 N-Arylated NH125 analogue 1 rapidly eradicates a diverse panel of drug-resistant bacteria and fungi (≥99.9% kill in 1 to 10 minutes).
N-Arylated NH125 analogue 1 rapidly eradicates a diverse panel of drug-resistant bacteria and fungi (≥99.9% kill in 1 to 10 minutes).
Abstract
While a number of disinfection techniques are employed in healthcare units, the eradication of drug-resistant microorganisms remains a challenge. We recently reported N-arylated NH125 analogue 1, which demonstrated potent biofilm eradication and antibacterial activities against a panel of drug-resistant pathogens. The broad-spectrum activities observed for 1 along with its rapid eradication of MRSA persister cells suggested that this agent, and related analogues, can serve as disinfectants for antibiotic resistant pathogens in healthcare settings. Here, we report the rapid bactericidal activities of 1 against a panel of exponentially-growing, drug-resistant pathogens. Against MRSA, MRSE, VRE and MDR A. baumannii, 1 eradicated bacterial cells after five minutes when tested at 50 μM (3- to 6-log reduction of CFU per mL). We highlighted the rapid killing activities by demonstrating that 1 eradicates 99.99% of viable MRSA 1707 cells in one minute (50 μM, 4-log reduction of CFU per mL). In addition, 1 rapidly eradicated fungal pathogen C. neoformans in kill kinetic experiments. A solution of 1 demonstrated similar shelf stability to known disinfectant BAC-16 when tested up to 111 days after being stored. Collectively, our data highlights the potential of 1 to be used as a disinfecting agent to prevent healthcare-associated, drug-resistant infections.
Introduction
Healthcare facilities are designed to be safe places for individuals in need of treatment and healing. Contaminated surfaces within hospital settings contribute to the transmission of healthcare-associated pathogens, including drug-resistant microorganisms.1–9 With the continuing rise of microbial resistance, we face significant challenges regarding the prevention and eradication of infectious agents.2–4,6,10 Bacterial pathogens such as Staphylococcus aureus, Staphylococcus epidermidis, Acinetobacter baumannii and Enterococcus are notorious for antibiotic resistance and long survival times (up to months), increasing the risk of acquiring infections due to contamination in healthcare facilities.11–13 It is estimated that 2 million patients are affected by healthcare-associated infections, resulting in 100 000 deaths each year.14
Biofilms are surface-attached bacterial communities embedded within an extracellular matrix of biomolecules that house metabolically-dormant persister cells.15–17 Over the past several decades, biofilms have been recognized as a significant source of bacterial infection in hospitals as they are able to persist on surfaces for extended periods of time. In fact, S. aureus and S. epidermidis biofilms are the primary source of medical device-related infections.14 In addition, bacterial biofilms formed by multidrug resistant Gram-negative pathogens have become a widespread problem in healthcare facilities.14,18 Without question, there is a considerable need for new and effective disinfectant agents to rapidly eradicate drug-resistant bacteria.
