Abstract
This paper explores cutting-edge deep learning methods for information extraction from medical imaging free text reports at a multi-institutional scale and compares them to the state-of-the-art domain-specific rule-based system - PEFinder and traditional machine learning methods - SVM and Adaboost. We proposed two distinct deep learning models - (i) CNN Word - Glove, and (ii) Domain phrase attention-based hierarchical recurrent neural network (DPA-HNN), for synthesizing information on pulmonary emboli (PE) from over 7,370 clin-ical thoracic computed tomography (CT) free-text radiology reports collected from four major healthcare centers. Our proposed DPA-HNN model encodes domain-dependent phrases into an attention mechanism and represents a radiology report through a hierarchical RNN structure composed of word-level, sentence-level and document-level representations. Experimental results suggest that the performance of the deep learning models that are trained on a single institutional dataset, are better than rule-based PEFinder on our multi-institutional test sets. The best F1 score for the presence of PE in an adult patient population was 0.99 (DPA-HNN) and for a pediatrics population was 0.99 (HNN) which shows that the deep learning models being trained on adult data, demonstrated generalizability to pediatrics population with comparable accuracy. Our work suggests feasibility of broader usage of neural network models in automated classification of multi-institutional imaging text reports for a variety of applications including evaluation of imaging utilization, imaging yield, clinical decision support tools, and as part of automated classification of large corpus for medical imaging deep learning work.
Keywords: Convolutional Neural Network (CNN), Recurrent Neural Network (RNN), pulmonary embolism, text report classification, radiology report analysis
1. Introduction
Diagnostic imaging accounts for 10 percent (100 billion dollars) of annual health care costs [1]. Due to market and patient care related pressures there is a constant demand for maintaining consistent clinical diagnostic excellence in the setting of rising imaging volumes, greater imaging complexity, and a demand by clinicians for rapid results. As a result the highest demands for new technology are around solutions that drastically improve workflow while maintaining or improving quality of care, building in effciencies for the clinician, health system, and ultimately for the patient. Consequently, technologies that can aid in automating the medical imaging workflow are in high demand and have inspired advances in machine learning methods which show promise in assisting radiologists to analyze complex imaging and text data [2, 3, 4, 5, 6]. Yet, despite the rapid exploration around new machine learning tools for use in medical imaging diagnostic tasks, a significant barrier remains for this technology to be applied at scale: the diagnostic information for the imaging studies are contained within unstructured clinician-created free text reports. So while it may be possible to acquire millions of medical images for machine learning applications, extracting the diagnostic information in those images as structured labels for machine learning model training requires a highly specialized understanding of diagnostic imaging report context, syntax, structure, and specific terminologies all unique to the radiology enterprise. One example of an important life-threatening medical condition is pulmonary embolism (PE) which is diagnosed with computed tomography imaging (CT) and relevant details stored in the medical record as free-text. CT exam for PE is an expensive test with risks of radiation exposure, contrast injection reactions, and incidental findings leading to further testing [7]. To identify imaging studies that are positive for PE, like many other radiology diagnoses, information extraction from the free-text radiology reports is needed in order to perform large scale analyses. If the hospital was able to leverage the computerized tools, it would be possible to rapidly and accurately account for the percentage of positive and negative studies, per provider/service/specialty, and use that data over time to identify opportunities for implementing or reeducating ordering providers about decision support tools for PE evaluation and reduce the number of negative examinations in a targeted approach [8].
Natural Language Processing (NLP) techniques can be a key to successfully analyzed the radiology reports to extract clinically important findings and recommendations [9, 10]. NLP has already shown potential to automate the task of classifying imaging reports in a way that could inform decisions regarding medical imaging utilization and appropriateness [11, 12, 13]. Chapman et al. [14] developed an application called PEFinder based on an extension of NegEx to detect lexical cues other than negation and defined how each cue modifies a preceding or succeeding concept. Yu et al. [15] used an NLP system called Narrative Information Linear Extraction (NILE) that combines linguistic and machine learning approaches to improve identification of pulmonary embolism location and also demonstrated promising results. Well-studied early programs that have also leveraged machine learning techniques include MedLEE (Medical Language Extraction and Encoding System), which relies on controlled vocabulary and grammatical rules in order to convert free-text into a structured database [16] [17]. Dang et al. [9] processed 1059 radiology reports with Lexicon Mediated Entropy Reduction (LEXIMER) to identify the reports that include clinically important findings and recommendations for subsequent action. More recently, Sohn et al. [18] used machine learning to identify patients with abdominal aortic aneurysms. Other work has used various machine learning techniques such as support vector machines to classify MRI free text reports [11].
Recent advances in computing power and machine learning techniques have prompted the rise of a new generation of machine learning technique called convolutional neural networks (CNNs) that have successfully been applied to image recognition tasks, including facial recognition, object detection, and image classification [6, 19]. CNNs and other similar deep learning architectures have significant implications for diagnostic imaging [5, 20, 4, 21, 22], but rather than being limited to image recognition tasks, it is possible that CNNs can also be applied to text data. Character-based and very deep CNN models are generally applied for text classification tasks with very large datasets [23, 24], where complex neural network architectures play a major role to capture all generic features of the data while disregarding domain-dependent word-level features. Instead, we used a shallow CNN model proposed by Kim [25] as it has been shown to obtain state-of-the-art results on multiple benchmark datasets using pre-trained word vectors. The adaptation of this CNN model combined with GloVe word embeddings [26] served as a strong baseline for our experiments.
Another class of neural networks architectures, recurrent neural networks (RNNs), has recently gained a lot of attention from researchers for modeling sequences and have been shown to perform well in solving various NLP tasks because of their ability to deal with variable-length input and output [27]. RNNs have also been used to address tasks in the medical domain [28]. To the best of our knowledge, there is no existing work that applies RNN-based models to classify free text medical imaging reports based on PE categorical measures and analyze the comparative performance against the CNN model.
In this study, we present a comparative analysis of distinct deep learning techniques (CNN and RNN) in classifying a database of free text chest (contrast enhanced) CT reports based on the presence of pulmonary embolism. We train the models on a small sub-set of Stanford training set (2512 reports) and present the performance on four distinct institutional datasets. Our results show that deep learning models can be transferable to completely different institutional dataset only being trained on a single institutional data. The main contributions of this paper can be summarized as follows:
- Methodological contributions
-
(a)We proposed a novel domain phrase attention-based hierarchical recurrent neural network model (DPA-HNN) to classify the radiology reports. The model encodes radiology reports through an intuitive hierarchical structure composed of word-level, sentence-level and document-level representations. Compared to a simple word-level attention mechanism, domain phrase attention suits the radiology domain better as the radiologists traditionally follow a domain-specific note writing style and some domain phrases occur frequently in reports.
-
(b)We also proposed the use of a well-known state-of-the-art convo-lutional neural network model [25] for classifying radiology reports corpus based on the presence of PE factors. We integrated the CNN model with pre-trained Glove vectors: (CNN Word - Glove) [26] for learning the semantics of the whole radiology reports from a relatively small annotated training set.
-
(a)
- Comparative analysis
-
(a)We analyzed the comparative performance of the two proposed deep learning architectures (CNN Word - Glove and DPA-HNN) in the context of free-text radiology reports labeling according to the presence of PE factors.
