV capsid interactors shown, 21 were identified in DENV or ZIKV, and 10 of these are known to be nucleolar (Fig. 3g; Supplementary Table 6). These findings are consistent with a recently published report identifying several nucleolar proteins as interaction partners of ZIKV capsid 29. We focused on the nucleolar dead-box helicase protein, DDX55, and confirmed an interaction with WNV capsid via co-immunoprecipitation (Fig. 3h). We also show the nucleolar localization of DDX55 by immunofluorescence (Supplementary Fig. 4b), as well as colocalization of native DDX55 and capsid in WNV-infected cells (Supplementary Fig. 4c). We find that DDX55 is antiviral, as depletion of DDX55 using two distinct siRNAs results in an increase in viral titer for all three flaviviruses (Fig. 3i; Supplementary Fig. 4d).
RNAi Screening uncovers host factors that interact with WNV proteins and impact flavivirus infection
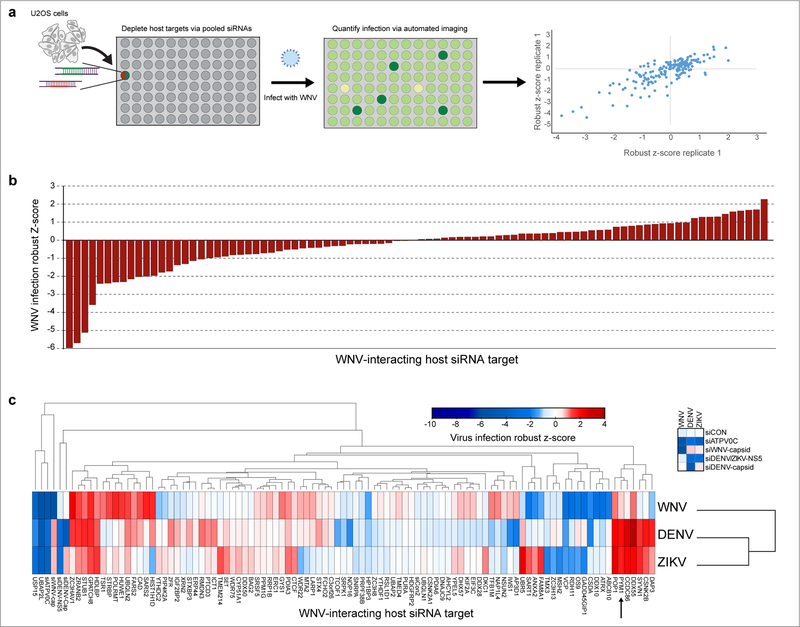
Next, we set out to determine which WNV-host interactions are most relevant by identifying those that influence WNV replication. We selected 122 WNV-interacting factors, including the 21 WNV-capsid interacting proteins shared between WNV and DENV or ZIKV (Supplementary Table 8). We pooled two siRNAs targeting each host factor in a 384-well plate, performed each screen in duplicate and analyzed the results with automated immunofluorescence microscopy (Fig. 4a). We used positive controls (siWNV-capsid, siATPV0C) as well as a scramble siRNA negative control (siCON). First, we quantified the total cell number and excluded those wells that had less than 70% viability, compared to the plate average. Next, we calculated robust z-scores for the percent infection (Fig. 4b). Of the 88 factors that were non-toxic, 26 (~30%) had an effect on WNV infection (Fig. 4b; Supplementary Table 8). We identified 12 anti-viral and 14 pro-viral genes, including previously identified factors influencing flaviviral infection (Supplementary Table 4) 2,3,5,8,30–32.
Fig. 4. RNAi screening reveals important roles for host proteins that interact with WNV proteins.
a, Schematic of the WNV infection siRNA screen. 122 WNV-interacting host proteins were depleted using a pool of 2 siRNAs for each target (x-axis). The screen was performed with two independent biological replicates and robust z-scores of WNV infection are plotted for each individual replicate. b, The average robust Z-score for each of the 88 (non-toxic) genes calculated from two independent biological replicates is shown. c, siRNA screening against WNV, DENV, and ZIKV performed with two independent biological experiments. The colors in the heatmap correspond to the average robust z-score of infection and are indicated in the accompanying scale. Columns (viruses) were clustered using One minus Pearson correlation and rows (targets) were clustered by Euclidean distance. The WNV capsid-interacting host protein PYM1 is indicated with an arrow. The complete screening dataset is provided in Supplementary Table 8.
We next determined which interactors impact the replication of the related flaviviruses, DENV and ZIKV (Fig. 4c). We confirmed an effect with the positive controls as appropriate (siDENV/ZIKV NS5, siDENV-capsid, siWNV-capsid, siATPV0C) and not the negative control (siCON) (Fig. 4c). Of the non-toxic genes (88 genes), we identified host factors that are specific to WNV infection (13, ~15%), as well as those that had an effect on WNV and either DENV or ZIKV (13, ~15%), and those influencing infection of all three flaviviruses (8, 9%). Notably, of the non-toxic interactors tested, 40 (45%) had an effect on at least one flavivirus, including host factors previously implicated in flavivirus infection (VCP, FAM8A1) (Fig. 4c; Supplementary Table 8) 9,32. Of the 48 capsid interactors tested, 18 (~ 38%) had a phenotype in infection with one or more flaviviruses (Fig. 4c, Supplementary Table 8). Interestingly, several of the nucleolar capsid-interacting proteins were shown to be antiviral across multiple flaviviruses, including DDX55 (Fig. 3i), CCDC86 and POP1, suggesting that the nucleolus may be important in host antiviral defense.