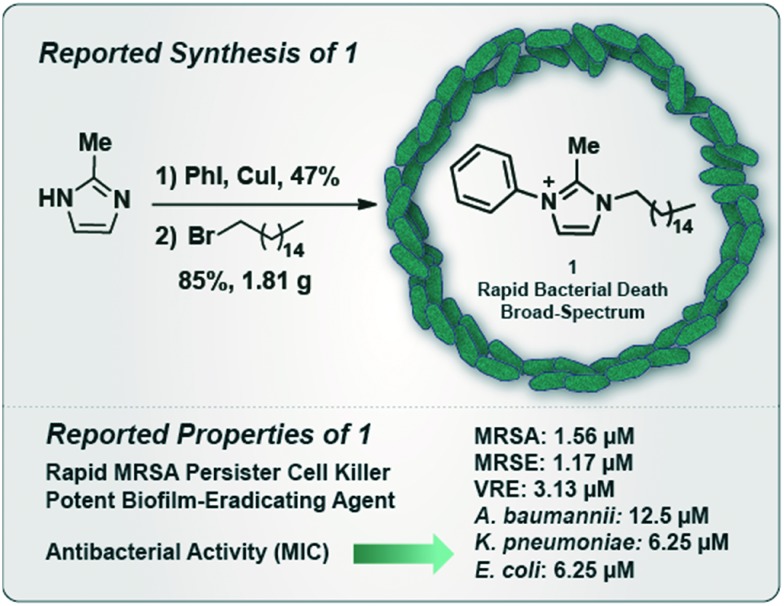
We recently reported N-arylated NH125 analogue 1 as a broad spectrum antibacterial agent that demonstrates potent biofilm-eradicating activities and kills methicillin-resistant S. aureus (MRSA) persister cells in stationary cultures (Fig. 1).19,20 We found that 1 elicits antibacterial activities through membrane lysis, which is a result of its polar head group (charged, imidazole) and long aliphatic tail. These structural features allow 1 to penetrate and disrupt the amphipathic nature of lipid bilayers, leading to cell lysis and bacterial death. Interestingly, the amphipathic structural features of 1 correspond to quaternary ammonium cations (QACs)21–23 agents, which are utilized in multiple applications to eradicate microorganisms. Additionally, we reported that 1 can be readily synthesized in two steps from 2-methylimidazole via 1.) copper(i)-catalyzed C–N coupling with iodobenzene (47% yield), and 2.) SN2 reaction involving the resulting N-phenylimidazole with 1-bromohexadecane (85% yield of 1, multigram scale; Fig. 1).19,20
Fig. 1. Reported chemical synthesis and antibacterial activities of N-arylated NH125 analogue 1.
Due to the significant antibacterial activities that 1 demonstrates against drug-resistant pathogens, we felt this compound could serve as an outstanding disinfectant for clinical settings. However, our previous kinetic kill experiments were carried out solely against stationary MRSA cultures and required extended exposure times (e.g., 30 to 90 minutes).19,20 In these studies, we assessed the rapid kill potential of 1 against a panel of exponentially-growing, drug-resistant pathogens. To do this, we subjected various microbial pathogens to 1 for short time exposures (e.g., 1 to 10 minutes), then determined kill efficacy via colony count assessment by comparing treated versus untreated (vehicle control only) cultures. Our experimental results are reported in colony forming units (CFU) per milliliter (mL).
Results and discussion
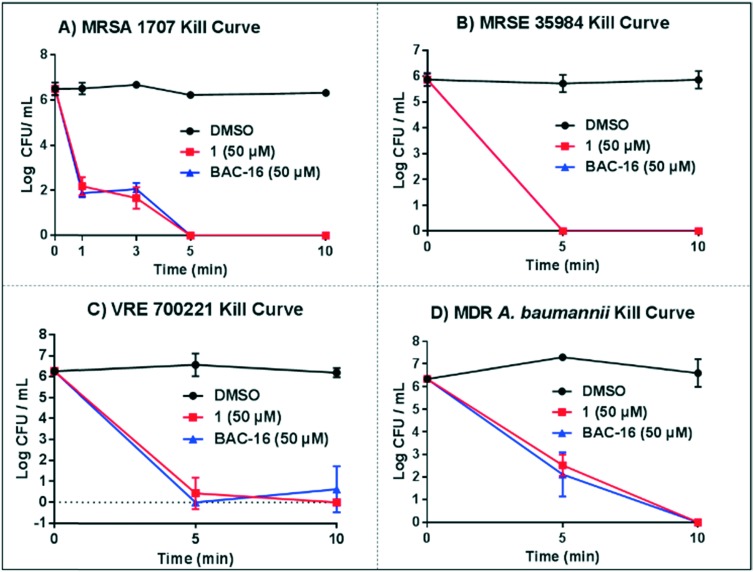
We started these investigations with MRSA 1707 due to the clinical importance of this Gram-positive human pathogen in healthcare-associated infections.11 In kill kinetic experiments against exponentially-growing cultures, we found 1 to rapidly kill MRSA 1707 cells after 1 minute at 50 μM (4-log reduction of CFU per mL, 99.99% kill; Fig. 2A). Similar results were observed with 1 following a 3 minute treatment of MRSA 1707 cultures; however, after 5 and 10 minutes of exposure to 1, we observed a complete eradication of MRSA 1707 cultures (no viable bacterial colonies were found at either time point; >6-log reduction in CFU per mL). We tested benzyldimethylhexadecyl ammonium chloride (BAC-16), a quaternary ammonium cation disinfectant, as a comparator in these experiments which demonstrated the same rapid kill kinetic profile as 1.
