Abstract
Site-specific fluorescence probes can be used to measure distances within proteins when used as part of a Förster resonance energy transfer (FRET) pair. Here we report the synthesis of a coumarin maleimide (Mcm-Mal) that is fluorogenic upon reaction with cysteine. We demonstrate that cysteine, acridonylalanine (Acd) double mutant proteins can be produced by unnatural amino acid mutagenesis and reacted with Mcm-Mal to generate Mcm/Acd labeled proteins for FRET studies. The Mcm/Acd FRET pair is minimally-perturbing, easy to install, and well-suited to studying protein distances in the 15-40 Å range. Furthermore, Mcm/Acd labeling can be combined with tryptophan fluorescence in three color FRET to monitor multiple interactions in one experiment.
Fluorescence spectroscopy methods can be valuable ways of studying protein folding and dynamics, as they allow one to observe protein motions in real time under physiological conditions.1, 2 If two probes can be attached to the protein, one can obtain structural information using distance-dependent interactions such as Förster resonance energy transfer (FRET) and quenching by photo-induced electron transfer.3 The development of probes that can be easily installed for these applications is an important area of bioorganic chemistry. Here, we report the synthesis of methoxycoumarin maleimide (Mcm-Mal, Fig. 1), a probe that can be used as part of a FRET pair with acridonylalanine (Acd). We further demonstrate that Mcm/Acd labeling can be combined with a single Trp mutant for three color FRET experiments, which can be used to simultaneously monitor two interactions, such as a protein/protein interaction and a conformational change within one of the proteins.
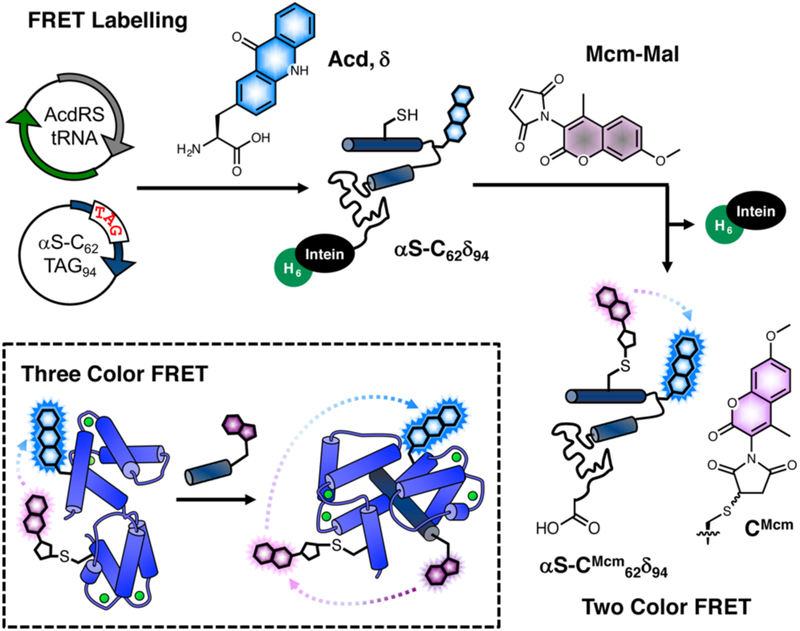
Fig. 1.
Mcm labelling for FRET experiments. Protein expressed with Cys and acridonylalanine (Acd, δ) mutants is harvested as an intein fusion and reacted with methoxycoumarin maleimide (Mcm-Mal) for two color FRET experiments. Inset: Three color FRET between Trp (purple), Mcm (pink), and Acd (cyan) can be used to detect binding (Trp excitation) and conformational change (Mcm excitation) simultaneously.