Flavivirus Capsid proteins interact with PYM1 and flavivirus infection inhibits NMD
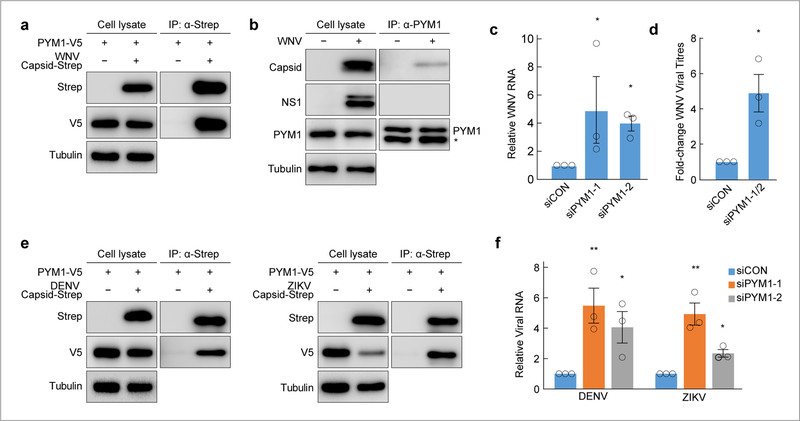
PYM1 interacted with all three flavivirus capsids and this interaction was observed in a recent ZIKV-human interaction study (Fig. 3g; Supplementary Table 5)29. Previously, we identified PYM1 as an HCV capsid (core)-interacting protein with a pro-viral role in infection14. In contrast, our data indicates that PYM1 is antiviral against flaviviruses (Fig. 4c, Supplementary Table 8). We confirmed the WNV capsid-PYM1 interaction by co-immunoprecipitation with strep-tagged WNV capsid and V5-tagged PYM1 (Fig. 5a). We demonstrated this interaction in the context of viral infection by immunoprecipitating native PYM1 from WNV-infected cells and detecting the interaction with native WNV capsid, but not with another viral protein, NS1 (Fig. 5b). Moreover, we show co-localization of WNV capsid and PYM1 in infected cells by immunofluorescence (Supplementary Fig. 6a). We confirmed the antiviral phenotype of PYM1 outside of the context of screening with two distinct siRNAs targeting PYM1, measuring WNV RNA by RT-qPCR (Fig. 5c; Supplementary Fig. 6b) and viral titers from control and PYM1-depleted cells (Fig. 5d). We validated the interaction of V5-tagged PYM1 with strep-tagged DENV and ZIKV capsid (Fig. 5e). We also validated the antiviral phenotype for both DENV and ZIKV upon PYM1 depletion with two distinct siRNAs (Fig. 5f).
Fig. 5. PYM1 is antiviral against flaviviruses and interacts with flavivirus capsid proteins.
a, Immunoprecipitations in HEK293 cells expressing the indicated constructs (WNV capsid-2X strep, PYM1-V5) were performed with Strep-Tactin resin (IBA) and blotted with the indicated antibodies. Results are representative of two biologically independent experiments. b, Immunoprecipitation of PYM1 from WNV-infected U2OS cells show a specific interaction between PYM1 with WNV capsid, but not with another WNV protein, NS1. Mouse IgG light chain is detected below PYM1 in the PYM1-IP fraction and is indicated by (*). Results are representative of two biologically independent experiments. c, Relative WNV RNA (WNV/18S) following infection upon siRNA depletion of PYM1 with two independent siRNAs (siPYM1–1, siPYM1–2). Infections are normalized to cells treated with siControl (siCON). Shown is the mean±SE, n = three biologically independent experiments. Significance was calculated using an unpaired, two-tailed student’s t-test and is indicated by * p < 0.05. d, Control (siCON) or PYM1-depleted (siPYM1-1/2) U2OS cells were infected with WNV Kunjin. TCID50 values were measured from cell supernatants. Shown is the mean±SE, n = three biologically independent experiments. Significance was calculated using an unpaired, one-tailed student’s t-test and is indicated by * p < 0.05. e, Immunoprecipitations in HEK293 cells expressing the indicated constructs (DENV or ZIKV capsid-2X strep, PYM1-V5) were performed with Strep-Tactin resin (IBA) and blotted with the indicated antibodies. Results are representative of two biologically independent experiments. f, Relative viral RNA (DENV/18S or ZIKV/18S) following infection upon siRNA depletion of PYM1 with two independent siRNAs (siPYM1–1, siPYM1–2). Infections are normalized to cells treated with siControl (siCON). Shown is the mean±SE, n = three biologically independent experiments. Significance was calculated using an unpaired, two-tailed student’s t-test and is indicated by * p < 0.05, ** p < 0.005.
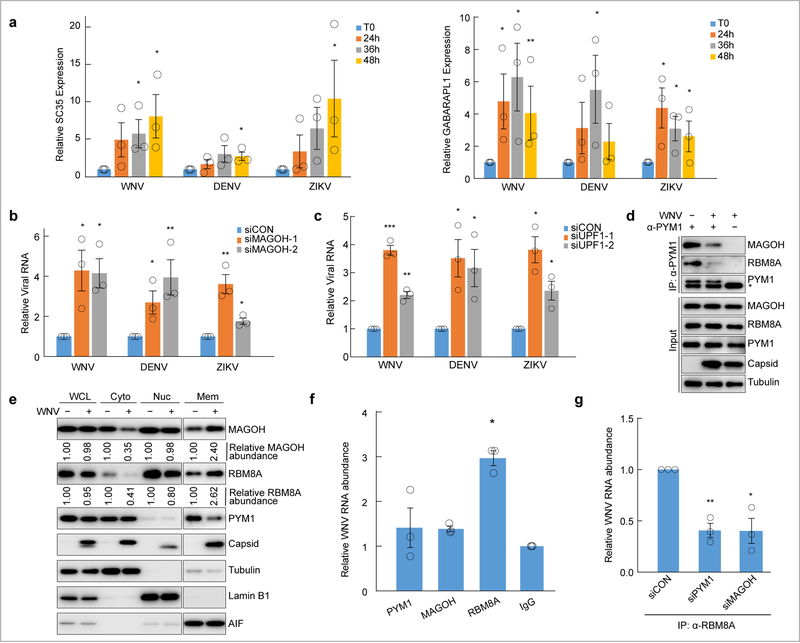
PYM1 is implicated in RNA processes, including the exon-junction complex, translation and nonsense-mediated decay (NMD) 16–21. NMD is an RNA decay pathway that can target diverse RNAs for degradation and is known to impact infection of multiple RNA viruses14,33,34. However, it is unknown whether WNV infection interferes with NMD activity. Therefore, we first determined if WNV, DENV or ZIKV infection alters NMD by monitoring endogenous targets. These include an NMD target generated through alternative splicing (SC35), a long 3’ UTR containing-target (GABARAPL1), a 5’ uORF-containing target (ASNS) and CARS, which has an unknown NMD-inducing feature35–37. We measured the abundance of these NMD targets during infection and found an increase in SC35, GABARAPL1, CARS and ASNS over 48 hours with all three infections (Fig. 6a, Supplementary Fig. 7a) 36. This accumulation is more pronounced and appears to be induced at earlier time points during WNV infection. We observed a less-pronounced, but statistically significant, increase in NMD targets during DENV infection. As a control, we measured the housekeeping gene, LDHA, and observed no accumulation upon infection (Supplementary Fig. 7b). To establish a role for PYM1 in NMD during flaviviral infection, we show that depletion of PYM1 in WNV-infected cells results in a further accumulation of the long 3’ UTR-containing NMD target, GABARAPL1, but not SC35 (Supplementary Fig. 7c).
