Abstract
Background
The etiology of cleft lip with or without cleft palate (CL/P), a common congenital birth defect, is complex and involves the contribution of genetic and environmental factors. Although many candidate genes have been identified, the regulation and interaction of these genes in CL/P remain unclear. In addition, the contribution of microRNAs (miRNAs), non-coding RNAs that regulate the expression of multiple genes, to the etiology of CL/P is largely unknown.
Methods
To identify the signatures of causative biological pathways for human CL/P, we conducted a systematic literature review for human CL/P candidate genes and subsequent bioinformatics analyses. Functional enrichment analyses of the candidate CL/P genes were conducted using the pathway databases GO and KEGG. The miRNA-mediated post-transcriptional regulation of the CL/P candidate genes was analyzed with miRanda, PITA, and TargetScan, and miRTarbase. Genotype-phenotype association analysis was conducted using GWAS. The functional significance of the candidate miRNAs was evaluated experimentally in cell proliferation and target gene regulation assays in human lip fibroblasts.
Results
Through an extensive search of the main biomedical databases, we mined 177 genes with mutations or association/linkage reported in individuals with CL/P, and considered them as candidate genes for human CL/P. The genotype-phenotype association study revealed that mutations in 12 genes (ABCA4, ADAM3A, FOXE1, IRF6, MSX2, MTHFR, NTN1, PAX7, TP63, TPM1, VAX1, and WNT9B) were significantly associated with CL/P. In addition, our bioinformatics analysis predicted 16 microRNAs (miRNAs) to be post-transcriptional regulators of CL/P genes. To validate the bioinformatics results, the top six candidate miRNAs (miR-124-3p, miR-369-3p, miR-374a-5p, miR-374b-5p, miR-497-5p, and miR-655-3p) were evaluated by cell proliferation/survival assays and miRNA-gene regulation assays in cultured human lip fibroblasts. We found that miR-497-5p and miR-655-3p significantly suppressed cell proliferation in these cells. Furthermore, the expression of the predicted miRNA-target genes was significantly downregulated by either miR-497-5p or miR-655-3p mimic.
Conclusion
Expression of miR-497-5p and miR-655-3p suppresses cell proliferation through the regulation of human CL/P-candidate genes. This study provides insights into the role of miRNAs in the etiology of CL/P and suggests possible strategies for the diagnosis of CL/P.
Electronic supplementary material
The online version of this article (10.1186/s12920-019-0535-2) contains supplementary material, which is available to authorized users.
Keywords: Cleft lip, Gene mutation, MicroRNA, Systematic review, Bioinformatics analysis
Background
Cleft lip with or without cleft palate (CL/P) is a common congenital malformation affecting speech, hearing, feeding, among others functions [1]. Individuals with CL/P require a comprehensive treatment, including multiple plastic and maxillofacial surgeries from birth to adulthood, and speech therapy [2]. It has an average worldwide birth prevalence of 1 in 1000, ranging from 1 in 500 in the Asian population to 1 in 2500 in the African population, with wide variability per geographic origin, ethnicity, and socioeconomic status [3, 4]. The complexity of its etiology seems to result from various genetic and environmental risk factors along with gene- environment interactions (e.g., approximately 70% cases are non-syndromic) [5]. Genome-wide association studies (GWAS) and linkage studies have identified genetic susceptibility to CL/P and differences in such susceptibility in various populations and ethnic groups [6]. However, as seen in many complex diseases or traits, most of the mutations and loci identified from such studies are in the noncoding genomic regions and do not have specific functional annotations [7, 8]. Although much progress has been made in identifying genes whose mutations are associated with CL/P, little is known about the mechanisms by which environmental and epigenetic factors adversely influence gene expression during lip development. Recent studies indicate that environmental factors contribute to changes in phenotype or gene expression at post-transcriptional level through the regulation of noncoding RNAs, including microRNAs (miRNAs) [9]. miRNAs are short (~ 22 nucleotides) noncoding RNA molecules that regulate gene expression at the post- transcriptional level, and they fine-tune the expression of ~ 30% of all mammalian protein-encoding genes [10]. miRNAs were first reported in mammalian systems in 2001 [11], but the latest release of the miRNA database [miRBase (ver. 20): more than 24,000 miRs annotated] highlights the rapid growth of this field of research; however, the expression patterns and functions of most miRNAs still remain to be discovered. The miRNA-gene regulatory mechanisms contribute to the pathogenesis in various diseases [12, 13]. Nonetheless, a limited number of miRNAs (e.g. miRNA- 140, miRNA-17-92 cluster, miRNA-200b, miRNA-133b) have been reported as miRNAs involved in craniofacial deformities in zebrafish and mouse models, as well as in humans [14]. Therefore, an investigation of the functional regulation, at the level of biological pathways and post- transcriptional regulation mechanism, will improve our understanding of genetic susceptibility to CL/P [7, 15].
In this study, in order to identify signatures of causative pathways in the complex etiology of CL/P, we performed a systematic review and subsequent bioinformatics analysis for CL/P-candidate genes. In addition, we analyzed miRNA-gene regulation by bioinformatics analyses. The function in cell proliferation/survival for six candidate miRNAs was experimentally evaluated by cell proliferation assays. Our findings will help us understand the role of genes and miRNAs that are associated with CL/P, in biological pathways and networks. Such knowledge will provide the basis for the diagnosis, prevention, and treatment of craniofacial anomalies such as CL/P.
Methods
Data sources
The publishing guidelines set forth by PRISMA (Preferred Reporting Items for Systematic Reviews and Meta- Analyses) were followed during the literature search and review. The search was conducted using three main literature databases: Medline (Ovid), PubMed (NLM), and Embase (Ovid). In addition, related citations were searched in Scopus (Elsevier, Inc.) to check whether any unique studies were missed from the regular database searches.
Inclusion and exclusion criteria
The articles meeting the following eligibility criteria were included in our systematic review:
described genes causing or potentially associated with CL/P in humans;
referred to studies of either syndromic or non-syndromic CL/P;
were published as original articles (not as review articles, editorials, dissertations, conference proceedings or comments);
included cross-sectional, case-control, cohort studies, or clinical trials;
were published in English.
After screening for articles using the criteria above, we manually excluded those articles in which:
gene mutations were not described in the original articles;
CL/P was not specifically described;
CL/P resulted from environmental factors;
treatments for CL/P were described;
were case reports;
the articles failed to fit in any of the above criteria but did not have CL/P candidate genes or related information.
Search methods to identify studies
Concepts included in the search were: CL/P, genetics (gene mutations), and humans. A combination of Medical Subject Headings (MeSH) terms and titles, abstracts, and keywords was used to develop the initial Medline search string, and then adapted to search the other databases. The last search date was May 28, 2018.
Study selection and data collection
All the citations found in the search process were stored in RefWorks (ProQuest) and any duplicates were excluded. Search strategies and results were tracked using the Primary Excel Workbook designed for systematic reviews (http://libguides.sph.uth.tmc.edu/excel_SR_workbook). To check the reliability of study selection between the screeners, Cohen’s Kappa test was performed using a randomly selected sample of 66 citations screened for CL/P-candidate genes by titles and abstracts. After achieving a > 90% score for the Cohen’s Kappa, all the titles and abstracts of the articles found through the database search were independently examined by two screeners. The full text of the articles not excluded in the above process was manually reviewed, and all results from the screening were recorded in the Primary Excel Workbook. The data collected were displayed as a descriptive narrative. A codebook for data extraction from eligible articles was developed, as described previously [16]. The data elements extracted for the codebook included citation information, study level information (characteristics and results), and quality level information. The quality assessment of each study identified was performed using the Newcastle- Ottawa Scale (NOS), considering the selection criteria of pre- and post- fortification samples, comparability of these groups, and the ascertainment of either the exposure or outcome. NOS assigns a maximum score of 9 points where studies showing < 5 points have high risk of bias and limitations, with these being excluded from a meta-analysis [17].
Bioinformatics analysis of CL/P genes
Functional enrichment analyses of the candidate genes were conducted using the pathway databases Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) through tools of the database for annotation, visualization, and integrated discovery (DAVID, version 6.8, http://david.ncifcrf.gov) [18]. The enrichment of CL/P genes in a pathway or GO term was tested by the hypergeometric test implemented in DAVID. The p-values were adjusted by false discovery rate (FDR, q-value). The significant pathways were obtained by q < 0.05 and at least four genes from the list of input genes (CL/P genes) in each pathway included. Hierarchical level 4 was used as the cutoff in order to avoid too general GO terms. The miRNA- mediated post-transcriptional regulation of the CL/P candidate genes was analyzed by using the following method: first, the miRNA-gene pairs were identified from three computationally predicted miRNA-target gene interaction databases, miRanda, PITA, and TargetScan [19–22]; then, the miRNA-gene pairs that were experimentally proofed were selected through miRTarbase analysis [13]; finally, for each miRNA, the enrichment of CL/P candidate genes was examined using Fisher’s Exact Test. To investigate which human phenotype terms are strongly enriched for the candidate genes, the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) (http://webgestalt.org) was used to perform the overrepresentation enrichment analysis (OEA) using the Human Phenotype Ontology (HPO) annotation database. The top 30 results from the OEA were retrieved on March 15, 2019. The minimum number of genes per category was set to 5 (default). The Benjamini–Hochberg procedure was used for multiple test correction [23]. FDR values were used for all statistical analyses.
Genotype-phenotype association analysis
Genotype-phenotype association analysis was conducted using GWAS available from dbGaP (dbGaP accession phs000774.v1.p1, https://www.ncbi.nlm.nih.gov/gap). This dbGaP repository includes 11,925 individuals from 4058 families, mostly from trios. Out of 11,925 individuals, 5327 were reported as Caucasians, 2221 as Asians, 473 as Africans, and 3904 were reported as having more than one ancestry. A total of 5008 individuals were reported as of Hispanic ethnicity. Not including the parents, 52% of children were males and 48% were females. Array-based genotypes from Illumina Infinium HumanCore Beadchips, including GWAS markers from HumanCore v1 with additional exome and custom contents from the Center for Craniofacial and Dental Genetics (CCDG) consortium, were analyzed with the PLINK software (version 1.90b) to test the association of each directly genotyped variant in the genes listed through the systematic review. Transmission disequilibrium test (TDT) was applied for maximizing analytical power for trios and for minimizing artifacts from population stratification.
Cell culture
Human lip fibroblasts were obtained from JCRB Cell Bank (#JCRB9103KD) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin, and L-glutamine. Human lip fibroblasts were plated onto 96-well cell culture plates at a density of 5000/well and treated with a mimic for either negative control, miR-124-3p, miR- 369-3p, miR- 374a-5p, miR-374b-5p, miR-497-5p, and miR-655-3p (mirVana miRNA mimic, ThermoFisher Scientific) using TransIT-X2 system (Mirus Bio LLC, Madison, WI), following the manufacturer’s protocol (3 pmol mimic and 0.3 μl transfection reagent in 100 μl DMEM per well). Cell proliferation was determined using a cell counting kit 8 (Dojindo Molecular Technologies, Gaithersburg, MD) (n = 6 per group).