The fluorescent labeling of proteins for FRET studies has been greatly enabled by recent advances in site-specific protein modification, particularly through incorporation of unnatural amino acids (Uaas) and biorthogonal reactions.4 However, there are still limitations to commonly used fluorophores such as fluorescein and rhodamine that can hamper FRET experiments. Their large size can be disruptive to protein folding, and their working range for FRET may not be suited to distance changes in the proteins. The fluorescein/ rhodamine FRET pair has a Förster radius (R0, the distance of half-maximal energy transfer) of ~50 Å, making it useful for studying distances in the 30-90 Å range. Since many important intraprotein distances are shorter than 30 Å, small short-range FRET pairs are better suited to studying these protein regions.
Our laboratory has worked to develop non-perturbing, short/medium-range probe pairs that can be selectively excited in proteins. These include fluorescent Uaas such as Acd.5, 6 Acd is excited at 380-400 nm with emission at 420-450 nm. We have shown that Acd can be a valuable FRET acceptor from methoxycoumarin (Mcm, excited at 325 nm) with a working range of 15-40 Å.5 Mcm is an excellent FRET donor to Acd since it has a high extinction coefficient with a peak in the absorption spectrum that coincides with a minimum in the Acd absorption spectrum (see ESI, Fig. S13). However, in previous studies, Mcm was introduced as 7-methoxycoumarin-4-yl-alanine, which must be incorporated by solid phase peptide synthesis.5 We wished to introduce Mcm through selective Cys modification in order to more easily generate Mcm/Acd labelled proteins.
We considered two ways of installing Mcm, either through conjugate addition of a 3-maleimide derivative (Mcm-Mal) or through an SNAr reaction of a 3-bromo derivative (Mcm-Br, see ESI, Scheme S1). Mcm-Br was synthesized from 7-methoxy-4-methylcoumarin by bromination with NBS (yield 78%). Mcm-Mal was also synthesized from 7-methoxy-4-methylcoumarin. After nitration with HNO3 and Ac2O, the nitrocoumarin derivative was reduced by hydrogenation in the presence of Pd/C (32% yield of the desired 3-amino isomer). The 3-aminocoumarin derivative obtained was converted to Mcm-Mal by condensation with maleic anhydride (84% yield).
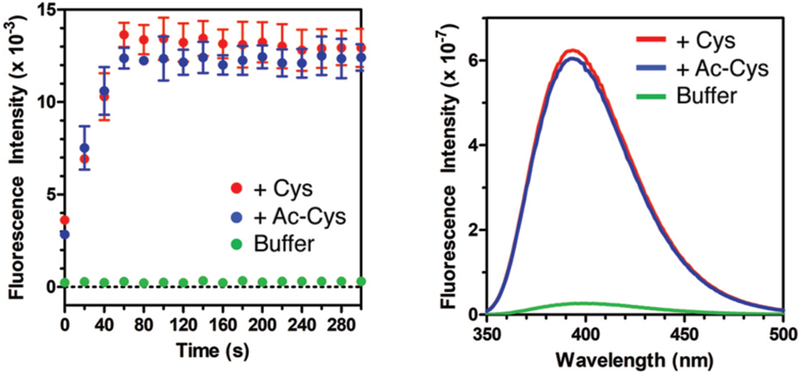
We tested the reaction of Mcm-Mal and Mcm-Br with Cys in aqueous buffer and found that reactions of Mcm-Mal were much faster (complete within a few seconds at 10 μM), with a >20-fold increase in fluorescence at 390 nm (see ESI, Figs. S2 and S4). High performance liquid chromatography (HPLC) and mass spectrometry (MS) analysis indicated that the very rapid reaction initially yielded the conjugate addition product shown in Fig. 1, without maleimide ring opening (see ESI, Figs. S3 and S5). This product then isomerizes on a timescale of hours to a compound of identical mass, which we attribute to the lactam form S5 (see ESI, Scheme 2 and Fig. S5). This is similar to reactions recently reported for detection of Cys in its amino acid form by Tong et al.7 The authors ascribed the high levels of fluorogenicity that they observed to relief of two quenching mechanisms, one by conjugate addition, and the other by opening of the maleimide ring. Since this second step involves the Cys α-amine, we also tested the reaction of Mcm-Mal with N-acetyl cysteine (Ac-Cys), which is more representative of Cys in proteins. The resulting >20-fold turn-on of fluorescence was comparable to that seen in reactions with Cys (Fig. 2) These Ac-Cys/Mcm-Mal data, together with our observations of the faster fluorescence turn-on relative to ring-opening in Cys/Mcm-Mal reactions, imply that the conjugate addition is the primary determinant of fluorescence turn-on.
