Abstract
Human rhinoviruses (RVs) are picornaviruses that can cause a variety of upper and lower respiratory tract illnesses, including the common cold, bronchitis, pneumonia, and exacerbations of chronic respiratory diseases such as asthma. There are currently > 160 known types of RVs classified into three species (A, B, and C) that use three different cellular membrane glycoproteins expressed in the respiratory epithelium to enter the host cell. These viral receptors are intercellular adhesion molecule 1 (used by the majority of RV-A and all RV-B types), low-density lipoprotein receptor family members (used by 12 RV-A types), and cadherin-related family member 3 (CDHR3; used by RV-C). RV-A and RV-B interactions with intercellular adhesion molecule 1 and low-density lipoprotein receptor glycoproteins are well defined and their cellular functions have been described, whereas the mechanisms of the RV-C interaction with CDHR3 and its cellular functions are being studied. A single nucleotide polymorphism (rs6967330) in CDHR3 increases cell surface expression of this protein and, as a result, also promotes RV-C infections and illnesses. There are currently no approved vaccines or antiviral therapies available to treat or prevent RV infections, which is a major unmet medical need. Understanding interactions between RV and cellular receptors could lead to new insights into the pathogenesis of respiratory illnesses as well as lead to new approaches to control respiratory illnesses caused by RV infections.
Key Words: antiviral, cadherin-related family member 3, cellular receptor, intercellular adhesion molecule 1, low-density lipoprotein receptor, rhinovirus
Abbreviations: CDHR3, cadherin-related family member 3; ICAM-1, intercellular adhesion molecule 1; LDLR, low-density lipoprotein receptor; RV, rhinovirus
The Picornaviridae family of viruses includes some of the most common human pathogens, including enteroviruses and rhinoviruses (RVs). The RVs are classified in the Enterovirus genus and have historically been identified as specific to humans, but they were recently discovered also in wild chimps in Uganda.1 There are > 160 known genotypes of RVs classified into A, B, and C species according to their phylogenetic sequence and distinct genomic features.2 RV-A and RV-B types have been recognized for > 50 years, whereas RV-C types (which do not grow in standard tissue culture cell lines) were first discovered in 2006 with the use of molecular techniques.3, 4 Although RVs can bind to a number of cell types, the airway epithelium provides the primary host cells for each of the RV species.
Viruses in all three RV species frequently cause upper respiratory illnesses in children and adults.5 RV infections also contribute significantly to otitis media, sinusitis, and lower respiratory illnesses.6 RV-A and RV-C are more likely than RV-B to be associated with severe respiratory illnesses such as pneumonia, bronchiolitis, influenza-like illness, chronic rhinosinusitis, and exacerbations of asthma, COPD, and cystic fibrosis.7, 8, 9 Thus, RV infections are both common and associated with significant respiratory morbidity.5 Specific treatments are lacking, and the search for therapeutic targets has prompted renewed efforts to understand the molecular virology and pathogenesis of RV infections and illnesses. An important first step in the viral replication cycle is specific binding to host cells followed by entry into the cells. The following sections highlight recent discoveries related to RV structure and molecular virology, which emphasize the importance of studies on RV cellular receptors.