Quantitative RT-PCR
Total RNAs were isolated from cultured human lip fibroblasts treated with a mimic for target miRNAs for 1 day (n = 6 per group), using the QIAshredder and RNeasy mini extraction kit (QIAGEN), as previously described [24]. The PCR primers used for quantitative RT-PCR are listed in Additional file 1: Table S1.
Statistical analysis
Two-tailed student’s t tests were applied for the statistical analysis. A p value < 0.05 was considered statistically significant. For all graphs, data are represented as mean ± standard deviation (SD).
Results
Literature search
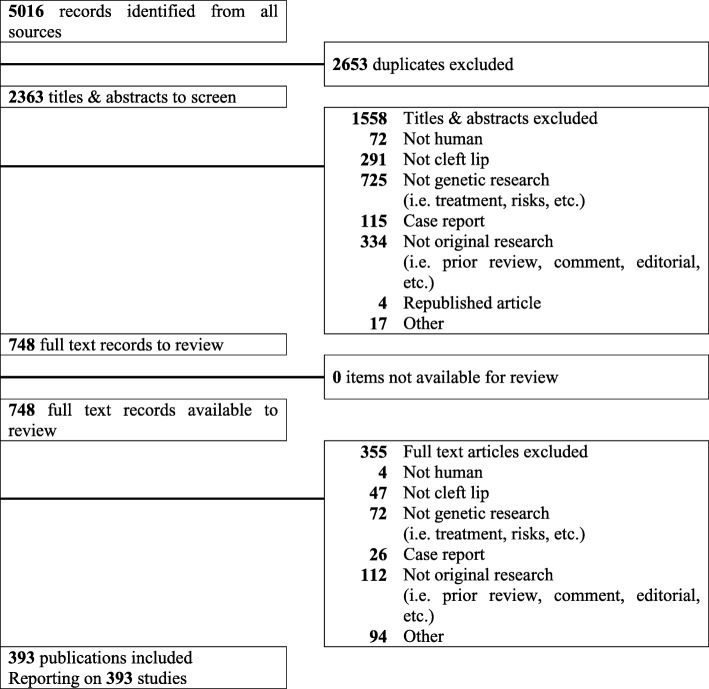
Our systematic search identified a total of 5016 publications. After eliminating 2653 duplicates from the list, the remaining 2363 articles were further screened, using the titles and abstracts, independently by the two screeners, which resulted in 1558 publications being further excluded based on reasons such as referring to non-genetic studies and case reports. The remaining 748 articles were further assessed for eligibility through manual full-text review. Through this process, 393 articles satisfying all inclusion criteria were selected while 355 articles were excluded. These selected 393 studies were used for collection of CL/P-candidate genes and in the follow-up analyses (Fig. 1 and Additional file 1: Table S2).
Fig. 1.
PRISMA flowchart for study selection. A graphical representation of the flow of citations reviewed in the course of the systematic review was provided using a PRISMA flow diagram
Summary of human CL/P genes
We identified 172 CL/P-candidate genes from the qualified studies above (Table 1 and Additional file 1: Tables S3–S8). For the bioinformatics analyses, we excluded phenotypic markers and genes with unknown genomic location. Among the CL/P-candidate genes, 10 genes were studied at least five times in different populations: IRF6 (52 studies, encoding interferon regulatory factor 6, located at genomic locus 1q32.2), MTHFR (26 studies, encoding methylenetetrahydrofolate reductase, at 1p36.2), TGFA (18 studies, encoding transforming growth factor alpha, at 2q13.3), MSX1 (25 studies, encoding msh homeobox 1, at 4p16.2), TGFB3 (16 studies, encoding transforming growth factor beta 3, at 14q24.3), NECTIN1 (10 studies, encoding Nectin cell adhesion molecule 1, at 11q23.3), BMP4 (10 studies, encoding bone morphogenetic protein 4, at 14q22.2), FOXE1 (6 studies, encoding forkhead box E1, at 9q22.33), BCL3 (6 studies, encoding B-cell CLL/lymphoma 3, at 19q13.32), and CRISPLD2 (5 studies, encoding cysteine rich secretory protein LCCL domain containing 2, at 16q24.1). Most of the gene mutations (168/177 = 94.9%) were reported only in non-syndromic CL/P while mutations in nine genes (ADH1C, FGFR1, IRF6, MID1, NECTIN1, PHF8, SOX9, TGFA, and TP63) were also reported in syndromic CL/P (9/177 = 5.1%). Several genes were reported only in syndromic cases; CHD7 (CHARGE syndrome), CR1 (van der Woude syndrome), EFNB1 (craniofrontonasal syndrome), KISS1R (Kallmann syndrome), MID1 (Opitz G/BBB syndrome), REN (van der Woude syndrome), and SOX9 (Pierre-Robin syndrome). Mutations in eight genes (BCL3, BMP4, IRF6, MSX1, MTHFR, NECTIN1, TGFA, and TGFB3) were reported across different ethnic groups such as Caucasians, African Americans, Hispanics, and Asians, including but not limited to Chinese, Japanese, Indian, Turkish, Polish, Finnish, Brazilian, American, and European. By contrast, a total of 75 genes were reported as insignificant in some populations. For example, while mutations in TGFA were significant in the Iranian, Korean, Caucasian and Chilean population, they were not significant in the South American, Italian, Malaysian or Indian populations. Mutations in IRF6 were not significant in some studies, but they were significant in larger studies in various populations (Additional file 1: Tables S3–S8). Recent advances in the discovery of genetic variants at whole genome level and the design of genome-wide approaches enable investigators to identify the involvement of multiple genes and loci in a single study (e.g., GWAS). Mutations in multiple genes and loci were reported in 87 and 12 studies, respectively. A significant association of IRF6, MSX1 and NECTIN1 with CL/P has been reported in several GWAS in different populations (Additional file 1: Table S4). However, no significant association was reported in several studies conducted in Danish, Mesoamerican, and African populations for IRF6, American and Latvian populations for MSX1, and Taiwanese population for NECTIN1 (Additional file 1: Table S6).
Table 1.
Summary of genes associated with cleft lip with/without cleft palate in humans
| Gene symbol | Gene name | Loci | Syndromic/ Nonsyndromic |
|---|---|---|---|
| ABCA4 | ATP binding cassette subfamily A member 4 | 1p22.1 | Nonsyndromic |
| ABCB1 | ATP binding cassette subfamily B member 1 | 7q21.12 | Nonsyndromic |
| ADAM3A | ADAM metallopeptidase domain 3A | 8p11.22 | Nonsyndromic |
| ACSS2 | Acyl-coenzyme A synthetase short-chain family member 2 | 20q11.22 | Nonsyndromic |
| ADAM5 | ADAM metallopeptidase domain 5 | 8p11.22 | Nonsyndromic |
| ADAMTS20 | ADAM metallopeptidase with thrombospondin type 1 motif 20 | 12q12 | Nonsyndromic |
| ADGRL2 | Adhesion G protein-coupled receptor L2 | 1p31.1 | Nonsyndromic |
| ADH1C | Alcohol dehydrogenase 1C (class I), gamma polypeptide | 4q23 | Nonsyndromic & Syndromic: Fetal alcohol syndrome |
| AQP7 | Aquaporin 7 | 9p13.3 | Nonsyndromic |
| ARHGAP29 | Rho GTPase activating protein 29 | 1p22.1 | Nonsyndromic |
| ASS1 | Argininosuccinate synthase 1 | 9q34.11 | Nonsyndromic |
| AXIN2 | Axin 2 | 17q24.1 | Nonsyndromic |
| BAG4 | BCL2 associated athanogene 4 | 8p11.23 | Nonsyndromic |
| BCL3 | B-cell CLL/lymphoma 3 | 19q13.32 | Nonsyndromic |
| BHMT2 | Betaine-homocysteine S-methyltransferase 2 | 5q14.1 | Nonsyndromic |
| BLM | Bloom syndrome RecQ like helicase | 15q26.1 | Nonsyndromic |
| BMP4 | Bone morphogenetic protein 4 | 14q22.2 | Nonsyndromic |
| BMPR1B | Bone morphogenetic protein receptor type 1B | 4q22.3 | Nonsyndromic |
| BRIP1 | BRCA1 interacting protein C-terminal helicase 1 | 17q23.2 | Nonsyndromic |
| CBS | Cystathionine-beta-synthase | 21q22.3 | Nonsyndromic |
| CDH1 | Cadherin 1 | 16q22.1 | Nonsyndromic |
| CDH2 | Cadherin 2 | 18q12.1 | Nonsyndromic |
| CENPJ | Centromere protein J | 13q12.12-q12.13 | Nonsyndromic |
| CHD7 | Chromodomain helicase DNA binding protein 7 | 8q12.2 | Syndromic: CHARGE syndrome |
| CLPTM1 | CLPTM1, transmembrane protein | 19q13.32 | Nonsyndromic |
| COL4A2 | Collagen type IV alpha 2 chain | 13q34 | Nonsyndromic |
| COL4A3 | Collagen type IV alpha 3 chain | 2q36.3 | Nonsyndromic |
| COL4A4 | Collagen type IV alpha 4 chain | 2q36.3 | Nonsyndromic |
| COL21A1 | Collagen type XXI alpha 1 chain | 6p12.1; 6p12.3-p11.2 | Nonsyndromic |
| CR1 | Complement C3b/C4b receptor 1 (Knops blood group) | 1q32.2 | Syndromic: Van der Woude syndrome |
| CRISPLD2 | Cysteine rich secretory protein LCCL domain containing 2 | 16q24.1 | Nonsyndromic |
| CYP1A1 | Cytochrome P450 family 1 subfamily A member 1 | 15q24.1 | Nonsyndromic |
| CYP2E1 | Cytochrome P450 family 2 subfamily E member 1 | 10q26.3 | Nonsyndromic |
| DCAF7 | DDB1 and CUL4 associated factor 7 | 17q23.3 | Nonsyndromic |
| DHFR | Dihydrofolate reductase | 5q14.1 | Nonsyndromic |
| DICER1 | Dicer 1, ribonuclease III | 14q32.13 | Nonsyndromic |
| DLX1 | Distal-less homeobox 1 | 2q31.1 | Nonsyndromic |
| DMD | Dystrophin | Xp21.2-p21.1 | Nonsyndromic |
| DVL2 | Dishevelled segment polarity protein 2 | 17p13.1 | Nonsyndromic |
| E2F1 | E2F transcription factor 1 | 20q11.22 | Nonsyndromic |
| EFNB1 | Ephrin B1 | Xq13.1 | Syndromic: Craniofrontonasal syndrome |
| EIF2B3 | Eukaryotic translation initiation factor 2B subunit gamma | 1p34.1 | Nonsyndromic |
| EGF61 | Epidermal growth factor 61 | 4q25 | Nonsyndromic |
| ESRRG | Estrogen related receptor gamma | 1q41 | Nonsyndromic |
| EVC | EvC ciliary complex subunit 1 | 4p16.2 | Nonsyndromic |
| EVC2 | EvC ciliary complex subunit 2 | 4p16.2 | Nonsyndromic |
| EYA1 | EYA transcriptional coactivator and phosphatase 1 | 8q13.3 | Nonsyndromic |
| F13A1 | Coagulation factor XIII A chain | 6p25.1 | Nonsyndromic |
| FAM49A | Family with sequence similarity 49 member A | 2p24.2 | Nonsyndromic |
| FGF1 | Fibroblast growth factor 1 | 5q31.3 | Nonsyndromic |
| FGF2 | Fibroblast growth factor 2 | 4q28.1 | Nonsyndromic |
| FGF3 | Fibroblast growth factor 3 | 11q13.3 | Nonsyndromic |
| FGF10 | Fibroblast growth factor 10 | 5p12 | Nonsyndromic |
| FGF18 | Fibroblast growth factor 18 | 5q35.1 | Nonsyndromic |
| FGFR1 | Fibroblast growth factor receptor 1 | 8p11.23 | Nonsyndromic & Syndromic: Kallmann syndrome |
| FGFR2 | Fibroblast growth factor receptor 2 | 10q26.13 | Nonsyndromic |
| FOXE1 | Forkhead box E1 | 9q22.33 | Nonsyndromic |