Fig. 6. NMD is inhibited by flaviviruses through targeting of the EJC and the EJC protein RBM8A binds to viral RNA.
a, U2OS cells were infected with the indicated viruses at MOI 10 and analyzed at the indicated time points. Two endogenous targets of the NMD mRNA surveillance pathway (SC35, left panel; GABARAPL1, right panel) were analyzed by quantitative RT-PCR. Gene expression data (gene/18S) are normalized to uninfected controls. Shown is the mean±SE; n = three biologically independent experiments. Significance was calculated using the ratios of the target gene to the control gene and is indicated by * p < 0.05, ** p < 0.005 using an unpaired, two-tailed student’s t-test. b, Relative WNV, DENV and ZIKV viral RNA (viral RNA/GAPDH) was measured in control cells (siCON) and MAGOH-depleted cells using two independent siRNAs (siMAGOH-1, siMAGOH-2) at 24h post-infection. Infections are normalized to cells treated with siControl (siCON). Shown is the mean±SE, n = three biologically independent experiments. c, Relative WNV, DENV and ZIKV viral RNA was measured in control and UPF1-depleted cells using two independent siRNAs (siUPF1–1, siUPF1–2) at 24h post-infection. Infections are normalized to cells treated with siControl (siCON). Shown is the mean±SE, n = three biologically independent experiments. For all experiments significance was calculated using the ratios of the target gene to the control gene and is indicated by * p < 0.05, ** p < 0.005, *** p < 0.0005 using an unpaired, two-tailed student’s t-test. d, Immunoprecipitation of PYM1 from uninfected or WNV Kunjin-infected U2OS cells. Results are representative of two biologically independent experiments. Mouse IgG light chain is detected below PYM1 in the PYM1-IP fraction and is indicated by (*). e, Cellular fractionation experiments show the relative abundance of MAGOH, RBM8A, PYM1 and WNV capsid in the whole cell lysate (WCL), cytoplasmic (Cyto), nuclear (Nuc) and membrane/organelle (Mem) fractions between uninfected and WNV Kunjin-infected U2OS cells at 24 hours post-infection. Tubulin (cytoplasm), Lamin B1 (nucleus), and AIF (mitochondria) are shown as fractionation markers and loading controls. The relative abundance of MAGOH and RBM8A were calculated by quantifying the bands using Quantity One software (Bio-Rad). For each compartment, the relative protein abundance in the uninfected sample was set to 1. Results are representative of two biologically independent experiments repeated with similar results. f, Antibodies specific for PYM1, MAGOH and RBM8A were used to immunoprecipitate native proteins from WNV Kunjin-infected cells. RNA was extracted from the eluates and associated viral RNA was measured by quantitative RT-PCR (viral RNA/18S RNA). The viral RNA associated with the immunoprecipitated proteins was normalized to an IgG control. Shown is the mean±SE; n = three biologically independent experiments. Significance was calculated using the ratios of the viral RNA to 18S RNA using an unpaired, one-tailed student’s t-test and is indicated by * p < 0.05. g, An antibody specific for RBM8A was used to immunoprecipitate native proteins from control (siCON), PYM1-depleted (siPYM1) or MAGOH-depleted (siMAGOH), WNV-infected cells. RNA was extracted from the eluates and associated viral RNA was measured by quantitative RT-PCR (viral RNA/18S RNA). The viral RNA associated with the immunoprecipitated proteins was normalized to an IgG control and to the relative input viral RNA. Shown is the mean±SE; n = three biologically independent experiments. Significance was calculated using an unpaired, one-tailed student’s t-test and is indicated by * p < 0.05, ** p < 0.005.
PYM1 (Partner of Y14 and MAGOH) interacts with and promotes the recycling of the exon junction complex (EJC) proteins, MAGOH and RBM8A (Y14), which can target transcripts to NMD 16,18. Indeed, tethering PYM1, MAGOH or RBM8A to an RNA target leads to NMD-dependent degradation 16,38. To determine if the antiviral role of PYM1 is related to its EJC function, we tested the role of MAGOH in viral infection. We validated knock-down of MAGOH and confirmed that MAGOH depletion inhibits NMD (Supplementary Fig. 8a) 39,40. We observed an increase in WNV, DENV and ZIKV infection in MAGOH-depleted cells, as compared to control cells (Fig. 6b). Given that PYM1 and the EJC complex are involved in NMD, we next determined if NMD itself is antiviral against flaviviruses. Therefore, we depleted a canonical and essential component of NMD, UPF1, and confirmed both knock-down (Supplementary Fig. 8b, left panel) and inhibition of NMD by accumulation of the NMD target SC35 using two distinct siRNAs (Supplementary Fig. 8b, right panel). We monitored infection in UPF1-depleted cells via RT-qPCR and observed an increase in WNV, DENV and ZIKV RNA (Fig. 6c). These data suggest that NMD is antiviral and that the interaction between WNV capsid and PYM1 may subvert this process.
We next tested whether the WNV capsid-PYM1 interaction interferes with PYM1 function. We immunoprecipitated PYM1 from uninfected cells and observed interactions with the EJC complex proteins MAGOH and RBM8A. However, these interactions are diminished upon WNV infection, suggesting disruption of these associations (Fig. 6d). We did not observe a significant change in abundance of MAGOH and RBM8A during WNV infection (Fig. 6d), but reasoned that disruption of their interactions with PYM1 may alter the localization of these proteins. Therefore, we performed fractionation experiments in uninfected and WNV-infected cells and monitored the relative abundance of MAGOH and RBM8A in the whole cell lysate, cytoplasmic, nuclear and membrane/organelle fractions (Fig. 6e). While the total abundance of MAGOH and RBM8A is unchanged in infected whole cell lysates, we observed decreased abundance in the cytoplasmic fraction, with a concomitant increase in the membrane/organelle fraction upon infection. This suggests that WNV infection leads to sequestration of PYM1, and disruption of EJC protein function. Interestingly, we observed a change in the localization of PYM1 upon infection (Fig. 6e). Given that WNV capsid binds and sequesters PYM1, attenuating its interaction with MAGOH and RBM8A, we hypothesized that depletion of PYM1 may induce this mislocalization. Indeed, we found that depletion of PYM1 results in a similar redistribution of these EJC proteins (Supplementary Fig. 8c). These data suggest that WNV capsid inhibits the function of MAGOH and RBM8A by altering the localization of these proteins.
