Abstract
Pseudomonas syringae produces highly efficient biological ice nuclei (IN) that were proposed to influence precipitation by freezing water in clouds. This bacterium may be capable of dispersing through the atmosphere, having been reported in rain, snow, and cloud water samples. This study assesses its survival and maintenance of IN activity under stressing conditions present at high altitudes, such as UV radiation within clouds. Strains of the pathovars syringae and garcae were compared to Escherichia coli. While UV-C effectively inactivated these cells, the Pseudomonas were much more tolerant to UV-B. The P. syringae strains were also more resistant to radiation from a solar simulator, composed of UV-A and UV-B, while only one of them suffered a decline in IN activity at −5 °C after long exposures. Desiccation at different relative humidity values also affected the IN, but some activity at −5 °C was always maintained. The pathovar garcae tended to be more resistant than the pathovar syringae, particularly to desiccation, though its IN were found to be generally more sensitive. Compared to E. coli, the P. syringae strains appear to be better adapted to survival under conditions present at high altitudes and in clouds.
Subject terms: Atmospheric science, Air microbiology
Introduction
The Gram-negative bacterium Pseudomonas syringae is a common member of epiphytic communities and an important phytopathogen in diverse crops1. It was the first organism found to produce biological ice nuclei (IN), being able to freeze supercooled water with exceptional efficiency at temperatures above −10 °C2. The IN activity of this organism originates from a large protein situated at the cell’s outer membrane, which forms multimeric clusters that structure water into an ice-like array promoting its phase change3,4. Homologs of this protein, called InaZ, have since been identified in other ecologically similar species also active as IN. This trait has been linked to the increased susceptibility to frost damage above −5 °C of plants harboring populations of these bacteria on their leaves, a significant concern to agriculture5.
The particular ability of P. syringae and similar organisms to form efficient IN has been long suggested to possibly influence atmospheric processes6,7. Glaciation, the freezing of the droplets that compose clouds, is an important mechanism leading to precipitation (including hail formation), and is largely determined by IN particles present in suspension in the air. Due to the reduced vapor pressure over ice crystals than supercooled liquid water, frozen particles can accumulate water and grow to sizes large enough to start the precipitation process inside the cloud8,9. This is known as the Wegener-Bergeron-Findeisen process, and its significance is evidenced by observations such as that the ice phase of clouds is the main source of rain in continental areas across the globe10. An additional mechanism that can amplify the influence of IN is the Hallett-Mossop process, the rapid multiplication by orders of magnitude of secondary ice crystal fragments caused by the riming and splintering of primary ice surfaces11. Since this occurs predominantly between −3 and −8 °C, temperatures where biological IN are the major active nuclei present in the environment8, this has been proposed as another potential contribution that organisms like P. syringae can have for the precipitation cycle6.
In addition to cloud glaciation, microbial cells can also exhibit activity as cloud condensation nuclei (CCN) in warm clouds12. These aerosol particles are essential for the condensation of water vapor into the liquid droplets that make up clouds. Interestingly, besides their IN activity, studies with bacteria of the Pseudomonas genus have also shown their ability to produce biosurfactants that can act as highly efficient CCN13–15. Further research of microbial life in the atmosphere also includes the effects of cells on cloud chemistry, particularly in relation to the metabolism of organic compounds in its aqueous phase16.
Multiple works have reported the presence of cultivable P. syringae and other ice nucleating bacteria in rain and snow samples (Morris, et al.17 and Šantl-Temkiv, et al.18, for example). Recently, Stopelli, et al.19 isolated with selective culture media IN-active P. syringae from snow collected at an altitude of 3,580 m at Jungfraujoch, Switzerland. Furthermore, these organisms have also been isolated directly from clouds20,21. Members of the Pseudomonas genus were the most frequently identified bacterial isolates from cloud water samples collected at an altitude of 1,465 m at the puy de Dôme summit in France between 2007 and 201022. A number of Pseudomonas strains were also isolated from clouds and rain at the Outer Hebrides, Scotland, although those did not present IN activity13. These evidences point to the widespread distribution of these bacteria in the atmosphere and support their relationship with clouds and the precipitation cycle.
Besides Pseudomonas, a concentration of about 104 total bacterial cell numbers per cubic meter of air is estimated to be found typically over land, though this figure may significantly change with the altitude, weather, season, and the underlying ecosystem23,24. Particles with the size of bacteria have a relatively long residence time in the air, on the order of days, during which they have the potential to cross long distances24,25. Effective dispersal trough this medium is, however, conditioned to cell survival as aerosols, which is a considerable challenge in this situation26.
The viability of aerosolized bacteria can be severely limited by atmospheric factors. Particularly, dehydration and exposure to ultraviolet (UV) light have been found to be the major barriers for successful aerial transportation of microorganisms26. Even inside clouds, cells are still subjected to UV, and additionally to low temperatures, freezing, and chemical stresses such as low pH and oxidizing species16,27. An important means of escape from this situation may be through precipitation, which can be facilitated by the IN activity of the biological particle, as mentioned above. Both field measurements and laboratory studies have shown that cells with this activity can be preferentially precipitated from clouds in this manner, more so than non-nucleating particles19,28,29. In this way, ice nucleation could be a valuable feature for the surviving airborne bacteria to return to the ground and again be able to multiply and propagate.
A few previous studies explored the response of P. syringae to simulated solar radiation in the context of atmospheric survival. Attard, et al.30 reported a reduction in viability and a somewhat small impact in IN activity of Pseudomonas strains after prolonged exposure to UV-A light. In contrast, Joly, et al.27 measured no loss of viability for two P. syringae strains exposed to a source emitting UV-B, but with a much reduced UV-A output compared to the proportion in which this wavelength range is present in the solar spectrum. IN were not evaluated in this work. Here, we improve upon these findings with a more detailed investigation of the effects of different UV ranges at a series of fluences on cell survival and ice nucleation.
