Abstract
In June 2016, a disease characterised by intestinal haemorrhage with a mortality rate of approximately 5% was observed in a duck farm in Shandong province, China. Here, we report the isolation and characterisation of a reovirus from duck tissue samples by inoculating duck embryos and duck embryo fibroblasts (DEF). The isolate replicated in DEF and Vero cells and formed syncytia. Sequence analysis revealed that the viral genome was 23,434 nt in length with typical structure organization, consisting of 10 dsRNA segments ranging from 3998 nt (L1) to 1190 nt (S4) in size, and was genetically distinct from previous Chinese duck-origin reoviruses. Phylogenetic analyses showed that the isolate was most closely related to the recently reported duck reovirus D2533/6/1-10 isolated in Germany, forming a monophyletic branch different from known reference avian reoviruses. Experimental infection results indicated that the isolate replicated transiently in ducklings and was shed via faeces. Infection with the isolate caused epithelial cell damage and lymphocyte apoptotic death in the bursa of Fabricius, which may result in immunosuppression in infected ducklings. The role of the isolate in current duck haemorrhage enteritis remains to be determined, but its damage to the bursa warrants further investigation of the duck immune response.
Subject terms: Viral pathogenesis, Viral epidemiology, Viral pathogenesis, Viral epidemiology
Introduction
Avian reoviruses (ARVs) are widely distributed in a variety of avian species, including wild birds1–4. They are members of genus Orthoreovirus in the family Reoviridae. The viral double stranded RNA genome contains 10 segments divided into three size classes based on their electrophoretic motility on a sodium dodecyl sulfate-polyacrylamide gel: large (L1, L2 and L3), medium (M1, M2 and M3) and small (S1–S4)5. The genome is predicted to encode eight structural proteins (λA, λB, λC, μA, μB, σA, σB and σC) and several nonstructural proteins (μNS, σNS, p10, etc). ARVs have been associated with a variety of diseases in domestic fowl, including chickens (viral arthritis/tenosynovitis, malabsorption syndrome, gastroenteritis and respiratory disease)6, turkeys (infectious enteritis)7, ducks and geese (fatal infection and spleen necrosis)8–10. In wild birds, reoviruses have been detected in white stork, grey heron, rock pigeon and so on4. ARV isolates were traditionally classified using standard serological procedures, but more recently have been characterised by molecular methods based on genome sequences and deduced amino acid analyses. However, for pathogenic isolates, it is difficult to assign a new isolate to a pathogenic group or pathotype according to its genetic background due to the lack of clearly defined virulence determinant(s) of the avian reovirus.
In China, Muscovy duck reovirus (MDRV) infection was first recognised in domestic Muscovy ducks in 200111, and the disease was seen only in Muscovy ducks (Carina moschata) and mule ducks, an infertile hybrid cross between male Muscovy and female Pekin ducks (Anas platyrhynchos). Genetic analysis showed that the virus was closely related to MDRV isolated in Europe12. Since 2006, outbreaks of disease characterised by spleen necrosis in Pekin ducklings were observed, and the aetiological agent was identified to be a reovirus distinct from previous MDRV isolates10. Currently, the reovirus has been tentatively classified as a “novel” duck reovirus (NDRV) to distinguish it from the “classical” MDRV13,14. Virus infections were subsequently diagnosed widely in mainland China, affecting almost all domestic ducks and geese13,15–17. In June 2016, an outbreak of disease characterised by intestinal haemorrhage was observed in a commercial duck farm in Shandong Province, with a mortality rate of around 5% before the age of 4 weeks. In this study, we conducted an aetiological investigation and isolated a reovirus genetically distinct from previous duck reoviruses identified in China. The pathogenicity of the isolate was evaluated in ducklings.
Results
Virus isolation and characterisation
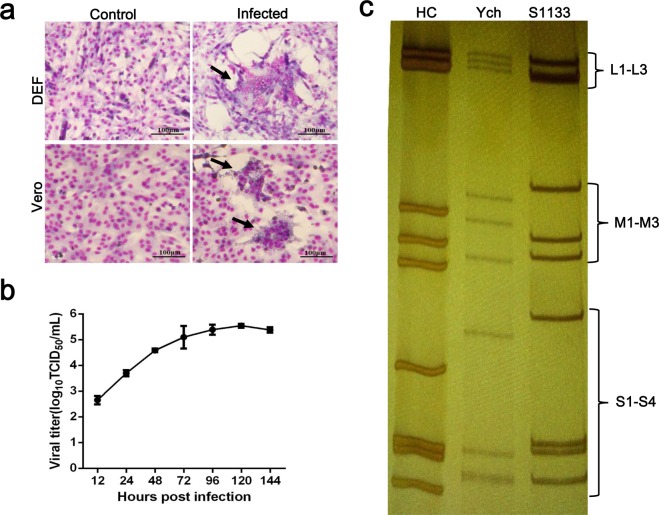
Bacterial infection was first ruled out since no typical pathogenic bacterium was isolated from tissue samples. Attempts were then made for virus isolation. The mixed tissue homogenate was inoculated into duck embryos and sequentially passaged eight times. No consistent mortality was observed for inoculated embryos, but irregular necrotic foci were found in the livers and spleens of some embryos at necropsy on 5 dpi. We then infected DEFs with embryo allantoic fluid of the fourth passage and observable CPE, characterised by the formation of a few syncytia and cell detachment appeared on 4 dpi after four successive passages. Further passage in DEFs resulted in the rapid formation of syncytia and the virus titre peaked at 3.47 × 105 TCID50/mL (Fig. 1a,b). Thus, the isolate was designated as Pekin duck/China/Ych/2016 (Ych hereafter). When DEF-adapted Ych was used to infect fresh Vero cell monolayers, syncytium formation was observed (Fig. 1a), suggesting the isolate replicated in Vero cells. Infectious virus quantitation in cell culture showed that Ych was resistant to chloroform treatment and virus replication in DEFs was not significantly affected by addition of 5-bromo-2′-deoxyuridine in the maintenance medium. These results indicated that the isolate was a nonenveloped RNA virus.
Figure 1.
Characteristics of the isolated Ych strain. (a) Syncytium formation induced by Ych in DEF and Vero cells at 48 hours-post-infection (Giemsa stain). (b) Growth kinetics of Ych in DEF cells. Cells were infected with virus at a multiplicity of infection of 0.01. The titers represent the means ± SD (n = 3) from one of the three independent experiments. (c) SDS-PAGE analysis of the genomic segment mobility of HC (a duck reovirus), Ych and S1133 (a chicken reovirus). Full-length gel is presented in Supplementary Fig. S1.