PYM1, MAGOH and RBM8A are RNA binding proteins involved in NMD and tethering any of these proteins to an RNA target leads to NMD-dependent degradation 16,38. Therefore, we hypothesized that one of these proteins may serve that function by directly binding WNV RNA. To test this, we used a modified CLIP protocol in which we cross-linked RNA binding proteins to RNA in WNV-infected cells, immunoprecipitated native PYM1, MAGOH or RBM8A and determined the abundance of viral RNA using RT-qPCR (Fig. 6f), and immunoprecipitated protein by immunoblot (Supplementary Fig. 8d) in the eluates. We normalized the viral RNA to a nonspecific control (18S RNA) and compared the relative amount of bound viral RNA to an IgG control. These experiments revealed a specific interaction between viral RNA and RBM8A (Fig. 6f). Next, we tested whether loss of PYM1 or MAGOH might impact RBM8A binding to viral RNA. We found that depletion of either PYM1 or MAGOH led to decreased interaction of RBM8A and viral RNA (Fig. 6g). Depletion of MAGOH also leads to reduced RBM8A which may impact RBM8A binding to viral RNA (Supplementary Fig. 8e). Altogether, these data suggest that the EJC targets flavivirus RNA for NMD-mediated decay and that flavivirus capsids interact with PYM1 to interfere with this pathway.
Discussion
Here, we provide the first complete AP-MS-generated, WNV-host protein-protein interaction (PPI) map. Our subsequent RNAi screen targeting a subset of factors in WNV, DENV and ZIKV infection to identify 40 proteins that interact with viral proteins and influence flavivirus infection (Fig. 2, 4c; Supplementary Tables 2, 8). We identified eight proteins, five pro-viral (CSDA, RDH11, UBAP2L, VCP, UBP15) and three antiviral (DDX55, GPATCH8, and PYM1), that both interacted with WNV proteins and had an effect on infection of all three flaviviruses. Of these eight proteins, five are RNA-binding proteins interacting with flavivirus capsids. These data underscore the importance of both flavivirus capsid-host protein interactions and RNA processing in flaviviral infection.
We focused on the interaction between PYM1 and WNV, DENV and ZIKV capsids and show that PYM1 is antiviral (Fig. 4c, 5a-f; Supplementary Table 8)14. PYM1 interacts with the EJC proteins MAGOH and RBM8A and plays a role in NMD. In addition to PYM1, the EJC protein, MAGOH, and the canonical NMD factor UPF1 are antiviral against WNV, DENV and ZIKV (Fig. 6b, c). NMD targets accumulate during flaviviral infection (Fig. 6a; Supplementary Fig. 7a); altogether, these data suggest that NMD is an antiviral pathway that is antagonized by WNV. Our studies show that WNV infection interferes with the interaction between PYM1 and the EJC proteins MAGOH and RBM8A (Fig. 6d), resulting in their mislocalization (Fig. 6e). This suggests that the interaction of WNV capsid with PYM1 during infection prevents its association with MAGOH and RBM8A, thereby attenuating NMD.
Previous reports have indicated that tethering EJC proteins to RNA substrates results in degradation by NMD16,38. We show that RBM8A binds viral RNA and that depletion of PYM1 diminishes this interaction, suggesting that PYM1 is required for this association (Fig. 6f, g). These data support a model in which NMD targets viral RNA through the EJC protein RBM8A, and that WNV capsid subverts this response through interaction with PYM1. This prevents the recycling of the EJC and results in the sequestration of EJC proteins to diminish the interaction between the EJC proteins and viral RNA, as well as between EJC proteins and host mRNA NMD targets, resulting in the stabilization of both (Supplementary Fig. 9). Previous studies have shown that haploinsufficiency of the EJC proteins MAGOH, EIF4A3, or RBM8A within the developing brain resulted in severe microcephaly41–43. ZIKV infection has been implicated in the development of microcephaly in fetuses and emerging research suggests that other neurotropic flaviviruses, including WNV, can result in similar fetal damage44,45. Further studies are required to determine if the sequestration and altered localization of MAGOH and RBM8A during flaviviral infection contributes to the neurological defects in developing fetuses.
NMD is known to restrict some RNA viruses33,34,46. In some cases, viruses counteract the restrictive mechanism of NMD through interaction with the NMD protein UPF147,34. It is unknown in these cases if the EJC plays a role in viral RNA recognition or degradation. Nevertheless, these data suggest a conserved antiviral role for this pathway 33,34,46. Interestingly, other studies have shown that some viruses can hijack NMD or EJC factors to aid in viral translation, replication or infectious viral particle production 14,48–50. Our ongoing studies will allow us to further define the relationship between flaviviruses and NMD, as well as explore additional mechanisms by which flaviviruses manipulate host cell processes.
Methods
Cells and viruses
HEK293 and U2OS cell lines were obtained from the American Type Culture Collections. All cultures were grown under standard conditions in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum, 100 units pencillin, 100 µg streptomycin and 20 mM L-alanyl-L-glutamine dipeptide (GlutaMax, Gibco). HEK293 cells were transfected using X-treme GENE 9 (Roche Applied Science) according to the manufacturer’s instructions. U2OS cells were transfected using Fugene HD (Promega) according to the manufacturer’s instructions. BHK cells were maintained as previously described 9. The WNV-KUNV isolate (CH16532) was a generous gift of R. Tesh (World Reference Center of Emerging Viruses and Arboviruses, Galveston, TX) was propagated using the same protocol as WNV. DENV-2 (NGC from BEI), and ZIKV Mexico 2016 (Mex2–81; a generous gift from R. Tesh) were grown as previously described 51. Viral titers were determined in BHK-21 cells by plaque assay.