-
(b)We compared the performance of the proposed deep learning models with a state-of-the-art representative of well feature engineered NLP approaches - PEFinder [14], as well as with traditional machine learning models - Support Vector Machine (SVM) and Adaboost with bag-of-words features which represent the class of traditional methods with limited feature engineering.
- (c)
-
(a)
In the following sections, we present a detailed description of the dataset, materials and method, followed by the experimental setup, results and discussion. Finally, we discuss some limitations in Section 6 and conclude the paper in Section 7.
2. Dataset
Corpora
- We obtained 227,809 radiology reports from Duke University Medical Center, 117,816 reports from Stanford University Medical Center, and 12,091 reports from Colorado Childrens Hospital for contrast-enhanced CT examinations of the chest performed between January 1, 1998 and January 1, 2016.1
The Impression sections were extracted from the original reports by searching for the “impression” keyword, signifying the beginning of the impression section, and using simple heuristics to determine the end of the section. We structured our heuristics to extract more details on the impressions rather than filtering out the noise, which inevitably resulted in some parses being more verbose than necessary.
Training Set and Test Set
- The model training was performed using the Stanford University radiology report corpus of 117,816 reports of adult patients; from these, 4,512 reports were randomly selected and annotated by three experienced radiologists who read the whole report and assigned binary class labels. Two binary labels were assigned to individual reports - these were defined according to two categorical measures of PE: (1) PE present/absent; (ii) PE acute/chronic. The annotators assigned binary labels based on the following assumption; If a PE was definitely present in the report it was annotated as positive, or else annotated as negative. Chronicity was labeled as either acute or chronic based on the text description. In the setting of acute on chronic, or “mixed” chronicity, the report was to be labeled as acute to reduce the false negative rate. 4512 annotated reports were randomly divided into training (2512 reports), validation (1000 reports), and testing (1000 reports) sets.
External Test Set
- External test data sets were created from the Duke and Colorado reports: 1000 reports from the Duke medical center corpus and 1000 reports from the Colorado Children’s corpus were randomly selected and annotated by two experienced radiologists following the same annotation protocol as adopted during the Stanford dataset annotation (above).
Inter-rater reliability among the three raters for annotation of Stanford, Duke and Colorado datasets was calculated using Fleiss’ kappa [31] - a generalization of Cohen’s Kappa for more than two raters. The raters were highly consistent for two categories, “PE positive” and “PE Acute”, with kappa scores of 0.959 and 0.969 respectively. The kappa metric reflects the initial ratings, following which a single radiologist resolved all conflicting cases manually for preparing the ground truth labels.
External Validation Set (UPMC dataset)
We obtained 858 reports from University of Pittsburgh medical center (UPMC) that were originally used to develop PEFinder classifiers. The reports were all de-identified in a fully HIPAA-compliant manner. Three medical students independently annotated the reports with five distinct states and binary annotations for each document were obtained from the user annotations as follows: “probably positive” and “definitely positive” were collapsed to positive; and “probably negative”, “indeterminate”, and “definitely negative” were considered negative. After collapsing annotations to binary values, the authors generated labels for each report by a majority vote of the annotators. We refer the reader to the original paper [14] for a detailed description of the UPMC dataset.
3. Materials and Methods
3.1. CNN Word - Glove Architecture
CNNs have been applied to natural language modeling by convolving a filter across a fixed length word representation as input into the model, and final classification is obtained through a series of matrix multiplications and non-linear function mappings [25]. Such a model can automatically learn the semantics of local structure of text, such as phrases, while avoiding having to keep memory over the entire sequence of words for a given sequence. However these models have empirically been shown to require larger training sets. To the down-stream performance of a CNN, the effcient vectorized representation of words and/or phrases are important factors. Neural network based word embedding approach [32] can learn a vector which enables a model to perform well on the task of relating a word to its context in a given window. For example, Glove [26] combines components of skip-gram and continuous bag of words model by incorporating local window based information with document level statistics in their word vectors.
The CNN Word - Glove model used for this task was initially proposed by Kim et. al. [25]. The CNN model with only one layer of convolution was trained on top of GloVe vectors obtained from an unsupervised neural network model trained on 42 billion words from text extracted from crawling web pages. Since the GloVe vector is created on very generic documents, it is expected that some specific clinical terms never occur in the vocabulary. All words that do not match the Glove corpus are converted to an unknown token, which is initialized to a random vector. The input embeddings are then padded with zeros at the end to ensure that all training examples have the same size. For this study, we choose a input length of 300 words, which is the 99.5 percentile for all impressions in our dataset. The examples containing longer sequences are truncated at the tail end. Such truncation of the report length has a minor effect on the model performance as radiologists usually document the key findings earlier in the impression section (within the first 2 or 3 sentences).
Figure 1 shows the high level schema of the CNN model. First, we generated the vector representations of all the words that belong to the Impression section of the Stanford corpus using the pre-trained GloVe model. The 115,816 Stanford report (excluding 2000 validation and test reports) impressions each contained 124 words and symbols on average with 31,470 unique tokens across all impressions. A preprocessing step was included to treat all punctuation as separate word tokens. The vector representation of the words are processed through a convolutional layer with 200 filters, a window size of 10 words, and a rectified linear unit as the non-linearity. This layer is followed by a max pooling layer which takes the max across all values of a given filter. Dropout, a regularization method to prevent overfitting of the data, is then used on the output. The result is fed into a fully connected output layer followed by a sigmoid layer to convert the raw scores to probabilities. The two prediction tasks are trained on two different CNN models −1) {PEPositive, PENegative} and 2){PEAcute, PEChronic}.
Figure 1:
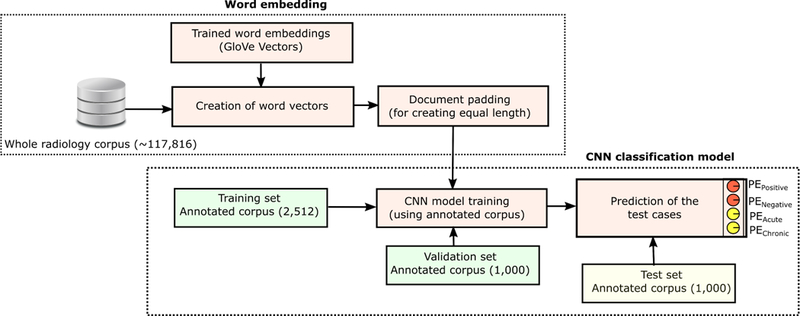
CNN Word - Glove Architecture, showing the training, validation and testing situation for Stanford dataset.
3.2. Domain Phrase Attention-based Hierarchical Recurrent Neural Network (DPA-HNN) Model
The semantics of a radiology report can be modeled through a hierarchical structure composed of word-level, sentence-level and document-level representations. Recent work on document-level sentiment classification showed promising performance by taking similar hierarchical structures into consideration [29, 30].