Virion Structure
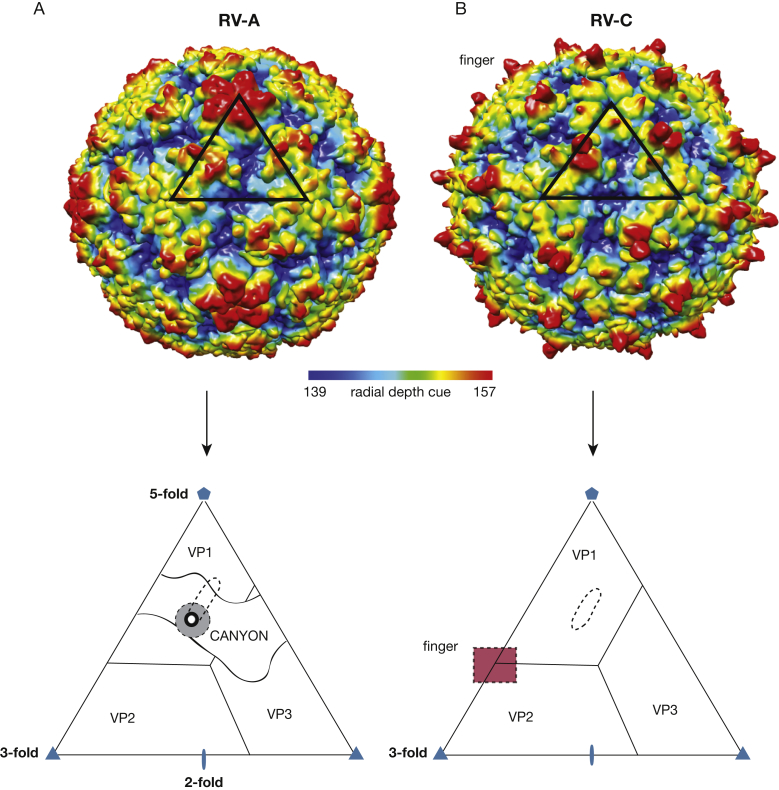
Several RV protein capsid structures have been resolved to atomic-level resolution by using X-ray crystallography and cryo-electron microscopy techniques. All RV genomes encode four capsid structural proteins (VP1, VP2, VP3, and VP4) and seven nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) that mediate viral polyprotein processing and genome replication. The capsid has icosahedral symmetry and is constructed of 60 copies each of VP1, VP2, VP3, and VP4 (Fig 1). The linear, single-stranded, positive-sense viral RNA is the template for viral protein synthesis, a process driven by an internal ribosome entry site element in the 5′-proximal sequence of the genome RNA. Initiation of internal ribosome entry site-driven translation involves recruitment of the cellular translation machinery to a location about 700 bases into the approximately 7,200 base in the viral RNA. Translation results in single long polyprotein of about 2,200 amino acids.10 Co-translational and posttranslational polyprotein processing releases the various mature components, of which the capsid precursors (VP0, VP1, and VP3) then directly assemble around nascent new genomes to form a tightly packed progeny virion.10, 11 Subsequent maturation steps include RNA-catalyzed cleavage of the precursor VP0 protein into the final VP2 (N-terminus) and VP4 (C-terminus) capsid units. The external surface of the capsid exposes elements of VP1, VP2, and VP3 proteins, whereas the VP4 protein lines the interior surface of the particle, in contact with the encapsidated viral genome.
Figure 1.
Structures of RV-A and RV-C colored by radial distance (Å) to the virus center. A black triangle indicates one icosahedral asymmetrical unit on each of the two viruses. A, Structure of RV-A with an enlarged triangle to show outline of the canyon, receptor pocket (dotted oval), receptor footprint (dotted circle, shaded gray), and pore (dark circle). B, Structure of RV-C with an enlarged triangle to show outline of binding pocket (dotted oval) and region of finger-like projection (dotted square, shaded red). A continuous canyon is absent in RV-C. RV = rhinovirus.
The overall surfaces of RV-A and RV-B have uneven topographies dominated by prominent, star-shaped plateaus at the 12 fivefold axes of symmetry, each of which is encircled by a cleft-like depression (“canyon”). Proteins at each threefold axis of symmetry coalesce into another smaller protrusion (Fig 1A). Underneath each canyon floor and within each VP1 core of every RV-A and RV-B is a small pore leading to a nonsurface hydrophobic pocket. In RV-A, this pocket is partially occupied by bulky cell-derived lipids called “pocket factors.” In the RV-B, the analogous pocket, although structurally present, is left unoccupied.12, 13, 14 This pocket has been exploited in the development of certain antiviral compounds that can also bind here, filling the cavity and/or displacing the native factors.15
The capsid structure of RV-C differs from that of RV-A and RV-B. Not only is the fivefold plateau much smaller, there are 60 RV-C unique spiky protrusions (fingers) caused by a VP1 loop insertion and located on the particle surface at the protomer junctions of VP1, VP2, and VP3 (Fig 1B). The spikes are formed by specific residues on VP1 and VP2, and these surface elements are likely to be immunogenic.12 The RV-C canyon is narrow and noncontinuous, unlike the RV-A and RV-B canyons. The VP1 proteins have small internal pockets, but these lack external pores and are therefore inaccessible to drugs or pocket factors.