Affinity Purification and Sample preparation for Mass Spectrometry
Viral bait proteins encoding a C-terminal 2X Strep II affinity tag were used for affinity purification and were derived from the following strains: WNV 2000-crow 3356, DENV serotype 2 16681, ZIKV French Polynesia 2013 H/FP/2013. Affinity purifications were performed as previously described12. Purified protein eluates were digested with trypsin for LC-MS/MS analysis. Samples were denatured and reduced in 2M urea, 10 mM NH4HCO3, 2 mM DTT for 30 min at 60C, then alkylated with 2 mM iodoacetamide for 45 min at room temperature. Trypsin (Promega) was added at a 1:100 enzyme:substrate ratio and digested overnight at 37C. Following digestion, samples were concentrated using C18 ZipTips (Millipore) according to the manufacturer’s specifications. Desalted samples were evaporated to dryness and resuspended in 0.1% formic acid for mass spectrometry analysis.
Mass Spectrometry
Digested peptide mixtures were analyzed by LC-MS/MS on a Thermo Scientific Velos Pro ion trap mass spectrometry system equipped with a Proxeon Easy nLC 1000 ultra high pressure liquid chromatography and autosampler system. Sample were injected onto a pre-column (2 cm x 100 um I.D. packed with ReproSil Pur C18 AQ 5um particles) in 0.1% formic acid and then separated with a two-hour gradient from 5% to 30% ACN in 0.1% formic acid on an analytical column (10 cm x 75 um I.D. packed with ReproSil Pur C18 AQ 3 um particles). The mass spectrometer collected data in a data-dependent fashion, collecting one full scan followed by 20 collision-induced dissociation MS/MS scans of the 20 most intense peaks from the full scan. Dynamic exclusion was enabled for 30 seconds with a repeat count of 1. The results raw data was matched to protein sequences by the Protein Prospector algorithm. Data were searched against the SwissProt Human protein sequence database (downloaded March 6, 2012) concatenated to a decoy database where each sequence was randomized in order to estimate the false positive rate. The searches considered a precursor mass tolerance of 1 Da and fragment ion tolerances of 0.8 da, and considered variable modifications for protein N-terminal acetylation, protein N-terminal acetylation and oxidation, glutamine to pyroglutamate conversion for peptide N-terminal glutamine residues, protein N-terminal methionine loss, protein N-terminal acetylation and methionine loss, and methionine oxidation, and constant modification for carbamidomethyl cysteine. Prospector data was filtered using a maximum protein expectation value of 0.01 and a maximum peptide expectation value of 0.05.
Interactome scoring and visualization
The WNV interactome was compiled through selecting bait-prey pairs with a MIST score > .68, computed with the weights we previously establish for HCV-host interactome: reproducibility (.36), abundance (.09) and specificity (.55) 14,52. The network was also scored with the complementary CompPASS scoring algorithm 15. Only the top 1% CompPASS WD scores per bait were included in the final iteration of the interactome. Any WNV protein for which we did not detect the bait protein in all the replicates analyzed was excluded from further study. To simplify the interactome and analyses, we manually removed all ribosomal proteins. We used a database of known human-human protein interactions to overlay additional connections between host proteins to identify multiprotein complexes (CORUM) 53. The final iteration of the WNV-interactome was visualized as a network representation using Cytoscape, version 2.8.3 54. The complete details of the scoring for the set of identified prey proteins, including the individual scores for the abundance, reproducibility, and specificity and the final interactome can be found in Supplementary Tables 1, 2.
Enrichment Analysis
For each given gene list, pathway and process enrichment analysis was carried out with one or more of the following ontology sources: KEGG Pathway, GO Biological Processes, GO Molecular Function, Reactome Gene Sets, Canonical Pathways and CORUM. All genes in the human genome were used as the enrichment background. Terms with p-value < 0.01, minimum count 3, and enrichment factor > 1.5 (enrichment factor is the ratio between observed count and the count expected by chance) were collected and grouped into clusters based on their membership similarities. More specifically, p-values were calculated based on accumulative hypergeometric distribution and q-values were calculated using the Benjamini-Hochberg procedure to account for multiple testing 55. Kappa scores were used as the similarity metric when performing hierarchical clustering on the enriched terms; sub-trees with similarity > 0.3 are considered a cluster. The most statistically significant term within a cluster was chosen as the representative category for each cluster. All analyses and visualization were carried out using Metascape (http://metascape.org/gp/index.html#/main/step1). A detailed list of all terms, results and statistics for each analysis are provided in the supplementary data (Supplementary Tables 3, 6, 7).
siRNA Screening
U2OS cells were seeded at 2000 cells/well in 50 µl media per well in 384-well for siRNA knockdown. Two siRNAs for each selected host target (Ambion Silencer Select) were transfected with HiPerFect (Qiagen) according to manufacturer recommendations at a final siRNA concentration of 20nM. Cells were infected three days after knockdown, at a density of 16,000 cells/well. WNV, DENV and ZIKV infections were done at an MOI of 0.01. Infectivity was measured 24 hours post infection by immunostaining and automated microscopy. Cells were fixed with 4% formaldehyde for 10 minutes, permeabilized with 0.1% Triton X-100 for 15 minutes, washed three times with PBS and blocked with 2% BSA in PBS-T. Cells were stained overnight with the mouse 4G2 (anti-envelope) antibody (1:4000 for DENV and ZIKV, 1:12000 for WNV Kunjin). Cells were washed three times with 2% BSA in PBS-T and incubated with 5 μg/mL Hoechst 33432 to identify nuclei and with an Alexa Fluor-488-conjugated α-mouse secondary antibody (1:1000) for one hour at room temperature. Cells were washed three times with 2% BSA in PBS-T and imaged in 50 ul PBS. Four sites per well were imaged at 20X magnification (ImageXpress Micro; Molecular Devices), and two wells were analyzed for each condition for each replicate. Each screen was performed in duplicate. Automated image analysis (MetaXpress; Molecular Devices) was used to segment the images and determine the number of DAPI-positive and 488-positive cells. The percentage of infected cells was calculated, averaged for the four sites in each well, and log transformed. The plate median and interquartile range were calculated and used to calculate a robust Z score for each well using the following equation: [(log10(%infection) − log10(median)/(IQR × 0.74)] 56. The robust z-scores for the experimental replicates were average and candidates were identified as positive if the averaged robust Z score was <−1.3 or >1.3. Cytotoxic candidates were identified based on nuclei counts and those with a >30% decrease in cell number as compared to the average cell count were excluded from further study. The complete screening results and a complete list of siRNAs used in this study can be found in Supplementary Tables 8, 9.