To identify the isolate, genomic RNA was extracted from concentrated virus particles and constructed RNA library was subjected to next generation sequencing (NGS). Analysis of the NGS data revealed that Ych was a reovirus distinct from the known China-origin duck reovirus isolates. To confirm the isolate, we designed a primer pair specific for the Ych λC gene on the basis of the NGS sequence data, and reverse transcription-polymerase chain reaction (RT-PCR) was performed using the RNA template extracted from the original duck tissue sample. A gene fragment identical to that revealed by deep sequencing was obtained. We further analysed the electrophoretic mobilities of the viral double-strand RNA segments and revealed that the ten genomic segments displayed clearly distinguishable migration patterns from those of duck reovirus DRV-HC and ARV-S1133 (Fig. 1c). It was noticed that the S3 segment of Ych migrated more closely to S4 rather than the S2 segment in comparison with ARV-S1133 and DRV-HC. Differences were also evident in other segments with the exception of L1–L3.
Genomic analysis of the Ych isolate
Based on the NGS data, a draft genome sequence was first aligned and then verified by Sanger sequencing using designed primer pairs (Table 1). The resulting genome sequence of Ych was 23,434 nucleotides (nt) in length, and the genome organisation was similar to reference ARVs, consisting of 10 segments from 1190 nt (S4) to 3998 nt (L1) (GenBank accession numbers MK173029–MK173038) (Table 2). The 5′ untranslated region (UTR) of the positive-sense strand ranged in length from 12 to 30 nt, and the 3′-UTR ranged in length from 32 to 98 nt, containing the same 5′and 3′terminal nucleotide motifs (5′-GCUUUU…UAUUCAUC-3′) as described for other avian reoviruses16,18. Proteins encoded by representative avian reovirus homologous segments were identified in the genome (Table 2). The polycistronic S1 segment was predicted to encode putative proteins p10, p13 and σC, homologous to chicken ARV p10, p17 and σC, respectively19.
Table 1.
Primers used for amplification and sequencing of the isolate Ych.
| Gene segment | Primer name | Sequence (5′ → 3′) | Location | Product size (bp) |
|---|---|---|---|---|
| S1 | Ych-S1F | GCATGCAATGGTGGTACAGTG | 35–55 | 1506 |
| Ych-S1R | CTTACTGCGTGACATGGACC | 1521–1540 | ||
| S2 | Ych-S2F | GTACGAGTTTTTCTCTGTGCC | 30–50 | 1238 |
| Ych-S2R | CCTAATTGGTGAAAGTGGCC | 1248–1267 | ||
| S3 | Ych-S3F | CAATGGAGGTGCGTATGCC | 29–47 | 1049 |
| Ych-S3R | CTGAAGGTAGTGGGTCGTGTC | 1057–1077 | ||
| S4 | Ych-S4F | CAACACTTCTGCTGCTGCCG | 56–75 | 1055 |
| Ych-S4R | GGGAAACAGACAATAAGACG | 1091–1110 | ||
| M1 | Ych-M1-1F | CTATCTAGCCACACCCGTG | 18–36 | 1313 |
| Ych-M1-1R | CGTCACTATCCATAATAGTG | 1311–1330 | ||
| Ych-M1-2F | GATATCAGATGCTTCGGGAAG | 1074–1094 | 965 | |
| Ych-M1-2R | CAGCTACGATGCGAAATTCG | 2019–2038 | ||
| M2 | Ych-M2-1F | TATCGCTCACCATGGGCAAC | 20–39 | 1187 |
| Ych-M2-1R | CTCATTCGGATTGAACGAGCC | 1186–1206 | ||
| Ych-M2-2F | CCTCGCACTTACAACATCCG | 973–992 | 1145 | |
| Ych-M2-2R | CTGGCGTGGATTCAGCTTAAC | 2097–2117 | ||
| M3 | Ych-M3-1F | ATGATGGCGTCCACTAAGTGG | 22–42 | 1388 |
| Ych-M3-1R | CGATTCATACGTTGCAGATCC | 1389–1409 | ||
| Ych-M3-2F | CAGATATGGTAGCGTGTCGAC | 1242–1262 | 730 | |
| Ych-M3-2R | CGTCCATGATCCACGTTGAG | 1952–1971 | ||
| L1 | Ych-L1-1F | GCTCCAGTTTCTGAGAAGAAAG | 60–81 | 1249 |
| Ych-L1-1R | CTCTCGAGGGATACATGACC | 1289–1308 | ||
| Ych-L1-2F | TCTTGAGCAACTTGCACCTC | 1165–1184 | 1292 | |
| Ych-L1-2R | CATCGACGCTCTAATCGATTG | 2436–2456 | ||
| Ych-L1-3F | CCTATACGGATCACTAATCCG | 2316–2336 | 1589 | |
| Ych-L1-3R | CAATCGATTAGAGCGTCGATG | 3885–3904 | ||
| L2 | Ych-L2-1F | CAGTCAAAGGTGTTTTGGCC | 19–38 | 1337 |
| Ych-L2-1R | CTGAAATAGAGGGTCCCAAG | 1336–1355 | ||
| Ych-L2-2F | CACCCTATTGGTTCCTTACG | 1227–1246 | 1336 | |
| Ych-L2-2R | CTGAATAGGCTAGAGAAAGC | 2543–2562 | ||
| Ych-L2-3F | GTGATCGCTTGGAGATGTGG | 2434–2453 | 1324 | |
| Ych-L2-3R | CGCACAAAGTTCTGCATTCC | 3738–3757 | ||
| L3 | Ych-L3-1F | CAGATTCGAGGTTTGCGCTTG | 19–39 | 1344 |
| Ych-L3-1R | CAAGTCCAATGGATAACCAGC | 1342–1362 | ||
| Ych-L3-2F | ATTCCCTTTGCTGGCATGC | 1218–1236 | 1400 | |
| Ych-L3-2R | GGTAGTCCAACTGCATGTAG | 2598–2617 | ||
| Ych-L3-3F | CTTCCACTGGCTGGATTGTG | 2466–2485 | 1375 | |
| Ych-L3-3R | TGGAGGCACGTAGAAAGACG | 3821–3840 |
Table 2.