Cellular Receptors
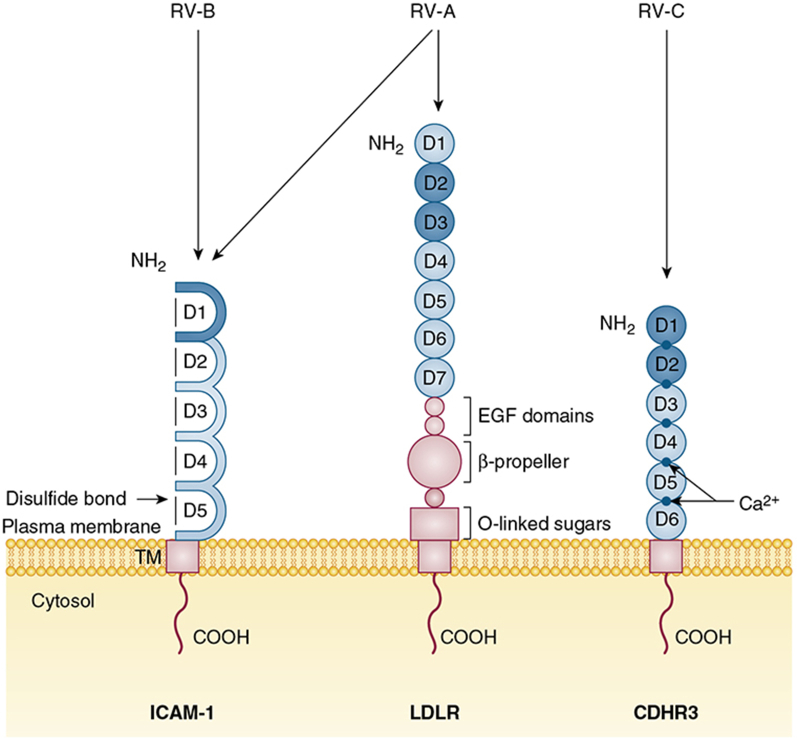
Collectively, all RVs use one of three major types of plasma membrane glycoproteins to bind and enter into host epithelial cells (Fig 2). The RV-A and RV-B types subdivide into major and minor infectivity groups based on their receptor specificity. The major group viruses, which constitute all RV-B and most RV-A types, use intercellular adhesion molecule 1 (ICAM-1) as their receptor.16 Some major group RVs, especially once they are cell culture adapted, can also use heparan sulfate proteoglycans as an alternative receptor.17 The minor group viruses comprise those 12 RV-A types that bind to protein members of the low-density lipoprotein receptor (LDLR) family instead of ICAM-1. All RV-C, in contrast, react with cadherin-related family member 3 (CDHR3), a protein expressed by ciliated airway epithelial cells in vivo.18, 19 As with RV-A, some RV-C can adapt to bind proteoglycans, such as heparin, on the surface of HeLa cells if they undergo repeated passages in vitro.20 RV-A and RV-B binding to their ICAM-1 or LDLR cell receptor initiates the process of receptor-mediated endocytosis, which allows particle entry and genome uncoating into the host cell.21 A similar process likely occurs for RV-C when it binds CDHR3.
Figure 2.
Cellular receptors of RV-A, RV-B, and RV-C. ICAM-1 has five consecutive extracellular ligand binding immunoglobulin domains linked by disulfide bonds. The NH2 domain (D1) interacts with major group RV-A and all RV-B. The LDLR family of glycoproteins (receptors LDLR, VLDLR, and LDLR-related protein) are characterized by distinct repeats of ligand binding domains, EGF domains, and β-propeller modules. The LDLR receptor has seven consecutive extracellular ligand-binding domains followed by three EGF domains, a β-propeller module, and O-linked sugar domain. The NH2 second and third domains (D2 and D3) interact with minor group RV-A. CDHR3 has six extracellular ligand-binding domains structurally supported by obligate Ca2+ ions at the domain junctions. The first two NH2 domains (D1 and D2) are predicted to interact with RV-C. All RV receptors have a TM and COOH. Their virus contact domains are shaded. CDHR3 = cadherin-related family member 3; COOH = C-terminal cytoplasmic tail; EGF = epidermal growth factor; ICAM-1 = intercellular adhesion molecule 1; LDLR = low-density lipoprotein receptor; NH2 = N-terminal; TM = transmembrane region. See Figure 1 legend for expansion of other abbreviation.