Fig. 2.
Mcm-Mal turn-on experiments. Fluorescence intensity of 10 μM Mcm-Mal in Tris buffer (100 mM, pH 7.0, DMSO 1%) alone or mixed with 1 equiv Cys or Ac-Cys. Fluorescence excitation at 325 nm, emission monitored at 400nm; background corrected data are shown, see raw data in ESI Fig. S6. Inset: Fluorescence spectra acquired after 6 h of incubation.
Following these trial reactions with small molecules, we evaluated the turn-on of Mcm-Mal fluorescence in reactions at a Cys residue in two proteins, calmodulin (CaM) and α-synuclein (αS). CaM is a calcium sensor protein that undergoes a dramatic conformational change in the presence of calcium and helical peptide binding partners.8, 9 αS is a disordered protein that contributes to the pathogenesis of Parkinson’s disease.10, 11 These provide examples of the value of FRET to monitor conformational change and protein/protein interactions. Cys mutants of CaM and αS (CaM-C12, αS-C62, αS-C114) were reacted with Mcm-Mal, and fluorescence emission was monitored at 390 nm. In all cases, we observed a turn-on of fluorescence, but this was significantly less than what was observed with Cys or Ac-Cys, due in part to higher background fluorescence in protein reactions because of the need to keep the protein Cys reduced. Matrix-assisted laser desorption ionization (MALDI) MS analysis of whole proteins and trypsin digests confirmed that modification took place exclusively at the Cys, without ring-opening. (see ESI, Fig. S8-S12)
In order to perform FRET measurements, one must doubly label the protein with a donor/acceptor probe pair. We installed the Mcm/Acd FRET pair by using site-directed mutagenesis and amber codon suppression to generate constructs with single Cys and Acd mutations, followed by reaction of Cys with Mcm-Mal. (Fig. 1) The reaction progress was monitored using the turn-on of Mcm-Mal fluorescence and MALDI MS. (see ESI, Figs. S8 and S10) We commonly use C-terminal intein-His6 tags to facilitate isolation of full-length proteins containing unnatural amino acids from truncated proteins.12 His6 tags can be cleaved with β-mercaptoethanol either before or after Mcm-Mal labelling. Due to the highly sensitive nature of FRET measurements, we typically perform a second purification pass by FPLC or HPLC.