Genetic characteristics of duck reovirus strain Ych.
| Genome segment | Size (nt) | Length (nt) of | Encoded protein and size (aa) | Strain with highest amino acid similarity (%) | ||
|---|---|---|---|---|---|---|
| 5′UTR | ORF | 3′UTR | ||||
| L1 | 3998 | 20 | 3921 | 57 | λA (1306) | D2533/6/1-10 (98.1) |
| L2 | 3826 | 14 | 3780 | 32 | λB (1259) | D2533/6/1-10 (97.4) |
| L3 | 3899 | 12 | 3855 | 32 | λC (1284) | D2533/6/1-10 (94.9) |
| M1 | 2282 | 12 | 2199 | 71 | μA (732) | D2533/6/1-10 (93.3) |
| M2 | 2150 | 30 | 2022 | 98 | μB (673) | D2533/6/1-10 (98.5) |
| M3 | 1990 | 21 | 1908 | 61 | μNS (635) | D2533/6/1-10 (95.8) |
| S1 | 1573 | 22 | 282 | 34 | p10 (93) | D2533/6/1-10 (94.7) |
| 369 | p13 (122) | D2533/6/1-10 (92.7) | ||||
| 1014 | σC (337) | D2533/6/1-10 (91.7) | ||||
| S2 | 1325 | 15 | 1251 | 59 | σA (416) | D2533/6/1-10 (96.4) |
| S3 | 1201 | 30 | 1104 | 67 | σB (367) | D2533/6/1-10 (97.6) |
| S4 | 1190 | 23 | 1104 | 63 | σNS (367) | D2533/6/1-10 (97.0) |
Nucleotide and amino acid (aa) sequences of all segments of Ych were compared with representative members of Orthoreovirus species (Table 3 & Tables S1–S10 in Supplementary Materials). Moderate sequence identities were seen for the relatively conserved fragments encoding inner core proteins of ARVs, including the λA encoding gene (71.5–72.7% nt, 83.8–84.5% aa), λB (66.0–67.4% nt, 75.5–76.1% aa), λC (55.7–56.5% nt, 55.4–56.0% aa), μA (58.4–60.7% nt, 60.4–61.1% aa) and σA (63.6–64.3% nt, 66.7–67.4% aa). In terms of more divergent outer capsid proteins, Ych shared sequence identity values with those of the representative isolates of ARVs: μB (62.5–65.9% nt, 66.5–72.1% aa), σB (55.8–61.2% nt, 52.4–60.3% aa), and σC (37.3–43.6% nt, 22.2–26.7% aa). The nt and aa identity could not absolutely satisfy the species demarcation criteria in the genus Orthorovirus18, but fell into the grey zone of the cut-off values, with the exception of σC. Interestingly, Ych shared the highest nt/aa identity with the newly-described reovirus D2533/6/1-10 genome segments at a level ranging from 85.5–94.1% (nt) and 91.7–98.5% (aa), which was isolated from Pekin ducklings in Germany20.
Table 3.
Comparison of nucleotide and amino acid sequence identities of Ych with representative members of Orthoreovirus species (%).
| Strains | λA | λB | λC | μA | μB | μNS | σC | σA | σB | σNS | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ARV-D2533/6/1-10 | nt | 94.1 | 91.7 | 87.7 | 93.4 | 89.6 | 92.1 | 85.5 | 89.6 | 91.8 | 90.3 |
| aa | 98.1 | 97.4 | 94.9 | 93.3 | 98.5 | 95.8 | 91.7 | 96.4 | 97.6 | 97.0 | |
| ARV-Ch/Tua | nt | 72.0–72.8 | 66.6–67.3 | 55.2–56.1 | 59.4–60.3 | 64.2–65.7 | 57.6–58.9 | 33.6–37.0 | 63.6–64.7 | 54.8–58.7 | 61.2–62.9 |
| aa | 84.3–85.0 | 75.4–76.3 | 54.2–55.2 | 60.4–61.7 | 69.6–72.4 | 55.8–57.6 | 21.6–24.4 | 66.7–67.6 | 51.9–55.7 | 62.8–64.9 | |
| ARV-Wa Classicalb | nt | 71.3–72.3 | 65.8–66.5 | 56.0–56.4 | 57.6–60.6 | 62.4–62.7 | 57.7–58.5 | 36.0–38.0 | 63.3–64.8 | 59.1–60.2 | 61.3–62.7 |
| aa | 83.8–84.2 | 75.5–76.4 | 55.3–55.6 | 60.0–61.1 | 66.5–66.9 | 55.8–57.4 | 24.1–25.2 | 66.2–67.1 | 55.2–57.6 | 63.3–64.9 | |
| ARV-Wa Novelc | nt | 71.4–72.1 | 65.8–66.3 | 56.0–56.4 | 60.3–60.7 | 65.7–66.0 | 57.3–57.8 | 39.6–44.5 | 63.8–64.9 | 60.6–61.6 | 60.7–62.0 |
| aa | 83.8–84.6 | 75.6–75.9 | 55.8–56.0 | 60.7–61.3 | 71.2–72.0 | 55.5–56.6 | 26.7–27.3 | 66.7–67.4 | 59.2–60.6 | 63.0–64.1 | |
| NBVd | nt | 65.3–66.0 | 36.5–56.1 | 45.8–46.7 | 49.5–50.9 | 62.7–63.1 | 28.4–48.4 | 25.8–31.1 | 56.8–58.1 | 43.2–44.2 | 55.2–55.4 |
| aa | 72.2–72.6 | 36.7–40.1 | 36.4–40.8 | 45.3–46.2 | 68.3–69.2 | 7.6–38.3 | 6.5–17.7 | 56.1–56.6 | 30.0–30.3 | 51.1–51.6 | |
| BRVe | nt | 36.7 | 53.2 | 39.8 | 44 | 47.6 | 39.8 | — | 41.6 | 35.4 | 41.4 |
| aa | 8.4 | 49.4 | 27.9 | 31.9 | 39 | 25.8 | — | 29.9 | 16.9 | 25.6 | |
| RRVf | nt | 54.4 | 55.3 | 40.7 | 46.6 | 56.5 | 39.5 | 34.2 | 42.9 | 39.5–39.9 | 44 |
| aa | 49.7 | 56.3 | 28 | 38.7 | 53 | 24.8 | 16.1 | 33.8 | 20.4–21.3 | 31 | |
| MRVg | nt | 51.0–51.3 | 53.9–54.6 | 41.0–41.8 | 40.9–41.4 | 50.4–51.0 | 38.4–40.1 | 25.3–28.5 | 41.8–43.5 | 35.4–36.4 | 40.4–41.4 |
| aa | 42.9–43.5 | 52.2–52.7 | 28.2–28.6 | 26.8–27.4 | 44.9–45.7 | 20.9–21.7 | 11.6–15.4 | 27.2–28.4 | 14.7–16.4 | 23.1–24.0 | |
aChicken (S1133, 138, 176, AVS-B, GX/2010/1, T1781) and turkey (19831M09, 22342/13, D1246) origin ARVs.