ICAM-1 Glycoprotein
The transmembrane glycoprotein ICAM-1 was identified independently by three research groups as the cellular receptor for major group RVs.16, 22, 23 ICAM-1 is a member of the immunoglobulin superfamily of cell adhesion proteins, and it is typically expressed by epithelial cells, endothelial cells, and leukocytes when they are activated by inflammatory stimuli, or physical or chemical stress. The ICAM-1 structure has five consecutively linked extracellular immunoglobulin domains, a carboxyl-proximal transmembrane region, and a short C-terminal cytoplasmic domain (Fig 2).24, 25 The N-terminal immunoglobulin domain (D1) is captured during interactions with the RVs by a binding pocket composed of the VP1 and VP2 capsid proteins (Fig 1A), deep within the canyon feature.26 As part of its normal cell function, ICAM-1 is a co-stimulatory molecule for cell activation, and it helps to regulate leukocyte migration from blood to the tissues in inflammatory sites when it binds to two integrin receptors, leukocyte function-associated antigen and macrophage-1 antigen.27, 28
For major group RVs, excluding the RV-B, the hydrophobic pocket of VP1 can assume two different conformations, with the pocket filled or empty.29 Binding to ICAM-1, for all major group viruses, occurs only when the pocket is empty, because contact residues of the VP1 and VP2 capsid proteins, located at the base of the canyon and in proximity to the pocket pore, are not otherwise in an appropriate conformation. Once ICAM-1 has captured a virus, endocytosis is triggered, and during this process, the low pH of the subsequent endosome allows conformational changes in the capsid that enable viral genome RNA translocation into the cytosol.30
LDLR Family
The discovery of ICAM-1 as an RV receptor was later followed by the discovery of LDLR family members as the additional receptors for the remaining, minor group RV.31 There are at least three members of the LDLR family of glycoproteins that can bind and internalize RVs. These include low-density lipoprotein receptor (LDLR), the very-low-density lipoprotein receptor (VLDLR), and the LDLR-related protein (LRP). Receptors in this family are characterized by distinctive arrangement of multiple structural modules in their extracellular regions that include ligand-binding repeats, epidermal growth factor precursor repeats, and YMTD spacer domains (β-propeller modules). The LDLR and VLDLR receptors have an additional O-linked sugar domain (Fig 2). All of them have a single C-proximal transmembrane domain and a short cytoplasmic tail.32 In its natural role in cells, LDLR mediates the uptake of its natural ligand, cholesterol-carrying lipoprotein particles, by the process of endocytosis mediated by clathrin-coated pits. The captured cargos are then released due to the low pH environment of the endosome.33 The cytoplasmic tail of LDLR family members contains motifs used for interactions with a number of cytoplasmic adaptor and scaffold proteins, as part of the signal transduction pathways.34
To initiate an RV infection, two domains (D2 and D3) of VLDLR interact with VP1 residues at the apex of the fivefold plateau of the virion icosahedron.35 As with ICAM-1 binding, these events initiate endocytosis, viral entry, and release of viral RNA into the cytoplasm.
CDHR3 Glycoprotein
CDHR3 is a transmembrane glycoprotein that is currently found to be the only known receptor for RV-C. The protein is classified in the cadherin superfamily of proteins, many of which participate in calcium-dependent cell-cell adhesion or other related cell functions such as signaling pathway triggering cellular development and homeostasis. All cadherins have linear tandem repeats consisting of variable numbers of structurally similar extracellular domains, preceded by an N-terminal signal sequence and then terminated with a C-proximal transmembrane domain, linked to a short cytoplasmic tail.36 The extracellular domains arrange themselves like a stiff, linear rod, with the help of obligate Ca2+ ions at the domain junctions.37 The sequence of CDHR3 shows this protein to have six extracellular domains. The native function of CDHR3 on the surface of cells is currently unknown, but its gene is highly expressed in airway epithelia, fallopian tubes, and the brain.38 The gene is maintained with a high degree of sequence conservation among all known human and animal genomes.39, 40
Although the specific contacts between CDHR3 and RV-C have not yet been definitively mapped, protein modeling suggested that the first two extracellular domain repeats of CDHR3 are important,19, 40 and these most likely bind viruses in a location other than the diminished canyon, perhaps even at the twofold axes. More recently, studies of recombinant CDHR3 have shown that the first extracellular domain contacts RV-C.41 The hydrophobic pocket within the VP1 of RV-C is present, but the pore is absent, and the pocket cannot accommodate any known pocket factor.12 The “collapsed” structure of the pocket is physically similar to the empty pockets observed in the RV-B.
Of the two human alleles of CDHR3, the rs6967330-A allele encoding the Y529 protein variant is associated with higher RV-C binding and cellular replication capacity compared with the rs6967330-G allele encoding the C529 protein variant.19 The reason for this dichotomy is unknown but may involve differences in domain structure and cell surface stability of the two CDHR3 protein variants that consequently produce the different levels of RV-C accessibility on expressing cells.40 In clinical and biochemical studies, the rs6967330-A allele compared with the rs6967330-G allele is associated with higher overall CDHR3 protein expression on cell surfaces, which leads directly to increased RV-C binding and higher levels of viral replication.19, 40, 42
Opportunities for Antiviral Agents
Current over-the-counter treatments for RV infections are limited to those that can help relieve symptoms. Ongoing efforts to develop antiviral agents or vaccines that target RVs have been hampered by technical factors that include the large number of RV serotypes and their considerable genetic diversity. The RNA polymerase of RV exacerbates this problem because it is error prone, and the observed high natural mutation rates increase the likelihood of drug resistance. Moreover, upper and lower respiratory symptoms can be caused by many types of respiratory viruses in addition to RV, and any specific diagnosis of RV infections cannot be distinguished according to symptoms alone. Given that antiviral agents are only typically effective if administered during the early stages of respiratory infection, lack of an accurate point-of-care diagnostic test has been a hurdle. However, despite these obstacles, modern efforts to develop new RV vaccines and antiviral therapies are ongoing. The latest therapeutic strategies include the development of synthetic compounds or neutralizing antibodies that might potentially inhibit these viruses at different phases of their life cycles; for example, by blocking cellular receptors used by RVs or by augmenting antiviral immune responses.