| FOXF2 | Forkhead box F2 | 6p25.3 | Nonsyndromic |
| FOXG1 | Forkhead box G1 | 14q12 | Nonsyndromic |
| FOXP2 | Forkhead box protein P2 | 7q31.1 | Nonsyndromic |
| FZD6 | Frizzled class receptor 6 | 8q22.3 | Nonsyndromic |
| GABRB3 | Gamma-aminobutyric acid type A receptor beta 3 subunit | 15q12 | Nonsyndromic |
| GAD1 | Glutamate decarboxylase 1 | 2q31.1 | Nonsyndromic |
| GCH1 | GTP cyclohydrolase 1 | 14q22.2 | Nonsyndromic |
| GLI2 | GLI family zinc finger 2 | 2q14.2 | Nonsyndromic |
| GREM1 | Gremlin 1, DNA family BMP antagonist | 15q13.3 | Nonsyndromic |
| GSTM1 | Glutathione S-transferase mu 1 | 1p13.3 | Nonsyndromic |
| GSTP1 | Glutathione S-transferase pi 1 | 11q13.2 | Nonsyndromic |
| GSTT1 | Glutathione S-transferase theta 1 | 22q11.23 | Nonsyndromic |
| HECTD1 | HECT domain E3 Ubiquitin protein ligase 1 | 14q12 | Nonsyndromic |
| HKDC1 | Hexokinase domain containing 1 | 10q22.1 | Nonsyndromic |
| IRF6 | Interferon regulatory factor 6 | 1q32.2 | Nonsyndromic |
| JAG2 | Jagged2 | 14q32.33 | Nonsyndromic |
| JARID2 | Jummonji and AT-rich interaction domain containing 2 | 6p22.3 | Nonsyndromic |
| KIF2A | Kinesin family member 2A | 5q12.1 | Nonsyndromic |
| KIF7 | Kinesin family member 7 | 15q26.1 | Nonsyndromic |
| KISS1R | KISS1 receptor | 19p13.3 | Syndromic: Kallmann syndrome |
| KRT18 | Keratin 18 | 12q13.13 | Nonsyndromic |
| LACTB | Lactamase beta | 15q22.2 | Nonsyndromic |
| LHX8 | LIM homeobox 8 | 1p31.1 | Nonsyndromic |
| LPHN2 | Adhesion G protein-coupled receptor L2 | 1p31.1 | Nonsyndromic |
| MAFB | MAF bZIP transcription factor B | 20q12 | Nonsyndromic |
| MGAM | Maltase-glucoamylase | 7q34 | Nonsyndromic |
| MID1 | Midline 1 | Xp22.2 | Nonsyndromic & Syndromic: Opitz G/BBB syndrome |
| MMP3 | Matrix metallopeptidase 3 | 11q22.2 | Nonsyndromic |
| MMP9 | Matrix metallopeptidase 9 | 20q13.12 | Nonsyndromic |
| MROH7 | Maestro heat like repeat family member 7 | 1p32.3 | Nonsyndromic |
| MRPL53 | Mitochondrial ribosomal protein L53 | 2p13.1 | Nonsyndromic |
| MSX1 | msh homeobox 1 | 4p16.2 | Nonsyndromic |
| MSX2 | msh homeobox 2 | 5q35.2 | Nonsyndromic |
| MTR | 5-methyltetrahydrofolate-homocysteine methyltransferase | 1q43 | Nonsyndromic |
| MTRR | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 5p15.31 | Nonsyndromic |
| MYH9 | Myosin heavy chain 9 | 22q12.3 | Nonsyndromic |
| MTHFD1 | Methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 | 14q23.3 | Nonsyndromic |
| MTHFR | Methylenetetrahydrofolate reductase | 1p36.22 | Nonsyndromic |
| NAT1 | N-acetyltransferase 1 | 8p22 | Nonsyndromic |
| NAT2 | N-acetyltransferase 2 | 8p22 | Nonsyndromic |
| NECTIN1 | Nectin cell adhesion molecule 1 | 11q23.3 | Nonsyndromic |
| NECTIN2 | Nectin cell adhesion molecule 2 | 19q13.32 | Nonsyndromic |
| NECTIN3 | Nectin cell adhesion molecule 3 | 3q13.13 | Nonsyndromic |
| NOG | Noggin | 17q22 | Nonsyndromic |
| NTN1 | Netrin 1 | 17p13.1 | Nonsyndromic |
| OFC1 | Orofacial cleft 1 | 6p24 | Nonsyndromic |
| OFC2 | Orofacial cleft 2 | 2p14-p13 | Nonsyndromic |
| PAH | Phenylalanine hydroxylase | 12q23.2 | Nonsyndromic |
| PARVA | Parvin alpha | 11p15.3 | Nonsyndromic |
| PTCH1 | Patched 1 | 9q22.32 | Nonsyndromic |
| PAX3 | Paired box 3 | 2q36.1 | Nonsyndromic |
| PAX7 | Paired box 7 | 1p36.13 | Nonsyndromic |
| PAX9 | Paired box 9 | 14q13.3 | Nonsyndromic |
| PCYT1A | Phosphate cytidylyltransferase 1, choline, alpha | 3q29 | Nonsyndromic |
| PDGFC | Platelet derived growth factor C | 4q32.1 | Nonsyndromic |
| PEMT | Phosphatidylethanolamine N-methyltransferase | 17p11.2 | Nonsyndromic |
| PKP1 | Plakophilin 1 | 1q32.1 | Nonsyndromic |
| PHF8 | PHD finger protein 8 | Xp11.22 | Nonsyndromic |
| PHYH | Phytanoyl-CoA 2-hydroxylase | 10p13 | Nonsyndromic |
| RPS26 | Ribosomal Protein S26 | 12q13.2 | Nonsyndromic |
| PRSS1 | Protease, serine 1 | 7q34 | Nonsyndromic |
| PRSS35 | Protease, serine 35 | 6q14.2 | Nonsyndromic |
| RAD51 | RAD51 recombinase | 15q15.1 | Nonsyndromic |
| RAD54B | RAD54 homolog B | 8q22.1 | Nonsyndromic |
| RARA | Retinoic Acid Receptor Alpha | 17q21.2 | Nonsyndromic |
| RECQL5 | RecQ like helicase 5 | 17q25.1 | Nonsyndromic |
| REG3A | Regenerating family member 3 alpha | 2p12 | Nonsyndromic |
| REN | Renin | 1q32.1 | Syndromic: Van der Woude syndrome |
| RFC1 | Replication factor C subunit 1 | 4p14 | Nonsyndromic |
| RHPN2 | Rhophilin Rho GTPase binding protein 2 | 19q13.11 | Nonsyndromic |
| RUNX2 | Runt related transcription factor 2 | 6p21.1 | Nonsyndromic |
| RYK | Receptor-like tyrosine kinase | 3q22.2 | Nonsyndromic |
| SATB2 | SATB homeobox 2 | 2q33.1 | Nonsyndromic |
| SEC16A | SEC 16 homolog A, endoplasmic reticulum export factor | 9q34.3 | Nonsyndromic |
| SERPINA6 | Serpin family A member 6 | 14q32.13 | Nonsyndromic |
| SLC6A4 | Solute carrier family 6 member 4 | 17q11.2 | Nonsyndromic |
| SMAD1 | SMAD family member 1 | 4q31.21 | Nonsyndromic |
| SMAD2 | SMAD family member 2 | 18q21.1 | Nonsyndromic |
| SOX9 | SRY-box 9 | 17q24.3 | Syndromic: Pierre-Robin Syndrome |
| SPRY1 | Sprouty RTK signaling antagonist 1 | 4q28.1 | Nonsyndromic |
| SUMO1 | Small ubiquitin-like modifier 1 | 2q33.1 | Nonsyndromic |
| STK32B | Serine/threonine kinase 32B | 4p16.2 | Nonsyndromic |
| SYNE3 | Spectrin repeat containing nuclear envelop family member 3 | 14q32.13 | Nonsyndromic |
| TAF1B | TATA-box binding protein associated factor, RNA polymerase I subunit B | 2p25.1 | Nonsyndromic |
| TANC2 | Tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | 17q23.2-q23.3 | Nonsyndromic |
| TBX22 | T-box 22 | Xq21.1 | Nonsyndromic |
| TCN2 | Transcobalamin 2 | 22q12.2 | Nonsyndromic |
| TEX11 | Testis expressed 11 | Xq13.1 | Nonsyndromic |
| TFAP2A | Transcription factor AP-2 alpha | 6p24.3 | Nonsyndromic |
| TGFA | Transforming growth factor alpha | 2p13.3 | Nonsyndromic |
| TGFB1 | Transforming growth factor beta 1 | 2p13.3 | Nonsyndromic |
| TGFB2 | Transforming growth factor beta 2 | 1q41 | Nonsyndromic |
| TGFB3 | Transforming growth factor beta 3 | 14q24.3 | Nonsyndromic |
| TIMP2 | TIMP metallopeptidase inhibitor 2 | 17q25.3 | Nonsyndromic |
| TMEM19 | transmembrane protein 19 | 12q21.1 | Nonsyndromic |
| TNS1 | Tensin 1 | 2q35 | Nonsyndromic |
| TOX3 | TOX high mobility group box family member 3 | 16q12.1 | Nonsyndromic |
| TP63 | Tumor protein p63 | 3q28 | Syndromic: Hay wells or AEC syndrome |
| TPH2 | Tryptophan hydroxylase 2 | 12q21.1 | Nonsyndromic |
| TPM1 | Tropomyosin 1 (alpha) | 15q22.2 | Nonsyndromic |
| TULP4 | Tubby like protein 4 | 6q25.3 | Nonsyndromic |
| TYMS | Thymidylate synthase | 18p11.32 | Nonsyndromic |
| VAX1 | Ventral anterior homeobox 1 | 10q25.3 | Nonsyndromic |
| VAX2 | Ventral anterior homeobox 2 | 2p13.3 | Nonsyndromic |
| VWA8 | Von Willebrand factor A domain-containing protein 8 | 13q14.11 | Nonsyndromic |
| WNT3 | Wnt family member 3 | 17q21.31-q21.32 | Nonsyndromic |
| WNT3A | Wnt family member 3A | 1q42.13 | Nonsyndromic |
| WNT5A | Wnt family member 5A | 3p14.3 | Nonsyndromic |
| WNT5B | Wnt family member 5B | 12p13.33 | Nonsyndromic |
| WNT6 | Wnt family member 6 | 2q35 | Nonsyndromic |
| WNT9B | Wnt family member 9B | 17q21.32 | Nonsyndromic |
| WNT10A | Wnt family member 10A | 2q35 | Nonsyndromic |
| WNT11 | Wnt family member 11 | 11q13.5 | Nonsyndromic |
| YOD1 | TOD1 deubiquitinase | 1q32.1 | Nonsyndromic |
| ZNF385B | Zinc finger protein 385B | 2q31.2-q31.3 | Nonsyndromic |
Due to space limits, the details (e.g. publication ID) of each gene were not provided in this table
GO term enrichment analysis
We analyzed CL/P genes enriched in the GO terms to assess the functional features of CL/P genes (Additional file 1: Tables S9–S12). The most specific enriched terms among GO biological processes showed strong association with development and morphogenesis of other organs (Additional file 1: Tables S9 and S10). These results suggest that CL/P genes may potentially cause additional developmental disorders while 70% of CL/P cases are non-syndromic without any additional birth defects. In non-syndromic CL/P cases, molecules that are specifically expressed in the lip region during lip formation may help explain the molecular mechanism in regard to how only lip formation is affected. Lip formation involves the growth and fusion of maxillary and nasal processes during embryogenesis [25]. We identified a strong association with positive and negative regulators of several cellular processes, including cell proliferation, apoptosis, differentiation, and epithelial-to-mesenchymal transition. We also found that genes involved in folic acid metabolic process were significantly enriched (7 CL/P genes) (Additional file 1: Table S10). The current approach for the prevention of neural tube defects and CL/P is folic acid supplementation [26]. Our results suggest that some individuals with CL/P have defects in folic acid metabolism, and that a folic acid supplement can potentially prevent CL/P.