Two strains of P. syringae were used in this study: pv. syringae IBSBF 281T (pathovar type strain) and pv. garcae IBSBF 158, previously reported to be IN-active by Gonçalves and Massambani31. In addition to UV, the strains were also tested against desiccation at different relative humidity (RH) values (33% and <5%), a still much less understood factor for these bacteria in this context. In this manner, the aim of the experiments was to simulate particularly important26 individual conditions that Pseudomonas would be exposed to in the high atmosphere that could constrain their dispersion through the air. Nevertheless, it is worth mentioning that both tested stress factors, solar light and dehydration, can also be found at different degrees on their natural plant surface habitat. We compared the effects of distinct UV radiation sources: narrow-band UV-C and UV-B lamps, commonly used in photobiological studies, and UV-A + UV-B from a solar simulator. Previously, microorganisms have been shown to exhibit significantly different responses to these UV sources32, making this a novel, relevant exploration of P. syringae resilience. Additionally, a strain of Escherichia coli, another Gram-negative gamma-proteobacterium like Pseudomonas, but non-ice nucleation active and not a common inhabitant of plant surfaces, was used for comparison. With its status as a model organism, while not being expected to present substantial tolerance to environmental stresses present outside its usual habitat in the mammalian gut, E. coli is widely used for distinguishing the resistance of bacteria, often as a non-tolerant reference33,34. As follows, our study provides new information about microorganisms in the atmospheric environment testing varied conditions with different strains, and as can be seen next, a broad range of responses were measured.
Results
Ice nucleation activity of the P. syringae strains
Ice nucleation profiles across a range of temperatures were determined by the droplet freezing assay35 for the P. syringae strains (Fig. 1). Cells were grown in nutrient-limited LNP medium36 and at cultivation conditions chosen to allow for maximum expression of the IN phenotype. Different to what was previously reported by Gonçalves and Massambani31, the pv. syringae strain (281) had a stronger measured IN activity than the pv. garcae strain (158). No IN active at −3 °C could be reliably detected for 158, while 9 ± 5 × 10−6 nuclei per cell could be measured at this temperature for 281. Despite that, the overall IN spectrum of both strains was mostly similar for the remaining temperatures tested. For the other experiments, only the nuclei concentration at −5 °C was chosen to be studied. This temperature was used for being at the lower range for maximum environmental significance of the IN activity, be it for causing frost damage on leaves5 or affecting cloud glaciation6,11.
Figure 1.

Ice nucleation spectrum of the P. syringae strains. Cultures of P. syringae pv. syringae 281 and P. syringae pv. garcae 158 grown in LNP medium at 15 °C were tested with the droplet freezing assay to quantify their ice nuclei (IN) concentration at decreasing temperatures. The data points at warmer temperatures represent the initial temperature at which frozen droplets could be detected for each strain. Error bars denote standard deviations of the means (n = 5).
P. syringae survival and ice nucleation activity following exposure to different UV ranges
The UV resistance of the microorganisms was first tested with common laboratory narrow-band (monochromatic) UV-C and UV-B sources. Spectra of all the lamps used for the experiments can be observed in Fig. 2, and fluence values and intensities were measured by a Vilber Loumart radiometer (RMX-3W, Marne-la-Vallée, France). The survival curve to UV-C radiation (254 nm) of P. syringae pv. syringae 281 remained within 1 standard deviation of E. coli, while P. syringae pv. garcae 158 presented a generally slightly higher mean survival but still overlapped with E. coli at the final 120 J/m2 point (Fig. 3). Under our tested conditions, at a fluence of 60 J/m2, the CFU counts of 281 were reduced to 0.3 ± 0.1% of the initial population, which was of about 5 × 106 cells/ml for both strains for all experiments. This can be compared to 158, for which this UV-C fluence caused just over a 1 log decrease in viability (6 ± 3% survival). In any case, both Pseudomonas can be considered sensitive to this radiation range. It is important to note, though, that solar UV-C is mostly absent below the highest reaches of the atmosphere.
Figure 2.
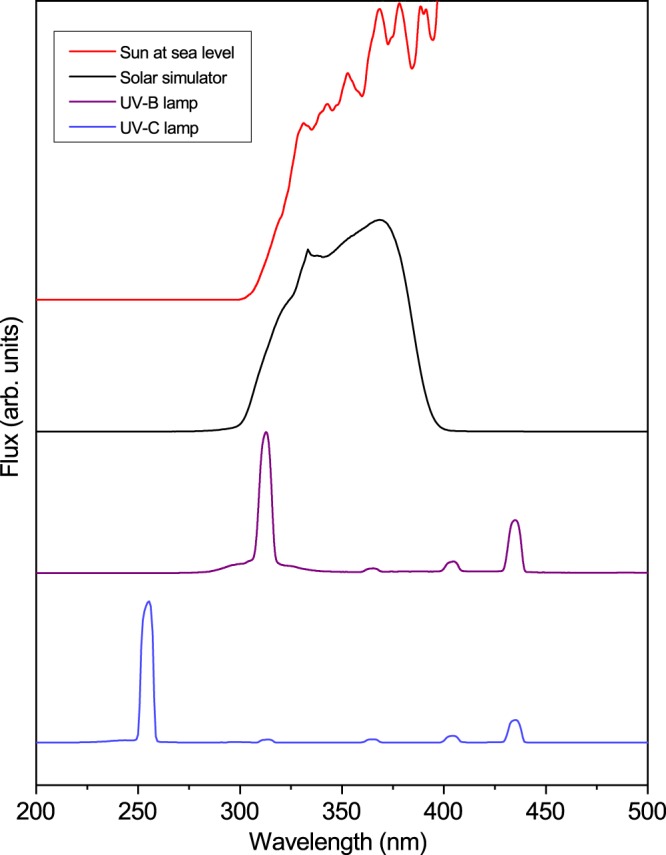
Comparison of the spectra of the lamps used for the experiments. From top to bottom (in arbitrary flux unities): the Sun over Earth’s surface (ASTM G173-03, smoothed), the Oriel Sol-UV-2 solar simulator, and the UV-B and UV-C lamps, measured with an Ocean Optics QE65000 spectrometer. It is apparent that the commonly used UV-B and UV-C narrow-band lamps largely differ from the UV in the real environment.
Figure 3.

Survival curves to UV-C (254 nm) radiation. P. syringae strains were exposed as 5 × 106 cells/ml suspensions, and E. coli as 5 × 107 cells/ml. P. syringae pv. syringae 281 and P. syringae pv. garcae 158 are shown to be as sensitive as E. coli to this UV range often used in laboratory experiments but absent on Earth’s surface. Error bars denote standard deviations of the means (n = 3).
For the UV-B (312 nm) assays, the observed survival of the P. syringae strains was considerably greater than E. coli (Fig. 4). At a fluence of 5,000 J/m2, E. coli was inactivated by nearly 3 orders of magnitude, while both 281 and 158 still retained 31 ± 10% and 67 ± 15% of viability, respectively. Treatment with higher fluences (up to 20,000 J/m2) evidenced a larger UV tolerance of 158 compared to 281, which was inactivated by over 4 logs at this final point (not being reliably recovered for proper quantification). Despite being present in the environment, the UV-B of the narrow-band lamps poorly represents the real solar spectrum. For a more faithful recreation of UV in Earth’s atmosphere, an Oriel solar simulator (Sol-UV-2, Bozeman, USA) was used (Fig. 2).