Capsid-binding Agents
Antiviral approaches have focused heavily on capsid-binding agents, which are small molecules that can react directly with the RV-A and RV-B to inhibit receptor binding and, therefore, entry into the host cell.43, 44 Most of the tested inhibitors work by insertion into the VP1 hydrophobic pocket underneath the floor of the canyon. These drugs can stabilize RV-A and RV-B virions by displacing the floor of the canyon and preventing consequent changes in capsid conformation that are necessary for virus uncoating and/or release of viral RNA.45, 46 In actual clinical trials, however, most such capsid-binding agents worked well in vitro but only had limited efficacy. Pleconaril, vapendavir (BTA798), and pirodavir each successfully reduced the measured viral load using a human viral challenge model but provided limited clinical efficacy.47 Furthermore, it is now known that the RV-C lack an accessible hydrophobic pocket and are not susceptible to similar capsid-binding compounds.12 The initial failures to account for the reactiveness of the RV-C within clinical trials likely contributed to the limited efficacy in intervention studies.
Replication Inhibitors
In contrast to capsid binders, which work outside the cells, protease inhibitors prevent cleavage of viral proteins required for replication inside the infected cells. Protease inhibitors such as rupintrivir have been tested in RV challenge experiments, but to date, have exhibited only low efficacy against panels of RV-A and RV-B types in vitro.48 Other replication-inhibitory molecules that have been tested with equivalent limited efficacy include SG85 (3C protease inhibitor) and guanidine and TBZE029 (2C protease inhibitors).49, 50 Future strategies for anti-RV medications may include: inhibition of the RV polymerase51 or the 2A, 3A, or 3AB nonstructural proteins52, 53; or even interference with capsid assembly processes.54
Vaccines
The diversity of RV types and the expectation of only limited cross-protection have been key obstacles to the development of RV vaccines, although these features have prompted multiple studies to identify putatively shared epitopes among the RVs. For example, proposed subunit vaccines might consist of small but immunogenic regions of the virus proteins (VP2, VP4, and viral polymerase) representing subunits of broadly conserved regions across RV types.55 Preclinical testing of this approach across multiple RV types has had limited success.56 Likewise, addition of adjuvants such as AS03, MF59, and AS04 to inactivated RV vaccines has improved immunogenicity.57
Recently, the concept of a polyvalent inactivated RV vaccine has been revisited through studies involving mice and rhesus macaque models. Broadly neutralizing responses were induced in these animal models; these responses were dependent on high valency of the included virus types (25-valent in mice, 50-valent in rhesus macaques) and high antigen dose.58 Multivalent RV vaccine approaches have yet to be tested in humans.
In addition, new discoveries related to antiviral immunity have renewed interest in the use of biologic response modifiers to augment innate immune responses to this and other respiratory viruses. For example, interferon beta was tested in clinical trials and showed some promise for preventing virus-induced exacerbations of asthma despite not meeting prespecified outcomes.59 There are many other candidate strategies for boosting host innate immunity to respiratory viruses, and this general approach is exciting because it has the potential to induce resistance to multiple species of viral pathogens.
Receptor Blockers
Another potential antiviral approach is to block cell surface RV receptors on host cells with antibodies or small molecules. Monoclonal antibodies specific for ICAM-1, or even soluble recombinant versions of the receptor, can inhibit RV replication in vitro. However, this approach was costly, required multiple doses per day, and had limited efficacy in clinical trials.60 As alternatives, synthetic compounds such as multivalent Fab fusion proteins were developed to block RV-A and RV-B receptors.61 For example, a study showed that an ICAM-1 domain 1-specific antibody (14C11) could inhibit multiple major group RV types in vitro and prevent major group RV infections in a mouse model without interfering with ICAM-dependent cellular trafficking.62 Parallel approaches to block LDLR or CDHR3 have not yet been tested.
Conclusions
The discovery of RV-C lagged behind that of other RVs by 50 years because standard cell lines used by viral diagnostic laboratories lack expression of CDHR3. Modern diagnostic methods led to the discovery of RV-C viruses, and 10 years following that discovery, CDHR3 was identified as the first and probably the only cellular receptor for RV-C. As analysis of RV-C/receptor interactions progresses, what might these discoveries mean with respect to curing or preventing the common cold, a goal that is especially important to children and adults with chronic lung diseases such as asthma?