We have used our doubly labeled αS and CaM constructs in proof-of-principle FRET experiments to demonstrate the value of the Mcm/Acd pair. For αS, we monitored a conformational change of the monomer induced by addition of the compacting osmolyte trimethylamine N-oxide (TMAO). TMAO is a naturally occuring osmolyte that is abundant in aquatic organisms, and counteracts the denaturing effects of urea required for osmotic pressure regulation.13, 14 Previous work has demonstrated that αS undergoes successive compaction with exposure to increasing amounts of TMAO.15-17
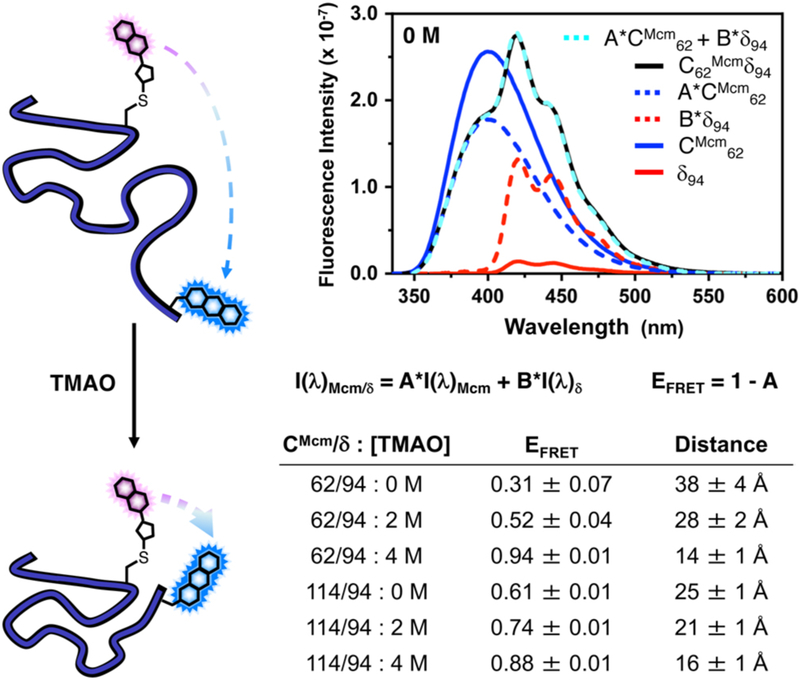
Here, we chose to monitor TMAO-induced conformational changes in αS by introducing Cys residues at positions 62 or 114 and Acd at position 94. Single-labelled constructs containing either Mcm or Acd and the double-labelled constructs were prepared as described above. Fluorescence spectra for all of the single and double-labelled constructs were collected in 0, 2, and 4 M TMAO. Singly-labelled spectra were used to analyse the double-labelled spectra to determine FRET efficiency (EFRET). This was performed by linearly weighting the singly-labelled spectra and minimizing the squared difference between the doubly-labelled spectrum and the sum of the linearly weighted single-labelled spectra at each wavelength. (Fig. 3) This allows one to correct for changes in quantum yield or spectral overlap when the chromophore changes environment, and directly affords the relative Mcm quenching (A in Fig. 3) and EFRET (EFRET = 1-A) to determine the FRET distance through application of an appropriate form of the Förster equation. (see ESI for details)
Fig. 3.
αS TMAO FRET Experiments. Left: Incubation of αS with TMAO causes protein compaction, which can be monitored by FRET. Top Right: Fluorescence emission spectra (325 nm excitation) of 1 μM concentrations of αS-CMcm62δ94 and the corresponding singly-labelled αS-CMcm62δ94 and αS-CMcm62δ94 constructs in 0 M TMAO. Deconvolution of the doubly labelled spectrum by fitting to a weighted sum of the singly-labelled spectrum are shown. Bottom Right: EFRET and corresponding inter-fluorophore distances for αS constructs under varying conditions determined using spectral deconvolution. See ESI for details.
The 0 M data are consistent with Flory scaling models of inter-residue distances in disordered proteins. (ESI, Fig. S24) Moreover, we observe compaction for both label positions with increasing amounts of TMAO, as previously observed in this system with other FRET pairs.16, 18 These results were corroborated by measuring the change in Mcm fluorescence lifetime for the two double-labelled pairs relative to the analogous Mcm single-labelled proteins. (see ESI)
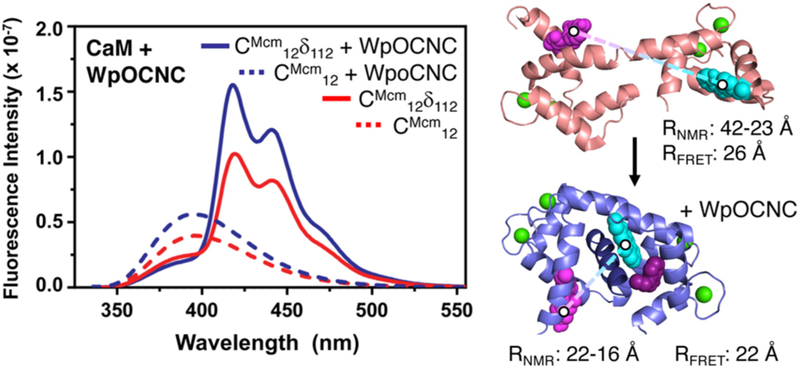
For CaM, we monitored the conformational change induced by peptide binding. Acd-labelled CaM was expressed and reacted with Mcm-Mal to give CaM-CMcm12δ112. Based on existing crystal and NMR structures of CaM, we expected that a significant change in EFRET would occur due to a decrease in distance between positions 12 and 112 upon peptide binding. (Fig. 4) We note that in several CaM crystal structures, an extended conformation is observed, making the 12/112 interresidue distance >50 Å. However, NMR studies have shown that the N- and C-terminal lobes of CaM are dynamic, and that the conformations sampled in solution are more compact.19
Fig. 4.