Among the enriched GO terms in its domain Molecular Function (MF) (Additional file 1: Tables S9 and S11), we observed enrichment of several terms related to molecular binding: transcription factor activity, sequence-specific binding (28 CL/P genes), sequence-specific DNA binding (23 CL/P genes), chromatin binding (16 CL/P genes), and binding to Frizzled, a family of G protein-coupled receptors for WNT ligands (11 CL/P genes). The remaining enriched terms in the MF domain included: protein homodimerization activity (26 CL/P genes); growth factor activity (13 CL/P genes), which is induced by FGF, BMP, TGFβ and PDGF; protein tyrosine kinase activity (10 CL/P genes), 1- phosphatidylinositol-3 kinase activity (8 CL/P genes), and phosphatidylinositol-4,5-bisphosphate 3-kinase activity (8 CL/P genes) induced by FGF signaling; and others. Thus, the GO MF term analysis highlighted the contribution of morphologic factors involved in WNT, FGF, BMP, TGFβ, and PDGF signaling pathways.
Among GO terms representing cellular components (Additional file 1: Tables S9 and S12), most terms were enriched in extracellular components: extracellular region (48 CL/P genes), extracellular space (36 CL/P genes), cell surface (27 CL/P genes), and proteinaceous extracellular matrix (ECM) (22 CL/P genes). In GO terms, many growth factors including WNT, FGF, BMP, TGFβ, and PDGF and their extracellular regulators (e.g. ECM) were identified in these groups. These findings are in agreement with the fact that growth factors are activated in the extracellular space.
Human phenotype ontology enrichment analysis
We used the HPO database to investigate which human phenotype terms were enriched for the identified set of CL/P candidate genes. The most enriched phenotypic term was abnormal number of teeth (28 genes), followed by CL (23 genes), cleft upper lip (22 genes), reduced number of teeth (25 genes), and oral cleft (36 genes) (Additional file 1: Table S13). Among the top 30 results, most phenotypes were directly related to CL/P, congenital dental disorders, or congenital abnormalities of the fingers.
KEGG pathway analysis
We hypothesized that CL/P genes share common features among wide arrays of biological functions and pathways. We therefore examined which biological pathways were enriched with CL/P genes by using the DAVID bioinformatics tool and the canonical pathways from the KEGG database (Additional file 1: Table S14). The regulatory pathway annotation was performed based on the score and visualization of the pathways used in the KEGG database. Among the cellular functions in KEGG pathways, 19 pathways were statistically significant in the enrichment of CL/P genes (FDR < 5%). Seven of these pathways were related to cancer: pathways in cancer (43 CL/P genes), basal cell carcinoma (16 CL/P genes), proteoglycans in cancer (16 CL/P genes), melanoma (10 CL/P genes), pancreatic cancer (8 CL/P genes), bladder cancer (6 CL/P genes), and colorectal cancer (7 CL/P genes). Previous population-based studies suggest that individuals with CL/P have a higher risk of cancer in the breast, brain, and lung later in life [27–29]. Among them, molecules related to Hippo, FGF, WNT, and TGFβ signaling are highlighted in these cancer-related pathways. These cellular signaling pathways were significantly involved in CL/P: Hippo signaling pathway (22 CL/P genes), signaling pathways regulating pluripotency of stem cells (21 CL/P genes), phosphoinositide 3-kinase (PI3K)- Akt signaling pathway (15 CL/P genes), mitogen-activated protein kinase (MAPK) signaling pathway (13 CL/P genes), WNT signaling pathway (11 CL/P genes), and TGF-β signaling pathway (9 CL/P genes).
Genotype-phenotype association analysis
To determine the contribution of genetic variations to CL/P, a genotype-phenotype association analysis was conducted using GWAS for the genetics of orofacial clefts and related phenotypes (e.g. CL/P) available from dbGaP (dbGaP accession phs000774.v1.p1). By PLINK analysis, we identified 2975 cases vs 8751 controls out of 11,925 individuals. We investigated whether single nucleotide polymorphisms (SNPs) mapped to the CL/P-candidate genes are associated with human CL/P phenotypes using TDT of The Genetics of Orofacial Clefts and Related Phenotypes GWAS dataset. This method is appropriate for statistical imbalances between transmitted and non-transmitted alleles in parent-child trios. We investigated all directly genotyped (i.e. not imputed) variants within the genes found in the systematic review. A total of 5437 variants from 179 genes were included in the association analysis, while some genes did not have the genotyped SNPs in this dataset and some SNPs did not present any variations in the analyzed samples. Because most SNPs in the same gene have strong linkage disequilibrium, we set our candidate-wise significance threshold at 2.79 × 10− 4 by using the Bonferroni level with the number of genes tested (0.05/179). We identified 56 SNPs from 12 genes with p-values smaller than 2.79 × 10− 4 (Table 2). The top association signals were from IRF6 and NTN1 (Netrin 1), which also showed nominal genome-wide significance in the GWAS dataset (p < 5 × 10− 8). We also observed gene-wide significant signals (p < 2.5 × 10− 6) from ABCA4 (ATP-binding cassette, sub-family A, member 4) and PAX7 (paired box 7). Although ADAM3A (ADAM metallopeptidase domain 3A), FOXE1 (forkhead box E1), MSX2 (msh homeobox 2), MTHFR (methylenetetrahydrofolate reductase), TP63 (tumor protein p63), TPM1 (tropomyosin 1), VAX1 (ventral anterior homeobox 1), and WNT9B (wingless-type MMTV integration site family, member 9B) did not reach the genome-wide nor gene-wide significance thresholds, these genes reached candidate-wise significance level (p < 2.79 × 10− 4).
Table 2.
Genotype-phenotype association
| Top marker | Type | Gene | p-value |
|---|---|---|---|
| rs1044516 | UTR3 | IRF6 | 2.66 × 10−9 |
| rs1880646 | Intron | NTN1 | 3.02 × 10−9 |
| rs4147827 | Intron | ABCA4 | 8.51 × 10−7 |
| rs742071 | Intron | PAX7 | 2.09 × 10−6 |
| rs6474148 | Intron | ADAM3A | 3.26 × 10−6 |
| rs1443435 | UTR3 | FOXE1 | 1.39 × 10−5 |
| rs6585429 | Intron | VAX1 | 2.28 × 10−5 |
| rs2066462 | Silent | MTHFR | 7.38 × 10−5 |
| rs3803499 | Intron | TPM1 | 1.91 × 10−4 |
| rs1515497 | Intron | TP63 | 2.19 × 10−4 |
| rs14459 | UTR3 | MSX2 | 2.34 × 10−4 |
| rs4968282 | Intron | WNT9B | 2.65 × 10−4 |
Environmental and epigenetic factors
In addition to gene mutations, genetic background such as ethnicity, population and gender have substantial influence on the birth prevalence of CL/P. Environmental factors such as maternal age, smoking, alcohol consumption, obesity, and micronutrient deficiencies are known to be strong susceptibility risk factors. Recent studies suggest that environmental factors can interact with the epigenetic system and control gene expression at the post- transcriptional level [30–32]. To explore the degree to which miRNAs regulate the expression of CL/P genes, we performed a bioinformatics analysis to identify miRNAs whose target genes are statistically enriched with CL/P genes (Table 3). By applying the adjusted p-value (FDR) < 0.1, a total of 16 miRNAs were significantly enriched with the targeted CL/P genes. These 16 miRNAs were categorized into 10 known and one unknown miRNA families: the miR-27 family (hsa-miR- 27b- 3p, 23 CL/P genes), miR-124 family (hsa- miR- 124-3p, 29 CL/P genes), miR-154 family (hsa-miR-369-3p, 13 CL/P genes; hsa- miR-655-3p, 14 CL/P genes; hsa-miR-300, 18 CL/P genes; hsa-miR-381-3p, 18 CL/P genes), miR-203 family (hsa-miR-203a-3p, 9 CL/P genes), miR-368 family (hsa-miR-376b-3p, 10 CL/P genes), miR-374 family (hsa-miR- 374a-5p, 22 CL/P genes; hsa-miR-374b-5p, 22 CL/P genes), miR-497 family (hsa-miR-497-5p, 25 CL/P genes), miR-503 family (hsa-miR-503-5p, 10 CL/P genes), miR-550 family (hsa-miR-550a-5p, 5 CL/P genes; hsa-miR-550a-3-5p, 5 CL/P genes), miR-1271 family (hsa-miR-1271-3p, 5 CL/P genes), and an unknown family (hsa-miR-3678-3p, 5 CL/P genes). Notably, except for hsa-miR-27b-3p [33], these miRNAs have not yet been reported in CL/P. In the enrichment analysis, CL/P genes regulated by multiple miRNAs were: EN2 (targeted by 6 miRNA families), FZD6 (targeted by 5 miRNA families), HECTD1 (targeted by 5 miRNA families), and YOD1 (targeted by 5 miRNA families) (Table 4). Because gene expression of EN2, FZD6, HECTD1, and YOD1 is regulated by several miRNAs, expression of these genes may be more susceptible to environmental factors during lip formation.