Now that RV receptors are known, it is worth considering whether blocking all three receptors would be an effective and feasible treatment. Given that all three known receptor proteins have other key functions in cells, topical administration of antiviral agents aimed at these proteins would be preferred to minimize side effects and possible toxicity. When considering prophylaxis vs treatment, prophylaxis for RVs has the potential for greater success given that the efficacy of antiviral drugs typically wanes quickly as the illness progresses. In addition, point-of-care diagnostics for RVs, which might be necessary for a treatment strategy, are still experimental and not available in the clinic. Other challenges with this approach could include both the cost and regulatory concerns of producing a medication with three active ingredients.
Because the RVs cause more respiratory infections in humans than any other pathogens, new therapeutic approaches are needed. All RVs infect airway epithelial cells by receptor-mediated endocytosis after recognition of one of only three main receptors: LDLR, ICAM-1, or CDHR3. Molecular definition of this first step in the replication cycle for all three RV species has enabled comprehensive studies of RV biology in vitro and offers major new opportunities toward identifying novel strategies for prevention and treatment.
Acknowledgments
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: J. E. G. has received grants from the National Institutes of Health/National Institute of Allergy and Infectious Diseases and consulting fees from PrEP Biopharm Inc, Regeneron, Meissa Vaccines Inc, and MedImmune, outside the submitted work. In addition, J. E. G. and A. C. P. have two patents pending (Methods of Propagating Rhinovirus C in Previously Unsusceptible Cell Lines and Adapted Rhinovirus C). None declared (S. B.).
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Footnotes
FUNDING/SUPPORT: This study was funded by the National Institutes of Health/National Institute of Allergy and Infectious Diseases [U19 AI104317] and National Heart, Lung, and Blood Institute [P01 HL070831].
References
- 1.Scully E.J., Basnet S., Wrangham R.W. Lethal respiratory disease associated with human rhinovirus C in wild chimpanzees, Uganda, 2013. Emerg Infect Dis. 2018;24(2):267–274. doi: 10.3201/eid2402.170778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmenberg A.C., Spiro D., Kuzmickas R. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324(5923):55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochkov Y.A., Palmenberg A.C., Lee W.M. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17(3):627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78(9):1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox D.W., Khoo S.K., Zhang G. Rhinovirus is the most common virus and rhinovirus-C is the most common species in paediatric intensive care respiratory admissions. Eur Respir J. 2018;52(2) doi: 10.1183/13993003.00207-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy J.L., Turner R.B., Braciale T., Heymann P.W., Borish L. Pathogenesis of rhinovirus infection. Curr Opin Virol. 2012;2(3):287–293. doi: 10.1016/j.coviro.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwane M.K., Prill M.M., Lu X. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis. 2011;204(11):1702–1710. doi: 10.1093/infdis/jir634. [DOI] [PubMed] [Google Scholar]
- 8.Annamalay A.A., Jroundi I., Bizzintino J. Rhinovirus C is associated with wheezing and rhinovirus A is associated with pneumonia in hospitalized children in Morocco. J Med Virol. 2017;89(4):582–588. doi: 10.1002/jmv.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox D.W., Bizzintino J., Ferrari G. Human rhinovirus species C infection in young children with acute wheeze is associated with increased acute respiratory hospital admissions. Am J Respir Crit Care Med. 2013;188(11):1358–1364. doi: 10.1164/rccm.201303-0498OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fry E.E., Stuart D.I. Virion structure. In: Ehrenfeld E.D.E., Roos R.P., editors. The Picronaviruses. ASM Press; Washington, DC: 2010. pp. 59–71. [Google Scholar]
- 11.Martinez-Salas E, Ryan MD. Translation and protein processing. In: Ehrenfeld E, Domingo E, Roos RP, eds. The Picornaviruses. Washington, DC: ASM Press; 2010:141-161.
- 12.Liu Y., Hill M.G., Klose T. Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc Natl Acad Sci U S A. 2016;113(32):8997–9002. doi: 10.1073/pnas.1606595113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gern JE, Palmenberg AC. Rhinoviruses. In: Knipe DM, Howley PM, eds. Fields Virology, 6th edition. Philadelphia, PA: Lippincott Williams & Wilkins; 2013:531-549.