b“Classical” waterfowl origin ARVs: Muscovy duck (ZJ2000M, 815-12, D1546, D2044) and goose (D20/99).
c“Novel” waterfowl origin ARVs: Muscovy duck (ZJ00M, NP03, J18), Pekin duck (TH11, HC, 091), wild Mallard duck (SD-12) and goose (03G).
dNBVs (AF059718.1, AF059722.1, AF059726.1, AF218360.1, JF342672.1-JF342677.1, JF342666.1-JF342671.1, AY357730.1-AY357733.3, JF811580.1-JF81153.1).
eBRVs (AF059719.1, AF059723.1, AF059727.1, HQ847903.1-HQ847908.1).
fRRVs (AY238886.1, KT696547.1-KT696556.1).
gMRVs: MRV-1 (B/03), MRV-2 (BYD1), MRV-3 (T3D, ZJ2013), MRV-4 (Ndelle).
“—”: no equivalent sequence.
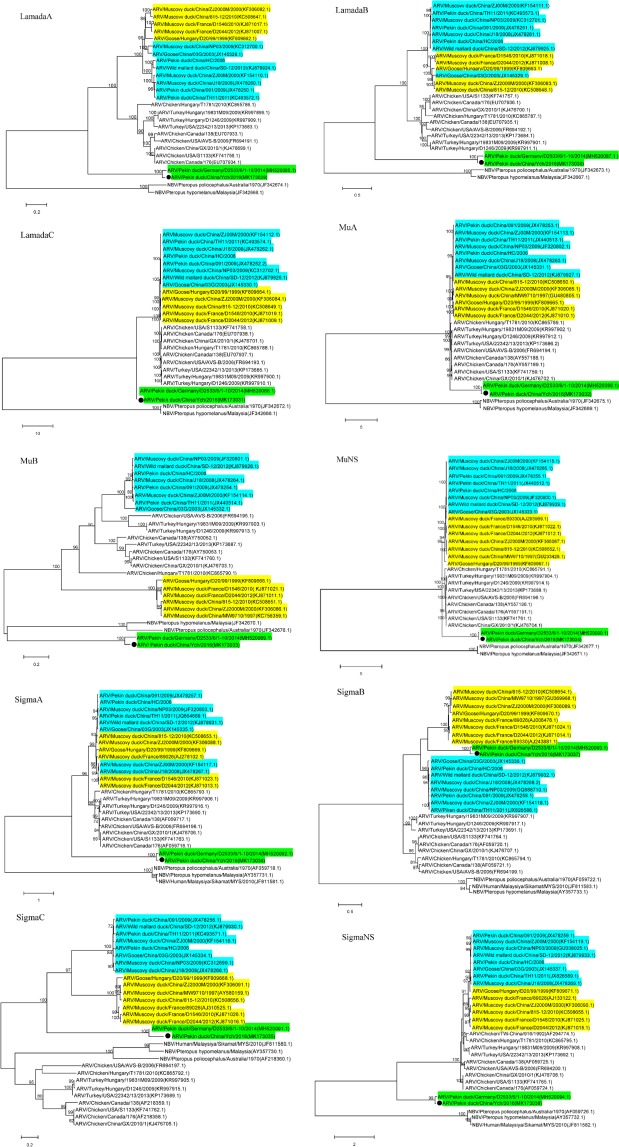
The open reading frames (ORFs)of the nucleotide sequences of ten segments were used to construct the phylogenetic trees (Fig. 2). Phylogenetic analyses revealed a strong evolutionary relationship with strain D2533/6/1-10, and Ych always appeared on the same monophyletic branch with the German isolate, which was distinct from other known orthoreovirus isolates. Based on our analyses, the waterfowl-origin reoviruses could be divided into three genogroups, (i) the “classical” Muscovy duck reovirus strains isolated from Muscovy ducks and geese, (ii) the “novel” duck reovirus strains, (iii) the newly-emerging duck reovirus strains Ych and D2533/6/1-10.
Figure 2.
Phylogenetic relationship of Ych to selected orthoreovirus species based on the nucleotide sequences of ten ORFs. Maximum likelihood trees were constructed using a General Time Reversible model (MEGA 7.0.14 program) with bootstrap values calculated from 1000 replicates. Bootstrap values lower than 0.7 were hidden. GenBank accession numbers of reference strains appear next to the virus names. The “classical” waterfowl origin ARVs, “novel” waterfowl origin ARVs and newly emergent duck reovirus are marked with yellow, blue and green backgrounds, respectively.
Serological characterisation
The serotype relationship of Ych with duck reovirus DRV-HC and ARV-S1133 was determined by virus neutralisation test (Table 4). The three strains were neutralised by homologous antiserum, however, they could not be cross neutralised by antibody against a heterologous strain. The result demonstrated that Ych was serologically different from DRV-HC and S1133.
Table 4.
Serological relationships between Pekin duck and chicken origin reoviruses studied by cross-neutralisation tests.
| Virus | Neutralisation titre of antisera to* | ||
|---|---|---|---|
| Ych | HC | S1133 | |
| Ycha | 4 | <1 | <1 |
| HCa | <1 | 5 | <1 |
| S1133b | <1 | <1 | 4 |
*(−log2).
aPekin duck origin ARVs.
bChicken origin ARV.
Results of experimental infection
Compared with uninfected controls, no abnormal clinical signs were noted in ducklings infected by oral/intranasal or subcutaneous inoculation during the experimental period, and neutralising antibody was not detected in sera collected from the infected ducks on 9, 12 and 16 dpi. However, virus-specific RNA could be sequentially detected from cloacal swabs of some infected ducklings from 1–4 dpi (Table 5), suggesting the virus replicated in ducklings. As 12-day-old ducklings were infected by simultaneous oral and subcutaneous inoculation, virus specific RNA was detected from caecal tonsil (1/5) and bursa of Fabricius (3/5) when ducklings were euthanised on 3 dpi. Virus could also be recovered after inoculation of the RT-PCR-positive samples into DEF monolayers. These data further indicate that the isolate replicated in the intestinal lymphoid tissue and bursa of Fabricius of infected ducklings. However, the virus was not detected in tissue samples collected on 5 dpi and thereafter, suggesting that the virus infection of the bursa was transient.
Table 5.
