 6-Thiopurine (6TP) is a potent cytotoxic agent that is a clinically prescribed anti-metabolite employed in the treatment of numerous blood cancers since 1952.
6-Thiopurine (6TP) is a potent cytotoxic agent that is a clinically prescribed anti-metabolite employed in the treatment of numerous blood cancers since 1952.
Abstract
6-Thiopurine (6TP) is a potent cytotoxic agent that is a clinically prescribed anti-metabolite employed in the treatment of numerous blood cancers since 1952. However, its reported severe toxicities limit its general usage in the clinic. We previously have undertaken investigations into identifying the mode of toxicity for 6TP, and have found that the oxidative metabolites of 6TP, specifically 6-thiouric acid (6TU), is responsible for the in vitro inhibition of UDP-glucose dehydrogenase (UDPGDH) in a UV-vis method. In this method, inhibition was quantified through the quantification of NADH production, however, purines absorb at the same wavelength and thereby can interfere with the NADH detection. Herein, we report a HPLC method that allows for direct quantification of UDP-glucuronic acid, product from UDPGDH, for the assessment of inhibition towards UDPGDH with no interference from purines. In this method it was revealed that 6TP possesses a greater inhibitory properties than previously observed; 111 vs. 288 μM. Building upon the data collected from a previously performed rat hepatocyte study, which correlated our in vitro to in vivo inhibition theories about UDPGDH, we have developed a bio-mimic in vitro assay to aid in the inhibitory assessment of 6TP and analogs. In our efforts to expand the use of 6TP, and analogs constructed, our laboratory has undertaken a screening campaign to identify small molecule potentiators that work in synergy with 6TP in other types of cancers. Three chalcone-based compounds have been discovered through our total synthesis campaign of uvaretin, and it has been found that 11c has strong synergism with 6TP in the pancreatic cancer cell line MIA PaCa-2. Through the work presented herein, we reveal new methods to assess toxicity of 6TP and future analogs and new small molecules that work in synergy to expand the therapeutic applications of this neglected cytotoxic agent.
1. Introduction
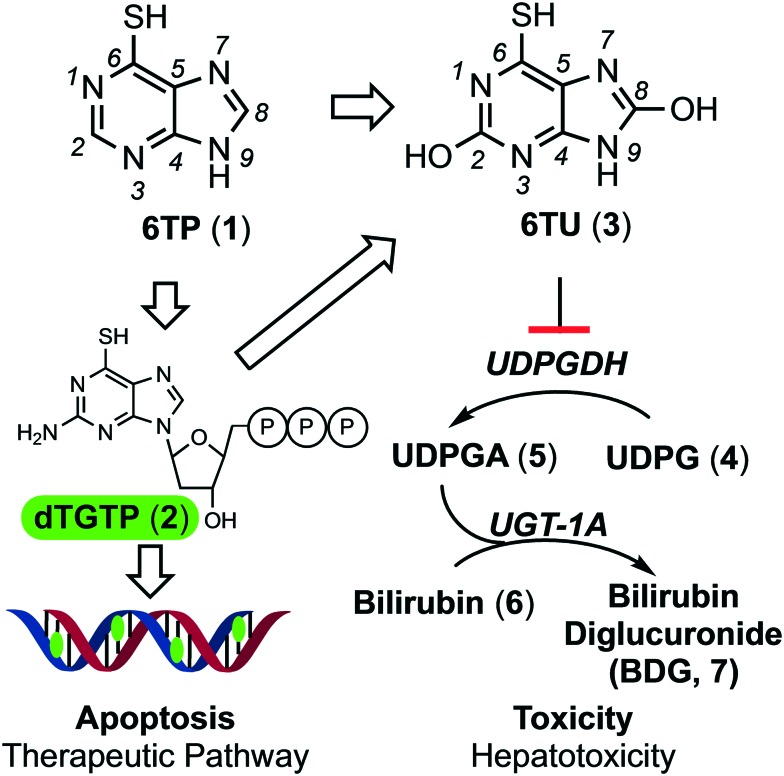
6-Thiopurine (6TP, 1, Fig. 1) has been a continually prescribed anticancer agent since 1952, two years following its discovery by Gertrude Elion and George Hitchings at Burroughs Wellcome Laboratories in 1950.1 This agent serves as a treatment option for numerous diseases, such as, but not limited to: acute lymphocytic leukemia (ALL),2,3 non-Hodgkin's leukemia,4,5 Crohn's disease,6–8 and inflammatory bowel disease.9,10 The therapeutic mode of action for 6TP comes from its biochemical transformation into deoxythioguanosine triphosphate (2), which is subsequently incorporated into DNA as a dGMP mimic that results in the cell undergoing apoptosis upon checkpoint activation.11–14 Numerous enzymatic transformations are required to convert 6TP into 2, some of the thiol methylated intermediate species serves as checkpoint activator.15–18 In addition to 6TPs therapeutic pathway, an excretion pathway is occurring simultaneously. Xanthine oxidase (XO) oxidizes both the C2 and C8 positions of 6TP giving rise to 6-thiouric acid (6TU, 3), an excretable species, however 3 is retained by the body beyond 24 hours post-6TP treatment.19 Many of the intermediates within the therapeutic pathway are also susceptible to enzymatic transformations/oxidations leading to the formation of additional 6TU. As such, the prescribed dosage of 6TP is increased to compensate for the loss from such transformations.