- 14.Oliveira M.A., Zhao R., Lee W.M. The structure of human rhinovirus 16. Structure. 1993;1(1):51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 15.Hadfield A.T., Diana G.D., Rossmann M.G. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc Natl Acad Sci U S A. 1999;96(26):14730–14735. doi: 10.1073/pnas.96.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greve J.M., Davis G., Meyer A.M. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56(5):839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 17.Vlasak M., Goesler I., Blaas D. Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J Virol. 2005;79(10):5963–5970. doi: 10.1128/JVI.79.10.5963-5970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griggs T.F., Bochkov Y.A., Basnet S. Rhinovirus C targets ciliated airway epithelial cells. Respir Res. 2017;18(1):84. doi: 10.1186/s12931-017-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bochkov Y.A., Watters K., Ashraf S. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015;112(17):5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochkov Y.A., Watters K., Basnet S. Mutations in VP1 and 3A proteins improve binding and replication of rhinovirus C15 in HeLa-E8 cells. Virology. 2016;499:350–360. doi: 10.1016/j.virol.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs R., Blaas D. Uncoating of human rhinoviruses. Rev Med Virol. 2010;20(5):281–297. doi: 10.1002/rmv.654. [DOI] [PubMed] [Google Scholar]
- 22.Staunton D.E., Merluzzi V.J., Rothlein R., Barton R., Marlin S.D., Springer T.A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56(5):849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 23.Tomassini J.E., Graham D., DeWitt C.M., Lineberger D.W., Rodkey J.A., Colonno R.J. cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc Natl Acad Sci U S A. 1989;86(13):4907–4911. doi: 10.1073/pnas.86.13.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staunton D.E., Marlin S.D., Stratowa C., Dustin M.L., Springer T.A. Primary structure of ICAM-1 demonstrates interaction between members of the immunoglobulin and integrin supergene families. Cell. 1988;52(6):925–933. doi: 10.1016/0092-8674(88)90434-5. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard A.K., Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med. 2000;28(9):1379–1386. doi: 10.1016/s0891-5849(00)00223-9. [DOI] [PubMed] [Google Scholar]
- 26.Kolatkar P.R., Bella J., Olson N.H., Bator C.M., Baker T.S., Rossmann M.G. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18(22):6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bella J., Kolatkar P.R., Marlor C.W., Greve J.M., Rossmann M.G. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci U S A. 1998;95(8):4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond M.S., Staunton D.E., de Fougerolles A.R. ICAM-1 (CD54): a counter-receptor for Mac-1 (CD11b/CD18) J Cell Biol. 1990;111(6 Pt 2):3129–3139. doi: 10.1083/jcb.111.6.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadfield A.T., Lee W., Zhao R. The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle. Structure. 1997;5(3):427–441. doi: 10.1016/s0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
- 30.Nurani G., Lindqvist B., Casasnovas J.M. Receptor priming of major group human rhinoviruses for uncoating and entry at mild low-pH environments. J Virol. 2003;77(22):11985–11991. doi: 10.1128/JVI.77.22.11985-11991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofer F., Gruenberger M., Kowalski H. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci U S A. 1994;91(5):1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon H., Blacklow S.C. Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem. 2005;74:535–562. doi: 10.1146/annurev.biochem.74.082803.133354. [DOI] [PubMed] [Google Scholar]
- 33.Beglova N., Blacklow S.C. The LDL receptor: how acid pulls the trigger. Trends Biochem Sci. 2005;30(6):309–317. doi: 10.1016/j.tibs.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Cam J., Bu G. Low-density lipoprotein receptor family: endocytosis and signal transduction. Mol Neurobiol. 2001;23(1):53–67. doi: 10.1385/MN:23:1:53. [DOI] [PubMed] [Google Scholar]
- 35.Verdaguer N., Fita I., Reithmayer M., Moser R., Blaas D. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat Struct Mol Biol. 2004;11(5):429–434. doi: 10.1038/nsmb753. [DOI] [PubMed] [Google Scholar]
- 36.Niessen C.M., Leckband D., Yap A.S. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91(2):691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sotomayor M., Schulten K. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys J. 2008;94(12):4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Broad Institute of MIT and Harvard, GTEx Portal: CDHR3 gene expression. https://gtexportal.org/home/gene/CDHR3. Accessed December 10, 2018.