Table 3.
MicroRNA (miRNA) enrichment analysis of 161 human CL/P genes (FDR < 0.1)
| miRNA family ID | miRNA name | Gene symbol | FDR (BH) |
|---|---|---|---|
| miR-154 | hsa-miR-369-3p | CHD7, DMD, EYA1, GABRB3, GREM1, HOXB3, NOG, PAX6, TIMP2, TULP4, BAG4, FZD6, YOD1 | 0.03 |
| miR-154 | hsa-miR-655-3p | BCL2, DMD, EN2, GREM1, HOXB3, MAFB, MID1, NTN1, PAX6, SATB2, TULP4, CYP1A1, FZD6, YOD1 | 0.03 |
| miR-374 | hsa-miR-374a-5p | CHD7, DMD, EYA1, FGFR2, FOXG1, HECTD1, HOXB3, JARID2, MSX1, NOG, NTN1, PAX6, RAD51, RHPN2, RUNX2, TGFA, TNS1, WNT5A, WNT5B, YOD1, EN2, FZD6 | 0.07 |
| miR-374 | hsa-miR-374b-5p | CHD7, DMD, EYA1, FGFR2, FOXG1, HECTD1, HOXB3, JARID2, MSX1, NOG, NTN1, PAX6, RAD51, RHPN2, RUNX2, TGFA, TNS1, WNT5A, WNT5B, YOD1, EN2, FZD6 | 0.07 |
| miR-497 | hsa-miR-497-5p | AXIN2, BAG4, BCL2, CHD7, CRISPLD2, EN2, EYA1, FGF1, FGF2, FGFR1, FGFR2, FOXP2, FZD6, HECTD1, JARID2, PAX7, RHPN2, SATB2, SLC6A4, WNT3A, YOD1, MTHFR, RUNX2, TFAP2A, TPM1 | 0.07 |
| miR-124 | hsa-miR-124-3p | GABRB3, GCH1, JAG2, KIF2A, MTR, MYH9, ROR2, RYK, TANC2, ARHGAP29, COL4A4, DVL2, EFNB1, FGF1, FGFR1, FOXF2, GREM1, HOXB3, KIF7, PDGFC, PEMT, PKP1, RAD51, RUNX2, SOX9, TBX22, TP63, TPM1, WNT5B | 0.09 |
| miR-1271 | hsa-miR-1271-3p | CBS, DCAF7, FZD6, RAD51, YOD1 | 0.09 |
| miR-203 | hsa-miR-203a-3p | E2F1, EN2, GREM1, HECTD1, KIF2A, PAX6, RUNX2, SMAD2, TP63 | 0.09 |
| miR-27 | hsa-miR-27b-3p | BCL3, CHD7, COL21A1, DVL2, EN2, EYA1, FGF1, FOXP2, GABRB3, GCH1, GREM1, HOXB3, PAX9, RARA, SATB2, SMAD1, STK32B, SMAD2, DCAF7, PAX3, PAX7, SEC16A, WNT9B | 0.09 |
| miR-154 | hsa-miR-300 | CR1, DVL2, FGF1, FGFR2, FOXF2, FOXP2, GABRB3, GAD1, HECTD1, JAG2, MID1, PDGFC, TANC2, TGFB3, WNT5A, CRISPLD2, GREM1, PHF8 | 0.09 |
| NA | hsa-miR-3678-3p | E2F1, GABRB3, JARID2, SLC6A4, YOD1 | 0.09 |
| miR-368 | hsa-miR-376b-3p | CRISPLD2, EN2, FGFR1, MYH9, PAX3, PAX6, PRSS35, TANC2, FGF2, FOXG1 | 0.09 |
| miR-154 | hsa-miR-381-3p | CR1, DVL2, FGF1, FGFR2, FOXF2, FOXP2, GABRB3, GAD1, HECTD1, JAG2, MID1, PDGFC, TANC2, TGFB3, WNT5A, CRISPLD2, GREM1, PHF8 | 0.09 |
| miR-503 | hsa-miR-503-5p | HECTD1, JARID2, MAFB, WNT3A, BCL2, DHFR, FGF2, FGFR1, SMAD2, TFAP2A | 0.09 |
| miR-550 | hsa-miR-550a-3-5p | CBS, DCAF7, FZD6, RAD51, YOD1 | 0.09 |
| miR-550 | hsa-miR-550a-5p | CBS, DCAF7, FZD6, RAD51, YOD1 | 0.09 |
BH Benjamini–Hochberg adjustment for multiple test correction [23]
Table 4.
Human CL/P genes targeted by at least two microRNA (miRNA) families
| Gene | # miRNA families | miRNA family |
|---|---|---|
| EN2 | 6 | miR-154, miR-203, miR-27, miR-368, miR-374, miR-497 |
| YOD1 | 5 | miR-1271, miR-154, miR-374, miR-497, miR-550 |
| FZD6 | 5 | miR-1271, miR-154, miR-374, miR-497, miR-550 |
| HECTD1 | 5 | miR-154, miR-203, miR-374, miR-497, miR-503 |
| GREM1 | 4 | miR-124, miR-154, miR-203, miR-27 |
| HOXB3 | 4 | miR-124, miR-154, miR-27, miR-374 |
| PAX6 | 4 | miR-154, miR-203, miR-368, miR-374 |
| RAD51 | 4 | miR-124, miR-1271, miR-374, miR-550 |
| CHD7 | 4 | miR-154, miR-27, miR-374, miR-497 |
| EYA1 | 4 | miR-154, miR-27, miR-374, miR-497 |
| RUNX2 | 4 | miR-124, miR-203, miR-374, miR-497 |
| FGF1 | 4 | miR-124, miR-154, miR-27, miR-497 |
| FGFR1 | 4 | miR-124, miR-368, miR-497, miR-503 |
| GABRB3 | 3 | miR-124, miR-154, miR-27 |
| FGFR2 | 3 | miR-154, miR-374, miR-497 |
| JARID2 | 3 | miR-374, miR-497, miR-503 |
| CRISPLD2 | 3 | miR-154, miR-368, miR-497 |
| FOXP2 | 3 | miR-154, miR-27, miR-497 |
| TANC2 | 3 | miR-124, miR-154, miR-368 |
| DVL2 | 3 | miR-124, miR-154, miR-27 |
| DCAF7 | 3 | miR-1271, miR-27, miR-550 |
| BCL2 | 3 | miR-154, miR-497, miR-503 |
| SATB2 | 3 | miR-154, miR-27, miR-497 |
| FGF2 | 3 | miR-368, miR-497, miR-503 |
| SMAD2 | 3 | miR-203, miR-27, miR-503 |
| DMD | 2 | miR-154, miR-374 |
| WNT5A | 2 | miR-154, miR-374 |
| NOG | 2 | miR-154, miR-374 |
| NTN1 | 2 | miR-154, miR-374 |
| FOXG1 | 2 | miR-368, miR-374 |
| RHPN2 | 2 | miR-374, miR-497 |
| WNT5B | 2 | miR-124, miR-374 |
| JAG2 | 2 | miR-124, miR-154 |
| FOXF2 | 2 | miR-124, miR-154 |
| PDGFC | 2 | miR-124, miR-154 |
| CBS | 2 | miR-1271, miR-550 |
| BAG4 | 2 | miR-154, miR-497 |
| MAFB | 2 | miR-154, miR-503 |
| PAX7 | 2 | miR-27, miR-497 |
| WNT3A | 2 | miR-497, miR-503 |
| TFAP2A | 2 | miR-497, miR-503 |
| TPM1 | 2 | miR-124, miR-497 |
| GCH1 | 2 | miR-124, miR-27 |
| KIF2A | 2 | miR-124, miR-203 |
| MYH9 | 2 | miR-124, miR-368 |
| TP63 | 2 | miR-124, miR-203 |
| PAX3 | 2 | miR-27, miR-368 |
Experimental validation
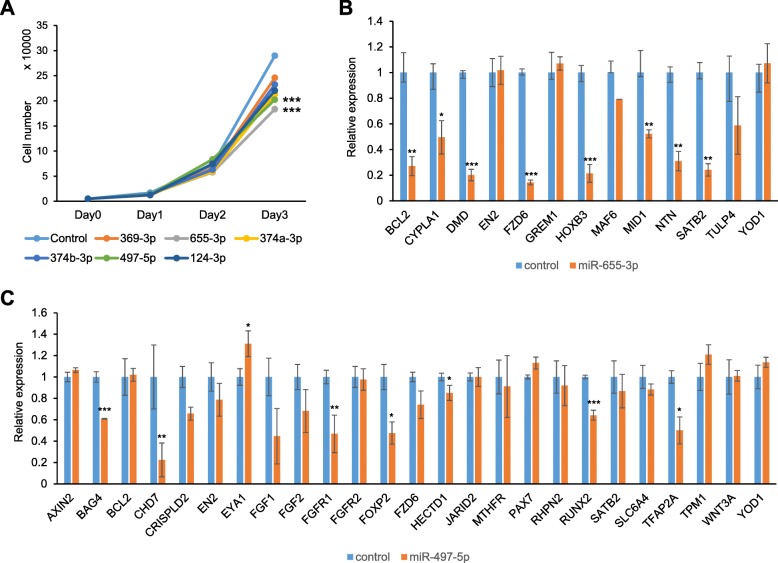
miRNA regulates expression of its genes anti- correlationally [34]. To test whether induction of potential CL/P-candidate miRNAs caused proliferation defects through the suppression of target gene expression, we treated cultured human lip fibroblasts with each miRNA mimic. The miR-497-5p and miR-655-3p mimics were most significantly inhibited cell proliferation in human lip fibroblasts (Fig. 2a). To identify target genes regulated by either miR-497-5p or miR-655-3p, we performed quantitative RT-PCR analyses for the predicted target genes (AXIN2, BAG4, BCL2, CHD7, CRISPLD2, EN2, EYA1, FGF1, FGF2, FGFR1, FGFR2, FOXP2, FZD6, HECTD1, JARID2, MTHFR, PAX7, RHPN2, RUNX2, SATB2, SLC6A4, TFAP2A, TPM1, WNT3A, and YOD1 for miR-497-5p; and BCL2, CYP1A1, DMD, EN2, FZD6, GREM1, HOXB3, MAFB, MID1, NTN1, PAX6, SATB2, TULP4, and YOD1 for miR-655-3p) in human lip fibroblasts after treatment with mimics of either miR-497-5p or miR-655-3p. The expression of almost target genes except EN2, GREM1, MAF6, TULP4 and YOD1 was significantly downregulated by treatment with miR- 655-3p mimic (Fig. 2b). The expression of target genes (BAG4, CHD7, CRISPLD2, FGFR1, FOXP2, HECTD1, RUNX2, and TFAP2A) was significantly downregulated by treatment with miR-497-5p mimic (Fig. 2c). PAX6 was excluded because its expression is restricted in the anterior ectoderm during early embryogenesis and the ectoderm of craniofacial surface during craniofacial development [35–37]. EYA1 expression was unexpectedly increased after treatment with miR-497-5p mimic.