Fig. 1. Illustration of the therapeutic and excretion, leading to reported off-target toxicities, pathways of 6-thiopurine. Atom number has been included on 6TP and 6TU for reference.
While 6TP has been used in the treatment of leukemia for over 60 years, its general application has been on the decline due to toxicities associated with its use. The most predominate toxic side effects are jaundice and hepatotoxicity, corresponding to the reported increase in bilirubin (6) levels within patients.13,16 The bilirubin pathway is responsible for the excretion of bilirubin, post red-blood cell senescence.20 Excretion of this non-polar species is achieved through the conjugation of two UDP-glucuronic acids (UDPGA, 5) by UDP-glucuronosyltransferase (UGT-1A), forming an excretable polar species. UDPGA is formed from the oxidation of UDP-glucose (UDPG, 4) by UDP-glucose dehydrogenase (UDPGDH).21,22 While 6TP is a currently prescribed antileukemic agent, its toxic side effects are so potent and prevalent that the administration of 6TP is given in an on/off strategy, allowing time for the toxic species to be cleared by the body. Unfortunately, this greatly reduces its therapeutic efficacy, restricts the quality of life of the patients taking the drug, and has ultimately limited its use as an anticancer therapy.
Our laboratory is interested in investigating drugs that either have been removed from clinical use due to fatal toxicities or ones currently used in limited applications due to associated toxicities, such as 6TP. To accomplish this, we investigate to identify the target that results in the reported toxicity from the agent and then explore for possible substrate to target correlations. Analogs are constructed based upon these observations in the aims of reducing the reported toxicity, so that the agent can be returned to clinical use, and if possible, in new expanded therapeutic capacities. For 6TP, we have previously disclosed our investigations into the role that the excretion metabolites, oxidized at either or both the C2/C8 positions, have upon UDPGDH.23 From this, we have hypothesized that the hepatotoxicity, and its related side effects, caused by 6TP administration arises from inhibition of UDPGDH that prevent proper bilirubin conjugation and excretion. Our laboratory has shown that 6TP has no significant inhibition towards UDPGDH with a Ki of 288 μM, however, its main excretion metabolite 6TU has a 41-fold increase in inhibition with a Ki of 7 μM. The method that was developed to assess inhibition of UDPGDH by these excretion metabolites is a UV/vis based procedure in which the rate of enzymatic turn-over was measured by NADH production.
Based upon our mode of toxicity onset investigations, in which the C8 position was shown to play a vital role in UDPGDH inhibition, C8-halogen bearing analogs of 6TP were synthesized.24 Inhibition towards UDPGDH was greatly reduced with these C8-analogs, and furthermore, they were shown to retain cytotoxicity towards the acute lymphocytic leukemia cell line Reh. In our continuing efforts to redesign 6TP into a form with reduced toxicity, while retaining its therapeutic mode of action, further enhanced methods to assess 6TP and any analogs constructed is required. In these efforts, a more accurate correlation of the inhibitory properties of 6TP, excretion metabolites, and analogs constructed is necessary. However, our previous method for inhibition assessment has a critical flaw, purines interfere with the NADH signal quantification. As such, this requires the development of a new method of inhibition assessment that does not depend upon NADH quantification. Herein is disclosed our HPLC method that was developed to assess the inhibitory properties of these species via direct quantification of UDPGA, in which purines possess no interfering signals. Additionally, the construction of a bio-mimic system, based upon previously findings from our rat hepatocyte study, has been accomplished. From these new models that assess toxicity of 6TP, new avenues of investigation for newly synthesized analogs can be performed. Lastly, research has also been undertaken, and disclosed in this work, in the discovery of small molecules that work in synergy with 6TP to expands is therapeutic usefulness to pancreatic cancer.