Figure 4.

Survival curves to UV-B (312 nm) radiation. P. syringae strains were exposed as 5 × 106 cells/ml suspensions, and E. coli as 5 × 107 cells/ml. P. syringae pv. syringae 281 and P. syringae pv. garcae 158 are shown to tolerate much larger UV-B fluences than E. coli, while 158 presented the highest resistance. Error bars denote standard deviations of the means (n = 4).
For the “environmental” UV experiments with the solar simulator, cell suspensions were exposed placed on top of a frozen foam block to avoid evaporation and excessive heating. The intensities used were 75.5 W/m² for the UV-A and 48.7 W/m² for the UV-B at the ranges measured by the radiometer. This UV-B value, which can be taken as the more biologically relevant region of the spectrum, is equivalent to about 5.2 times the intensity at an altitude of 850 m in São Paulo, Brazil (9.3 W/m²) and 3.1 times the value at an altitude of 5091 m at the Atacama Desert, Chile (15.6 W/m²), as reported in a previous paper32. The Pseudomonas strains were found to be significantly more tolerant than E. coli to this “environmental” UV range (Fig. 5). A 60 minute exposure, equivalent to up to several hours in the real environment, did not reduce by more than 1 log the viability of 281 (32 ± 16% survival) or 158 (25 ± 14%), while E. coli survived at only 0.2 ± 0.1%. Interestingly, the response of both P. syringae strains, for which survival curves were nearly overlapping, was much more similar to the UV-A + UV-B used for these experiments than for the monochromatic UV-B (Fig. 4), again evidencing the biological relevance of differing spectra.
Figure 5.

Survival curves to “environmental” UV radiation (UV-A + UV-B). For this more accurate representation of the solar UV found in the troposphere, P. syringae pv. syringae 281 and P. syringae pv. garcae 158 suffered a decrease in viability of less than 90% after 60 minutes of exposure, from an initial population of ~5 × 106 cells/ml. Meanwhile, LB-grown E. coli presented a 1,000-fold reduction in survival from its initial numbers of ~5 × 107 cells/ml. When grown in LNP , the minimal medium used for the P. syringae strains, E. coli was even more sensitive, with a drop in viability after the full exposures of almost 5 logs (from about 107 cells/ml in this case). Error bars denote standard deviations of the means (n = 3).
To test if the Pseudomonas culture conditions somehow favored the greater survival of these cells in relation to LB-grown E. coli, further experiments were done. E. coli cultivated in nutrient-poor LNP medium, the same used for the P. syringae strains, was tested at the solar simulator. Remarkably, its survival after one hour of irradiation, 0.003 ± 0.002%, was even inferior to cells grown in rich medium at the same fluences (0.2 ± 0.1%), a difference of almost 2 logs (Fig. 5). In this manner, a poorer growth medium was not observed to contribute to UV tolerance, and, in fact, actually resulted in more sensitive cells.
The cumulative IN activity of the cells at −5 °C was measured after irradiation for a much longer period (120 minutes) of simulated “environmental” UV (Fig. 6). The strain 281 exhibited no significative difference from its initial cumulative IN concentration when assayed after the exposure. Meanwhile, 158 presented an up to 10-fold decrease from the typical 10−2–10−1 nuclei per cell of this strain at this same temperature (seen in the “0 min” control and also in Fig. 1). Despite that, the Pseudomonas ice nuclei endured UV even better than the cells themselves. Experiments with very large UV-C fluences (10,000 J/m2), far beyond the point where no surviving cell could be expected (see Fig. 3), also yielded no difference in 281’s IN at −5 °C, and a reduction by only about 20 times for 158 (Fig. 6), further confirming this observation.
Figure 6.
IN activity following prolonged exposures to “environmental” UV radiation (UV-A + UV-B) (right panel) and to UV-C (254 nm) radiation (left panel). For the “environmental” UV, when compared to non-irradiated controls (“0 min”), the concentration of cumulative ice nuclei (IN) per cell at −5 °C of P. syringae pv. syringae 281 exposed for two hours suffered no significant change, while P. syringae pv. garcae 158 samples exhibited an up to 10-fold reduction. The IN of P. syringae pv. syringae 281 exposed to 10,000 J/m2 of UV-C remained stable when compared to non-irradiated controls (“0 J/m2”). P. syringae pv. garcae 158, instead, was reduced by about 20-fold. Error bars denote standard deviations of the means (n = 7 for 281 and n = 5 for 158 for the “environmental” UV experiments, n = 6 for 281 and n = 7 for 158 for the UV-C experiments). Based on the Mann-Whitney U test, significantly different values are marked as *(p < 0.05) and non-significant differences as “ns”.
Survival and ice nucleation activity of desiccated cells
For the desiccation assays, cells were dried inside microcentrifuge tubes for 6 days at 20 °C, or kept hydrated in NaCl 0.9% solution as controls. During this period, the tubes were stored inside containers with controlled relative humidity (RH), using MgCl2 for 33% RH and silica gel for <5% RH. Those results are presented in Fig. 7. The largest tolerance was exhibited by the 158 strain at <5% RH, in which 22 ± 8% of its initial population of 5 × 106 cells survived. At the same treatment, the viability of 281 was reduced by 103–104 times (also from 5 × 106 cells). At 33% RH, the surviving percentage of 158 was 4 ± 2%, about 10 times more than 281. For both tested RH (33% and <5%), E. coli mean survival was around 3 to 4% (from 5 × 107 cells). As can be observed, 158 presented some degree of tolerance to desiccation, while 281 was much more sensitive. Comparing between organisms, no clear tendency is apparent in relation to RH and cell survival. Hydrated control samples remained mostly at the same initial CFU concentration, except for 281 for which the number of cells increased slightly. Again, for comparison, E. coli grown in LNP minimal medium was tested. It presented a far reduced survival, with a decrease of over 3 orders of magnitude in relation LB-grown cells (from about 107 cells, Fig. 7).
Figure 7.

Desiccation tolerance at different relative humidity (RH) values. Survival of hydrated controls and samples desiccated at 33% and <5% RH for 6 days at 20 °C of P. syringae pv. syringae 281, P. syringae pv. garcae 158, and E. coli grown either on LB or LNP are presented. The 281 strain is shown to be sensitive to dehydration, while 158 is considerably more tolerant to this treatment. E. coli tolerated relatively well the desiccation, but when grown in LNP minimal medium these cells were inactivated by additional 3 logs. Error bars denote standard deviations of the means (n = 4).