In contrast to the existing work [30] that encodes global user preference and product characteristics via a word-level user-product attention mechanism for a product-oriented sentiment classification task, we propose a novel Domain Phrase Attention-based Hierarchical Neural Network (DPA-HNN) model (Figure 2) by encoding clinical domain-dependent phrases into a sentence-level attention mechanism and representing a radiology report through a hierarchical structure composed of word-level, sentence-level and document-level representations. Compared to a general word-level attention mechanism, our domain phrase attention applied at the sentence-level plays a more important role in classifying radiology reports as radiologists traditionally follow a domain-specific note writing style. Moreover, some domain phrases occur frequently in radiology documents, justifying the need to propose our DPA-HNN model with a domain phrase attention mechanism.
Figure 2:
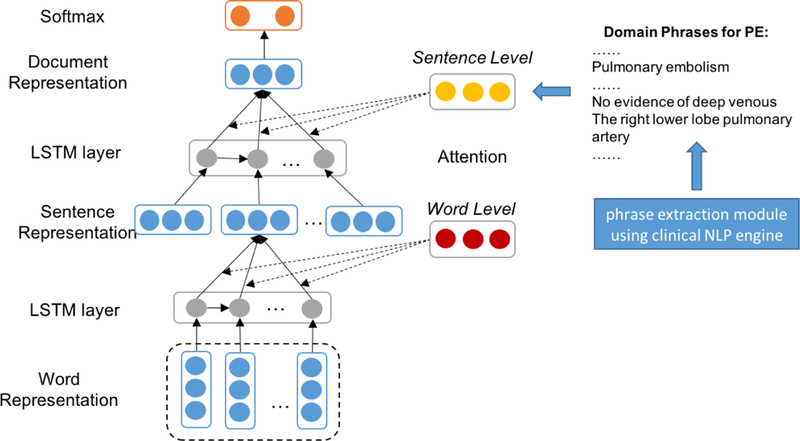
Domain Phrase Attention-based Hierarchical Neural Network (DPA-HNN) architecture.
Although RNN is theoretically a powerful model to encode sequential information, in practice it often suffers from the vanishing/exploding gradient problems while learning long-range dependencies [33]. LSTM [34] and GRU [35] networks are known to be successful remedies to these problems. We chose LSTMs for our experiments because there had not been shown any distinguishable difference between the performance of a LSTM unit and a GRU unit in the literature [36, 37]. We use LSTM as our hidden layer activation unit to model the semantic representations of sentences and documents. Typically, each cell in a LSTM is computed as follows:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
where are the weight matrices and are the biases of LSTM to be learned during training, parameterization, and transformations of the input, forget and output gates, respectively. σ is the sigmoid function and ⊙ stands for element-wise multiplication. xt is the input of a LSTM cell unit and ht represents the hidden state at time t.
Assume that a document has L number of sentences, where each sentence si contains Ti words. wit with t ∈ [1, T ] represents the words in the ith sentence. For word-level computations, xt represents the word embedding vectors wt. The first hidden layer vectors hit with t ∈ [1, T ] are used to represent a sentence. For sentence-level computations, xt represents the sentence embedding vectors si. The hidden layer vectors hi with i ∈ [1, L] are used to represent a document in this case.
We regard the last hidden layer as the representation of a document and place a softmax layer on top of it to predict the class labels e.g. {PEPositive, PENegative} or {PEAcute, PEChronic} for the radiology report. In other words, the output layer (softmax) of DPA-HNN considers two possible labels. As such, we build two separate models to classify PE positive/negative and PE acute/chronic. Considering h∗ as the final representation of a radiology report, the softmax layer can be formulated as:
| (7) |
where Ws and bs are the parameters of the softmax layer. We use the negative log likelihood of the correct labels as our training loss function:
| (8) |
where j is the label of a document d.
Domain Phrase Attention Mechanism:
In the settings of hierarchical neural networks without attention, the hidden states are fed to an average pooling layer to obtain the sentence representation and the final document representation. For example, the final feature representation of a radiology report can be computed as:
| (9) |
We propose a domain phrase attention mechanism to capture the most important part of a document by considering domain phrases at the sentence level. It is reasonable to reward sentences that are clues to correctly classify a document as indicated in [29]. Hence, we pay extra attention to domain phrases if they are present in a sentence. We encode each domain phrase as continuous and real-valued vectors which are randomly initialized. This yields:
| (10) |
| (11) |
| (12) |
where Ws and Wdp are projection parameters and bs is the bias parameter to be learned during training.
Our domain phrase (DP) generation algorithm works as follows. First, we extract clinical concepts from the radiology reports based on SNOMED-CT ontology [38] using a hybrid clinical NLP engine [39]. Then, our algorithm uses the extracted clinical concepts to form the DPs based on two heuristics: (1) we combine the consecutive clinical concepts occurred in the same sentence as one domain phrase. For example, in the sentence, “three small low attenuation lesions within the liver, which are too small to characterize”, “low attenuation” and “lesions” are tagged as two separate clinical concepts by the clinical NLP engine. Since they are consecutive words in the sentence, we regard them as the domain phrase “low attenuation lesions”. In other words, the clinical concepts should be consecutively present in a sentence in order to be the part of a domain phrase, and (2) we obtain shallow parsing annotation for the text. If one token is tagged as “B-NP”, then together with its following tokens tagged as “I-NP”, “B-PP”, and “B-NP”, the entire chunk will be considered as one phrase. If any token in this phrase is annotated as a clinical concept by the clinical NLP engine, we regard the phrase as a domain phrase. For example, the shallow parsing annotation for “The right lower lobe pulmonary artery” is “The[B-NP] right[I-NP] lower[I-NP] lobe[I-NP] pulmonary[I-NP] artery[I-NP]”. Here, the token “The” is tagged as “B-NP”, and its following tokens are tagged as “I-NP” while “right lower lobe pulmonary artery” is tagged as a clinical concept. Therefore, the whole chunk “The right lower lobe pulmonary artery” is considered as one domain phrase. The list of Domain Phrases (DPs) is generated from the Stanford training set. The total number of DPs in the list is 343. The average number of words in DPs is ≈ 4. We display the frequency distribution of word counts in DPs with few examples in Figure 3.
Figure 3:
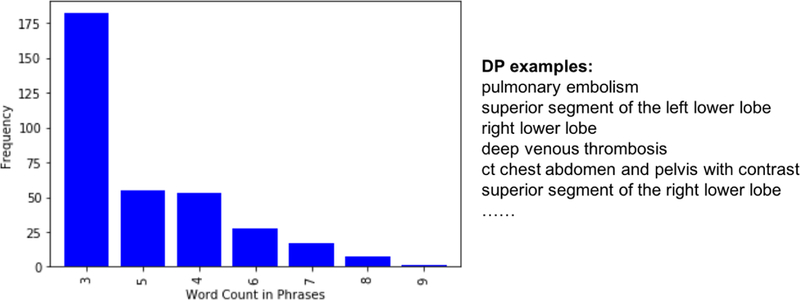
The Frequency Distribution of Word Count in DPs and DP Examples.
4. Experiments and Evaluation
4.1. Models for Comparison
For legitimate comparison, we experimented with the following seven models.
PEFinder: a state-of-the-art rule-based method for PE radiology text report classification [14].
Support Vector Machine with radial basis kernel function (SVM): a popular machine learning method for binary classification; we used sparse bag-of-words as feature vector.
Adaptive Boosting (Adaboost): a machine learning algorithm that uses selective boosting on top of the bag-of-words feature vector.