2. Results and discussion
2.1. Development of an HPLC method for direct UDPGA quantification for UDPGDH inhibition assessment
Our laboratory has previously reported the inhibitory effects of the excretion metabolites of 6TP, in which 6TU was found to be the most inhibitory, towards UDPGDH through a UV-vis based method through NADH quantification. While a minimal concentration of the purines were employed in this method, there was interfering signals between the purine and NADH via absorption quantification. The development of a new method to assess inhibition, in which the interfering signal of the purines cannot bias absorption, was undertaken to further corroborate our initial findings of the effects of the hydroxylated C2/8 positions towards UDPGDH. To assess the inhibitory effects of the excretion metabolites upon UDPGDH, a HPLC method was developed that allows for baseline separation of the substrates (UDPG and NAD+) and products (UDPGA and NADH). Baseline separation and column regeneration, allowing for reproducible back-to-back runs, is accomplished over a 23 minute period with a dual-solvent gradient method (HPLC chromatogram and mobile phase gradient profile, and the UDPGA standard curve constructed is presented in the ESI†). This HPLC method requires no derivatization of UDPGA, nor multiple mobile phases as previously employed in other HPLC methodologies.25–27
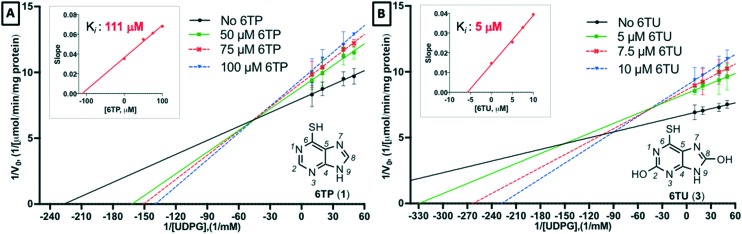
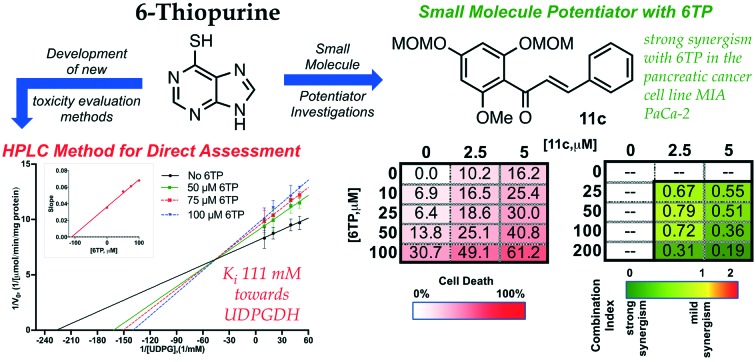
With an enhanced HPLC method developed for the quantification of UDPGDH activity by monitoring UDPGA formation established, efforts were directed towards assessing inhibition of 6TP and 6TU. Following the protocol previously employed by our laboratory,23 kinetic studies were performed against UDPGDH with four varying concentrations of 6TP and 6TU employed, and the determination of Ki values was accomplished by plotting the slope of from each independent analysis set verses the concentration of the purine; calculations of the Ki was performed by taking the negative-inverse of the x-intercept. The Ki for 6TU was found to be 5 μM via direct analysis, showing only a marginal difference of the 7 μM observed in our in-direct method of assessment (Fig. 2B). However, 6TP was shown to have considerably more inhibitory properties towards UDPGDH when assessed in this direct method. The Ki for 6TP via direct assessment was found to be 111 μM, where previously we reported 288 μM through our in-direct NADH detection method (Fig. 2A).
Fig. 2. Assessment of inhibition of UDP-glucose dehydrogenase by 6TP and 6TU via direct quantification of UDP-glucuronic acid by HPLC separation and quantification. A) Concentration of 6TP, varying UDPG, screened were 0, 50, 75, 100 μM with the calculated slopes of each line equaling 0.0353, 0.0549, 0.0612, 0.0680, respectively. Plotting slopes versus concentration gave a regression line of y = 0.000328x + 0.0364 with a 0.9881 R-square. B) Concentration of 6TU, varying UDPG, screened were 0, 5, 7.5, 10 μM with the calculated slopes of each line equaling 0.0147, 0.0254, 0.0328, 0.0394, respectively. Plotting slopes versus concentration gave a regression line of y = 0.002474x + 0.01416 with a 0.9946 R-square. Each data point for Lineweaver Burk plots are an average of three independent runs with standard error bars shown.
Through the development of a non-interfering method to assess the inhibition of UDPGDH by 6TP and 6TU, it was revealed that 6TP has a two-fold greater inhibitory effect than previously reported. No significant change in the Ki for 6TU was observed in this method. We have previously reported our hypothesis that 6TU, as well as 8-OH-6TP are the main excretion metabolites of 6TP responsible for UDPGDH inhibition, which correlates to the reported toxic side effects from 6TP administration. While the current investigation has revealed a Ki of 111 μM for 6TP towards UDPGDH, levels that could be potentially impact enzymatic activity, we believe that our hypothesis of inhibition by C8-hydroxylated species remains unaffected and that inhibition of UDPGDH by 6TP is not significantly impairing for multiple reasons, such as: 1) the oxidation of 6TP to 6TU by xanthine oxidase is rapid, 2) the inhibition by 6TP is 21-folds lower than 6TU, and 3) the biotransformation into its therapeutic nucleotide mimic species occurs concurrently to oxidation by xanthine oxidase thereby leaving a lower available pool of 6TP to elicit any inhibition towards UDPGDH.