The IN activities of the desiccated P. syringae strains were strikingly different (Fig. 8). After being resuspended from the RH 33% and RH <5% treatments, the 281 strain presented a reduced mean cumulative nuclei concentration of about 3–4 × 10−3 per cell at −5 °C. Probably, this can be attributed to the disruption of the cells’ outer membranes where the ice nucleation protein clusters lie. Its hydrated controls remained at typical values for these cultures, at 1.1 ± 1.0 × 10−1. However, the IN concentrations for the 158 strain, which are normally similar to those of 281 (see Fig. 1), were lower by 103–104 times, even for hydrated cells that kept most of their viability. Its mean measured IN activities (concentration per cell at −5 °C) ranged from 1 × 10−4 to 9 × 10−6, after the 6-day period at 20 °C. Since Pseudomonas IN are known to be heat-sensitive, the temperature was suspected to play a role in this decline, so new experiments were performed storing the 158 strain samples at 4 °C. Survival was very similar at these conditions, and a substantially increased maintenance of IN at the hydrated control and RH 33% samples was observed, while the RH <5% treatment remained mostly identical (Supplementary Fig. S1). Yet, these values were still lower than the typical IN concentration for this strain (Fig. 1).
Figure 8.

IN activity following desiccation. The concentration of cumulative ice nuclei per cell at −5 °C of P. syringae pv. syringae 281 presented a significant (p < 0.01) decrease after desiccation at 33% and < 5% RH for 6 days at 20 °C compared to its hydrated controls. Surprisingly, P. syringae pv. garcae 158 suffered a large reduction in IN in relation to its typical values (repeatedly measured as 10−2–10−1 per cell at −5 °C, Fig. 1) even while hydrated, as indicated by the downwards grey arrow. Error bars denote standard deviations of the means (n = 6). Based on Bonferroni-corrected Mann-Whitney U tests, significantly different values are marked as *(p < 0.05) or as **(p < 0.01), and non-significant differences as “ns”.
Discussion
Both P. syringae strains were found to be very sensitive to UV-C, exhibiting survival comparable to the non-UV tolerant bacterium E. coli (Fig. 3). This range of Solar radiation (<280 nm) does not reach Earth’s lower atmosphere (the troposphere), being completely absorbed at the stratosphere. Despite that, the 254 nm wavelength produced by low-pressure mercury lamps is widely used to assess the effects of UV on bacteria in laboratory studies. It efficiently damages DNA and inactivates microorganisms, and, as such, is commonly referred to as “germicidal UV”37. Nevertheless, some bacteria, such as Deinococcus radiodurans, show extreme tolerance to UV-C38. For comparison, a UV-C fluence of about 1,000 J/m2 is required to inactivate 90% of its population32, while the highest fluence tested in this work was just 120 J/m2 (Fig. 3). The biological effects of the different UV radiation ranges and known UV tolerance systems are presented in the Supplementary Discussion.
Covering the wavelengths between 280 and 320 nm, UV-B is the most energetic, and potentially damaging, range of solar radiation that reaches the ground. At higher elevations, the UV flux is increased in relation to lower altitudes, more so at the UV-B region39. In agreement to that, Wang, et al.40 measured a more severe DNA damage in plants at an altitude of 1,700 m than at 300 m. In contrast to the UV-C assays, the P. syringae strains were significantly more resistant to this UV range (at 312 nm) than E. coli (Fig. 4). Even then, it must be recognized that these organisms are comparatively much less tolerant than D. radiodurans, for which over 120,000 J/m2 of UV-B is required to inactivate about 90% of its population at identical experimental conditions32. Still, these results point to some degree of resistance of P. syringae to UV-B, and it is important to notice that these cells are naturally exposed to a much different spectrum which effects considerably different responses32.
The strain 158 (P. syringae pv. garcae) was more tolerant than 281 (P. syringae pv. syringae) to the UV-B at the higher fluences tested. This evidences substantial differences between the strains which can also be observed for the other experiments discussed further below, despite both being classified as the same species. In fact, P. syringae pv. garcae can be distinguished as belonging to a separate, discrete genomospecies than P. syringae pv. syringae on the basis of DNA differences, though a lack of discerning phenotypic characteristics has prevented its reclassification with other strains within the “P. coronafaciens” taxon41. This fact may explain the observations made in this study, where the distinct measured phenotypes probably reflect the underlying dissimilar genotypes.
For a more accurate, relevant representation of the environmental UV than the narrow-band lamps, a solar simulator emitting UV-A + UV-B was used (Fig. 2). Under these conditions, the P. syringae strains were shown to be much more resistant than E. coli (Fig. 5). This can be partially seen as a consequence of the observed higher tolerance of Pseudomonas to the UV-B (Fig. 4), though the presence of UV-A (320–400 nm) can significantly affect some organisms. For example, D. radiodurans is surprisingly sensitive to this higher wavelength fraction of the environmental UV32,38. This confirms that P. syringae can be considered truly tolerant to solar radiation. Additionally, it is important to note that, due to differences between the UV-B lamps’ line emission at 312 nm and the solar simulator broad spectrum (~290–400 nm, Fig. 2), the biological response to both sources is not exactly equivalent. Thus, the survival curves presented for UV-B and “environmental” UV cannot be directly compared on the basis of the measured fluences, considering the Vilber Loumart radiometer’s probes read a spectral range, not a single wavelength.
Joly, et al.27 exposed two P. syringae isolated from cloud water for 10 hours to total final fluences of 85.7 kJ/m2 of UV-A and 27 kJ/m2 of UV-B at 5 °C. After this period, the isolates suffered virtually no viability loss. In the results presented in Fig. 5, it is revealed that Pseudomonas strains actually tolerate acute expositions to considerably larger fluences than the ones used for this previous study. Still, these authors used a lamp with a much different spectrum from the solar simulator, with proportionally less UV-A and a large amount of visible light. Attard, et al.30 used a 1,000 Watt xenon lamp filtered by a glass lid with a total UV-A intensity of 33 W/m2. Interestingly, exposure for 42 hours in distilled water at 17 °C, only reduced the viability of three different P. syringae isolates by about 90% in relation to non-irradiated controls. UV-B measurements (if significant) were not provided, but the final calculated UV-A exposure was of almost 5,000 kJ/m2. Possibly, this less acute exposition could have improved the tolerance of the strains, considering the lower intensity and absence of the more damaging UV-B wavelengths.