CNN Word-Glove: CNN model built on top of GloVe vectors (Section 3.1).
Hierarchical Recurrent Neural Network (HNN): RNN baseline model without any attention mechanism (Section 3.2).
Attention-based Hierarchical Neural Network (A-HNN): RNN-based model with the general word-level attention mechanism using random initialization (Section 3.2).
Domain Phrase Attention-based Hierarchical Neural Network (DPA-HNN): our proposed model with Domain Phrase (DP) attention mechanism (Section 3.2).
4.2. Model Training
The CNN model has been trained on the vector embedding of 2512 reports from the Stanford corpus. We used dropout rate (p) of 0.5, l2 constraint (s) of 3, and mini-batch size of 50. The network weight update has been done through stochastic gradient descent (SGD) over shuffled mini-batches with the Adadelta update rule. Weights for the model were initialized randomly using the Xavier initialization method to roughly keep the scale of the gradients the same across all layers.
For RNN-based models, we used the same pre-trained word embeddings as used by CNN model to represent the words. Each RNN was built with 300 hidden units (i.e. LSTMs as discussed in Section 3.2). Similar to the CNN models, our RNN-based models were also trained with the SGD algorithm with update direction computed using a batch size of 32 via the Adadelta update rule. We trained these models for 200 epochs and 400 iterations of validation. The vector size for each granularity i.e. word, sentence, and document were 300 dimensions.
Using the Stanford validation set (1000 reports), we optimized the hyperpa-rameters such as dropout rate, number of epochs, and batch size by selecting a best trade-off between the validation accuracy and the computational memory requirement.
4.3. Classification Performance
We evaluate the performance of our models on all the data sets (Stanford, UPMC, Colorado Childrens and Duke) using the following metrics: Precision, Recall, F1 value, and Area Under the Curve (AUC). In order to convert the predicted probability values of neural models to binary class labels, we determine an optimal cutoff threshold of the probability of the positive class by maxi-mizing: Precision(ti)+Recall(ti) for all the thresholds (ti) between 0 to 1.The classification results from all methods are displayed in Table 2.
Table 2:
Comparative performance measures. Boldface numbers represent column-wise superior performance achievement on a particular dataset.
| PE Positive/Negative | PE Acute/Chronic | |||||||
|---|---|---|---|---|---|---|---|---|
| P | R | F1 | AUC | P | R | F1 | AUC | |
| Stanford test set | ||||||||
| PEFinder | 0.87 | 0.90 | 0.89 | N/A | 0.91 | 0.91 | 0.91 | N/A |
| SVM | 0.96 | 0.96 | 0.95 | 0.85 | 0.93 | 0.97 | 0.95 | 0.93 |
| Adaboost | 0.98 | 0.98 | 0.98 | 0.95 | 0.97 | 0.97 | 0.97 | 0.92 |
| CNN | 0.92 | 0.97 | 0.95 | 0.99 | 0.91 | 0.91 | 0.91 | 0.99 |
| HNN | 0.94 | 0.96 | 0.95 | 0.98 | 0.92 | 0.97 | 0.94 | 0.95 |
| A-HNN | 0.99 | 0.96 | 0.97 | 0.98 | 0.97 | 0.96 | 0.97 | 0.99 |
| DPA-HNN | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.97 | 0.98 | 0.99 |
| UPMC dataset | ||||||||
| PEFinder | 0.87 | 0.96 | 0.91 | N/A | 0.91 | 0.95 | 0.93 | N/A |
| SVM | 0.71 | 0.70 | 0.58 | 0.83 | 0.49 | 0.70 | 0.57 | 0.86 |
| Adaboost | 0.83 | 0.84 | 0.83 | 0.90 | 0.84 | 0.83 | 0.82 | 0.91 |
| CNN | 0.76 | 0.95 | 0.85 | 0.97 | 0.82 | 0.92 | 0.87 | 0.97 |
| HNN | 0.82 | 0.74 | 0.77 | 0.90 | 0.88 | 0.73 | 0.75 | 0.88 |
| A-HNN | 0.82 | 0.74 | 0.77 | 0.90 | 0.88 | 0.72 | 0.75 | 0.88 |
| DPA-HNN | 0.87 | 0.87 | 0.87 | 0.95 | 0.91 | 0.90 | 0.90 | 0.94 |
| Duke test set | ||||||||
| PEFinder | 0.84 | 0.99 | 0.90 | N/A | 0.86 | 0.99 | 0.92 | N/A |
| SVM | 0.95 | 0.98 | 0.96 | 0.94 | 0.96 | 0.98 | 0.97 | 0.95 |
| Adaboost | 0.97 | 0.96 | 0.97 | 0.85 | 0.97 | 0.97 | 0.97 | 0.77 |
| CNN | 0.98 | 0.97 | 0.97 | 0.90 | 0.98 | 0.98 | 0.98 | 0.91 |
| HNN | 0.93 | 0.79 | 0.85 | 0.92 | 0.95 | 0.74 | 0.81 | 0.91 |
| A-HNN | 0.90 | 0.83 | 0.86 | 0.91 | 0.89 | 0.79 | 0.83 | 0.88 |
| DPA-HNN | 0.94 | 0.81 | 0.86 | 0.94 | 0.90 | 0.95 | 0.92 | 0.99 |
| Colorado Childrens test set | ||||||||
| PEFinder | 0.69 | 0.79 | 0.73 | N/A | 0.66 | 0.87 | 0.72 | N/A |
| SVM | 0.98 | 0.99 | 0.99 | 0.91 | 0.99 | 0.99 | 0.99 | 0.94 |
| Adaboost | 0.99 | 0.98 | 0.99 | 0.97 | 0.99 | 0.99 | 0.99 | 0.98 |
| CNN | 0.99 | 0.95 | 0.96 | 0.97 | 0.99 | 0.98 | 0.98 | 0.99 |
| HNN | 0.99 | 0.99 | 0.99 | 0.99 | 0.56 | 0.74 | 0.60 | 0.81 |
| A-HNN | 0.87 | 0.80 | 0.83 | 0.98 | 0.60 | 0.87 | 0.66 | 0.77 |
| DPA-HNN | 0.80 | 0.80 | 0.80 | 0.93 | 0.71 | 0.87 | 0.77 | 0.98 |
For Stanford test set, DPA-HNN has the best scores on all evaluation metrics for both PE Positive/Negative and PE Acute/Chronic classifications. Compared with HNN and A-HNN, DPA-HNN encodes domain phrase attention and improves the performance. All the improvements of DPA-HNN model over HNN model and A-HNN model were found to be statistically significant (p < 0.05). From the results we can see that, overall, neural network-based methods have better performance than the classic PEFinder, SVM and Adaboost methods in term of F1 and AUC scores on Stanford test set.
On UPMC dataset, DPA-HNN has the best precision scores for both tasks, while CNN has the best AUC scores. On Duke test set, DPA-HNN has the best AUC scores for both tasks, while CNN has the best precision and F1 scores. On Colorado Childrens test set, HNN has the best scores on all evaluation metrics for PE Positive/Negative classification, while not performing well on PE Acute/Chronic classification.