2.2. Generation of an in vitro bio-mimic UDPGDH/UGT-1A evaluation system
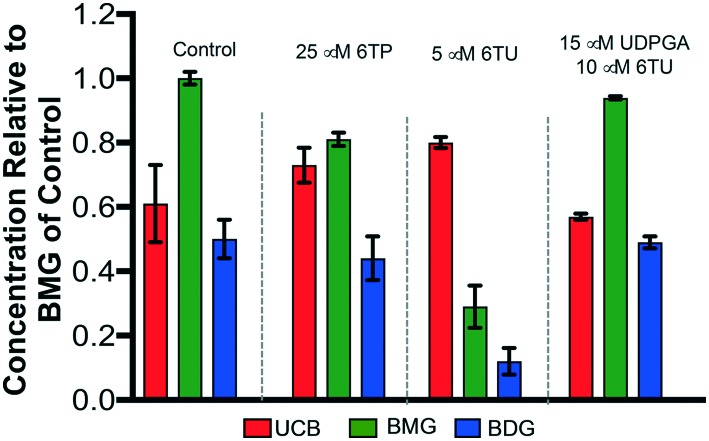
Recently, we reported a hepatocyte study that allowed for in vivo corroboration of our in vitro data regarding 6TP and 6TU inhibition of UDPGDH and UGT-1A. Highlights of the overall findings from this hepatocyte investigation are presented in Fig. 3.24 The treatment of 6TP resulted in a marginally lower level of BDG (bilirubin diglucuronide) formation, which we can now correlate to the recently observed greater inhibitory property of 6TP towards UDPGDH. This also results in a lower liable pool of UDPGA and the observed BMG (bilirubin monoglucuronide) depression. 6TU was shown to have a larger impact upon the formation levels of BDG, through the proposed inhibition of UDPGDH. Validation of this UDPGDH inhibition claim was revealed when external UDPGA was added to a system in the presence of 6TU, in which levels of BMG formation returned to those of the control group.
Fig. 3. Overview of the performed hepatocyte studies to correlate 6TP and 6TU inhibition of UDPGDH. Each group is an average of three independent runs with standard error bars shown. UCB: unconjugated bilirubin (free), BMG: bilirubin diglucuronide (one UDPGA attached), BDG: bilirubin diglucuronide.
In alignment with the overall objective of decreasing the toxic response from 6TP, while retaining its known therapeutic mode of action, efforts towards analog construction are underway. Evaluation of inhibition towards UDPGDH and UGT-1A by all synthesized analogs will be required. While we have previously successfully employed a hepatocyte model to confirm our working hypothesis of inhibition, the need for a more readily accessible toxicity assessment model is essential. Cellular levels of UDPGDH vary, and as such the development of a physiological model can be difficult.28,29 As such, the in vitro model was chosen to mimic the result from our hepatocyte in vivo investigations. Our bio-mimic in vitro model system was based upon the buffer composition employed in our UGT-1A assay, a phosphate buffer system, containing bilirubin and alamethicin (inhibitor of glucuronidase, present within the microsomes employed in this study).23 In addition, both NAD+ and UDPG were included to allow for the formation of UDPGA, which can be used to assess BDG formation.
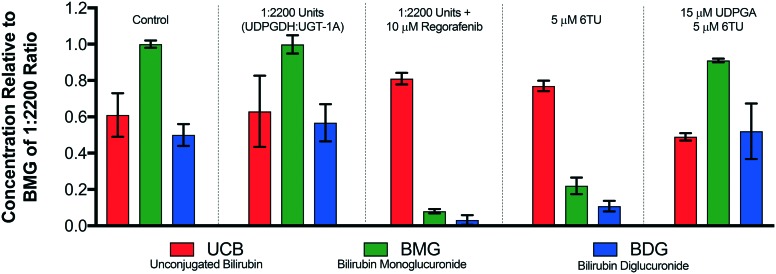
Efforts in the development of a bio-mimic in vitro system, to mimic the observed trends of the hepatocyte study (Fig. 3), commenced with investigations into determining the ratio of UDPGDH (6.8 units per mL) and microsomes (containing 2.2 units per mL of UGT-1A) was then undertaken. We decided to use 4.4 units per mL of UGT-1A in the presence of 2 mM bilirubin along with 10 mM NAD+ and 5 mM UDPG in a final volume of 500 μL as this mimicked our reported UGT-1A assay.23 The addition of a UDPGDH solution marked the start of the reaction. Employing 10 μM regorafenib, a UGT-1A inhibitor, 45 minutes into the enzymatic incubation will allow assessment to determine if the ratio of the two enzymes matches the hepatocyte model. The same concentration of regorafenib resulted in depression of BMG and BDG formation, not complete inhibition. As such, if the ratio of UDPGDH to microsomes is too high, the enhanced formation of UDPGA will complete with regorafenib resulting in bilirubin conjugation. If the levels are UDPGA are too low, this will result in a near complete inhibition of BDG and BMG formation post regorafenib treatment. Employing a 1 : 1 ratio of UDPGDH to UGT-1A resulted in a saturating amount of UDPGA formation, as did a 1 : 400, 1 : 800, and 1 : 1200 ratio (results are presented in the ESI†). Presented in Fig. 4 is the control group set from the hepatocyte study (Fig. 3). Employing a 1 : 2200 ratio of UDPGDH to UGT-1A afford a similar distribution of UCB, BMG, and BDG to the control set. When 10 μM of regorafenib was employed, the suppression of both BMG and BDG formation observed matched satisfactorily to the same investigational parameters within the hepatocyte study. This supports that the in vitro model mimics the hepatocyte model towards UGT-1A. Employing 5 μM of 6TU showed a decrease in the production of BMG and BDG; however not to the same extent as regorafenib. This result was expected, since 6TU is inhibiting UDPGDH and thereby limiting the amount of UDPGA formed, which corresponds to the decreased production of both BMG and BDG. To fully confirm that UDPGDH is inhibited in this in vitro model, subjecting the system to 5 μM of 6TU in the presence of 15 μM of UDPGA showed recovery of the system back to control levels. Employing a 1 : 2200 ratio of UDPGDH to microsomes has shown to provide data consistent with our hepatocyte model, and corrects correlates 6TU inhibition of UDPGDH and not UGT-1A.