RT-PCR detection of reovirus Ych virus gene in cloacal swab samples from experimentally infected ducklings.
| Days post-infection | Infection route | ||
|---|---|---|---|
| Oral and intranasal (n = 8) | Subcutaneous (n = 7) | Mock-infected control (n = 5) | |
| 1 | 5/8 | 0/7 | 0/5 |
| 2 | 8/8 | 0/7 | 0/5 |
| 3 | 8/8 | 2/7 | 0/5 |
| 4 | 2/8 | 5/7 | 0/5 |
| 5 | 0/8 | 0/7 | 0/5 |
| 6 | 0/8 | 0/7 | 0/5 |
Histopathological examination of the tissues of infected ducklings revealed inter-follicle oedema of the bursa of Fabricius (Fig. 3a). The epithelial cells of the bursa underwent vacuolar degeneration and debris or mucin were seen inside the vesicles on 3 dpi (Fig. 3b). On 5 dpi, marked sloughing of epithelial cells and apparent goblet cell metaplasia were seen in the mucosal layer (Fig. 3c,d). Monocyte infiltration and central eosinophilic focus formation were observed in the medullar area of the bursa follicles (Fig. 3c,d,e). Lymphocytes exhibited apoptotic images characterised by nuclei margination (Fig. 3f). No abnormality was observed in bursa of uninfected ducklings (Fig. 3g,h).TUNEL assay revealed an increase in the number of apoptotic cells compared with the mock control (Fig. 3i,j). These results suggested that Ych infection could induce damage to the bursa of Fabricius of infected ducklings although viral replication persisted for a short period only.
Figure 3.
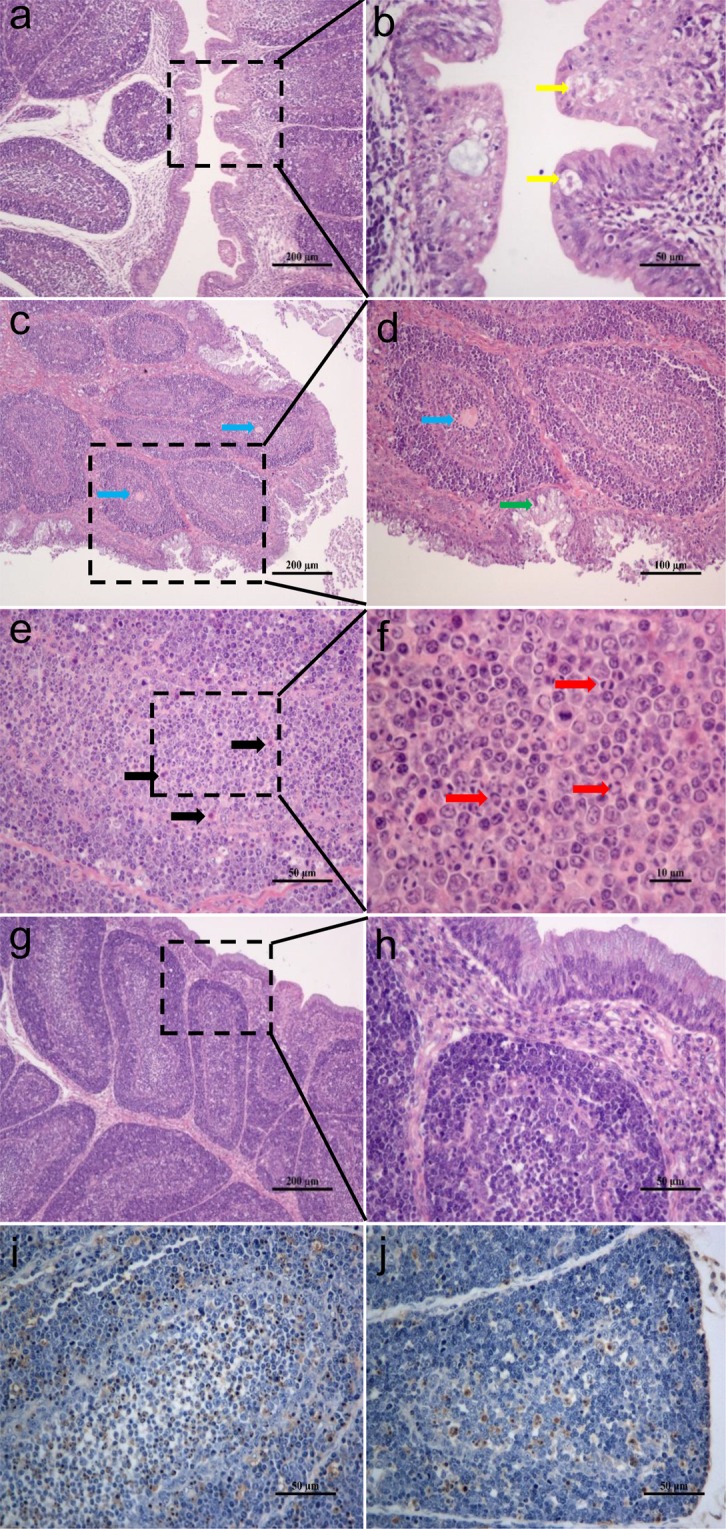
Microscopic lesions in the bursa of ducklings infected with reovirus Ych isolate. (a,b) showing the inter-follicle edema, vacuolation of epithelial cells (yellow arrow) (HE stain) (Scale bar = 200 & 50 μm, respectively). (c,d) showing the epithelial cell sloughing, goblet cell metaplasia in the mucosal layer (green arrow) and central eosinophilic focus (blue arrow) (HE stain) (Scale bar = 200 & 100 μm, respectively). (e,f) showing monocyte infiltration (black arrow) and cells with chromatin margination (red arrow) (HE stain) (Scale bar = 50 & 10 μm, respectively). (g,h) showing bursa sections of uninfected ducklings (HE stain) (Scale bar = 200 & 50 μm, respectively). (i,j) Marking of apoptotic cells of one infected duckling at 5 dpi (i) and uninfected control (j) with TUNEL assay. Haematoxylin stain was used as a cytoplasmatic contrast (Scale bar = 50 μm).