CaM Peptide binding FRET. Left: Fluorescence emission spectra of CaM-CMcm12δ112 (325 nm excitation) obtained in the presence and absence of the WpOCNC binding peptide. Note: The apparently small FRET change is the result of an opposing increase in Mcm emission upon binding. This can be accounted for by comparison to CaM-CMcm12 emission with and without WpOCNC. Full spectral deconvolution to obtain EFRET values is described in ESI. Right: Images of CaM in the presence an absence of the pOCNC peptide rendered from PDB entries 1X0220 and 1SY9,21 respectively. Distances obtained from EFRET measurements are consistent with the CaM conformations in these NMR studies when models of CMcm and Acd are included. See ESI for details.
Fluorescence spectra of CaM-CMcm12δ112 were obtained in Ca2+-buffered solutions, and indeed we observed significant Mcm-to-Acd FRET, evidence that positions 12 and 112 were closer than 40 Å. Upon addition of WpOCNC, a Trp derivative of a known high-affinity CaM binding peptide,21, 22 we observed an increase in FRET. As in our αS studies, we also obtained spectra for singly-labelled CaM-CMcm12 and CaM-δ112 under identical conditions, and fit a sum of these spectra to the CaM-CMcm12δ112 spectrum to quantitatively determine EFRET. The resulting interresidue distances of 26 Å and 22 Å in the absence and presence of the WpOCNC peptide, respectively, are in reasonable agreement with Mcm- and Acd-labelled CaM models based on existing NMR structures (Fig. 4 and ESI).20, 21 These studies demonstrate the value of Mcm/Acd FRET in monitoring a binding induced conformational change.
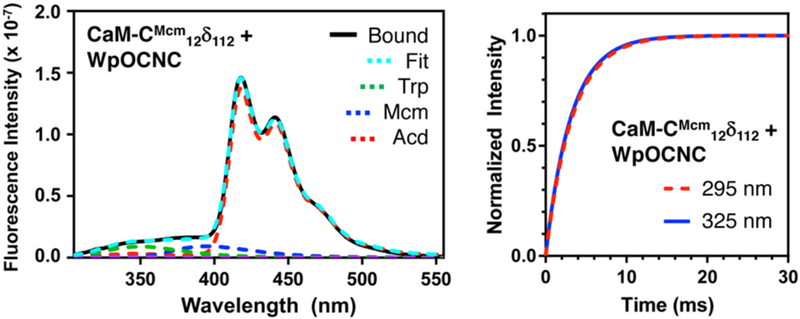
The longer wavelength excitation (325 nm) and emission of Mcm and Acd allowed us to selectively observe these changes in the presence of the Trp residue in WpOCNC. However, we can also take advantage of the spectral overlap of Trp emission with both Mcm and Acd to perform three color FRET experiments, monitoring binding and conformational change events in the same experiment. Although deconvolution to extract distance information is not reliable, FRET can still be used to measure binding kinetics. Using a stopped-flow fluorometer, we observed rapid binding of WpOCNC through 295 nm excitation, monitoring Acd emission at 420 nm. In the same experimental setup, we excited at 325 nm to monitor the conformational change in CaM alone. We found that the rates of binding (Ex295: 338 ± 93 s−1) and conformational change were nearly identical (Ex325: 306 ± 40 s−1), implying that the two processes are concerted. This is consistent with previous experiments showing that CaM populates compact structures even in the absence of peptide.19, 20 Our data support the idea that WpOCNC stabilizes existing compact CaM conformations rather than binding and then inducing a large conformational change. We note that the rates of binding and conformational change are also consistent with previous data for CaM binding of several peptides (~500 s−1 at 1 μM peptide and CaM).23-25