Fig. 4. Development of an in vitro model to assess UDPGDH and UGT-1A inhibition. Each group is an average of three independent runs with standard error bars shown.
2.3. Expansion of 6-thiopurine therapeutic applications
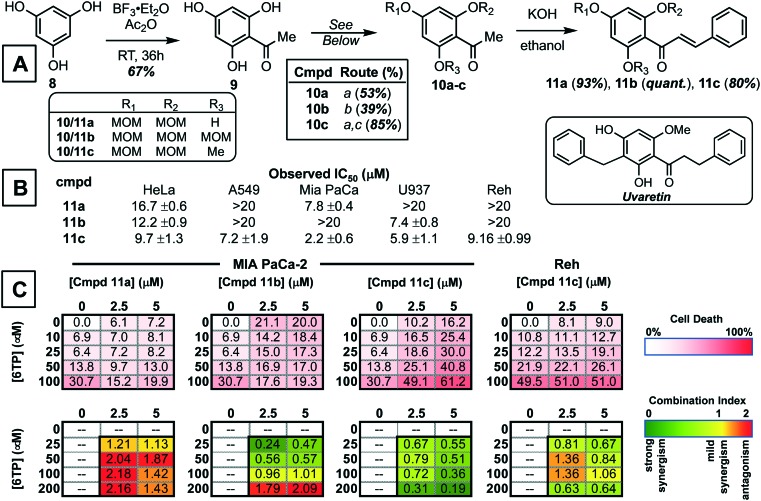
With the development of new methods to assess the inhibitory properties of any 6TP analogs constructed, three of which have already been reported by our laboratory,24 we turned our attention to expanding the therapeutic application of 6TP, and analogs. To accomplish this, we propose to investigate the effects of small molecules upon 6TP when used in combination, also referred to as small molecule potentiation. The action in which small molecules work in synergy with chemotherapeutic agents is variable. In some cases they help attenuate activity by modulating a key biochemical pathway in which the agents elicits is therapeutic action, and in others they aid in the transport of the chemotherapeutic agent, to mention a few possible modes of action. Discovering small molecules with the ability to potentiate the activity of chemotherapeutic agents is a difficult endeavor, most times their discovery is quite serendipitous. Our efforts to discover such small molecules lies within the intermediates accessed from total synthesis routes. We proposed that intermediates, whether fruitful or not towards the targeted natural product, can be used in general screens to determine if they have sole cytotoxic properties or possibly possess potentiating properties towards current chemotherapeutic agents, such as 6TP. Our efforts towards the total synthesis of uvaretin, shown in the insert of Fig. 5A, has allowed for the access of numerous small molecules possessing interesting and diverse biological activity. Illustrated in Fig. 5A is our basic synthetic route employed to gain access to the core structure of uvaretin. Starting from phloroglucinol (8), acetylation was performed under Lewis-acid conditions and acetic anhydride to afford the corresponding aromatic ketone (9). Details on the elaboration of 9 onto its three corresponding phenol protected counterparts 10a–c is presented within the ESI.† While numerous groups were attempted in route that accessed uvaretin, the methoxymethyl (MOM) group is highlighted in this work for the unique properties associated with the final chalcone product. Subjecting 10a–c to aldol condensation condition with benzaldehyde gave access to chalcones 11a–c.