Discussion
ARVs have been demonstrated to be involved in numerous diseases in commercial poultry and the pathogenicity of isolates differs considerably. Differences in pathogenicity have been described for a number of avian reoviruses6,7,21,22, and high morbidity and mortality with multifocal hepatic and spleen necrosis at necropsy have been recorded for the majority of duck and goose reovirus infections9–11,13,15,23–25. In this work, a reovirus Ych was isolated from tissue samples of diseased Pekin ducks, characterised by intestinal haemorrhage. The isolate could induce syncytia formation in infected cell cultures but did not cause duck embryo death as previously described for pathogenic duck reoviruses. Genomic analyses showed that Ych shared the common genome structural characteristics and the predicted polycistronic S1 genome segment, but it was genetically distant from known reovirus isolates identified in China10. It is interesting that the greatest sequence similarity values of the ten genome segments were seen with the recently described reovirus strain D2533/6/1-10, which was isolated from a bursa sample of Pekin ducks in Germany20. Phylogenetic analysis of all genes revealed that the two isolates form a distinct branch from reference ARVs, suggesting that they might have a common evolutionary origin. Based on the relatively small amount of ARV sequence data available in GenBank, no attributable genomic reassortment or intra-segment recombination was found for the evolutionary origin of the newly emergent duck reovirus. Cross neutralisation results demonstrated that Ych belongs to a serotype distinct from the “novel” duck reovirus and chicken reovirus S1133.
In this study, inoculating ducklings with Ych by oral, intranasal or subcutaneous routes, which are likely natural transmission routes for avian reoviruses, did not produce clinical disease and gross lesions, as were observed in the field cases. However, virus shedding from the infected ducklings was detected by RT-PCR, suggesting that the virus multiplied transiently in the infected duck intestine. However, virus shedding via faeces implies that infected ducklings may transmit virus laterally to pen mates as seen in other reoviruses in chickens and turkeys26. It was noticed that the German strain D2533/6/1-10 was isolated from a sample of duck bursa although its pathogenicity was not further evaluated20. Our experimental infection results demonstrate that the virus replicated in the bursa of Fabricius. These findings are also in agreement with previous reports that the intestine and bursa of Fabricius were the initial sites of replication of some chicken and turkey reovirus isolates27,28. Microscopic examination revealed damage to the bursa epithelial cells and increased number of apoptotic lymphocytes, suggesting that Ych infection might induce a transient and possibly permanent immunosuppression in infected ducklings. This might explain to some extent that no specific neutralising antibody against the virus was detected from the sera of infected ducklings until the end of observation period. However, the fact that virus was not detected in the spleen, liver, kidney or lung samples of the experimentally infected ducks suggests that Ych was noninvasive for these internal organs, thus affecting also the humoral immune response. This scenario has been reported in chickens for reoviruses possessing low pathogenicity29.
Describing the aetiological role of an ARV isolate can be complicated because some clinical disease states are often difficult to reproduce experimentally30. Haemorrhagic enteritis as seen in field cases was not reproduced in this study. Our experimental infection results demonstrate that the bursa of Fabricius may serve as a site of Ych replication in infected ducklings, and whether the increase of apoptotic lymphocytes could result in lymphoid cell deletion is under investigation. It was possible that under field conditions, the virus infection induced immunosuppression, predisposing the ducklings to infection with other pathogens, which would worsen the disease and lead to death.
In summary, diagnostic attempts at Pekin duck haemorrhagic enteritis led to the isolation of a new reovirus, Ych, genetically distinct and antigenically different from previous duck reovirus strains identified in China. Experimental infection of ducklings indicated that the virus was able to replicate in the bursa of Fabricius and induce epithelial cell damage and lymphoid cell apoptosis. The virus-induced damage to this immune organ implies that the newly emergent reovirus infection may serve as a priming or concurrent agent in the development of duck enteritis. However, the role of Ych in haemorrhagic enteritis needs to be further evaluated in combination with other agents or substances.
Materials and Methods
Virus isolation
Twenty-day-old Pekin ducks that died at a farm in Yucheng, Shandong Province were necropsied. Pathological examination revealed mild to moderate swollen liver and intestinal mucosa haemorrhagic plaques. Samples of liver, spleen and brain tissue were inoculated onto tryptic soy agar (BD, Sparks, MD, USA) plates containing 2% fetal bovine serum and incubated at 37 °C for 48 h for bacterial examination. Virus isolation was performed later. The intestinal mucus, liver and spleen samples were pooled and homogenised in sterile phosphate-buffered saline solution (pH 7.2) to give a 20% suspension (w/v). After centrifugation at 8,000 × g for 10 min, the supernatant was filtered through a 0.22 μm pore-size sterile filter and inoculated into five 9-day-old duck embryos (0.2 mL/embryo) by the chorioallantoic membrane route. Embryos were candled daily and were chilled at 4 °C to death on 5 days post-inoculation (dpi) to harvest allantoic fluid for subsequent passage. Since no consistent embryo death was observed during the serial embryo-passage, we inoculated the allantoic fluid of the fourth passage onto confluent monolayers of duck embryo fibroblasts. The syncytium formation of the infected cells was visualised by Giemsa staining. To test the growth of the isolate in Vero cells, the DEF-adapted virus suspension was inoculated onto the monolayer of Vero cells using the conventional method.
Virus characterisation
To determine whether the isolated virus had a lipid envelope, the DEF-adapted virus suspension was clarified by low speed centrifugation after two freeze/thaw cycles and treated with chloroform (5%, v/v) for 10 min at room temperature. The chloroform layer was then removed by centrifugation and the infectious virus titre of the supernatant was quantified in DEF culture after serial dilution. For nucleic acid type determination, virus replication ability was tested in DEF culture with or without addition of 5-bromo-2′-deoxyuridine (BrdU, 50 μg/mL) in the maintenance medium. Duck Tembusu virus and duck enteritis virus were used as RNA and DNA virus controls, respectively.
Virus RNA extraction and high-throughput sequencing
Based on the fact that the isolate was a non-enveloped RNA virus, we extracted viral RNA from the concentrated virus. Briefly, the virus-infected DEF cell suspension was centrifuged at 8,000 × g for 30 min at 4 °C to remove cell debris. The clarified supernatant was further centrifuged by ultracentrifugation at 100,000 × g for 2 h. After removing the supernatant, the pellet was resuspended in distilled water to 1/100 volume of the original supernatant and total RNA was extracted from the suspension with TRIzol (Invitrogen, Carlsbad, CA, USA). Using the extracted RNA as template, an RNA library was prepared with NEBNext® Ultra™ RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) and submitted for NGS with the Illumina HiSeq4000 (Shanghai Hanyu Bio-Tech Co., Ltd, Shanghai, China).