Fig. 2. Kill kinetic profiles of 1 against exponentially-growing bacteria, including: A) MRSA 1707, B) MRSE 35984, C) VRE 700221 and D) MDR A. baumannii 1794.
We then investigated the killing kinetics of 1 against exponentially-growing cultures of methicillin-resistant S. epidermidis (MRSE 35984). S. epidermidis is a major human pathogen and constitutes ∼80% of biofilms involved in material-associated infections.14 Therefore, it is important for disinfectants to demonstrate rapid killing activity profiles against S. epidermidis, especially drug-resistant strains. In kill curve experiments against MRSE 35984 cultures, compound 1 demonstrated total eradication to MRSE after 5 and 10 minutes when tested at 50 μM (∼6-log reduction of viable MRSE cells, ∼99.9999% kill; Fig. 2B).
Another Gram-positive pathogen important to human health and involved in healthcare-associated infections is vancomycin-resistant Enterococcus faecium (VRE). VRE can survive for extended periods of time in clinical environments, leading to human infection and resulting economic burden.11 One study reported that vancomycin-resistant E. faecium infections significantly increased hospital costs compared to vancomycin-sensitive E. faecium infections, suggesting the importance of control measures to prevent VRE transmission.24 We tested N-arylated NH125 analogue 1 at 50 μM against VRE 700221 cultures and, similar to our initial MRSA and MRSE findings, observed rapid and complete eradication of VRE 700221 cultures at 5 and 10 minutes (6-log reduction of viable VRE 700221 cells, 99.9999% kill; Fig. 2C).
We were also interested in evaluating 1 against the major Gram-negative human pathogen, A. baumannii. For this assessment, we selected the multi-drug resistant (MDR) strain A. baumannii 1794, which is resistant to several classes of conventional antibiotics. Following a 5 minute treatment, 1 eradicated 99.9% of the MDR A. baumannii culture at 50 μM (3-log reduction in CFU per mL). Complete eradication of A. baumannii 1794, was observed with 1 after a 10 minute treatment at 50 μM (>6-log reduction of viable cells, >99.9999% kill). Similar to our findings with the Gram-positive pathogens, BAC-16 reported near identical killing profiles.
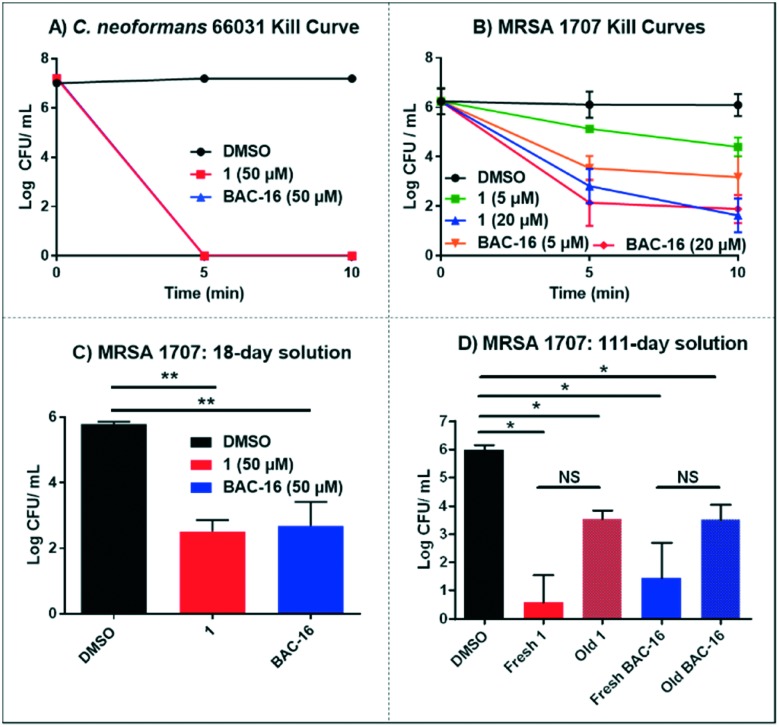
To expand the scope of these investigations, we tested 1 against the fungal pathogen Cryptococcus neoformans. C. neoformans infections are rare in healthy adults; however, this fungal pathogen can cause complications in those with compromised immune systems (i.e., HIV patients) and can be acquired in a healthcare setting.25 In kinetic kill experiments, N-arylated NH125 analogue 1 led to complete eradication of C. neoformans 66031 cells following 5 and 10 minute treatments at 50 μM (7-log reduction of viable fungal cells; Fig. 3A). The rapid killing activity profiles against diverse microorganisms, including both bacterial and fungal human pathogens, is an outstanding and promising feature displayed by 1.