As mentioned, the UV-B intensity used for the “environmental” UV experiments is much higher than the values found in some natural environments, even at high altitudes, as reported previously32. Those measurements were performed by the same probes used for the solar simulator with a Vilber Loumart radiometer, under clear sky conditions, at noon, during summer, and at similar latitudes. Taking the UV-B as the most biologically relevant radiation range for comparison, P. syringae strains can be expected to tolerate hours of direct sunlight exposure, even without attenuating factors such as association to cell clusters, mineral particles or organic fragments. Certainly, those factors can become increasingly important for survival at higher altitudes and for longer periods.
The IN activity of 281 was not reduced for cells irradiated for two hours under the solar simulator. The 158 strain, instead, suffered an up to 10-fold decrease at −5 °C, showing that UV can have at least some effect on this trait (Fig. 6). The P. syringae isolates irradiated with UV-A by Attard, et al.30 (mentioned above) presented either a non-significant difference in IN activity at −5 °C or suffered a small reduction. These authors discuss how dead bacteria could maintain their IN activity as long as cell integrity is not disrupted, preserving the large ice nucleation protein aggregates on the cells’ outer membranes. In fact, this is what was observed for cells completely inactivated by UV-C exposure, with again only a relatively small effect measured for 158 (Fig. 6).
Summarizing the UV results, our work reveals that the biological response to commonly used laboratory UV sources is a poor representation of what is found with the solar spectrum, and that while UV can be a serious limitation to the dispersal of P. syringae through the atmosphere, these bacteria are adapted to endure periods of complete exposure to sunlight. A relatively large subset of their population remained capable of influencing cloud nucleation, which, in turn, could favor their deposition back to the ground19,28,29. Perhaps, a greater challenge to these cells is desiccation, both in relation to survival and IN maintenance (see Figs 7 and 8).
Dehydration of cells causes membrane damage, DNA strand breakage, and an increased formation of reactive oxygen species from the cellular metabolism leading to protein oxidation42,43. Similarly to the UV (see Supplementary Discussion), DNA repair and antioxidant systems should be valuable features for bacterial survival under this condition. In the environment, tolerance to desiccation could possibly be achieved by activation of the aerosolized bacteria as cloud condensation nuclei (CCN), enabling hydration of the cells from water vapor harnessed from the air. Since Pseudomonas were shown to be potential efficient CCN due to biosurfactant production13–15, this could be another survival mechanism at the disposition of these cells.
Cell clustering could be an important factor contributing to P. syringae tolerance to desiccation when airborne. The experiments were conducted in a manner so as to minimize these artifacts, exposing cells as thin layers to rapidly equilibrate with the RH controlling agents, but masses of natural bacterial populations are probably better adapted to endure this stress. Monier and Lindow44 have previously shown that cells grown as aggregates over leaf surfaces present an increased desiccation tolerance with larger cells clusters, but artificially generated aggregates did not exhibit such effect. An explanation for that is the lack of locally-produced extracellular polymeric substances (EPS), which are formed by cells grown together as groups, but would not be extensively produced under the laboratory culture conditions used. In addition to this point, the osmotic shock of rehydration after desiccation under the tested laboratory conditions could be a significant stress by itself. As reported by Kosanke et al.45, a slow rehydration protocol can increase the survival of dried bacteria. Despite this effect being relatively minor when compared to the large range of survivals measured in this work, it could have also played a role in the results obtained.
Despite its low tolerance to desiccation, the IN activity of 281 was relatively well preserved after desiccation (Fig. 8). The measured decrease by roughly 30 times in IN concentration can possibly be attributed to cell membrane disruption during dehydration affecting the IN protein clusters. Its hydrated controls maintained the full typical IN concentration of this strain (about 10−1 per cell). In contrast, the IN activity of the more desiccation tolerant strain 158 was unexpectedly reduced even in the hydrated controls (Fig. 8) when compared to its typical IN concentration (Fig. 1). The large aggregates of InaZ proteins that form at the cells outer membranes, which are essential for forming efficient IN active at relatively high temperatures such as −5 °C, are known to be particularly heat sensitive36. Even though the temperature of 20 °C at which the hydrated and desiccated cells were kept at during these experiments were not expected to be detrimental to biological IN36, further tests with 158 were performed at 4 °C (Supplementary Fig. S1). This could not recover the typical IN concentration for this strain (Fig. 1), signifying some other mechanism contributes to the instability of the nuclei in this case where the cells are not actively growing and their metabolism, including protein synthesis, is probably reduced. Nevertheless, it can be summarized that one of our main findings is that desiccation can be a much more severe limitation to cells and IN than UV.
Aerial emission of P. syringae from plants was observed to be greater at dryer conditions on sunny days46,47. Despite the abundance of cells being lifted into the air, these desiccating circumstances may impart a significant limitation on living cell dispersion and IN activity right from the start. Alternatively, different studies48–50 found large increases of airborne IN evidenced to be to be of microbial origin during and following rainfall. The actual mechanism of aerosolization from raindrop impact has only recently began to be understood51, but these wet conditions may greatly enhance the efficiency of bacterial dissemination through the air within droplets (or at least at high humidity). This may be important for research on feedback effects between rain and IN. Precipitation events may increase further rain by seeding the atmosphere with suspended biological particles52, and the likely enhanced survival and IN preservation when compared to dry, desiccating conditions may be a key step enabling this cycle. Further studies will be necessary to make clear how the biological response of bacterial cells to environmental stresses can have further consequences to the ecosystem they inhabit.
Material and Methods
Strains, media, growth conditions, and survival quantification
The tested Pseudomonas syringae belonged to the strains IBSBF 281T (=NCPPB 281, ATCC 19310; pv. syringae type strain; isolated from Syringa vulgaris) and IBSBF 158 (pv. garcae; isolated from Coffea arabica, where it causes the brown spot disease on leaves). For maximum expression of the IN phenotype, cells were grown at 15 °C in LNP medium (3-(N-morpholino)propanesulfonic acid (MOPS), 10.46 g L−1; KCl, 1.86 g L−1; NH4Cl, 0.11 g L−1; Na2SO4, 1.42 g L−1; NaCl, 0.58 g L−1; MgCl2·6H2O, 0.20 g L−1; KH2PO4, 0.014 g L−1; CaCl2, 0.011 g L−1; FeCl3·6H2O, 0.0027 g L−1; sorbitol, 4 g L−1; pH 7.236) for 3 days to an OD of ~0.5. Colony-forming units (CFU) were enumerated on Difco Nutrient Agar added with 2.5% glycerol (NAG) plates incubated in the dark at 20 °C. E. coli BL21 was cultivated at 37 °C in Difco LB Broth. Cultures were grown overnight to an OD of ~5.0 in LB, and enumerated on LB agar plates incubated in the dark at 37 °C. For some experiments, E. coli was grown in LNP medium overnight at 37 °C to an OD of ~0.5. All cell suspensions were diluted in saline solution (NaCl 0.9% w/v) prepared with ultrapure Milli-Q water (Millipore, Molsheim, France). Survival is expressed as the fraction N/N0, where “N” is the dilution-corrected number of UFC recovered after each treatment and “N0” is the number of initial UFC from before the experiments. Survival fraction values are presented as means of the replicates, with error bars denoting standard deviations.