Using a combination of Cys labeling by Mcm-Mal and Acd incorporation allows one to easily introduce a FRET pair that is selectively excitable and minimally perturbing in proteins (WpOCNC Kds are nearly identical for CaM and CaM-CMcm12δ112, Fig. S17). Mcm/Acd FRET pairs can be used to monitor conformational changes of 15-40 Å, a useful scale for tracking motions within protein domains or among domains of moderately sized proteins. We have shown that spectra of Mcm- or Acd-only proteins can be used to correct for changes in quantum yield or spectral shape to accurately determine EFRET and inter-chromophore distance. While background signal from reducing agents hindered its demonstration here, the fluorescence turn-on of Mcm-Mal gives it the potential to be used for in situ FRET with Acd. Additionally, Trp FRET with both Mcm and Acd can be used in combination with Mcm/Acd FRET to monitor the kinetics of two processes simultaneously. We note that previous three color (or “two step”) FRET studies have primarily been single molecule experiments using bulky probes or even fluorescent proteins.26-29 An example using cyanophenylalanine, Trp, and 7-azatryptophan by Gai uses smaller probes, but these are all UV wavelength probes.30 The Trp/Mcm/Acd trio balances small probe size with selective excitation, and we are working to develop complementary red-shifted, minimalist fluorescence probes.
Supplementary Material
Fig. 5.
Three Color FRET and Rapid Mixing Measurements of Peptide Binding Kinetics Left: Emission spectrum of CaM-CMcm12δ112/WpOCNC complex (295 nm excitation) shown deconvoluted with Trp, Mcm, and Acd components. Right: Overlay of fits to stopped flow fluorescence data for binding of WpOCNC to 0.5 μM CaM-CMcm12δ112. Red: 295 nm excitation, 420 nm emission; Blue: 325 nm excitation, 420 nm emission. See ESI for raw data and additional steady state spectra for FRET experiments.
Acknowledgements
This work was supported by the University of Pennsylvania and the National Institutes of Health (NIH NS081033 to EJP). Instruments supported by the NIH and National Science Foundation include: MALDI MS (NSF MRI-0820996), LCMS and HRMS (NIH RR-023444), stopped flow fluorometer (NSF CHE-1337449), and NMR (NIH RR-022442). JJF, NI, CMH, CRW and IS respectively thank the following for fellowship support: NSF (DGE-1321851), Japan Society for the Promotion of Science, Age Related Neurodegenerative Disease Training Grant (NIH T32 AG000255), Structural Biology and Molecular Biophysics Training Grant (NIH T32 GM008275), and Royal Thai Foundation. JJF thanks Profs. Michele Vendruscolo, Christopher Dobson, Abhinav Nath, and Elizabeth Rhoades for sharing αS structural data.