Fig. 3. A) Kill kinetics of exponentially-growing C. neoformans 66031. B) Kill kinetics of exponentially-growing MRSA 1707 treated with variable concentrations of 1 and BAC-16. C) Efficacy of 18 day old solutions of 1 and BAC-16 (50 μM; CFU mL–1 in MRSA cultures after 10 minutes of exposure to compounds). D) Efficacy of new versus 111 day old solutions of 1 and BAC-16 (50 μM; CFU mL–1 in MRSA cultures after 10 minutes of exposure to compounds). *P-value <0.01, **P-value <0.0005. NS: not significant.
Following our initial assessment of 1 against a panel of diverse microorganisms at 50 μM, we investigated lower doses required for rapid killing. We performed kinetic kill experiments of 1 against MRSA 1707 at 5 and 20 μM. Our results demonstrate that at 20 μM, 1 causes a 3-log reduction of CFU per mL at 5 minutes (99.9% kill) and 4-log reduction of viable MRSA 1707 cells after 10 minute treatment (99.99% kill). However, when 1 was tested at 5 μM, minimal killing was observed at 5 minutes (<1-log) and moderate killing was observed at 10 minutes (1-log reduction of CFU per mL, 90% kill; Fig. 3B). BAC-16 provided near identical activities compared to 1 when tested at 20 μM; however, reported more rapid kill than 1 after a 5 minute treatment at 5 μM.
Shelf stability is crucial for the development of new disinfectants, we prepared a solution of 1 and BAC-16 (at 50 μM) in LB media and stored it on a shelf for extended periods of time. We tested the antibacterial activity of the compound solutions after 18 and 111 days of storage against MRSA 1707 by determining kill efficacies (in CFU mL–1) following 10 minutes exposure. After 18 days, both solutions remained active causing >3-log reduction of activity to MRSA 1707 cultures (Fig. 3C). On day 111, we compared the activity of a freshly-prepared solution against the 111 day stock of 1 and BAC-6 against MRSA 1707. We observed a slight reduction in activity for both solutions compared to fresh solutions of 1 and BAC-16 (Fig. 3D); however, we observed >99% kill against MRSA 1707. These data suggest that compound 1 and BAC-16 can be used in similar disinfectant applications against pathogenic microorganisms.
Despite this focused study with an eye towards the development of 1 as a disinfectant against drug-resistant microorganisms, NH125 and related analogues have potential use in multiple therapeutic applications. Based on previous reports, NH125 demonstrates efficacy in a C. elegans (live worm) model of S. aureus infection,26in vitro and in vivo anticancer27,28 and neuromodulatory activities.29,30 We assessed mammalian cytotoxicity of 1, NH125 and BAC-16 against HEK 293 (human embryonic kidney 293) cells in 48 hour MTT assays and found the half-maximal cytotoxic concentration (CC50) values to be <1.56, 2.41 and 6.84 μM, respectively (each of these compounds were completely toxic to HEK 293 cells at 25 μM). Alternative explorations are currently underway regarding our novel NH125 analogues20 and will be reported in due course.
Experimental
Kill kinetics of exponentially-growing bacterial cultures
An overnight culture of MRSA 1707 (ATCC, BAA-1707), S. epidermidis (ATCC, MRSE 35984), E. faecium (VRE 700221) or A. baumannii (ATCC, 1794) was diluted 1 : 1000 in 2 mL of LB media in test tubes containing test compounds at the desired, predetermined concentration, or vehicle (DMSO) alone. The cultures were then incubated at 37 °C with shaking at 250 rpm. At the desired time point (1, 3, 5 & 10 minutes), 100 μL aliquots were removed from the culture tubes, serially diluted and plated (LB, 1.5% agar petri dishes). The resulting petri dishes were then incubated at 37 °C overnight to allow growing of viable bacterial colonies. Following incubation, viable bacterial colonies were counted on each agar plate to determine colony forming units (CFU) per milliliter (mL). Plates containing between 30 and 300 colonies were used for analysis. Each experiment was conducted in three replicates from separate, individual bacterial colonies. Kill curves were plotted using Graphpad Prism 6.0.