Overall, DPA-HNN model shows performance improvement on Stanford test set, and partially on UPMC dataset and Duke test set. However, the performance on Colorado Childrens test set is not the best, which is reasonable because DPA-HNN and other neural network-based methods are trained on Stanford dataset which are mostly adult patients compared to the specific pediatrics population of Colorado Childrens. Further analyses revealed that the external datasets (UPMC dataset, Duke test set, Colorado Childrens test set) have varying distributions of average number of sentences and domain phrases in a document. The distributions and statistics are displayed in Figure 4 and Table 1. DPs play an important role in our proposed DPA-HNN model. For example, the Colorado data has an average number of 1.2 DPs in a document, which is much lower than the average number of 3.5 in Stanford test data, while the percentage of documents without DPs for Colorado data is much lower than the Stanford test data - that could be the reason why the DPA-HNN model trained on Stanford dataset does not work equally well on Colorado data. However, the average number of sentences in a document for this dataset is 6.2, which is very close to Stanford data of 6.4. Since the HNN model does not rely on DPs, it performed well on the Colorado test set, but the DPA-HNN model suffered due to lack of DPs in this test set.
Figure 4:
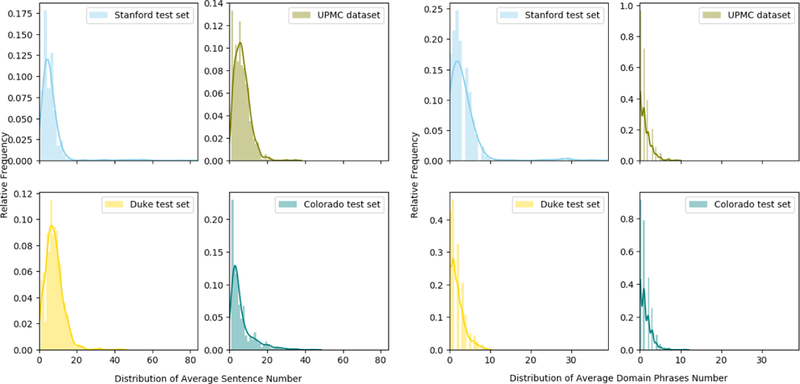
Distribution of Average Number of Sentences in a Document (left) and Distribution of Average Number of Domain Phrases in a Document (right).
Table 1:
Statistics of average number of sentences/DPs in a document across 4 datasets.
| Dataset | Stanford test set | UPMC dataset | Duke test set | Colorado test set |
|---|---|---|---|---|
| Average number of sentences in document | 6.4 | 6.7 | 8.0 | 6.2 |
| Average number of DPs in document | 3.5 | 1.2 | 1.8 | 1.2 |
| Percentage of Documents without DPs | 13.7% | 39.6% | 25.7% | 35.9% |
From the results, we can also observe that in general the evaluation scores of the PE Acute/Chronic classification task are lower than the PE Positive/Negative classification task denoting the complexity of the former compared to the later task.
4.4. Qualitative Analysis
To better understand what information a deep learning model in a natural language task is using to make its decisions, methods have been developed to visualize the impact of input words on the output decision [40]. For the CNN model, we used one such method called sensitivity analysis, which takes the partial derivative of the loss function with respect to each input variable. Since our input variables represent word vectors in this case, in order to get a importance score for a particular word we take the L1 norm of the vector of partial derivatives.
The heat maps (in Figure 5) represent result of sensitivity analysis of input for a positive example (left) and a negative example (right) where the CNN model predicted correctly from the Stanford test set. The result on the left shows the text of a report that is positive for all two prediction classes. We can see that there is little importance (light colored) placed on the long document with the exception of the relevant phrase (dark colored), which contains information on the PE status. Similarly on the right is an example of a report that is negative for PE and all other prediction classes. We can see in this case that the model places the most emphasis (dark color) on the first sentence which clearly states that there is no evidence of PE. From these qualitative results we note that the network is able to parse through large sequences of text to focus on phrases that are relevant to classification simply from the document level annotation.
Figure 5:
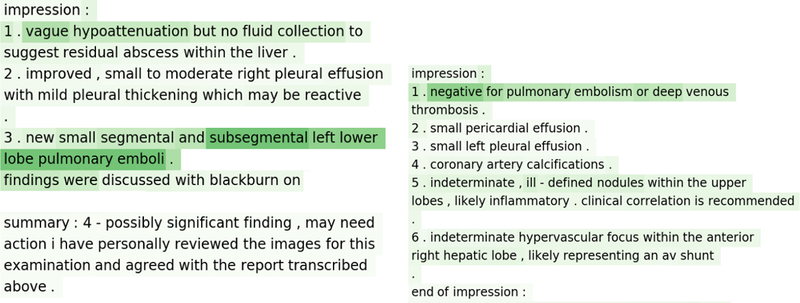
Results of sensitivity analysis of CNN model: cases with correct prediction -positive (left) and negative (right). The darker the color, the more weight there is.
In Figure 6, we present two examples of misclassified reports using the CNN model. In the first case (left) the model predicted PE negative, but the ground truth was PE positive. In the second case (right) the model predicted a PE positive, but the ground truth was PE negative. It is apparent in the example on the left that the CNN model was unable to infer that PE was present with the statement that there was decreased volume of clot. In the example on the right, the word “no” seemed to be separated by an abnormally long string of words prior to “pulmonary embolism” leading to a classification error. Such errors are reasonable as CNNs are only able to capture local information from the data to perform well in classification tasks [25].
Figure 6:

Results of sensitivity analysis of CNN model: cases with incorrect prediction - positive (left, CNN model predicted negative) and negative (right, CNN model predicted positive). The darker the color, the more weight there is.
However, our RNN based models A-HNN and DPA-HNN were able to predict the classes correctly for the same reports where CNN failed to classify (shown in Figure 6). This is shown via the hierarchical attention-based visualization results (Figure 7) where the weight value for each word in the sentence is obtained from the weights learned from word-level attention for the first LSTM layer. The weight value for each sentence in the report is obtained from the weights learned from sentence-level attention for the second LSTM layer. With the weights for both word-level and sentence-level attention, we can see that different sentences play different roles in a report and different words play different roles in each sentence toward the final classification of a radiology report. In the negative example (right) of Figure 7, the sentence 3: “there is no filling defect in the central pulmonary artery to indicate a pulmonary embolism” has the highest weight. The word “embolism” has the highest weight in this sentence, and the word “no” has the second highest weight in this sentence. In the positive example (left), the sentence 1: “impression 1 interval decrease in the volume of clot within the pulmonary arterial system” has the highest weight. The word “clot” has the highest weight in this sentence. The success of RNNs in classifying these reports correctly is understandable. Because, unlike CNNs, RNNs are able to capture global context from the data by considering long-term dependency among words [33].
Figure 7:
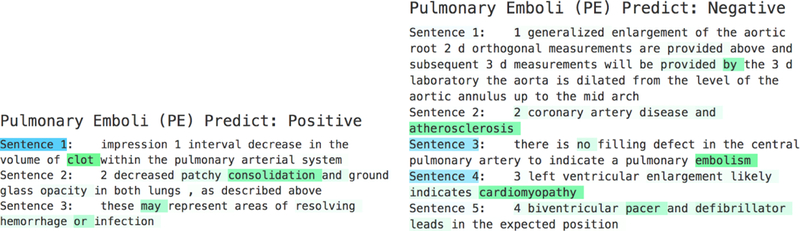
Sentence-level and word-level attention of the RNN model: cases where CNN failed to classify as shown in Figure 6. The blue color represents sentence-level importance and the green color stands for token-level importance; the darker the color, the more weight there is.