Fig. 2.
Experimental validation of predicted miRNAs. a Cell proliferation assays in human lip fibroblasts treated with the indicated mimics of miRNAs. Negative control (control, light blue), miR-369-3p (orange), miR-655-3p (gray), miR-374a-3p (yellow), miR-374b-3p (blue), miR-497-5p (light green), and miR-124-3p (dark blue). *** p < 0.001. b Quantitative RT-PCR for the indicated genes after treatment with negative control (light blue) or miR-655-3p mimic (orange). * p < 0.05, ** p < 0.01, *** p < 0.001. c Quantitative RT-PCR for the indicated genes after treatment with negative control (light blue) or miR-497-5p mimic (orange). * p < 0.05, ** p < 0.01, *** p < 0.001
Discussion
Orofacial cleft is described in approximately 400 known human syndromes [38–40]. Several factors have been implicated in clefting by studies of mouse models [25] and genetic screening in humans. Our literature search identified 177 genes as possible causative genes of CL/P. Our follow-up bioinformatics analysis could group genes by common features of CL/P genes in function, pathway, and miRNA regulation. As expected, the contribution of most of the highlighted pathways (e.g. FGF, Hippo, TGFβ, and WNT) to the growth and developmental process has been shown in previous mouse genetic studies for craniofacial development [25]. Cellular metabolic pathways such as one-carbon metabolism are also highlighted in KEGG pathway analysis. The one-carbon metabolism, mediated by the folate cofactor, is involved in multiple physiological processes including biosynthesis of purines and thymidine, amino acid homeostasis, epigenetic maintenance, and redox defense [41]. Mice with one-carbon metabolic aberrations (Mthfd1l−/− mice) are embryonic lethal by E12.5, with craniofacial deformities including midfacial cleft [41]. However, in human studies it is still controversial in the significance of genetic mutations in genes involved in one-carbon metabolism [42–45]; further studies are necessary to reach to the conclusive evidence.
Multiple processes synthesize miRNAs and then transcribe them as long primary transcripts that are cleaved by Dicer, a type 3 ribonuclease, to produce mature, functional miRNAs. In human genetic studies, the increasing number of studies show functional significance of single- nucleotide polymorphisms (SNPs) in genes related to CL/P [46–50]. These SNPs might alter the binding activity of miRNAs. For example, previous studies show that SNPs in the miRNA-binding sites of MSX1, FGF2, FGF5 and FGF9 are associated with the susceptibility of nonsyndromic orofacial clefts [51, 52]. A recent study shows that plasma miRNAs are differentially expressed in nonsyndromic CP and nonsyndromic CL/P [53]. The miRNAs may also systemically regulate gene expression during embryogenesis, while some miRNAs may uniquely regulate gene expression in some particular tissues with an interaction with mRNAs expressed in a tissue-specific manner. In mice, loss of Dicer results in absence of mature miRNAs and, therefore, the phenotype in Dicer knockout mice reflects how important miRNAs are for proper development. Interestingly, cranial neural crest (CNC) cell- specific Dicer knockout (DicerF/F;Wnt1-Cre) mice, but not epithelial-specific Dicer knockout (DicerF/F;K14-Cre) mice, exhibit severe midfacial deformities resulting from decreased cell proliferation and increased apoptosis in the developing craniofacial regions [54–57], indicating that miRNAs have crucial roles in the fate determination of CNC cells during midfacial development [54–57]. In this study, to evaluate the function of each miRNA in cell proliferation/survival, we employed human lip fibroblasts, CNC-derived cells, for our experimental validation. The functional significance of candidate miRNAs were tested in cell proliferation/survival assays in cultured human lip fibroblasts. We found that miR-369-3p, miR-655-3p, miR-374a-5p, miR-374b-5p, and miR-497-5p that were first identified in this study as candidates involved in human CL/P suppressed cell proliferation in cultured human lip fibroblasts. Top two candidate miRNAs, miR-655-3p and miR-497-5p, were further tested in the miRNA-gene regulation assay. We found that these miRNA mimics suppressed expression of genes associated with human CL/P. Thus, the miRNAs predicted were successfully validated in our experiments. Taken together, this study provides a better understanding of CL/P as well as data that will be available for the future research of genetic approaches used to characterize individual or cluster miRNAs identified in this study.
In this study, we validated that overexpression of miR-655-3p and miR-497-5p suppressed the expression of multiple genes (BCL2, CYPLA1, DMD, FZD6, HOXB3, MID1, NTN, and SATB2 by miR-655-3p; and BAG4, CHD7, FGFR1, FOXP2, HECTD1, RUNX2, and TFAP2A by miR-497-5p) that are associated with CL/P. Previous studies indicate that miR-655-3p is downregulated in several cancers through dysregulation of ADAM10 (a disintegrin and metalloproteinase domain-containing protein 10) and the WNT/β-catenin pathway [58, 59]. By contrast, it remains unknown how miR-655-3p expression is induced.
Previous studies also show that miR-497-5p is downregulated in several cancers [60–62]. Interestingly, miR-497-5p is upregulated during myofibroblast differentiation of lung resident mesenchymal stem cells and in the lung tissues of a pulmonary fibrosis mouse model [63]. While it remains unclear what factors and triggers induce miR-655-3p expression, increased miR-655-3p levels may result in accelerated tissue differentiation with less proliferation in various tissues. Several environmental risk factors for CL/P such as smoking, alcohol consumption and toxins [64] may cause CL/P through the upregulation of these CL/P-associated miRNAs. In addition, when the levels of these microRNAs are increased too much, multiple CL/P genes would be suppressed, as we demonstrated in this study. Our results may partially explain why individuals with CL/P show suppression of multiple CL/P genes without genetic mutations in the coding regions, which contributes to the complexity of the CL/P etiology.
In this study, we found that miR-497-5p suppressed CHD7 expression at maximum degree of 80% in cultured human lip fibroblasts. Mutations in CHD7 cause CHARGE syndrome, which is characterized by CL/P (in 20–36% of the cases), abnormal middle and external ear, coloboma, choanal atresia and hypoplastic semi-circular canals, rhombencephalic dysfunction, hypothalamo- hypophyseal dysfunction, mental retardation, and tracheoesophageal fistula, but do not contribute to nonsyndromic CL/P [65]. miR- 497-5p also suppressed RUNX2 expression; SNPs in RUNX2 are known to increase the risk of nonsyndromic CL/P. Although FOXP2 and BAG4 mutations are associated with nonsyndromic CL/P [66, 67], less is known about the role of these genes in CL/P development. Mouse genetic mutant models are useful tools to investigate the role of genes in lip formation, but the loss of these CL/P genes fails to cause CL/P in mice, although it has not been examined whether suppression of multiple CL/P-associated genes causes CL/P. However, mice deficient for Tfap2a, which is suppressed by miR-497-5p, exhibit midfacial cleft [68], suggesting that TFAP2A may be a principal downstream target of miR-497-5p in human lip fibroblasts. Mutations in TFAP2A cause Branchio-Oculo-Facial Syndrome (BOFS), which is characterized by CL/P, branchial arch defects, and ocular anomalies [69], and are associated with nonsyndromic CL/P in several populations. In addition, Tfap2a deficiency suppresses Fgf8 expression in craniofacial regions in mice [68], suggesting that FGF signaling is a downstream pathway of TFAP2A. In this study, we found that miR-497-5p suppressed FGFR1 gene expression in cultured human lip fibroblasts. Therefore, a combined downregulation of TFAP2A and FGFR1 may compromise this cascade. FGFR1 mutations are also associated with increased risk of nonsyndromic CL/P, as well as of Kallmann syndrome with CL/P [70] and Hartsfield syndrome, which is characterized by holoprosencephaly, ectrodactyly, and CL/P [71].
miR-655-3p also suppressed the expression of FZD6, a WNT receptor, with a maximum reduction of 85%, and of SATB2 in cultured human lip fibroblasts. Mutations in FZD6 are found in nonsyndromic CL/P individuals, and SNPs in SATB2 are associated with nonsyndromic CL/P in several populations. Importantly, mice deficient for Satb2 also show CL and CP, as seen in individuals with mutations in SATB2. Interestingly, wild-type mouse embryos maternally exposed to phenytoin, a drug used to prevent and control seizures but presenting congenital anomalies as common side effects, show decreased Satb2 expression in craniofacial tissues [72]. This suggests that miR-655-3p may be upregulated in this condition of CL/P. miR-655-3p also suppressed expression of DMD and MID1 in cultured human fibroblasts. These genes are located on the X chromosome so that their mutations are associated with X-linked nonsyndromic CL/P in men and women. Mutations in MID1 are associated with the X-linked Opitz G/BBB syndrome, characterized by midline defects such as CL and CP, hypertelorism, and laryngo-trachea-esophageal (LTE) abnormalities [73, 74]. CYP1A1 is involved in drug/agent metabolism and several of these metabolites are known to be carcinogens [75]; CYP1A1 mutations may be associated with CL/P induced with smoking and detoxification [76]. The role of other downstream targets (NTN1 and BCL2) of miR-655-3p in the CL/P etiology remains largely unknown, and mice deficient for either Ntn1 or Bcl2 fail to display CL/P and CP.
Our systematic data collection timely summarizes the CL/P-candidate genes; however, it has some limitations. For example, some genes are from syndromes that include CL/P as a feature, but often they do not. These causative genes may be more broadly related to development, but do not contribute to CL/P; CL/P may be secondary to other defects in these syndromes. The current genetic signature of CL/P or other complex genetic diseases may be due to the bias of the type of genes that have been studied. Unbiased genome sequencing approaches will likely overcome the limitations and help us discover genes associated with CL/P. Molecular profiling of CL/P at the genomic, transcriptomic, regulatory (e.g. enhancer), epigenomic, and proteomic levels in model organisms like mice will provide us with a more detailed and accurate view of how genetic changes cause CL/P.