Kill kinetics of exponentially-growing fungal cultures
Cryptococcus neoformans was purchased from ATCC (66031). C. neoformans first streaked onto YPD agar medium and incubated at 37 °C for two days. Single colonies were then inoculated into 3 mL of YPD liquid medium and cultured at 37 °C, 250 rpm overnight. Subsequently, the overnight cultures were diluted in fresh YPD medium, and test compounds dissolved in DMSO were added at a final concentration of 50 μM and the same volume of DMSO was used as control. After mixing well, the cultures were continuously incubated at 37 °C, 250 rpm. At different time points, aliquots were taken, diluted in fresh YPD medium, and plated on YPD agar medium. The plates were incubated at 37 °C and the number of colonies were recorded after one or two days. Each experiment was conducted in triplicate. Colony forming units (CFU) per milliliter data was plotted using Graphpad Prism 6.0.
Cytotoxicity assessment against HEK 293 cells
MTT assay was performed to determine compound cytotoxicity using human embryonic kidney 293 (HEK 293) cells. HEK 293 cells (104 cells in 100 μL) were seeded onto a 96-well plate containing DMEM supplemented with 10% FBS, 100 U mL–1 penicillin and streptomycin. The plate was then incubated at 37 °C in a humidified incubator under 5% CO2 overnight. The HEK 293 cells were then treated with serial concentrations of 1, NH125 and BAC-16 at 0, 1.56, 3.125, 6.25, 12.5, 25, 50, 100 and 200 μM. After further incubation at 37 °C for 48 hours, 10 μL of MTT (5 mg mL–1) in PBS was added to each well and incubated for 3 hours, followed by the aspiration of the medium. Dimethyl sulfoxide (DMSO, 100 μL) was added to each well to dissolve the MTT and the plate was agitated for 30 minutes. After this time, the optical density (OD) was measured at 570 nm using a UV/vis microplate spectrophotometer (BioTek). Six replications were performed per treatment and the percent kill was calculated as (1 – test OD570/non-treated OD570) × 100%. Data were analyzed by ORIGIN 8.0. From these experiments, the cytotoxic concentration 50 (CC50, the concentration found to be half-maximal toxic to HEK cells) for 1, NH125 and BAC-16 were determined to be <1.56, 2.41 and 6.84 μM, respectively. Note: HEK 293 cells were purchased from ATCC.
Conclusions
In conclusion, this study highlights the rapid killing profile of 1 against a range of exponentially-growing microbial pathogens. Impressively, 1 completely eradicated MRSA 1707, MRSE 35984, VRE 700221 and C. neoformans 66301 after 5 minutes and MDR A. baumannii 1794 after 10 minutes (6-log reduction to no detectable viable microbial cells). In addition, 1 displayed similar stability patterns to known disinfectant BAC-16. The rapid-killing activity displayed by 1 suggests that this microbicidal agent could be utilized as an effective disinfectant to eradicate human pathogens in healthcare settings.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We would like to acknowledge the University of Florida for start-up funds to support initial studies regarding N-arylated NH125 analogues. The National Institute of General Medical Sciences of the National Institutes of Health is acknowledged for providing financial support (R35GM128621 to RWH) to investigate biofilm-eradicating agents. We also thank Dr. Gina Parise Sloan for helpful discussions regarding disinfectant agents, and Hongfen Yang and Alejandra Chávez-Riveros for assistance with figures.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00613j
References