We further contextualize the results as confusion matrix for DPA-HNN on the Stanford test set in Figure 8. The confusion matrix is normalized class-wise to meaningfully represent the performance of the model. The X-axis represents gold-standard results, and the Y-axis represents predicted results from our DPA-HNN model. We can see that the false positive (top right part) and the false negative (down left part) rates are very low for both PE Positive/Negative classification and PE Acute/Chronic classification. In fact, for both classifications, only two cases are misclassified (sample shown in Figure 9). These errors were evaluated and found to be related to conflicting and skeptical language in the impression. For example, in Figure 9 (left), the impression clearly states “no definite pulmonary embolus”, however, shortly thereafter the report went on to suggest “artifact vs possible pulmonary embolus in the right upper lobe” and recommended an additional imaging test. In the other example in Figure 9 (right), the model focused on the word “subacute” to predict the report as chronic.
Figure 8:
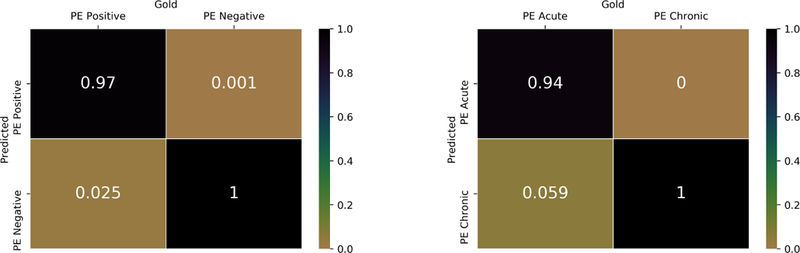
Normalized confusion matrices for results from DPA-HNN model. Darker color represents a higher value.
Figure 9:

DPA-HNN misclassified cases - PE negative but predicted positive (left) and PE Acute but predicted Chronic (right). The blue color represents sentence-level importance and the green color stands for token-level importance; the darker the color, the more weight there is.
5. Discussion
The increasing availability of computational methods to process vast amounts of unstructured information makes it possible to derive insights from large repositories of narrative medical data [41, 42]. However, the current state-of-the-art method to classify Radiology reports based on the presence or absence of a di-agnosis of PE, called PEFinder, is a purely rule-based system and may not be easily scaled to classify/annotate multi-institutional free-text reports with varying narrative styles. This restricts automated interpretation of large volumes of free text reports for creating solutions to support clinical processes such as image interpretation and auto-reporting in radiology. We report on the performance of machine learning models leveraging several neural network architectures to determine the PE status from unstructured CT reports. We compared the neural network models with the best available published feature-engineered model. To the best of our knowledge, our work is the first to compare several neural network techniques to evaluate classification performance on free text medical imaging reports on both intra-and inter-institutional data, and compare to the current NLP rule-based gold standard, PEFinder.
Our neural network models performed equivalently or better than PEFinder and existing machine learning models. Models such as MedLEE, cTAKES, NILE, and others require named entity recognition libraries, term definitions, parsed phrases, matching, etc., and a great deal of effort is required to create these resources - representing, in certain cases, decades of prior work. By contrast, our CNN and RNN based neural network models were mainly developed without hand-curating any semantic input. Instead, our neural network-based models were simply trained on a small sample of document-level classified reports and rapidly achieved the performance levels of the best available NLP tools in the field. This suggests that neural networks may be extremely powerful for classification/ annotation of large volumes of free text reports and can achieve optimal performance without any of the previously relied upon grammatical feature definitions, concept codes, or pre-defined terms [14, 15, 43].
More recently, the surge in medical imaging classification and computer vision in radiology has led to a demand for high-fidelity labeled medical images; neural network models, such as the ones demonstrated here, can serve to provide highly structured labels for medical images useful for deep learning techniques. One of the most exciting opportunities that deep learning classification of imaging text offers is the ability to aid in training and modeling in computer vision projects for which a large corpus of annotated imaging data is needed (and the free-text narrative report style is poorly suited) [44, 45]. Medical images are usually saved with accompanying radiology reports, and leveraging the natural language information for image analysis has great potential. For example, imaging studies from a clinical Picture Archiving and Communication Systems (PACS) can be automatically annotated by using our models to analyze the corresponding radiology reports. We can unleash the full capacity of deep learning for analyzing large volumes of medical images by automating the data collection and annotation process. Moreover, a sustainable system can be developed which allows the annotated image data to be continuously updated, shared, and integrated within a learning health care system.
Application of the neural network methodologies described in this work to other imaging free-text report annotation tasks would be both rapid and scalable using domain adaptation and transfer learning algorithms as shown in the literature [46, 47, 48]. To examine the generalization of our neural network models we also used imaging reports from other institutions as test sets. Our neural network models (trained on Stanford data) were tested on four different test sets. The rules of PEFinder were developed using reports from the UPMC test set, and so this provided an optimal test case. There is no significant difference in performance between our neural network models and PEFinder on the UPMC dataset. Our models were able to generalize to new reports not included in the training data and on reports from independent institutions, our models performed better than a rule-based NLP system and traditional machine learning models. The mild performance degradations noticed for the models applied on other institutional datasets except Stanford is still clinically acceptable with the lowest AUC of 0.93 for DPA-HNN. However, to achieve additional performance improvements, institutions can tune our trained models further based on their local datasets. Moreover, the hospitals can leverage the scalable deep learning tools demonstrated in this work on large volumes of data to identify trends and provide metrics around which to build performance and quality improvement programs, including the use of clinical decision support systems for PE imaging assessment, something that is mandated by congressional legislation beginning next year [49]. Moreover, the neural networks used for NLP tasks have tremendous value in many applications such as generating systems for radiology case prioritization based on report analysis, patient cohort generation, eligibility screening for clinical trials, and in radiology clinical decision support to manage imaging utilization or use yield as a metric. For example, understanding the rate of negative studies could serve as an indirect marker of utilization appropriateness and guide clinical decision making [50, 51, 8, 52, 53, 54, 55].
Overall, our work contributes to advancements in automated radiology report classification for PE leveraging a minimal amount of labeled examples, and offers opportunity to generate large volumes of annotated reports that would drive the development of clinical decision support systems towards improved imaging utilization, clinical research and automatic medical image interpretation for PE and other critical conditions.
6. Limitations
We acknowledge that our neural network classifiers are yet to achieve perfect scores. However, the accuracy measures are either superior or comparable to other published NLP studies. For instance, although there were 10 misclassifications out of the 1000 test cases by the CNN model, the classification errors are rather diffcult to explore in neural networks, as they are often a “black box”. However, the visualization we generated helped us understand the source of errors (Figure 6). The most common source of errors was the lack of a direct mention of the existence or absence of PE and limited documentation due to insuffcient image quality; instead, inference was needed based on context. As shown in Figure 7, our RNN based model correctly predicted the classes and located the most important sentences in the reports, but it is still hard to generalize how the model made the inference. We found only one example case for PE positive/negative classification, where CNN correctly predicted it positive, but RNN predicted it negative. However, it was not apparent how CNN was able to predict this case correctly based on the generated heat map. As such, many of these errors require subtle reasoning to reach the correct conclusion, which may be a limitation to the architecture of our models, in addition to training constraints posed by the size of our datasets.