Quantification of ice nucleation activity
Ice nucleation activity for each sample was quantified by the droplet freezing assay, using diluted cell suspensions placed as arrays of 32 drops of 10 μl on top of a paraffin-coated aluminum tray. This coating was previously applied as a 2% solution of paraffin in xylene, with the solvent removed by heat over a hot plate. The tray was covered with a transparent acrylic lid sealed on the borders by a ring made of EVA foam sheet and held in place by binder clips. This set was then positioned almost totally immersed in a low temperature circulating bath (Neslab LT-50, Newington, USA) filled with 96% ethanol. Temperature was monitored with a submerged mercury thermometer. From −2 °C, the bath temperature was reduced in 1 °C stages, which were held for at least 5 minutes. At each stage, the number of frozen drops was scored. IN concentration was calculated with the following equation adapted from Vali35, as commonly used for microbiological studies (e.g., Joly, et al.21): c(T) = [ln (N) − ln (N − N(T))]/A, where “c(T)” is the number of cumulative active IN per cell at temperature “T”, “N” is the number of drops tested, “N(T)” is the number of frozen drops at temperature “T”, and “A” is the number of cells per drop (determined by CFU counting from initial population). All experiments were replicated at least 4 times. Measured IN activity values are presented as means of the replicates, with error bars representing standard deviations. To determine significant differences, Mann-Whitney U tests and Bonferroni corrections were performed with PAST (v3.21) software53 (http://folk.uio.no/ohammer/past/).
UV irradiation experiments
UV-C irradiation was done with a Philips TUV-20W low-pressure mercury lamp with main emission line at 254 nm. UV-B was provided by a set of mercury lamps (two LightTech Narrow Band UV-B 20 W and one Philips TL 20 W/01 RS) with main emission line at 312 nm. A solar simulator (Oriel Sol-UV-2, Bozeman, USA) with a 1,000 Watt xenon arc lamp was used for the “environmental UV” irradiation experiments. This source’s output covers partially the UV-B and UV-A ranges with a spectrum similar to the one found at directly Sun-exposed environments on Earth (with negligible UV-C), except most of the visible light is removed by an optical filter (Fig. 2). Irradiation intensities and fluences were measured by a radiometer (Vilber Loumart RMX-3W, Marne-la-Vallée, France) with photocells specific to wavelength ranges centered in 254 nm at the UV-C (CX-254), 312 nm at the UV-B (CX-312), and 365 nm at the UV-A (CX-365).
For the UV-C and UV-B assays, cultures were diluted 100-fold in saline solution to a final volume of 8 ml in autoclaved 7 cm diameter glass petri dishes (without the lids). The samples were irradiated under orbital shaking while the fluences were monitored in real time by photocells placed next to the dishes. Aliquots taken at fluence intervals were diluted and plated for survival quantification by enumeration of CFU. For the “environmental” UV experiments, the samples were irradiated on top of a frozen foam block, also under shaking, so as to control their temperature to near 0 °C. This was done during these more prolonged exposures to avoid evaporation and to prevent excessive heating – which can itself affect the cells’ IN by leading to the disaggregation of the ice nucleation protein clusters on the cells’ outer membranes36. The intensities were measured beforehand as 75.5 W/m² for the UV-A and 48.7 W/m² for the UV-B, at the ranges read by the radiometer (centered on 365 and 312 nm). Aliquots from the exposed samples were then taken after determined time intervals, corresponding to UV-A and UV-B fluences which could be calculated afterwards from the intensity values.
Separate experiments were performed to test the effects of UV over the cells’ IN. Longer exposures were used in these assays since preliminary trials showed no effects of smaller fluences on IN activity. With the solar simulator, samples were exposed for 120 minutes at the same UV intensities as before, equivalent to a total 545 kJ/m² of UV-A and 348 kJ/m² of UV-B (at the ranges read by the radiometer), twice as much as the largest fluence of the survival tests. In this case, the diluted cultures were exposed as 2 ml volumes in 3 cm diameter dishes, allowing more samples to be placed below the source’s focus at the same time. The control samples, “0 min”, were aliquoted from the dishes before the beginning of the experiments and stored until the end of the irradiation when their IN activity was quantified parallel to the “120 min” samples.
Desiccation experiments
Desiccation assays were performed with 10 μl volumes taken directly from the cultures and deposited in the internal wall of horizontally positioned autoclaved 1.5 ml microcentrifuge tubes. The tubes were then placed inside sealed recipients containing either silica gel beads or water-saturated MgCl2. These treatments were used to provide controlled relative humidity (RH) values below 5% and of about 33%, respectively54. A <5% RH is typical for very high altitudes, considering water vapor sources at the ground surface. Hydrated controls were prepared by adding 10 μl from the cultures to a total 1 ml in tubes with saline solution (10−2 dilution). All samples were stored for 6 days inside an incubator at 20 °C. During this, the temperatures were monitored with mercury thermometers and were found to remain stable. At the end of this period, the dried samples were resuspended with 1 ml saline solution (10−2 dilution in relation to the cultures). Along with the hydrated control cell suspensions, these tubes were diluted for survival determination by plating and for IN quantification. Survival was calculated relative to initial controls aliquoted from the cultures, plated before the beginning of each experiment.
Supplementary information
Acknowledgements
This work was sponsored by the Brazilian Federal Agency for Support and Evaluation of Graduate Education – CAPES, the São Paulo Research Foundation – FAPESP (Projects 2016/06160-8 and 2012/18936-0), the Microsfera project (407816/2013-5) of the Brazilian Antarctic Program (ProAntar), the Brazilian National Council of Technological and Scientific Development – CNPq (CNPq 424367/2016-5), INCT-Criosfera (CNPq 028306/2009), and the University of São Paulo – USP, through the Brazilian Research Unity in Astrobiology – NAP/Astrobio. The authors would also like to thank Professor Josef Wilhelm Baader (IQ-USP) for lending the low temperature bath with which the ice nucleation experiments were performed.