Fig. 5. Synthesis of chalcone small molecules and investigations into possible synergy with 6-thiopurine for either increased cytotoxicity or expansion of therapeutic application scope. A) General synthesis route employed to gain access to the three chalcones 11a–c. Synthetic procedures: a. MOMCl (2.2 eq.), DIPEA, CH2Cl2, b. MOMCl (3.3 eq.), DIPEA, CH2Cl2, and c. Me2SO4, K2CO3, acetone. B) General cytotoxicity of chalcones 11a–c in HeLa (cervical), A549 (lung), MIA PaCa-2 (pancreatic), U937 (lymphoma), and Reh (acute lymphocyte leukemia) cancerous cell lines. C) Results of the average cell death induced by 11c & 6-thiopurine combinations evaluated in the 3 × 5 matrix and corresponding quantification of synergy with CI values (n = three biologic replicates).
Evaluation of 11a–c was then performed in a panel of cancerous cell lines: HeLa (cervical), A549 (lung), MIA PaCa-2 (pancreatic), U937 (lymphoma), and Reh (acute lymphocytic leukemia, ALL). Of these three chalcones, it was found that 11c possessed the most potent cytotoxicity towards the cell lines investigated as a sole agent (Fig. 5B). While 11a–b were observed to have lower potency in comparison to 11c, potentiating studies was performed with 6-thiopurine with all three chalcones. The assessment of the combinational properties of 11a–c with 6TP in various cancerous cell lines was performed in a matrix format (3 × 5 matrix) against five cancer types (HeLa-cervical, A549-lung, MIA PaCa-pancreatic, U937-lymphoma, and Reh-acute lymphocytic leukemia). The matrix was designed so that two concentrations of 11a–c would induce cell death between 10 and 20% within a 72 hour treatment, and the concentrations of 6TP would result in up to 50% cell death over the same time period. To quantity the effects of the two drugs in combination, as well as the combination index (CI) the Chou–Talalay method was employed.30 CI values were calculated for each combination, this resulted in 10 CI values per cell line investigated. Quantification of the combination experiments is shown in Fig. 5C, with the median CI values shown for each of the 6TP and 11a–c combinations. CI values of <1 indicate a strong synergy, with lower values possessing stronger synergy. Synergy was observed between 11c and 6TP in the pancreatic cell line MIA PaCa-2, giving low CI values. Given that 6TP has no reported toxicity within pancreatic cancer cell lines, this finding illustrates how the expansion of 6TP application can be accomplished via small molecule potentiation. Unexpectedly, no synergism was observed between 11a–b and 6TP in MIA PaCa-2 as observed with high CI values. Evaluation of potential synergism between 11c and 6TP in Reh, the cancer type that 6TP is prescribed for, showed mild synergy. No synergy was observed for 11a–b in Reh as well as for 11a–c in U937, A549, and HeLa cancerous cell lines.
Of the three chalcones synthesized, it was found that 11c possesses the best synergy with 6TP within the pancreatic cancer cell line MIA PaCa-2. Investigations are currently underway to elucidated the mode in which 11c potentiates the activity of 6TP. It is anticipated that these studies will give insight into how 11c works with 6TP in MIA PaCa-2, and also why 11c does not possess the same synergism with 6TP within the acute lymphocytic leukemia cell line Reh, the primary cancer type that 6TP is prescribed to treat. The discovery of small molecule potentiators is difficult, as the activity can differ between cancer types, as observed here with MIA PaCa-2 and Reh cell lines. We have shown that through the use of intermediates accessed within total synthesis campaigns that the discovery of such small molecules is possible. Furthermore, it has allowed for the discovery of 11c that expands the use of 6TP beyond acute lymphocytic leukemia to pancreatic cancers.
3. Conclusion
6-Thiopurine has been an approved chemotherapeutic drug since 1952, and is currently primarily used in the treatment of acute lymphocytic leukemia, in addition to other diseases. Unfortunately, its clinical application is limited due to reported toxicities associated with its application. Our laboratory has undertaken investigations into elucidating the mode of toxicity of 6TP in the aims of constructing new analogs with decreased, if not eliminated, toxicities. In addition, through screening campaigns of small molecules synthesized through our total synthesis campaigns, we seek to discover small molecules that work in synergy with 6TP to either enhance its activity or to expand the scope of therapeutic application. Through the new HPLC method developed for the assessment of UDPGDH inhibition, 6TP was shown to possess a two-fold greater inhibitory profile in compared to our previous method. Building upon this new method to assess the inhibition of UDPGDH, the found target of 6TP and its oxidative metabolites, and based upon a previously employed hepatocyte study, the construction of an in vitro bio-mimic inhibition model was undertaken. In this method, we are able to assess the inhibition of 6TP and 6TU towards UDPGDH and/or UGT-1A. Inhibitory assessment of 6TP analogs synthesized can now be done in a rapid and efficient process through this bio-mimic model system developed. Investigations into expanding the scope of 6TP application was successfully accomplished through the combinational screening of chalcones 11a–c with 6TP in the pancreatic (MIA PaCa-2) and acute lymphocytic leukemia (Reh) cancerous cell lines. Chalcone 11c was found to possess strong synergism with 6TP in the MIA PaCa-2, but not ReH. The enhanced cytotoxicity observed with the combination treatment of 6TP and 11c in MIA PaCa-2 relative to 6TP alone in the same cell line reveals an expansion of the application scope of 6TP to pancreatic cancer. Efforts are currently underway to explore the mode of action between 11c and 6TP and how it translated to this new synergist therapeutic effect. In summary, the development of new methods to assess inhibition about UDPGDH, as well as UGT-1A, has been performed and shown to be comparable to in vivo studies previously conducted. Furthermore, investigations into the discovery of small molecules to either potentiate the activity of 6TP or expand its therapeutic usefulness was performed and 11c was found to expand 6TP application into the pancreatic cancerous cell line MIA PaCa-2.