Genome sequence analysis
On the basis of the preliminary sequence data obtained from NGS, a set of gene-specific oligonucleotide primer pairs (Table 1) were designed. The genomic fragments were then amplified by RT-PCR and sequenced by the Sanger method to further verify the viral genomic sequence. The 5′- and 3′- ends of each segment were amplified using the 5′/3′ rapid amplification of cDNA end kit (Clontech, Mountain View, CA, USA) following the guidelines of the manufacturer.
The initial complete genome was assembled and manually edited using ContigExpress software (ContigExpress LLC, New York, NY, USA). ORFs were predicted using online software ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The putative protein functions of respective ORFs were then inferred by BLASTp in the non-redundant protein database. The nucleotide and amino acid sequence identity were analysed using the Clustal W method of MegAlign software (DNASTAR, Madison, WI, USA). Maximum likelihood phylogenetic analyses based on 10 ORFs nucleotide sequences were conducted using the General Time Reversible model using MEGA 7.0.14 software (www.megasoftware.net), and estimates based on bootstrap resampling were carried out with 1000 replicates.
Electrophoresis of virus RNA
To compare the electrophoretic mobilities of the genomic segments of the isolate with representative ARV strains, the virus was propagated in DEFs and total RNA was extracted from the ultracentrifugation-concentrated virus suspension using TRIzol as described above. The extracted RNA was then treated with DNase (NEB, Ipswich, MA, USA) and S1 Nuclease (Promega, Madison, WI, USA) to remove DNA and single-stranded RNA. The duck reovirus isolate HC and ARV S1133 were propagated in DEF and chicken fibroblasts, respectively, as described10, and RNA was extracted and treated individually in the same way then used as controls. Treated RNA samples were subjected to electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) at 180 V for 7 h and nucleic acid bands were stained using rapid sliver staining kit according to manufacture’s guideline (YEASEN Biotechnology Co., Ltd., Shanghai, China).
Preparation of antiserum and serological assays
Antisera against the isolated virus, duck reovirus HC and ARV S1133 were separately prepared in 3-week-old SPF chickens (Merial-Vital Laboratory Animal Technology Co., Ltd., Beijing, China) by four immunisations at 2-week intervals with inactivated virus suspension. Sera were collected 12 days after the last immunisation. For the cross-reactivity assay, heat-inactivated homologous or heterologous antiserum was 2-fold serially diluted and mixed with an equal volume of the virus suspension containing 200 TCID50/0.1 mL (50% tissue culture infective dose). After incubation at 37 °C for 1 h, the serum-virus mixture was inoculated into five wells of DEF monolayers in a 96-well plate. Cytopathic effect (CPE) was recorded for 5 days. The neutralising antibody titre was the highest serum dilution that resulted in the cells being completely protected.
Experimental infection of ducklings
One-day-old Pekin ducklings without maternal antibody against duck reovirus were purchased from Nankou Hatchery (Beijing Golden Star Duck Co., Ltd., Beijing, China). Ducklings were raised in negative pressured isolators with ad libitum access to feed and water. In experiment 1, twenty 2-day-old ducklings were divided into three groups and infected with the cell culture-prepared virus suspension as indicated in Table 5. Ducklings in group 1 were orally and intranasally infected with 0.5 mL cell culture suspension containing 7.35 × 104 TCID50 of the isolate. Group 2 were subcutaneously infected with the same dose, and group 3 were raised separately as mock-infected controls. Clinical signs were recorded for 16 days to evaluate the pathogenicity of the isolate for ducklings, and cloacal swabs were collected as indicated for detection of virus shedding. On 9, 12 and 16 dpi, serum samples were collected for antibody detection using the virus neutralisation test as described above. To evaluate the in vivo infectivity of the isolate, ten ducklings at 12 days old were infected by oral and subcutaneous inoculation with the same dose virus as described above. Five ducks were sacrificed on both 3 and 5 dpi, respectively, and liver, spleen, caecal tonsil, thymus and bursa of Fabricius samples were collected for virus detection by RT-PCR and re-isolation. Five uninfected control ducklings were examined in the same way at each sampling point.
Detection of viral RNA in tissue samples by RT-PCR
The viral RNA from tissue samples and cloacal swabs were extracted by viral RNA kit (Omega Bio-tek, Norcross, GA, USA) and converted to cDNA using a Reverse Transcription System (Promega, Madison, WI, USA) with specific primers following the manufacturer’s instructions. Specific primers based on the λC gene were designed and synthesised (Ych-4F: 5′-CTAAAGCTATTGACGTGGTGC-3′; Ych-4R: 5′-GGTAGTCCAACTGCATGTA G-3′). The length of RT-PCR product was 557 bp.
Histopathological examination
Histological sections were routinely prepared from the bursa after samples were fixed in 10% neutral buffered formalin solution and paraffin embedded. The sections were stained with haematoxylin and eosin (HE). For apoptosis detection, TUNEL assay (dUTP nick end labelling) was conducted using an In Situ Cell Detection kit (Roche, Mannheim, Germany) following the manufacturer’s instructions.
Ethics statement
Animal infection experiments were approved by the China Agricultural University Animal Ethics Committee, in accordance with the guidelines of the Review of Welfare and Ethics of Laboratory Animals approved by the Beijing Municipality Administration Office of Laboratory Animals. Experiments involving reovirus infections were conducted in the Biosafety Level 2 facilities in College of Veterinary Medicine, China Agricultural Unversity.
Supplementary information
Acknowledgements
This work was supported by National Key R&D Program under Grant 2016YFD0500800 and National Natural Science Foundation of China (NSFC) under Grant 31372461. We thank Professor Beiyu Jiang from Beijing Academy of Agricultural and Forestry Sciences for providing reovirus S1133 strain. We thank Gillian Campbell, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author Contributions
Y.C., X.H. and J.S. designed the experiments; Y.C., M.S. and J.W. performed the experiments; Y.C., J.S., X.H. and W.H. analyzed the results; Y.C. processed the images and graphs; Y.C. and J.S. wrote the paper. All authors amended and commented on the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xueying Hu, Email: hxying@mail.hzau.edu.cn.
Jingliang Su, Email: suzhang@cau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44178-3.