- Brown E. D., Wright G. D. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- Blair J. M. A., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J. V. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Rossiter S. E., Fletcher M. H., Wuest W. M. Chem. Rev. 2017;117:12415–12474. doi: 10.1021/acs.chemrev.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M. A., Dwyer D. J., Collins J. J. Nat. Rev. Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L. Drug Discovery Today: Technol. 2014;11:33–39. doi: 10.1016/j.ddtec.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Wright G. D. Chem. Commun. 2011;47:4055–4061. doi: 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- Doll M., Stevens M., Bearman G. Int. J. Infect. Dis. 2018;67:52–57. doi: 10.1016/j.ijid.2017.10.014. [DOI] [PubMed] [Google Scholar]
- Saka K., Akanbi II A., Obasa T., Raheem R., Oshodi A. J. Bacteriol. Parasitol. 2017;08:1–4. [Google Scholar]
- Mehta Y., Gupta A., Todi S., Myatra S., Samaddar D. P., Patil V., Bhattacharya P. K., Ramasubban S. Indian J. Crit. Care Med. 2014;18:149–163. doi: 10.4103/0972-5229.128705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin Zaman S., Hussain M. A., Nye R., Mehta V., Mamun K. T., Hossain N. Cureus. 2017;9:e1403. doi: 10.7759/cureus.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancer S. J. Clin. Microbiol. Rev. 2014;27:665–690. doi: 10.1128/CMR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu A. C., Tavares R. R., Borges A., Mergulhao F., Simoes M. J. Antimicrob. Chemother. 2013;68:2718–2732. doi: 10.1093/jac/dkt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell G., Russell A. D. Clin. Microbiol. Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival S. L., Suleman L., Vuotto C., Donelli G. J. Med. Microbiol. 2015;64:323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J., Stoodley P. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Wu H., Moser C., Wang H.-Z., Høiby N., Song Z.-J. Int. J. Oral Sci. 2015;7:1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D., Kemmerly S. A. Ochsner J. 2009;9:27–31. [PMC free article] [PubMed] [Google Scholar]
- Abouelhassan Y., Basak A., Yousaf H., Huigens III R. W. ChemBioChem. 2017;18:352–357. doi: 10.1002/cbic.201600622. [DOI] [PubMed] [Google Scholar]
- Basak A., Abouelhassan Y., Zuo R., Yousaf H., Ding Y., Huigens III R. W. Org. Biomol. Chem. 2017;15:5503–5512. doi: 10.1039/c7ob01028a. [DOI] [PubMed] [Google Scholar]
- Forman M. E., Jennings M. C., Wuest W. M., Minbiole K. P. C. ChemMedChem. 2016;11:1401–1405. doi: 10.1002/cmdc.201600176. [DOI] [PubMed] [Google Scholar]
- Jennings M. C., Ator L. E., Paniak T. J., Minbiole K. P. C., Wuest W. M. ChemBioChem. 2014;15:2211–2215. doi: 10.1002/cbic.201402254. [DOI] [PubMed] [Google Scholar]
- De Zoysa G. H., Cameron A. J., Hegde V. V., Raghothama S., Sarojini V. J. Med. Chem. 2015;58:625–639. doi: 10.1021/jm501084q. [DOI] [PubMed] [Google Scholar]
- Puchter L., Chaberny I. F., Schwab F., Vonberg R.-P., Bange F.-C., Ebadi E. Antimicrob. Resist. Infect. Control. 2018;7:1. doi: 10.1186/s13756-017-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni S., Haselow D., Lloyd S., Lockhart S., Moulton-Meissner H., Lester L., Wheeler G., Gladden L., Garner K., Derado G., Park B., Harris J. R. Emerging Infect. Dis. 2015;21:1719–1724. doi: 10.3201/eid2110.150249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong C., Eng S.-A., Lim M.-P., Nathan S. Front. Microbiol. 2016;7:1956. doi: 10.3389/fmicb.2016.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Yang J. M., Utsumi R., Okamoto T., Kitayama T., Hait W. N. Mol. Pharmacol. 2004;66:460–467. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- Liu L., Huang P., Wang Z., Chen N., Tang C., Lin Z., Peng P. BMC Cancer. 2016;16:813–820. doi: 10.1186/s12885-016-2853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry A. E., Adachi M., Nosyreva E., Na E. S., Los M. F., Cheng P., Kavalali E. T., Monteggia L. M. Nature. 2011;475:91–95. doi: 10.1038/nature10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Yang Q., Sun L., Liu S. J. Neuropharmacology. 2016;101:531–537. doi: 10.1016/j.neuropharm.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.