Conclusion
Our bioinformatics analysis results suggest that a disruption of extracellular cues and their signaling pathways might be a major cause of CL/P, and that miRNAs might play important roles in the CL/P etiology through the regulation of CL/P genes. In this study, we found that several miRNAs suppressed cell proliferation in cultured human lip fibroblasts. Among them, we confirmed that miR-655-3p and miR497-5p negatively regulated CL/P-candidate genes in the cultured cells. This study will have potential relevance to the miRNA-gene pathways and networks, not only in CL/P but also in other organogenesis processes. Therefore, this study will contribute to a better understanding of the mechanisms of CL/P and to future clinical interventions to prevent and diagnose CL/P.
Additional files
Table S1. PCR primer sets used in this study. Table S2. Summary of databases searched. Table S3. Genes with significant contribution to human CL/P (identified through single gene studies). Table S4. Genes with significant contribution to human CL/P (identified through multiple genes studies). Table S5. Genes with significant contribution to human CL/P (unknown coding genes). Table S6. Genes without significant contribution to human CL/P. Table S7. CL/P candidate genes with significant signals in GWAS. Table S8. CL/P candidate genes without significant signals in GWAS. Table S9. GO terms enriched with genes associated with cleft lip with/without cleft palate (CL/P) in humans. Table S10. GO Biological Process terms enriched with human CL/P genes (FDR < 0.005). Table S11. GO Molecular Function terms enriched with human CL/P genes (FDR < 0.005). Table S12. GO Cellular Component terms enriched with human CL/P genes (FDR < 0.05). Table S13. Top 30 Human Phenotype Ontology Categories. Table S14. KEGG pathways enriched with genes associated with cleft lip with/without cleft palate (CL/P) in humans. (ZIP 292 kb)
Acknowledgments
We thank Mrs. Helena VonVille for her valuable assistance with the systematic review and Dr. Guangchun Han and Dr. Hiroki Yoshioka for assistance with miRNA analysis.
Funding
This study was supported by grants from the NIH National Institute of Dental and Craniofacial Research (R03DE024759, R03DE026208, R01DE026767, and R03DE026509 to J.I.; R01LM012806, R21CA196508, R03DE027393, and R03DE028103 to Z.Z., and R03DE027711 to P.J.) and a faculty fund from UTHealth School of Dentistry to J.I. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All the data used in this study are from the public sources cited in our reference list. Additional files, which may be needed to reproduce the results presented in the manuscript, were made available as additional files.
Abbreviations
- CCDG
Center for Craniofacial and Dental Genetics
- CL/P
Cleft lip with or without cleft palate
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- ECM
Extracellular matrix
- FDR
False Discovery Rate
- GO
Gene Ontology
- GWAS
Genome-wide association study
- KEGG
Kyoto Encyclopedia of Genes and Genomes database
- MAPK
Mitogen-activated protein kinase
- MeSH
Medical Subject Headings
- MF
Molecular function
- miRNA
microRNA
- PI3K
Phosphoinositide 3-kinase
- SNP
Single nucleotide polymorphism
- TDT
Transmission Disequilibrium Test
Authors’ contributions
JI and ZZ designed the project, MG, ND, GJ, and PJ collected the data, MG, ND, AL, GJ, and PJ analyzed the data, AS and MZ carried out experiments, AS and JI drafted the manuscript, and JI and ZZ edited the manuscript. All authors approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- 2.Bender PL. Genetics of cleft lip and palate. J Pediatr Nurs. 2000;15(4):242–249. doi: 10.1053/jpdn.2000.8148. [DOI] [PubMed] [Google Scholar]
- 3.Bezerra J, Oliveira G, Soares C, Cardoso M, Ururahy M, Neto F, Lima-Neto L, Luchessi A, Silbiger V, Fajardo C. Genetic and non-genetic factors that increase the risk of non-syndromic cleft lip and/or palate development. Oral Dis. 2015;21(3):393–399. doi: 10.1111/odi.12292. [DOI] [PubMed] [Google Scholar]
- 4.Richards L, Plachez C, Ren T. Developmental biology: Frontiers for clinical genetics. Section editors: Roderick R McInnes, e-mail: Jacques Michaud, e-mail:. Mechanisms regulating the development of the corpus callosum and its. Clin Genet. 2004;66(4):276–289. doi: 10.1111/j.1399-0004.2004.00354.x. [DOI] [PubMed] [Google Scholar]
- 5.Ghazali N, Rahman NA, Kannan TP, Jaafar S. Screening of transforming growth factor Beta 3 and Jagged2 genes in the Malay population with nonsyndromic cleft lip with or without cleft palate. Cleft Palate Craniofac J. 2015;52(4):e88–e94. doi: 10.1597/14-024. [DOI] [PubMed] [Google Scholar]
- 6.do Rego Borges A, Sá J, Hoshi R, Viena CS, Mariano LC, de Castro Veiga P, Medrado AP, Machado RA, de Aquino SN, Messetti AC. Genetic risk factors for nonsyndromic cleft lip with or without cleft palate in a Brazilian population with high African ancestry. Am J Med Genet A. 2015;167(10):2344–2349. doi: 10.1002/ajmg.a.37181. [DOI] [PubMed] [Google Scholar]
- 7.Jia P, Zhao Z. Network.Assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum Genet. 2014;133(2):125–138. doi: 10.1007/s00439-013-1377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98(1):1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11(4):252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 10.Ross JS, Carlson JA, Brock G. miRNA: the new gene silencer. Am J Clin Pathol. 2007;128(5):830–836. doi: 10.1309/2JK279BU2G743MWJ. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Gong X, Purow B, Zhao Z. Uncovering MicroRNA and transcription factor mediated regulatory networks in glioblastoma. PLoS Comput Biol. 2012;8(7):e1002488. doi: 10.1371/journal.pcbi.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W, Mitra R, Lin CC, Wang Q, Cheng F, Zhao Z. Systematic dissection of dysregulated transcription factor-miRNA feed-forward loops across tumor types. Brief Bioinform. 2016;17(6):996–1008. doi: 10.1093/bib/bbv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoen C, Aschrafi A, Thonissen M, Poelmans G, Von den Hoff JW, Carels CEL. MicroRNAs in Palatogenesis and cleft palate. Front Physiol. 2017;8:165. doi: 10.3389/fphys.2017.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Jia P, Cuenco KT, Zeng Z, Feingold E, Marazita ML, Wang L, Zhao Z. Association signals unveiled by a comprehensive gene set enrichment analysis of dental caries genome-wide association studies. PLoS One. 2013;8(8):e72653. doi: 10.1371/journal.pone.0072653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sangani D, Suzuki A, VonVille H, Hixson JE, Iwata J. Gene mutations associated with temporomandibular joint disorders: a systematic review. OAlib. 2015;2(6). [DOI] [PMC free article] [PubMed]
- 17.Veronese N, Cereda E, Solmi M, Fowler SA, Manzato E, Maggi S, Manu P, Abe E, Hayashi K, Allard JP, et al. Inverse relationship between body mass index and mortality in older nursing home residents: a meta-analysis of 19,538 elderly subjects. Obes Rev. 2015;16(11):1001–1015. doi: 10.1111/obr.12309. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300.
- 24.Suzuki A, Pelikan RC, Iwata J. WNT/beta-catenin signaling regulates multiple steps of Myogenesis by regulating step-specific targets. Mol Cell Biol. 2015;35(10):1763–1776. doi: 10.1128/MCB.01180-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki A, Sangani DR, Ansari A, Iwata J. Molecular mechanisms of midfacial developmental defects. Dev Dyn. 2016;245(3):276–293. doi: 10.1002/dvdy.24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obican SG, Finnell RH, Mills JL, Shaw GM, Scialli AR. Folic acid in early pregnancy: a public health success story. FASEB J. 2010;24(11):4167–4174. doi: 10.1096/fj.10-165084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bille C, Winther JF, Bautz A, Murray JC, Olsen J, Christensen K. Cancer risk in persons with oral cleft--a population-based study of 8,093 cases. Am J Epidemiol. 2005;161(11):1047–1055. doi: 10.1093/aje/kwi132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunkhase E, Ludwig KU, Knapp M, Skibola CF, Figueiredo JC, Hosking FJ, Ellinghaus E, Landi MT, Ma H, Nakagawa H, et al. Nonsyndromic cleft lip with or without cleft palate and cancer: evaluation of a possible common genetic background through the analysis of GWAS data. Genom Data. 2016;10:22–29. doi: 10.1016/j.gdata.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taioli E, Ragin C, Robertson L, Linkov F, Thurman NE, Vieira AR. Cleft lip and palate in family members of cancer survivors. Cancer Investig. 2010;28(9):958–962. doi: 10.3109/07357907.2010.483510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang JB, Liu YL, Sun PW, Lv XD, Du M, Fan XM. Molecular mechanisms of congenital heart disease. Cardiovasc Pathol. 2010;19(5):e183–e193. doi: 10.1016/j.carpath.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Torano EG, Garcia MG, Fernandez-Morera JL, Nino-Garcia P, Fernandez AF. The impact of external factors on the epigenome: in utero and over lifetime. Biomed Res Int. 2016;2016:2568635. doi: 10.1155/2016/2568635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitsiou-Tzeli S, Tzetis M. Maternal epigenetics and fetal and neonatal growth. Curr Opin Endocrinol Diabetes Obes. 2017;24(1):43–46. doi: 10.1097/MED.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Sun C, Meng Y, Zhang B, Wang X, Su Y, Shi L, Zhao E. A pilot study: screening target miRNAs in tissue of nonsyndromic cleft lip with or without cleft palate. Exp Ther Med. 2017;13(5):2570–2576. doi: 10.3892/etm.2017.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 35.Compagnucci C, Fish JL, Schwark M, Tarabykin V, Depew MJ. Pax6 regulates craniofacial form through its control of an essential cephalic ectodermal patterning center. Genesis. 2011;49(4):307–325. doi: 10.1002/dvg.20724. [DOI] [PubMed] [Google Scholar]
- 36.Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121(5):1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 37.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113(4):1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 38.Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, Liu YE. Nonsyndromic cleft lip with or without cleft palate in China: assessment of candidate regions. Cleft Palate Craniofac J. 2002;39(2):149–156. doi: 10.1597/1545-1569_2002_039_0149_nclwow_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 39.Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, Liu YE. Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families. Am J Hum Genet. 2002;71(2):349–364. doi: 10.1086/341944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muenke M. The pit, the cleft and the web. Nat Genet. 2002;32(2):219–220. doi: 10.1038/ng1002-219. [DOI] [PubMed] [Google Scholar]