4. Experimental section
4.1. Reagents and equipment
All standard chemicals used in this study were the highest grades available and were purchased through Sigma-Aldrich (Saint Louis, MO, USA), VWR (Radnor, PA, USA), or Fisher Scientific (Denver, CO, USA). Specialized reagents were purchased through specific vendors. Glycylglycine (gly-gly: G1127), β-nicotinamide adenine dinucleotide (NAD+: N1636), uridine 5′-diphosphoglucose (UDPG disodium salt: U4625), uridine 5′-diphosophoglucuronic acid (UDPGA: U6751), uridine 5′-diphosphoglucose dehydrogenases (UDPGDH: U6885), 6-thiopurine monohydrate (6TP), bilirubin (including three mixed isomers, B4126), 4,5-diamino-6-hydroxypyrimidine hemisulfate salt (D19303), alamethicin (A5361), and pooled rat liver microsomes (M9066) were purchased from Sigma-Aldrich. 6-Mercaptopurine-2-ol (6-TX, QA-6668) was purchased from Combi-Blocks, and 6-thiouric acid (6-TU, SC-213040) from Santa Cruz Biotechnology. HPLC-grade water was obtained by passing distilled water through a reverse osmosis system followed by treatment with a Thermo Scientific Barnstead Smart2Pure 3UV purification system (Fisher, 10-451-045), herein referred to as nanopure water.
The cancerous cell line used in this investigation were purchased directly from the American Type Culture Collection: HeLa, U-937, A549, MIA PaCa-2, and Reh. Cells were grown in media supplemented with fetal bovine serum (FBS) and antibiotics (100 μg mL–1 penicillin and 100 U mL–1 streptomycin). Cells were incubated at 37 °C in a 5% CO2, 95% humidity atmosphere. Cellular viability was determined by quantification via Alamar blue.
All standard consumable supplies used in this study were purchased from VWR or Fisher Scientific. Specific equipment utilized in this work are: 1) Hewlett-Packard 8452 diode array UV/vis spectrophotometer (Palo Alto, CA, USA) equipped with a Lauda Brinkmann Ecoline RE 106 E100 circulating water bath purchased from VWR, 2) HPLC system consisting of an CBM-20A/20Alite system controller, SIL-20AHT auto sampler, SPD-20A, SPD-20AV UV-vis detector, LC-20AT Solvent delivery module, CTO-20A column oven, DGU-20A3R degassing unit and LC-20AD/20AT Gradient Valve Kit purchased from Shimadzu Scientific Instruments (Kyoto, Japan), and 3) all incubated reactions were performed with a Labcare America PRECISION water bath model 25 purchased from Fisher Scientific. All HPLC separations were performed on a Discovery C18 analytical column, 4.6 mm × 100 mm, 5 μm particle size (504955-30) along with the respective guard column (59576) purchased from Sigma-Aldrich. Data was processed and all figures and tables constructed via the program Prism 7.02 for Mac, GraphPad Software (La Jolla, CA, USA). All chemical structures were prepared with ChemDraw Professional 16.0 by PerkinElmer (Waltham, MA). All statistical calculations within this body of work was performed by the treatment of two-way factorials (positive and negative controls, design structure of RCBD, and T-tests) with Statistical Analysis System (SAS) software for Windows (Cary, NC, USA).
4.2. Biology
4.2.1. HPLC UDPGDH method for UDPGA detection and quantification
The substrates (UDP-glucose and NAD+) and products (UDPGA and NADH) of the enzymatic reaction carried out by UDPGDH was separated on a Discovery C18 analytical column, 4.5 mm × 100 mm, 5 μM particle size with guard column. A dual mobile phase was employed; the aqueous phase consisted of an 8 mM imidazole & 2.5 mM tetrabutylammonium hydrogen sulfate (TBAHS) buffer at a pH of 6.5 in nanopure water and methanol as the organic phase. A gradient elution profile was employed for full separation at a flow rate of 0.5 mL min–1, the method starts with a 90 : 10 (aqueous–organic percentage) mobile phase composition that is held for three-minutes, changed to 40 : 60 (aqueous–organic percentage) over one-minute, held for six-minutes, and then returned to initial composition over 2.7 minutes and held for 10 minutes to allow for column regeneration. The detection wavelength was 262 nm with a sample injection volume of 5 μL. Various concentrations of UDGPA were analyzed, and peak areas obtained were plotted relative to said concentrations to generate a working standard curve that was used to assess inhibition of UDPGDH by 6TP and 6TU as a function of UDPGA production.