- 39.Bonnelykke K., Sleiman P., Nielsen K. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46(1):51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 40.Palmenberg A.C. Rhinovirus C, asthma, and cell surface expression of virus receptor CDHR3. J Virol. 2017;91(7) doi: 10.1128/JVI.00072-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watters K.E., Palmenberg A.C. CHDR3 extracellular domains EC1-3 mediate rhinovirus C interactions with cells and as recombinant derivatives, are inhibitory to virus infection. PloS Pathogens. 2018;14(12):e1007477. doi: 10.1371/journal.ppat.1007477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonnelykke K., Coleman A.T., Evans M.D. Cadherin-related family member 3 genetics and rhinovirus C respiratory illnesses. Am J Respir Crit Care Med. 2018;197(5):589–594. doi: 10.1164/rccm.201705-1021OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacroix C., Laconi S., Angius F. In vitro characterisation of a pleconaril/pirodavir-like compound with potent activity against rhinoviruses. Virol J. 2015;12:106. doi: 10.1186/s12985-015-0330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J., Jung Y.K., Kim C. A novel series of highly potent small molecule inhibitors of rhinovirus replication. J Med Chem. 2017;60(13):5472–5492. doi: 10.1021/acs.jmedchem.7b00175. [DOI] [PubMed] [Google Scholar]
- 45.McKinlay M.A., Pevear D.C., Rossmann M.G. Treatment of the picornavirus common cold by inhibitors of viral uncoating and attachment. Annu Rev Microbiol. 1992;46:635–654. doi: 10.1146/annurev.mi.46.100192.003223. [DOI] [PubMed] [Google Scholar]
- 46.Valbuena A., Rodríguez-Huete A., Mateu M.G. Mechanical stiffening of human rhinovirus by cavity-filling antiviral drugs. Nanoscale. 2018;10(3):1440–1452. doi: 10.1039/c7nr08704g. [DOI] [PubMed] [Google Scholar]
- 47.Mirabelli C., Scheers E., Neyts J. Novel therapeutic approaches to simultaneously target rhinovirus infection and asthma/COPD pathogenesis. F1000Res. 2017;6:1860. doi: 10.12688/f1000research.11978.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binford S.L., Weady P.T., Maldonado F., Brothers M.A., Matthews D.A., Patick A.K. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother. 2007;51(12):4366–4373. doi: 10.1128/AAC.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lacroix C., George S., Leyssen P., Hilgenfeld R., Neyts J. The enterovirus 3C protease inhibitor SG85 efficiently blocks rhinovirus replication and is not cross-resistant with rupintrivir. Antimicrob Agents Chemother. 2015;59(9):5814–5818. doi: 10.1128/AAC.00534-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Palma A.M., Heggermont W., Lanke K. The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C. J Virol. 2008;82(10):4720–4730. doi: 10.1128/JVI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz B.A., Vance L.M. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J Virol. 1995;69(7):4189–4197. doi: 10.1128/jvi.69.7.4189-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falah N., Violot S., Décimo D. Ex vivo and in vivo inhibition of human rhinovirus replication by a new pseudosubstrate of viral 2A protease. J Virol. 2012;86(2):691–704. doi: 10.1128/JVI.05263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mousnier A., Bell A.S., Swieboda D.P. Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus. Nat Chem. 2018;10(6):599–606. doi: 10.1038/s41557-018-0039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glanville N., McLean G.R., Guy B. Cross-serotype immunity induced by immunization with a conserved rhinovirus capsid protein. PLoS Pathog. 2013;9(9):e1003669. doi: 10.1371/journal.ppat.1003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glanville N., Johnston S.L. Challenges in developing a cross-serotype rhinovirus vaccine. Curr Opin Virol. 2015;11:83–88. doi: 10.1016/j.coviro.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Williams G.R., Kubajewska I., Glanville N., Johnston S.L., Mclean G.R. The potential for a protective vaccine for rhinovirus infections. Expert Rev Vaccines. 2016;15(5):569–571. doi: 10.1586/14760584.2016.1142375. [DOI] [PubMed] [Google Scholar]
- 58.Lee S., Nguyen M.T., Currier M.G. A polyvalent inactivated rhinovirus vaccine is broadly immunogenic in rhesus macaques. Nat Commun. 2016;7:12838. doi: 10.1038/ncomms12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Djukanovic R., Harrison T., Johnston S.L. The effect of inhaled IFN-beta on worsening of asthma symptoms caused by viral infections. A randomized trial. Am J Respir Crit Care Med. 2014;190(2):145–154. doi: 10.1164/rccm.201312-2235OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLean G.R. Developing a vaccine for human rhinoviruses. J Vaccines Immun. 2014;2(3):16–20. doi: 10.14312/2053-1273.2014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang F., Yu M. Viral receptor blockage by multivalent recombinant antibody fusion proteins: inhibiting human rhinovirus (HRV) infection with CFY196. J Antimicrob Chemother. 2004;53(1):23–25. doi: 10.1093/jac/dkh019. [DOI] [PubMed] [Google Scholar]
- 62.Traub S., Nikonova A., Carruthers A. An anti-human ICAM-1 antibody inhibits rhinovirus-induced exacerbations of lung inflammation. PLoS Pathog. 2013;9(8):e1003520. doi: 10.1371/journal.ppat.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]