7. Conclusion
We proposed a novel domain phrase attention-based hierarchical recurrent neural network model (DPA-HNN) that can accurately classify free text chest CT reports into pre-defined PE related criteria. We adopted a well-known state-of-the-art convolutional neural network model [25] for radiology domain and integrated the model with pre-trained Glove vectors. For both models, we demonstrated both intra-and cross-institutional fidelity when compared to other more laborious NLP methods. Our results suggest feasibility of CNNs and RNNs in automated classification of imaging text reports and support the application of these techniques at scale in classifying free text imaging reports for various use cases including radiology patient prioritization, cohort generation for clinical research, eligibility screening for clinical trials, and assessing imaging utilization. These approaches may also have impact on other important research areas such as large scale generation of labeled data for generating computer vision models for automatic medical image interpretation.
Highlights.
Proposed two distinct deep learning models - (i) CNN Word - Glove, and (ii) Domain phrase attention-based hierarchical neural network (DPA-HNN), for synthesizing information on pulmonary emboli (PE) from clinical thoracic CT free-text radiology reports.
Visualization methods have been developed to identify the impact of input words on the output decision for both deep learning models.
Models are trained only on Stanford dataset (2512 reports) and are tested on four major healthcare centers dataset – Stanford (1000 reports), Duke (1000 reports), Colorado Children (1000 reports), and University of Pittsburg medical center (858 reports).
Comparative effectiveness of the deep learning models is judged against the current state-of-the-art –PEFinder as well as with traditional machine learning models – SVM and Adaboost with bag-of-words features.
This work proposed interesting experimental insight on the proficiency of CNN and RNN to automatize the analysis of unstructured imaging reports.
Acknowledgement
Financial support for this project was provided by grants from Philips Health-care and the Stanford Child Health Research Institute (Stanford NIH-NCATS-CTSA grant #UL1 TR001085).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All examination reports were de-identified in a fully HIPAA-compliant manner, and acquisition and processing of data was approved by the Institutional Review Board (IRB) of the institution where the reports were obtained.
References
- [1].Imaging utilization trends and reimbursement, http://www.diagnosticimaging.com/reimbursement/imaging-utilization-trends-and-reimbursement, accessed: 2018-03-30.
- [2].Pons E, Braun LM, Hunink MM, Kors JA, Natural language processing in radiology: a systematic review, Radiology 279 (2) (2016) 329–343. [DOI] [PubMed] [Google Scholar]
- [3].Xu Y, Tsujii J, Chang EI-C, Named entity recognition of follow-up and time information in 20 000 radiology reports, Journal of the American Medical Informatics Association 19 (5) (2012) 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hua K-L, Hsu C-H, Hidayati SC, Cheng W-H, Chen Y-J, Computer-aided classification of lung nodules on computed tomography images via deep learning technique, OncoTargets and therapy 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cho J, Lee K, Shin E, Choy G, Do S, Medical image deep learning with hospital pacs dataset, arXiv preprint arXiv:1511.06348
- [6].Bakthula R, Agarwal S, Automated human bone age assessment using image processing methods-survey, International Journal of Computer Applications 104 (13). [Google Scholar]
- [7].Lang K, Huang H, Lee DW, Federico V, Menzin J, National trends in advanced outpatient diagnostic imaging utilization: an analysis of the medical expenditure panel survey, 2000–2009, BMC medical imaging 13 (1) (2013) 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kilani RK, Paxton BE, Stinnett SS, Barnhart HX, Bindal V, Lungren MP, Self-referral in medical imaging: a meta-analysis of the literature, Journal of the American College of Radiology 8 (7) (2011) 469–476. [DOI] [PubMed] [Google Scholar]
- [9].Dreyer KJ, Kalra MK, Maher MM, Hurier AM, Asfaw BA, Schultz T, Halpern EF, Thrall JH, Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study 1, Radiology 234 (2) (2005) 323–329. [DOI] [PubMed] [Google Scholar]
- [10].Yetisgen-Yildiz M, Gunn ML, Xia F, Payne TH, A text processing pipeline to extract recommendations from radiology reports, Journal of biomedical informatics 46 (2) (2013) 354–362. [DOI] [PubMed] [Google Scholar]
- [11].Hassanpour S, Langlotz CP, Amrhein TJ, Befera NT, Lungren MP, Performance of a machine learning classifier of knee mri reports in two large academic radiology practices: A tool to estimate diagnostic yield, American Journal of Roentgenology (2017) 1–4. [DOI] [PubMed]
- [12].Hassanpour S, Langlotz CP, Predicting high imaging utilization based on initial radiology reports:: A feasibility study of machine learning, Academic radiology 23 (1) (2016) 84–89. [DOI] [PubMed] [Google Scholar]
- [13].Banerjee I, Madhavan S, Goldman RE, Rubin DL, Intelligent word embeddings of free-text radiology reports, in: AMIA 2017 Annual Symposium, Wash-ington, DC: (in press), 2017. [PMC free article] [PubMed] [Google Scholar]
- [14].Chapman BE, Lee S, Kang HP, Chapman WW, Document-level classification of ct pulmonary angiography reports based on an extension of the context algorithm, Journal of biomedical informatics 44 (5) (2011) 728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yu S, Kumamaru KK, George E, Dunne RM, Bedayat A, Neykov M, Hunsaker AR, Dill KE, Cai T, Rybicki FJ, Classification of ct pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing, Journal of biomedical informatics 52 (2014) 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB, A general natural-language text processor for clinical radiology, Journal of the American Medical Informatics Association 1 (2) (1994) 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hripcsak G, Austin JH, Alderson PO, Friedman C, Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports 1, Radiology 224 (1) (2002) 157–163. [DOI] [PubMed] [Google Scholar]
- [18].Sohn S, Ye Z, Liu H, Chute CG, Kullo IJ, Identifying abdominal aortic aneurysm cases and controls using natural language processing of radiology reports, AMIA Summits on Translational Science Proceedings 2013 (2013) 249. [PMC free article] [PubMed] [Google Scholar]
- [19].LeCun Y, Bengio Y, Hinton G, Deep learning, Nature 521 (7553) (2015) 436–444. [DOI] [PubMed] [Google Scholar]
- [20].Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM, Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning, IEEE transactions on medical imaging 35 (5) (2016) 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H, A comparative study for chest radiograph image retrieval using binary texture and deep learning classification, in: Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE, IEEE, 2015, pp. 2940–2943. [DOI] [PubMed] [Google Scholar]
- [22].Banerjee I, Crawley A, Bhethanabotla M, Daldrup-Link HE, Rubin DL, Transfer learning on fused multiparametric mr images for classifying histopathological subtypes of rhabdomyosarcoma, Computerized Medical Imaging and Graphics [DOI] [PubMed]
- [23].Conneau A, Schwenk H, Barrault L, LeCun Y, Very deep convolutional networks for text classification, in: Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics, EACL 2017, 2017, pp. 1107–1116. [Google Scholar]
- [24].Shin B, Chokshi FH, Lee T, Choi JD, Classification of radiology reports using neural attention models, in: International Joint Conference on Neural Networks (IJCNN), 2017, pp. 4363–4370. [Google Scholar]
- [25].Kim Y, Convolutional neural networks for sentence classification, arXiv preprint arXiv:1408.5882
- [26].Pennington J, Socher R, Manning CD, Glove: Global vectors for word representation., in: EMNLP, Vol. 14, 2014, pp. 1532–1543. [Google Scholar]
- [27].Sutskever I, Martens J, Hinton GE, Generating Text with Recurrent Neural Networks, in: Proceedings of ICML, 2011, pp. 1017–1024. [Google Scholar]
- [28].Choi E, Schuetz A, Stewart WF, Sun J, Using recurrent neural network models for early detection of heart failure onset, Journal of the American Medical Informatics Association 24 (2) (2016) 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yang Z, Yang D, Dyer C, He X, Smola AJ, Hovy EH, Hierarchical attention networks for document classification, 2016.