Author Contributions
G.G.A. planned and performed the experiments, analyzed the data and prepared the figures. F.R., F.L.T.G. and D.G. conceived the original idea and designed the experiments, acquired funding, and supervised the work. All authors contributed to critical interpretation of the data, writing and revising the manuscript. All authors approved the final version manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44283-3.
References
- 1.Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae - a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol R. 2000;64:624–653. doi: 10.1128/Mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maki LR, Galyan EL, Chang-Chien MM, Caldwell DR. Ice Nucleation Induced by Pseudomonas syringae. Appl Microbiol. 1974;28:456–459. doi: 10.1128/am.28.3.456-459.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green RL, Warren GJ. Physical and Functional Repetition in a Bacterial Ice Nucleation Gene. Nature. 1985;317:645–648. doi: 10.1038/317645a0. [DOI] [Google Scholar]
- 4.Govindarajan AG, Lindow SE. Size of Bacterial Ice-Nucleation Sites Measured In situ by Radiation Inactivation. Analysis. P Natl Acad Sci USA. 1988;85:1334–1338. doi: 10.1073/pnas.85.5.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindow SE, Arny DC, Upper CD. Bacterial Ice Nucleation - a Factor in Frost Injury to Plants. Plant Physiol. 1982;70:1084–1089. doi: 10.1104/Pp.70.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris CE, et al. Bioprecipitation: a feedback cycle linking Earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Global Change Biol. 2014;20:341–351. doi: 10.1111/gcb.12447. [DOI] [PubMed] [Google Scholar]
- 7.Sands D, Langhans V, Scharen A, De Smet G. The association between bacteria and rain and possible resultant meteorological implications. Időjárás. 1982;86:148–152. [Google Scholar]
- 8.Murray BJ, O’Sullivan D, Atkinson JD, Webb ME. Ice nucleation by particles immersed in supercooled cloud droplets. Chem Soc Rev. 2012;41:6519–6554. doi: 10.1039/c2cs35200a. [DOI] [PubMed] [Google Scholar]
- 9.Möhler O, DeMott PJ, Vali G, Levin Z. Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences. 2007;4:1059–1071. doi: 10.5194/bg-4-1059-2007. [DOI] [Google Scholar]
- 10.Mülmenstädt J, Sourdeval O, Delanoë J, Quaas J. Frequency of occurrence of rain from liquid-, mixed-, and ice-phase clouds derived from A-Train satellite retrievals. Geophys Res Lett. 2015;42:6502–6509. doi: 10.1002/2015GL064604. [DOI] [Google Scholar]
- 11.Hallett J, Mossop SC. Production of Secondary Ice Particles during Riming Process. Nature. 1974;249:26–28. doi: 10.1038/249026a0. [DOI] [Google Scholar]
- 12.Bauer, H. et al. Airborne bacteria as cloud condensation nuclei. J Geophys Res-Atmos108, 10.1029/2003jd003545 (2003).
- 13.Ahern HE, Walsh KA, Hill TCJ, Moffett BF. Fluorescent pseudomonads isolated from Hebridean cloud and rain water produce biosurfactants but do not cause ice nucleation. Biogeosciences. 2007;4:115–124. doi: 10.5194/bg-4-115-2007. [DOI] [Google Scholar]
- 14.Ekström S, et al. A possible role of ground-based microorganisms on cloud formation in the atmosphere. Biogeosciences. 2010;7:387–394. doi: 10.5194/bg-7-387-2010. [DOI] [Google Scholar]
- 15.Renard P, et al. Screening of cloud microorganisms isolated at the Puy de Dôme (France) station for the production of biosurfactants. Atmos Chem Phys. 2016;16:12347–12358. doi: 10.5194/acp-16-12347-2016. [DOI] [Google Scholar]
- 16.Delort, A. M. et al. In Microbial Ecology of Extreme Environments (eds C. Chénard & F. M. Lauro) 215–245 (Springer, 2017).
- 17.Morris CE, et al. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. The ISME Journal. 2008;2:321–334. doi: 10.1038/ismej.2007.113. [DOI] [PubMed] [Google Scholar]
- 18.Šantl-Temkiv T, et al. Characterization of airborne ice-nucleation-active bacteria and bacterial fragments. Atmos Environ. 2015;109:105–117. doi: 10.1016/j.atmosenv.2015.02.060. [DOI] [Google Scholar]
- 19.Stopelli E, et al. Ice nucleators, bacterial cells and Pseudomonas syringae in precipitation at Jungfraujoch. Biogeosciences. 2017;14:1189–1196. doi: 10.5194/bg-14-1189-2017. [DOI] [Google Scholar]
- 20.Amato P, et al. Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dôme: major groups and growth abilities at low temperatures. Fems Microbiol Ecol. 2007;59:242–254. doi: 10.1111/j.1574-6941.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 21.Joly M, et al. Ice nucleation activity of bacteria isolated from cloud water. Atmos Environ. 2013;70:392–400. doi: 10.1016/j.atmosenv.2013.01.027. [DOI] [Google Scholar]
- 22.Vaïtilingom M, et al. Long-term features of cloud microbiology at the puy de Dôme (France) Atmos Environ. 2012;56:88–100. doi: 10.1016/j.atmosenv.2012.03.072. [DOI] [Google Scholar]
- 23.Bauer H, et al. The contribution of bacteria and fungal spores to the organic carbon content of cloud water, precipitation and aerosols. Atmos Res. 2002;64:109–119. doi: 10.1016/S0169-8095(02)00084-4. [DOI] [Google Scholar]
- 24.Burrows SM, Elbert W, Lawrence MG, Pöschl U. Bacteria in the global atmosphere - Part 1: Review and synthesis of literature data for different ecosystems. Atmos Chem Phys. 2009;9:9263–9280. doi: 10.5194/acp-9-9263-2009. [DOI] [Google Scholar]
- 25.Wilkinson DM, Koumoutsaris S, Mitchell EAD, Bey I. Modelling the effect of size on the aerial dispersal of microorganisms. J Biogeogr. 2012;39:89–97. doi: 10.1111/j.1365-2699.2011.02569.x. [DOI] [Google Scholar]
- 26.Pulschen AA, et al. Survival of Extremophilic Yeasts in the Stratospheric Environment during Balloon Flights and in Laboratory Simulations. Appl Environ Microbiol. 2018;84:e01942–01918. doi: 10.1128/AEM.01942-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joly M, et al. Survival of microbial isolates from clouds toward simulated atmospheric stress factors. Atmos Environ. 2015;117:92–98. doi: 10.1016/j.atmosenv.2015.07.009. [DOI] [Google Scholar]