- 41.Momb J, Lewandowski JP, Bryant JD, Fitch R, Surman DR, Vokes SA, Appling DR. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci U S A. 2013;110(2):549–554. doi: 10.1073/pnas.1211199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanton SH, Henry RR, Yuan Q, Mulliken JB, Stal S, Finnell RH, Hecht JT. Folate pathway and nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol. 2011;91(1):50–60. doi: 10.1002/bdra.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munger RG, Tamura T, Johnston KE, Feldkamp ML, Pfister R, Cutler R, Murtaugh MA, Carey JC. Oral clefts and maternal biomarkers of folate-dependent one-carbon metabolism in Utah. Birth Defects Res A Clin Mol Teratol. 2011;91(3):153–161. doi: 10.1002/bdra.20762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyles AL, Wilcox AJ, Taylor JA, Shi M, Weinberg CR, Meyer K, Fredriksen A, Ueland PM, Johansen AM, Drevon CA, et al. Oral facial clefts and gene polymorphisms in metabolism of folate/one-carbon and vitamin a: a pathway-wide association study. Genet Epidemiol. 2009;33(3):247–255. doi: 10.1002/gepi.20376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyles AL, Wilcox AJ, Taylor JA, Meyer K, Fredriksen A, Ueland PM, Drevon CA, Vollset SE, Lie RT. Folate and one-carbon metabolism gene polymorphisms and their associations with oral facial clefts. Am J Med Genet A. 2008;146A(4):440–449. doi: 10.1002/ajmg.a.32162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee MK, Shaffer JR, Leslie EJ, Orlova E, Carlson JC, Feingold E, Marazita ML, Weinberg SM. Genome-wide association study of facial morphology reveals novel associations with FREM1 and PARK2. PLoS One. 2017;12(4):e0176566. doi: 10.1371/journal.pone.0176566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leslie EJ, Carlson JC, Shaffer JR, Butali A, Buxo CJ, Castilla EE, Christensen K, Deleyiannis FW, Leigh Field L, Hecht JT, et al. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum Genet. 2017;136(3):275–286. doi: 10.1007/s00439-016-1754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q, Carlson JC, Hetmanski JB, Wang H, Larson DE, et al. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am J Hum Genet. 2015;96(3):397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butali A, Suzuki S, Cooper ME, Mansilla AM, Cuenco K, Leslie EJ, Suzuki Y, Niimi T, Yamamoto M, Ayanga G, et al. Replication of genome wide association identified candidate genes confirm the role of common and rare variants in PAX7 and VAX1 in the etiology of nonsyndromic CL(P) Am J Med Genet A. 2013;161A(5):965–972. doi: 10.1002/ajmg.a.35749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma L, Xu M, Li D, Han Y, Wang Z, Yuan H, Ma J, Zhang W, Jiang H, Pan Y, et al. A miRNA-binding-site SNP of MSX1 is associated with NSOC susceptibility. J Dent Res. 2014;93(6):559–564. doi: 10.1177/0022034514527617. [DOI] [PubMed] [Google Scholar]
- 52.Li D, Zhang H, Ma L, Han Y, Xu M, Wang Z, Jiang H, Zhang W, Wang L, Pan Y. Associations between microRNA binding site SNPs in FGFs and FGFRs and the risk of non-syndromic orofacial cleft. Sci Rep. 2016;6:31054. doi: 10.1038/srep31054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Zou J, Li Q, Chen L, Gao Y, Yan H, Zhou B, Li J. Assessment of differentially expressed plasma microRNAs in nonsyndromic cleft palate and nonsyndromic cleft lip with cleft palate. Oncotarget. 2016;7(52):86266–86279. doi: 10.18632/oncotarget.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nie X, Wang Q, Jiao K. Dicer activity in neural crest cells is essential for craniofacial organogenesis and pharyngeal arch artery morphogenesis. Mech Dev. 2011;128(3–4):200–207. doi: 10.1016/j.mod.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2(3):152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 56.Zehir A, Hua LL, Maska EL, Morikawa Y, Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Dev Biol. 2010;340(2):459–467. doi: 10.1016/j.ydbio.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheehy NT, Cordes KR, White MP, Ivey KN, Srivastava D. The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development. 2010;137(24):4307–4316. doi: 10.1242/dev.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu G, Zheng K, Xia S, Wang Y, Meng X, Qin X, Cheng Y. MicroRNA-655-3p functions as a tumor suppressor by regulating ADAM10 and beta-catenin pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35(1):89. doi: 10.1186/s13046-016-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao XQ, Liang B, Jiang K, Zhang HY. Down-regulation of miR-655-3p predicts worse clinical outcome in patients suffering from hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017;21(4):748–752. [PubMed] [Google Scholar]
- 60.Chai L, Kang XJ, Sun ZZ, Zeng MF, Yu SR, Ding Y, Liang JQ, Li TT, Zhao J. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res. 2018;10:989–1003. doi: 10.2147/CMAR.S163335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, Kuang D, Zhao X, Chen D, Wang X, Yang Q, Wan J, Zhu Y, Wang Y, Zhang S, et al. miR-497-5p inhibits cell proliferation and invasion by targeting KCa3.1 in angiosarcoma. Oncotarget. 2016;7(36):58148–58161. doi: 10.18632/oncotarget.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Z, Li A, Yu Z, Li X, Guo X, Chen R. MicroRNA-497-5p Suppresses Tumor Cell Growth of Osteosarcoma by Targeting ADP Ribosylation Factor-Like Protein 2. Cancer Biother Radiopharm. 2017;32(10):371–378. doi: 10.1089/cbr.2017.2268. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Shi C, Wang C, Liu W, Chu Y, Xiang Z, Hu K, Dong P, Han X. The role of miR-497-5p in myofibroblast differentiation of LR-MSCs and pulmonary fibrogenesis. Sci Rep. 2017;7:40958. doi: 10.1038/srep40958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyszynski DF, Beaty TH. Review of the role of potential teratogens in the origin of human nonsyndromic oral clefts. Teratology. 1996;53(5):309–317. doi: 10.1002/(SICI)1096-9926(199605)53:5<309::AID-TERA5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 65.Felix TM, Hanshaw BC, Mueller R, Bitoun P, Murray JC. CHD7 gene and non-syndromic cleft lip and palate. Am J Med Genet A. 2006;140(19):2110–2114. doi: 10.1002/ajmg.a.31308. [DOI] [PubMed] [Google Scholar]
- 66.Mohamad Shah NS, Salahshourifar I, Sulong S, Wan Sulaiman WA, Halim AS. Discovery of candidate genes for nonsyndromic cleft lip palate through genome-wide linkage analysis of large extended families in the Malay population. BMC Genet. 2016;17:39. doi: 10.1186/s12863-016-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riley BM, Schultz RE, Cooper ME, Goldstein-McHenry T, Daack-Hirsch S, Lee KT, Dragan E, Vieira AR, Lidral AC, Marazita ML, et al. A genome-wide linkage scan for cleft lip and cleft palate identifies a novel locus on 8p11-23. Am J Med Genet A. 2007;143A(8):846–852. doi: 10.1002/ajmg.a.31673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Green RM, Feng W, Phang T, Fish JL, Li H, Spritz RA, Marcucio RS, Hooper J, Jamniczky H, Hallgrimsson B, et al. Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis Model Mech. 2015;8(1):31–43. doi: 10.1242/dmm.017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milunsky JM, Maher TA, Zhao G, Roberts AE, Stalker HJ, Zori RT, Burch MN, Clemens M, Mulliken JB, Smith R, et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet. 2008;82(5):1171–1177. doi: 10.1016/j.ajhg.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu H, Niu Y, Wang T, Liu S, Xu H, Wang S, Liu J, Ye Z. Novel FGFR1 and KISS1R mutations in Chinese Kallmann syndrome males with cleft lip/palate. Biomed Res Int. 2015;2015:649698. doi: 10.1155/2015/649698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simonis N, Migeotte I, Lambert N, Perazzolo C, de Silva DC, Dimitrov B, Heinrichs C, Janssens S, Kerr B, Mortier G, et al. FGFR1 mutations cause Hartsfield syndrome, the unique association of holoprosencephaly and ectrodactyly. J Med Genet. 2013;50(9):585–592. doi: 10.1136/jmedgenet-2013-101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mao XY, Tang SJ. Effects of phenytoin on Satb2 and Hoxa2 gene expressions in mouse embryonic craniofacial tissue. Biochem Cell Biol. 2010;88(4):731–735. doi: 10.1139/O10-013. [DOI] [PubMed] [Google Scholar]
- 73.Quaderi NA, Schweiger S, Gaudenz K, Franco B, Rugarli EI, Berger W, Feldman GJ, Volta M, Andolfi G, Gilgenkrantz S, et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17(3):285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 74.Hsieh EW, Vargervik K, Slavotinek AM. Clinical and molecular studies of patients with characteristics of Opitz G/BBB syndrome shows a novel MID1 mutation. Am J Med Genet A. 2008;146A(18):2337–2345. doi: 10.1002/ajmg.a.32368. [DOI] [PubMed] [Google Scholar]
- 75.Mescher M, Haarmann-Stemmann T. Modulation of CYP1A1 metabolism: from adverse health effects to chemoprevention and therapeutic options. Pharmacol Ther. 2018;187:71–87. doi: 10.1016/j.pharmthera.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 76.Shi M, Christensen K, Weinberg CR, Romitti P, Bathum L, Lozada A, Morris RW, Lovett M, Murray JC. Orofacial cleft risk is increased with maternal smoking and specific detoxification-gene variants. Am J Hum Genet. 2007;80(1):76–90. doi: 10.1086/510518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. PCR primer sets used in this study. Table S2. Summary of databases searched. Table S3. Genes with significant contribution to human CL/P (identified through single gene studies). Table S4. Genes with significant contribution to human CL/P (identified through multiple genes studies). Table S5. Genes with significant contribution to human CL/P (unknown coding genes). Table S6. Genes without significant contribution to human CL/P. Table S7. CL/P candidate genes with significant signals in GWAS. Table S8. CL/P candidate genes without significant signals in GWAS. Table S9. GO terms enriched with genes associated with cleft lip with/without cleft palate (CL/P) in humans. Table S10. GO Biological Process terms enriched with human CL/P genes (FDR < 0.005). Table S11. GO Molecular Function terms enriched with human CL/P genes (FDR < 0.005). Table S12. GO Cellular Component terms enriched with human CL/P genes (FDR < 0.05). Table S13. Top 30 Human Phenotype Ontology Categories. Table S14. KEGG pathways enriched with genes associated with cleft lip with/without cleft palate (CL/P) in humans. (ZIP 292 kb)
Data Availability Statement
All the data used in this study are from the public sources cited in our reference list. Additional files, which may be needed to reproduce the results presented in the manuscript, were made available as additional files.