RNA Isolation and Quantitative RT-PCR
Total cellular RNA was isolated using Trizol (ThermoFisher Scientific) purified and DNase-treated using RNA Clean & Concentrator (Zymo) per manufacturer’s instructions. Complimentary DNA (cDNA) was synthesized using 1 µg of input RNA with using random hexamer primers (Life Technologies) with MMLV reverse transcriptase (Invitrogen) in a total volume of 20 µl. cDNA reactions were diluted 1:5 and 20 µl of each diluted sample was used to make a pooled reference. The pooled reference was used for subsequent 10-fold dilutions to generate a standard curve for all targets being measured. cDNA reactions were further diluted 1:5 (1:25 total dilution) and SYBR green reactions contained 5 μl of 2x Maxima SYBR green/Rox qPCR Master Mix (Thermo), 5 μl of diluted cDNA, 5 pmol of both forward and reverse primers, analyzed by qPCR and the relative abundance of each target was calculated using the standard curve. The relative values for each transcript were normalized to a control RNA (18S rRNA or GAPDH) and compared between experimental conditions. A complete list of primers used for quantitative RT-PCR is provided in Supplementary Table 10.
Virus Titers
U2OS cells were treated with control or gene-specific siRNAs in 6-well plates and infected with WNV, DENV or ZIKV at 48 hours post-treatment with the indicated MOI. Media was aspirated, cells were washed three times in PBS and fresh media was added at four hours post-infection. At 24 hours post-infection, supernatants were collected from infected cells. Viruses were tittered by infecting BHK cells plated in 96-well plates with 10-fold dilutions of viral supernatant and infected for 48hrs. Cells were fixed with 4% formaldehyde for 10 minutes, permeabilized with 0.1% Triton X-100 for 15 minutes, washed three times with PBS and blocked with 2% BSA in PBS-T. Cells were stained overnight with the mouse 4G2 (anti-envelope) antibody (1:4000 for DENV and ZIKV, 1:12000 for WNV Kunjin). Cells were washed three times with 2% BSA in PBS-T and incubated with 5 μg/mL Hoechst 33432 to identify nuclei and with an Alexa-Fluor-488-conjugated α-mouse secondary antibody (1:1000) for one hour at room temperature. Cells were washed three times with 2% BSA in PBS-T and imaged in 50 ul PBS to calculate TCID50s.
Co-immunoprecipitations and Immunoblots
C-terminally Strep II-tagged WNV 2000-crow 3356, DENV serotype 2 16681 or ZIKV H/FP/2013 capsid and C-terminally V5-tagged PYM1 or DDX55 (from the Human Orfeome Collection 57) were ectopically expressed in HEK293 and cells were lysed in IP Buffer (150 mM NaCl, 50 mM Tris pH 7.4, 1 mM EDTA, 0.5% NP-40 substitute, supplemented with protease inhibitor cocktail (Sigma)). Lysates were clarified by centrifugation at 10,000 RPM at 4°C for 10 minutes. Clarified lysates were incubated with 20 µl Strep-Tactin resin (IBA) overnight at 4 °C to immunoprecipitate tagged viral proteins. Beads were washed 3X in IP Buffer and bound proteins were eluted with 1X Strep-Tactin Elution Buffer (IBA). Eluates were resuspended in 2X SDS loading buffer for SDS-PAGE and proteins were detected by western blotting using α-Strep (Abcam) and α-V5 (Bethyl) antibodies. To detect the interaction of WNV capsid and PYM1 in the context of viral infection and to test if WNV infection disrupts the interaction between PYM1 and MAGOH or RBM8A, U2OS cells were infected with WNV virus (MOI=5) for 24 hours, washed 3X with PBS and lysed in IP buffer. The cellular lysates were clarified and endogenous PYM1 was immunoprecipitated by α-PYM1 antibody (Novus) with Protein A/G Agarose beads (ThermoFisher Scientific) overnight at 4 °C. The beads and bound proteins were washed 3X in IP buffer and eluted with 0.2 M glycine pH 2.5 buffer, followed by neutralization with 1 M Tris pH 9 buffer. The eluates were resuspended in 2X SDS loading buffer and analyzed by western blotting with the indicated antibodies.
Cellular Fractionation Experiments
U2OS cells infected with WNV virus (MOI=5) were harvested at 12 hours, 24 hours and 36 hours respectively post infection. Cellular fractionation experiments were performed by using Cell Fractionation Kit (Cell Signaling) per manufacturer’s instruction. Briefly, cells were trypsinized and resuspended in 500 µl cold PBS. One of the aliquot (100 µl) was mixed with 6X SDS loading buffer, sonicated and then boiled as the Whole Cell Lysate (WCL). The remaining 400 µl cell suspension were pelleted and first lysed in Cytoplasm Isolation Buffer (CIB) supplemented with PMSF and protease inhibitors. After centrifuge for 5 min at 500 X g, the supernatant was saved as the Cytoplasmic Fraction (Cyto). The pellet was then resuspended in Membrane Isolation Buffer (MIB) and incubated on ice for 5 min. After centrifuge for 5 min at 8000 X g, the supernatant was removed and saved as the Membrane and Organelle Fraction (Mem). The pellet was incubated with Cytoskeleton/Nucleus Isolation Buffer (CyNIB) and sonicated as the Cytoskeletal and Nuclear Fraction (Nuc). Cell lysates from each fraction were loaded into SDS-PAGE and protein abundance was analyzed by Western Blotting.
RNA Immunoprecipitations
U2OS cells were infected with WNV at an MOI of 5. Twenty-four hours post-infection, cells were trypsinized and subjected to UV-crosslink. Cells were then lysed in Buffer A (30 mM Hepes at pH 7.4, 2 mM MgOAc, 0.1% NP40) and 150 mM KOAc supplemented with 1 mM DTT, PMSF, protease inhibitors and RNaseOUT Recombinant Ribonuclease Inhibitor (Invitrogen). Cell lysates were incubated with Protein A/G Agarose beads (ThermoFisher Scientific) at 4 °C for 1 hour with rotation. The precleared cell lysates were incubated with Protein A/G Agarose beads and specific antibodies or IgG control overnight at 4 °C. Following 5 times wash with Buffer A and 150 mM KOAc, the beads-bound proteins were eluted with 0.2 M glycine pH 2.5 buffer and then neutralized with 1 M Tris pH 9 buffer. Trizol reagent was added to eluates to extract RNA and the abundance of WNV RNA associated with PYM1, MAGOH, or RBM8A was analyzed by RT-qPCR. The beads were resuspended in 2X SDS loading buffer and boiled for Western Blotting.