References
- 1.Hollmén T, et al. Isolation and characterization of a reovirus from common eiders (Somateria mollissima) from Finland. Avian Diseases. 2002;46:478–484. doi: 10.1637/0005-2086(2002)046[0478:IACOAR]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 2.Sakai K, et al. Novel reovirus isolation from an Ostrich (Struthio camelus) in Japan. Vet Microbiol. 2009;134:227–232. doi: 10.1016/j.vetmic.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 3.Yu, K. et al. Complete genome sequence of an avian reovirus isolated from wild mallard ducks in china. Genome Announc2, 10.1128/genomeA.00813-14 (2014). [DOI] [PMC free article] [PubMed]
- 4.Stys-Fijol N, Kozdrun W, Czekaj H. Detection of Avian Reoviruses in Wild Birds in Poland. J Vet Res. 2017;61:239–245. doi: 10.1515/jvetres-2017-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benavente J, Martinez-Costas J. Avian reovirus: structure and biology. Virus Res. 2007;123:105–119. doi: 10.1016/j.virusres.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Olson NO, Weiss R. Similarity between arthritis virus and Fahey-Crawley virus. Avian Diseases. 1972;16:535–540. doi: 10.2307/1588670. [DOI] [PubMed] [Google Scholar]
- 7.Simmons DG, Colwell WM, Muse KE, Brewer CE. Isolation and characterization of an enteric reovirus causing high mortality in turkey poults. Avian Diseases. 1972;16:1094–1102. doi: 10.2307/1588834. [DOI] [PubMed] [Google Scholar]
- 8.Gaudry, D., Charles, J. M. & Tektoff, J. A new disease expressing itself by a viral pericarditis in Barbary ducks. C R Acad Sci Hebd Seances Acad Sci D274, 2916–2919 (1972). [PubMed]
- 9.Palya V, et al. Reovirus identified as cause of disease in young geese. Avian Pathol. 2003;32:129–138. doi: 10.1080/030794502100007187. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, et al. Isolation and characterization of a reovirus causing spleen necrosis in Pekin ducklings. Vet Microbiol. 2011;148:200–206. doi: 10.1016/j.vetmic.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Wu B, et al. Isolation and identification of Muscovy duck reovirus. Journal of Fujian Agricultural University. 2001;30:227–230. [Google Scholar]
- 12.Hu Q, et al. Cloning and sequence analysis of the muscovy duck reovirus MW9710 strain S1 gene fragment. Chinese Journal of Preventive Veterinary Medicine. 2004;26:22–25. [Google Scholar]
- 13.Chen S, et al. The isolation and identification of novel duck reovirus. Chinese journal of virology. 2012;28:224–230. doi: 10.1016/j.virol.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Farkas SL, et al. Detection of shared genes among Asian and European waterfowl reoviruses in the whole genome constellations. Infect Genet Evol. 2014;28:55–57. doi: 10.1016/j.meegid.2014.08.029. [DOI] [PubMed] [Google Scholar]
- 15.Yun T, et al. Complete genomic sequence of goose-origin reovirus from China. J Virol. 2012;86:10257. doi: 10.1128/JVI.01692-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun T, et al. Genomic characteristics of a novel reovirus from Muscovy duckling in China. Vet Microbiol. 2014;168:261–271. doi: 10.1016/j.vetmic.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Zhu YQ, et al. Molecular characterization of a novel reovirus isolated from Pekin ducklings in China. Arch Virol. 2015;160:365–369. doi: 10.1007/s00705-014-2241-x. [DOI] [PubMed] [Google Scholar]
- 18.Andrew, M. Q. King, M. J. A., Eric, B. C. & Elliot J. L. Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier, 521–528 (2011).
- 19.Bodelon G, Labrada L, Martinez-Costas J, Benavente J. The avian reovirus genome segment S1 is a functionally tricistronic gene that expresses one structural and two nonstructural proteins in infected cells. Virology. 2001;290:181–191. doi: 10.1006/viro.2001.1159. [DOI] [PubMed] [Google Scholar]
- 20.Farkas SL, et al. Genomic sequence and phylogenetic analyses of two novel orthoreovirus strains isolated from Pekin ducks in 2014 in Germany. Virus Res. 2018;257:57–62. doi: 10.1016/j.virusres.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Jones RC. Avian reovirus infections. Revue scientifique et technique (International Office of Epizootics) 2000;19:614–625. doi: 10.20506/rst.19.2.1237. [DOI] [PubMed] [Google Scholar]
- 22.Robertson MD, Wilcox GE, Kibenge FS. Prevalence of reoviruses in commercial chickens. Australian veterinary journal. 1984;61:319–322. doi: 10.1111/j.1751-0813.1984.tb07137.x. [DOI] [PubMed] [Google Scholar]
- 23.Banyai K, et al. The goose reovirus genome segment encoding the minor outer capsid protein, sigma1/sigmaC, is bicistronic and shares structural similarities with its counterpart in Muscovy duck reovirus. Virus genes. 2005;31:285–291. doi: 10.1007/s11262-005-3243-2. [DOI] [PubMed] [Google Scholar]
- 24.Malkinson M, Perk K, Weisman Y. Reovirus infection of young Muscovy ducks (Cairina moschata) Avian Pathol. 1981;10:433–440. doi: 10.1080/03079458108418493. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, et al. Sequence and phylogenetic analysis of M-class genome segments of novel duck reovirus NP03. The Canadian Journal of Veterinary Research. 2015;79:147–150. [PMC free article] [PubMed] [Google Scholar]
- 26.Al Afaleq AI, Jones RC. Localisation of avian reovirus in the hock joints of chicks after entry through broken skin. Res Vet Sci. 1990;48:381–382. doi: 10.1016/S0034-5288(18)31032-4. [DOI] [PubMed] [Google Scholar]
- 27.Jones RC, Islam MR, Kelly DF. Early pathogenesis of experimental reovirus infection in chickens. Avian Pathol. 1989;18:239–253. doi: 10.1080/03079458908418599. [DOI] [PubMed] [Google Scholar]
- 28.Pantin-Jackwood M. J., Spackman E., Day J. M. Pathology and Virus Tissue Distribution of Turkey Origin Reoviruses in Experimentally Infected Turkey Poults. Veterinary Pathology. 2007;44(2):185–195. doi: 10.1354/vp.44-2-185. [DOI] [PubMed] [Google Scholar]
- 29.Lenz SD, Hoerr FJ, Ellis AC, Toivio-Kinnucan MA, Yu M. Gastrointestinal pathogenicity of adenoviruses and reoviruses isolated from broiler chickens in Alabama. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 1998;10:145–151. doi: 10.1177/104063879801000205. [DOI] [PubMed] [Google Scholar]
- 30.Songserm Thaweesak, van Roozelaar Dirk, Kant Arie, Pol Jan, Pijpers Anton, Agnes ter Huurne Enteropathogenicity of Dutch and German avian reoviruses in SPF white leghorn chickens and broilers. Veterinary Research. 2003;34(3):285–295. doi: 10.1051/vetres:2003004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.