- [30].Chen H, Sun M, Tu C, Lin Y, Liu Z, Neural sentiment classification with user and product attention., in: EMNLP, 2016, pp. 1650–1659.
- [31].Fleiss JL, Measuring nominal scale agreement among many raters., Psychological bulletin 76 (5) (1971) 378. [Google Scholar]
- [32].Mikolov T, Chen K, Corrado G, Dean J, Efficient estimation of word representations in vector space, arXiv preprint arXiv:1301.3781
- [33].Bengio Y, Simard P, Frasconi P, Learning Long-Term Dependencies with Gradient Descent is Difficult, IEEE Transactions on Neural Networks 5 (2) (1994) 157–166. [DOI] [PubMed] [Google Scholar]
- [34].Hochreiter S, Schmidhuber J, Long Short-Term Memory, Neural Computation 9 (8) (1997) 1735–1780. [DOI] [PubMed] [Google Scholar]
- [35].Cho K, van Merrienboer B, Bahdanau D, Bengio Y, On the Properties of Neural Machine Translation: Encoder–Decoder Approaches, in: Proceedings of SSST-8, Eighth Workshop on Syntax, Semantics and Structure in Statistical Translation, 2014, pp. 103–111. [Google Scholar]
- [36].Chung J, Gulcehre C, Cho K, Bengio Y, Empirical evaluation of gated recurrent neural networks on sequence modeling, in: Proceedings of the Deep Learning and Representation Learning Workshop: NIPS, 2014. [Google Scholar]
- [37].Jozefowicz R, Zaremba W, Sutskever I, An empirical exploration of recurrent network architectures, in: Proceedings of the 32nd International Conference on International Conference on Machine Learning -Volume 37, 2015, pp. 2342–2350. [Google Scholar]
- [38].Stearns MQ, Price C, Spackman KA, Wang AY, Snomed clinical terms: overview of the development process and project status., in: Proceedings of the AMIA Symposium, American Medical Informatics Association, 2001, p. 662. [PMC free article] [PubMed] [Google Scholar]
- [39].Datla VV, Hasan SA, Qadir A, Lee K, Ling Y, Liu J, Farri O, Automated clinical diagnosis: The role of content in various sections of a clinical document, in: IEEE International Conference on Bioinformatics and Biomedicine, BIBM, 2017, pp. 1004–1011. [Google Scholar]
- [40].Arras L, Horn F, Montavon G, M¨uller K-R, Samek W, Explaining predictions of non-linear classifiers in nlp, arXiv preprint arXiv:1606.07298
- [41].Leeper NJ, Bauer-Mehren A, Iyer SV, LePendu P, Olson C, Shah NH, Practice-based evidence: profiling the safety of cilostazol by text-mining of clinical notes, PloS one 8 (5) (2013) e63499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gallego B, Dunn AG, Coiera E, Role of electronic health records in comparative effectiveness research, Journal of comparative effectiveness research 2 (6) (2013) 529–532. [DOI] [PubMed] [Google Scholar]
- [43].Cai T, Giannopoulos AA, Yu S, Kelil T, Ripley B, Kumamaru KK, Rybicki FJ, Mitsouras D, Natural language processing technologies in radiology research and clinical applications, RadioGraphics 36 (1) (2016) 176–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shen D, Wu G, Suk H-I, Deep learning in medical image analysis, Annual Review of Biomedical Engineering 19 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen C.-h., Computer vision in medical imaging, Vol. 2, World scientific, 2014. [Google Scholar]
- [46].Long M, Cao Y, Wang J, Jordan MI, Learning transferable features with deep adaptation networks, in: Proceedings of the 32nd International Conference on Machine Learning, ICML, 2015, pp. 97–105. [Google Scholar]
- [47].Long M, Zhu H, Wang J, Jordan MI, Unsupervised domain adaptation with residual transfer networks, in: Advances in Neural Information Processing Systems, 2016, pp. 136–144.
- [48].Long M, Zhu H, Wang J, Jordan MI, Deep transfer learning with joint adaptation networks, in: Proceedings of the 34th International Conference on Machine Learning, ICML, 2017, pp. 2208–2217. [Google Scholar]
- [49].Powell DK, Silberzweig JE, The use of acr appropriateness criteria: a survey of radiology residents and program directors, Clinical imaging 39 (2) (2015) 334– 338. [DOI] [PubMed] [Google Scholar]
- [50].Lungren MP, Amrhein TJ, Paxton BE, Srinivasan RC, Collins HR, Eastwood JD, Kilani RK, Physician self-referral: frequency of negative findings at mr imaging of the knee as a marker of appropriate utilization, Radiology 269 (3) (2013) 810–815. [DOI] [PubMed] [Google Scholar]
- [51].Lungren MP, Paxton BE, Kilani RK, Imaging self-referral: here we go again, American Journal of Roentgenology 201 (4) (2013) W658–W658. [DOI] [PubMed] [Google Scholar]
- [52].Paxton BE, Lungren MP, Srinivasan RC, Jung S-H, Yu M, East-wood JD, Kilani RK, Physician self-referral of lumbar spine mri with comparative analysis of negative study rates as a marker of utilization appropriateness, American Journal of Roentgenology 198 (6) (2012) 1375–1379. [DOI] [PubMed] [Google Scholar]
- [53].Amrhein T, Paxton B, Lungren M, Befera N, Collins H, Yurko C, East-wood J, Kilani R, Physician self-referral and imaging use appropriateness: negative cervical spine mri frequency as an assessment metric, American Journal of Neuroradiology 35 (12) (2014) 2248–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Raja AS, Ip IK, Prevedello LM, Sodickson AD, Farkas C, Zane RD, Hanson R, Goldhaber SZ, Gill RR, Khorasani R, Effect of computerized clinical decision support on the use and yield of ct pulmonary angiography in the emergency department, Radiology 262 (2) (2012) 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Frankovich J, Longhurst CA, Sutherland SM, Evidence-based medicine in the emr era, N Engl J Med 365 (19) (2011) 1758–1759. [DOI] [PubMed] [Google Scholar]