Confocal Microscopy
U2OS cells were plated into 12-well plates on glass coverslips at a density of 50,000 cells per well. After 24h, cells were infected with the indicated viruses at the indicated MOI for 24h. Cells were fixed with 4% formaldehyde, permeabilized for 10 min with 0.1% Triton X-100, and stained overnight at 4° C with the indicated antibodies. Coverslips were washed 3X in PBS-T and incubated with 5 μg/mL Hoechst 33432 to identify nuclei and with Alexa-Fluor-488 or 594-conjugated secondary antibodies (1:1000) for one hour at room temperature. Coverslips were washed three times with 2% BSA in PBS-T and mounted on glass slides using Vectashield (Vector Laboratories). Images were captured using oil immersion with a 63× objective with a Leica DM5500 Q confocal microscope.
RNAi, transfection, and Infection Studies
For RNAi experiments, 200,000 U2OS cells were plated in a 6-well plates and siRNAs were transfected using HiPerFect (Qiagen) according to manufacturer recommendations at a final concentration of 20nM and incubated for 48–72 hours. Cells were infected with WNV, DENV or ZIKV at the MOIs and timepoints indicated in the figure legends. Following infection, cells were collected in 1 ml Trizol for quantitative RT-PCR experiments or washed 3X in PBS and resuspended in 1 ml IP buffer for western blotting. For transfection experiments, 200,000 U2OS cells were plated in a 6-well plates and the indicate vectors were transfected using Fugene HD (Promega) and incubated for 48 hours. Following transfection, cells were collected in 1 ml Trizol for quantitative RT-PCR experiments or washed 3X in PBS and resuspended in 1 ml IP buffer for western blotting.
Antibodies
The following antibodies were used: α-strep tag (ab184224, abcam), α-FLAG tag (F7425, Sigma), α-V5 (A190–120A, Bethyl), α-tubulin (T6199, Sigma), α-WNV capsid (abcam, ab21673), α-WNV capsid (Genetex, GTX131947 ), α-WNV capsid (Sigma, SAB3500912), α-DENV capsid (Genetex, GTX103343), α-ZIKV capsid (Genetex, GTX133317), α-fibrillarin (Thermo Fisher Scientific, AMSA6771), α-fibrillarin (Cell Signaling Technologies, 2639S), α-DDX55 (Bethyl, A303–027A), α-PYM1 (Novus, NBP2–46366), α-MAGOH (Santa Cruz, sc-56724), α-RBM8A (Sigma, HPA018403), α-AIF (Cell Signaling, 5318S), α-Lamin B1 (Abcam, ab231282). The α-flavivirus glycoprotein (4G2) hybridoma was provided by Dr. M. Diamond (Washington University). Alexa-Fluor fluorescent secondary antibodies were from Life Technologies. HRP-conjugated secondary antibodies (α-mouse or α-rabbit) were from Amersham.
Plasmids
Open reading frames from strains WNV NY 2000-crow 3356 (capsid, prMEΔTM2, NS1, NS2A, NS2B, NS2B_3, NS3, NS4A, NS4B, NS5), DENV 16681 (capsid), ZIKV H/FP/2013 (capsid) were cloned into pCDNA4_TO with a C-terminal 2xStrep II affinity tag for expression in human cells (vectors provided by Nevan J. Krogan). Codon optimization for human codon usage was employed to improve expression of poorly expressing viral ORFs. The PYM1 and DDX55-expressing vectors with C-terminal V5 tags were obtained through the Human Orfeome Collection57
Data Availability Statement
Mass spectrometry data in this study are deposited in the PRIDE database (https://www.ebi.ac.uk/pride/archive/, Project Accession: PXD011728 ). A complete list of interaction scores are provided in Supplementary Tables 1, 2, 5. Gene Ontology enrichment analyses are provided in Supplementary Tables 3, 6, 7. Interactors found in previous flavivirus proteomic or genetic studies are detailed in Supplementary Table 4. RNAi screening data is provided in Supplementary Table 8. Additional supporting data are available from the corresponding authors upon request.
Supplementary Material
Acknowledgements
We thank members of the Cherry, Krogan and Ramage labs for discussion, advice and reagents. We thank the University of Pennsylvania High-Throughput Screening Core for providing reagents and technical expertise. We thank Julie To for virus preparations, Deidre Tatomer for RNA analyses and Mike Shales for graphical support. This work was supported by The American Liver Foundation Liver Scholar Award and the Creative and Novel Ideas in HIV Research Award to HR; NIH grants R01AI074951, RO1AI122749 and the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award to S.C.; NIH grants U19 AI118610, U19 AI135990, and P50 GM082250 to N.J.K.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Evans MV, Murdock CC & Drake JM Anticipating Emerging Mosquito-borne Flaviviruses in the USA: What Comes after Zika? Trends Parasitol 34, 544–547, doi: 10.1016/j.pt.2018.02.010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin DL et al. Dengue Virus Hijacks a Noncanonical Oxidoreductase Function of a Cellular Oligosaccharyltransferase Complex. MBio 8, doi: 10.1128/mBio.00939-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savidis G et al. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep 16, 232–246, doi: 10.1016/j.celrep.2016.06.028 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Richardson RB et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nature Microbiology, doi: 10.1038/s41564-018-0244-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H et al. A CRISPR-Based Screen Identifies Genes Essential for West-Nile-Virus-Induced Cell Death. Cell Rep 12, 673–683, doi: 10.1016/j.celrep.2015.06.049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marceau CD et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163, doi: 10.1038/nature18631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535, 164–168, doi: 10.1038/nature18625 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan MN et al. RNA interference screen for human genes associated with West Nile virus infection. Nature 455, 242–245, doi: 10.1038/nature07207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasunaga A et al. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog 10, e1003914, doi: 10.1371/journal.ppat.1003914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis ZH et al. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57, 349–360, doi: 10.1016/j.molcel.2014.11.026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heaton NS et al. Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity 44, 46–58, doi: 10.1016/j.immuni.2015.12.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jager S et al. Global landscape of HIV-human protein complexes. Nature 481, 365–370, doi: 10.1038/nature10719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jager S et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375, doi: 10.1038/nature10693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramage HR et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell 57, 329–340, doi: 10.1016/j.molcel.2014.12.028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sowa ME, Bennett EJ, Gygi SP & Harper JW Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403, doi: 10.1016/j.cell.2009.04.042 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bono F et al. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep 5, 304–310, doi: 10.1038/sj.embor.7400091 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diem MD, Chan CC, Younis I & Dreyfuss G PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol 14, 1173–1179, doi: 10.1038/nsmb1321 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Gehring NH, Lamprinaki S, Kulozik AE & Hentze MW Disassembly of exon junction complexes by PYM. Cell 137, 536–548, doi: 10.1016/j.cell.2009.02.042 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Hug N, Longman D & Caceres JF Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res 44, 1483–1495, doi: 10.1093/nar/gkw010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lykke-Andersen J, Shu MD & Steitz JA Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293, 1836–1839, doi: 10.1126/science.1062786 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Gehring NH, Neu-Yilik G, Schell T, Hentze MW & Kulozik AE Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11, 939–949 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Neufeldt CJ, Cortese M, Acosta EG & Bartenschlager R Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16, 125–142, doi: 10.1038/nrmicro.2017.170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agrawal T, Schu P & Medigeshi GR Adaptor protein complexes-1 and 3 are involved at distinct stages of flavivirus life-cycle. Sci Rep 3, 1813, doi: 10.1038/srep01813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Maio FA et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog 12, e1005841, doi: 10.1371/journal.ppat.1005841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatel-Chaix L et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe 20, 342–356, doi: 10.1016/j.chom.2016.07.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Denman AJ & Mackenzie JM The IMPORTance of the Nucleus during Flavivirus Replication. Viruses 9, doi: 10.3390/v9010014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colpitts TM, Barthel S, Wang P & Fikrig E Dengue virus capsid protein binds core histones and inhibits nucleosome formation in human liver cells. PLoS One 6, e24365, doi: 10.1371/journal.pone.0024365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balinsky CA et al. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol 87, 13094–13106, doi: 10.1128/JVI.00704-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coyaud E et al. Global interactomics uncovers extensive organellar targeting by Zika virus. Mol Cell Proteomics, doi: 10.1074/mcp.TIR118.000800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hafirassou ML et al. A Global Interactome Map of the Dengue Virus NS1 Identifies Virus Restriction and Dependency Host Factors. Cell Rep 22, 1364, doi: 10.1016/j.celrep.2018.01.038 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Scaturro P et al. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature, doi: 10.1038/s41586-018-0484-5 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Yasunaga A et al. Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection. PLoS Pathog 10, e1003914, doi: 10.1371/journal.ppat.1003914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balistreri G et al. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe 16, 403–411, doi: 10.1016/j.chom.2014.08.007 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Fontaine KA et al. The Cellular NMD Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein. MBio 9, doi: 10.1128/mBio.02126-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sureau A, Gattoni R, Dooghe Y, Stevenin J & Soret J SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J 20, 1785–1796, doi: 10.1093/emboj/20.7.1785 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F & Dietz HC Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36, 1073–1078, doi: 10.1038/ng1429 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Toma KG, Rebbapragada I, Durand S & Lykke-Andersen J Identification of elements in human long 3’ UTRs that inhibit nonsense-mediated decay. RNA 21, 887–897, doi: 10.1261/rna.048637.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gehring NH, Lamprinaki S, Hentze MW & Kulozik AE The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol 7, e1000120, doi: 10.1371/journal.pbio.1000120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baird TD et al. ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. Elife 7, doi: 10.7554/eLife.33178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh KK, Wachsmuth L, Kulozik AE & Gehring NH Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay. RNA Biol 10, 1291–1298, doi: 10.4161/rna.25827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao H et al. Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J Neurosci 35, 7003–7018, doi: 10.1523/JNEUROSCI.0018-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao H, McMahon JJ, Tsai YH, Wang Z & Silver DL Haploinsufficiency for Core Exon Junction Complex Components Disrupts Embryonic Neurogenesis and Causes p53-Mediated Microcephaly. PLoS Genet 12, e1006282, doi: 10.1371/journal.pgen.1006282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silver DL et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci 13, 551–558, doi: 10.1038/nn.2527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platt DJ et al. Zika virus-related neurotropic flaviviruses infect human placental explants and cause fetal demise in mice. Sci Transl Med 10, doi: 10.1126/scitranslmed.aao7090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miner JJ et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell 165, 1081–1091, doi: 10.1016/j.cell.2016.05.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia D, Garcia S & Voinnet O Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16, 391–402, doi: 10.1016/j.chom.2014.08.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mocquet V et al. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J Virol 86, 7530–7543, doi: 10.1128/JVI.07021-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyne JR, Jackson BR, Taylor A, Macnab SA & Whitehouse A Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J 29, 1851–1864, doi: 10.1038/emboj.2010.77 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serquina AK et al. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. J Virol 87, 8853–8861, doi: 10.1128/JVI.00925-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziehr B, Lenarcic E, Cecil C & Moorman NJ The eIF4AIII RNA helicase is a critical determinant of human cytomegalovirus replication. Virology 489, 194–201, doi: 10.1016/j.virol.2015.12.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rausch K et al. Screening Bioactives Reveals Nanchangmycin as a Broad Spectrum Antiviral Active against Zika Virus. Cell Rep 18, 804–815, doi: 10.1016/j.celrep.2016.12.068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verschueren E et al. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr Protoc Bioinformatics 49, 8 19 11–16, doi: 10.1002/0471250953.bi0819s49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giurgiu M et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res, doi: 10.1093/nar/gky973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smoot ME, Ono K, Ruscheinski J, Wang PL & Ideker T Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432, doi: 10.1093/bioinformatics/btq675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hochberg Y & Benjamini Y More powerful procedures for multiple significance testing. Stat Med 9, 811–818 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Birmingham A et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods 6, 569–575, doi: 10.1038/nmeth.1351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods 8, 659–661, doi: 10.1038/nmeth.1638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data in this study are deposited in the PRIDE database (https://www.ebi.ac.uk/pride/archive/, Project Accession: PXD011728 ). A complete list of interaction scores are provided in Supplementary Tables 1, 2, 5. Gene Ontology enrichment analyses are provided in Supplementary Tables 3, 6, 7. Interactors found in previous flavivirus proteomic or genetic studies are detailed in Supplementary Table 4. RNAi screening data is provided in Supplementary Table 8. Additional supporting data are available from the corresponding authors upon request.