- 28.Amato P, et al. Survival and ice nucleation activity of bacteria as aerosols in a cloud simulation chamber. Atmos Chem Phys. 2015;15:6455–6465. doi: 10.5194/acp-15-6455-2015. [DOI] [Google Scholar]
- 29.Stopelli E, et al. Ice nucleation active particles are efficiently removed by precipitating clouds. Sci Rep. 2015;5:16433. doi: 10.1038/Srep16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attard E, et al. Effects of atmospheric conditions on ice nucleation activity of Pseudomonas. Atmos Chem Phys. 2012;12:10667–10677. doi: 10.5194/acp-12-10667-2012. [DOI] [Google Scholar]
- 31.Gonçalves FLT, Massambani O. Bacteria and fungal spores as ice nuclei from Coffea arabica L. Rev Cien Nat. 2011;33:73–93. [Google Scholar]
- 32.Pulschen AA, et al. UV-resistant yeasts isolated from a high-altitude volcanic area on the Atacama Desert as eukaryotic models for astrobiology. Microbiologyopen. 2015;4:574–588. doi: 10.1002/mbo3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulino-Lima IG, et al. Isolation of UVC-Tolerant Bacteria from the Hyperarid Atacama Desert, Chile. Microb Ecol. 2013;65:325–335. doi: 10.1007/s00248-012-0121-z. [DOI] [PubMed] [Google Scholar]
- 34.Arrage AA, Phelps TJ, Benoit RE, White DC. Survival of Subsurface Microorganisms Exposed to Uv-Radiation and Hydrogen-Peroxide. Applied and Environmental Microbiology. 1993;59:3545–3550. doi: 10.1128/aem.59.11.3545-3550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vali G. Quantitative Evaluation of Experimental Results on Heterogeneous Freezing Nucleation of Supercooled Liquids. J Atmos Sci. 1971;28:402–409. doi: 10.1175/1520-0469(1971)028<0402:Qeoera>2.0.Co;2. [DOI] [Google Scholar]
- 36.Nemecek-Marshall M, Laduca R, Fall R. High-Level Expression of Ice Nuclei in a Pseudomonas syringae Strain Is Induced by Nutrient Limitation and Low-Temperature. J Bacteriol. 1993;175:4062–4070. doi: 10.1128/jb.175.13.4062-4070.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coohill TP, Sagripanti JL. Overview of the inactivation by 254 nm ultraviolet radiation of bacteria with particular relevance to biodefense. Photochem Photobiol. 2008;84:1084–1090. doi: 10.1111/j.1751-1097.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 38.Slade D, Radman M. Oxidative Stress Resistance in Deinococcus radiodurans. Microbiol Mol Biol R. 2011;75:133–191. doi: 10.1128/Mmbr.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blumthaler M, Ambach W, Rehwald W. Solar UV-A and UV-B Radiation Fluxes at 2 Alpine Stations at Different Altitudes. Theor Appl Climatol. 1992;46:39–44. doi: 10.1007/Bf00866446. [DOI] [Google Scholar]
- 40.Wang QW, Hidema J, Hikosaka K. Is UV-Induced DNA Damage Greater at Higher Elevation? Am J Bot. 2014;101:796–802. doi: 10.3732/ajb.1400010. [DOI] [PubMed] [Google Scholar]
- 41.Gardan L, et al. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959) Int J Syst Bacteriol. 1999;49:469–478. doi: 10.1099/00207713-49-2-469. [DOI] [PubMed] [Google Scholar]
- 42.Fredrickson JK, et al. Protein oxidation: key to bacterial desiccation resistance? The ISME Journal. 2008;2:393–403. doi: 10.1038/ismej.2007.116. [DOI] [PubMed] [Google Scholar]
- 43.Mattimore V, Battista JR. Radioresistance of Deinococcus radiodurans: Functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monier JM, Lindow SE. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. P Natl Acad Sci USA. 2003;100:15977–15982. doi: 10.1073/pnas.2436560100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kosanke JW, Osburn RM, Shuppe GI, Smith RS. Slow Rehydration Improves the Recovery of Dried Bacterial-Populations. Can J Microbiol. 1992;38:520–525. doi: 10.1139/M92-086. [DOI] [PubMed] [Google Scholar]
- 46.Lindemann J, Constantinidou HA, Barchet WR, Upper CD. Plants as Sources of Airborne Bacteria, Including Ice Nucleation-Active Bacteria. Appl Environ Microbiol. 1982;44:1059–1063. doi: 10.1128/aem.44.5.1059-1063.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindemann J, Upper CD. Aerial Dispersal of Epiphytic Bacteria over Bean Plants. Appl Environ Microbiol. 1985;50:1229–1232. doi: 10.1128/aem.50.5.1229-1232.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huffman JA, et al. High concentrations of biological aerosol particles and ice nuclei during and after rain. Atmos Chem Phys. 2013;13:6151–6164. doi: 10.5194/acp-13-6151-2013. [DOI] [Google Scholar]
- 49.Prenni AJ, et al. The impact of rain on ice nuclei populations at a forested site in Colorado. Geophys Res Lett. 2013;40:227–231. doi: 10.1029/2012GL053953. [DOI] [Google Scholar]
- 50.Constantinidou HA, Hirano SS, Baker LS, Upper CD. Atmospheric Dispersal of Ice Nucleation-Active Bacteria - the Role of Rain. Phytopathology. 1990;80:934–937. doi: 10.1094/Phyto-80-934. [DOI] [Google Scholar]
- 51.Joung YS, Ge Z, Buie CR. Bioaerosol generation by raindrops on soil. Nat Commun. 2017;8:14668. doi: 10.1038/ncomms14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bigg EK, Soubeyrand S, Morris CE. Persistent after-effects of heavy rain on concentrations of ice nuclei and rainfall suggest a biological cause. Atmos Chem Phys. 2015;15:2313–2326. doi: 10.5194/acp-15-2313-2015. [DOI] [Google Scholar]
- 53.Hammer Ø, Harper DA, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia electronica. 2001;4:9. [Google Scholar]
- 54.Winston PW, Bates DH. Saturated Solutions for the Control of Humidity in Biological Research. Ecology. 1960;41:232–237. doi: 10.2307/1931961. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