Notes and references
- 1.Haney CM, Wissner RF and Petersson EJ, Curr. Opin. Chem. Biol, 2015, 28, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giepmans BNG, Adams SR, Ellisman MH and Tsien RY, Science, 2006, 312, 217–224. [DOI] [PubMed] [Google Scholar]
- 3.Lakowicz JR, Principles of Fluorescence Spectroscopy, Springer; US, 3 edn., 2006. [Google Scholar]
- 4.Lang K and Chin JW, Chem. Rev, 2014, 114, 4764–4806. [DOI] [PubMed] [Google Scholar]
- 5.Speight LC, Muthusamy AK, Goldberg JM, Warner JB, Wissner RF, Willi TS, Woodman BF, Mehl RA and Petersson EJ, J. Am. Chem. Soc, 2013, 135, 18806–18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sungwienwong I, Hostetler ZM, Blizzard RJ, Porter JJ, Driggers CM, Mbengi LZ, Villegas JA, Speight LC, Saven JG, Perona JJ, Kohli RM, Mehl RA and Petersson EJ, Org. Biomol. Chem, 2017, 15, 3603–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong H, Zhao J, Li X, Zhang Y, Ma S, Lou K and Wang W, Chemical Communications, 2017, 53, 3583–3586. [DOI] [PubMed] [Google Scholar]
- 8.Chin D and Means AR, Trends Cell Biol, 2000, 10, 322–328. [DOI] [PubMed] [Google Scholar]
- 9.Vetter SW and Leclerc E, Eur. J. Biochem, 2003, 270, 404–414. [DOI] [PubMed] [Google Scholar]
- 10.Lashuel HA, Overk CR, Oueslati A and Masliah E, Nat. Rev. Neurosci, 2013, 14, 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R and Goedert M, Nature, 1997, 388, 839–840. [DOI] [PubMed] [Google Scholar]
- 12.Batjargal S, Walters CR and Petersson EJ, J. Am. Chem. Soc, 2015, 137, 1734–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang A and Bolen DW, Biochemistry, 1997, 36, 9101–9108. [DOI] [PubMed] [Google Scholar]
- 14.Canchi DR and García AE, Ann. Rev. Phys. Chem, 2013, 64, 273–293. [DOI] [PubMed] [Google Scholar]
- 15.Uversky VN, Li J and Fink AL, FEBS Lett, 2001, 509, 31–35. [DOI] [PubMed] [Google Scholar]
- 16.Moosa MM, Ferreon ACM and Deniz AA, ChemPhysChem, 2015, 16, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreon ACM, Moosa MM, Gambin Y and Deniz AA, Proc. Natl. Acad. Sci. USA, 2012, 109, 17826–17831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wissner RF, Batjargal S, Fadzen CM and Petersson EJ, J. Am. Chem. Soc, 2013, 135, 6529–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anthis NJ, Doucleff M and Clore GM, J. Am. Chem. Soc, 2011, 133, 18966–18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kainosho M, Torizawa T, Iwashita Y, Terauchi T, MeiOno A and Güntert P, Nature, 2006, 440, 52–57. [DOI] [PubMed] [Google Scholar]
- 21.Contessa GM, Orsale M, Melino S, Torre V, Paci M, Desideri A and Cicero DO, J. Biomol. NMR, 2005, 31, 185–199. [DOI] [PubMed] [Google Scholar]
- 22.Liu MY, Chen TY, Ahamed B, Li J and Yau KW, Science, 1994, 266, 1348–1354. [DOI] [PubMed] [Google Scholar]
- 23.Wu G, Berka V and Tsai A-L, J. Inorg. Biochem, 2011, 105, 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown SE, Martin SR and Bayley PM, J. Biol. Chem, 1997, 272, 3389–3397. [DOI] [PubMed] [Google Scholar]
- 25.Kovacs E, Tóth J, Vértessy BG and Liliom K, J. Biol. Chem, 2010, 285, 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK and Weiss S, Biophys. J, 2007, 92, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Lee S, Ragunathan K, Joo C, Ha T and Hohng S, Angew. Chem. Int. Ed, 2010, 49, 9922–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun YS, Wallrabe H, Booker CF, Day RN and Periasamy A, Biophys. J, 2010, 99, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voss S, Zhao L, Chen X, Gerhard F and Wu YW, J. Peptide Sci, 2014, 20, 115–120. [DOI] [PubMed] [Google Scholar]
- 30.Rogers JMG, Lippert LG and Gai F, Anal. Biochem, 2010, 399, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.