4.2.2. UDP-glucose dehydrogenase incubation
All protocols used for the formation of UDPGA via the enzymatic conversion by UDPGDH, and for purine inhibition, was performed by previously published procedures by our laboratory.23
4.2.3. UDP-glucuronosyltransferase activity assay
All protocols used for the quantification of UCB, BMG, BDG via the enzymatic conversion by UGT-1A, and for purine inhibition, was performed by previously published procedures by our laboratory.23,24
4.2.4. General cell death assessment via Alamar blue quantification
General procedure for single agent IC50 determination
Compounds were solubilized in DMSO (10 μM stock solutions) and added to a 96-well plate over a range of concentrations (31.6 nM to 200 μM) with media, and 40 μL was added to the 384-well plate in triplicate for each concentration of compound. Cells were then added to the plate (2000 cells per well for adherent and 3500 cells per well for suspension cells) in 10 μL of media. After 69 h of continuous exposure, 5 μL of Alamar Blue was added to each well, and the cells were allowed to incubate for an additional 3 h. The plates were then read for fluorescence intensity with an excitation of 560 nm and emission of 590 nm on a BioTek Synergy H1 plate reader. Doxorubicin and etoposide were both used as positive death controls, and wells with no compounds added as negative death controls. IC50 values were determined from three or more independent experiments using GraphPad Prism 7.0. (LaJolla, CA, USA).
Combination studies
All combinational cell death experiments were performed in 96-well plates with a total volume of 100 μL. To each well was added 49 μL of cell media, either 2.5 or 5 μM of 6TP (from a DMSO stock solution), and compounds 11a–c, independently. To each well on the plate was added two different concentration of 11a–c and five concentrations of 6TP (both prepared from 10 mM stock solutions in DMSO), cell media (adjusted to reach 50 μL volume) and 0.5 μL of DMSO to achieve a 1% DMSO concentration. To each well was then added 50 μL of a suspension of cells to obtain a final cell density of 4000 cells per well (adherent cells) and 7000 cells per well (suspension cells). Doxorubicin and etoposide were both used as positive death controls, and wells with no compounds added as negative death controls. Plates were incubated at 37 °C with 5% CO2 for 72 h, at which time they were assessed by Alamar blue. IC50 values were determined from three or more independent experiments using GraphPad Prism 7.0.
4.3. Chemistry
All reagents were commercially available and used without purification unless otherwise stated. NMR spectra were recorded with a Varian 400 MHz instrument. The chemical shifts are given in parts per million (ppm) relative to residual CHCl3 at δ 7.26 ppm or DMSO δ 2.50 ppm for proton spectra and relative to CDCl3 at δ 77.23 ppm or DMSO δ 39.52 ppm for carbon spectra, unless otherwise noted. Flash column chromatography was performed with silica gel grade 60 (230–400 mesh). Dichloromethane (CH2Cl2), and acetone were degassed with argon and passed through a solvent purification system containing alumina or molecular sieves. All commercially available reagents were used as received. All procedures including anhydrous solvents were performed with rigorously dried glassware under inert atmosphere.
4.3.1. 1-(2,4,6-Trihydroxyphenyl)ethan-1-one (9)
To a flame dried round bottom (RBF) under an argon atmosphere was added boron trifluoride etherate (8.81 mL, 71.37 mmol, 3 eq.), followed by phloroglucinol (8) (3.0 g, 23.79 mmol, 1 eq.), and acetic anhydride (2.25 mL, 23.79 mmol, 1 eq.). The solution was allowed to stir at room temperature for 16 h, at which time the reaction was quenched with the addition of a 10% sodium acetate solution (10× volume) and allowed to stir for an additional 24 h. The solution was filtered, the precipitate washed with washed to obtain 2,4,6-trihydroxyacetophenone (9) as a yellow solid in 67% yield (2.68 g). 1H NMR (400 MHz, methanol-d4) δ 5.79 (s, 2H), 2.59 (m, 3H). 13C NMR (101 MHz, methanol-d4) δ 204.56, 166.32, 165.90, 105.60, 95.59, 32.71.
4.3.2. 1-(2-Hydroxy-4,6-bis(methoxymethoxy)phenyl)ethan-1-one (10a)
To a RBF under an argon atmosphere was added 9 (500 mg, 2.97 mmol, 1 eq.) and 15 mL of CH2Cl2, which was then cooled to 0 °C and then diisopropylethylamine (1.6 mL, 8.91 mmol, 3 eq.) and MOMCl (0.5 mL, 6.5 mmol, 2.2 eq.) was added. The solution was then stirred overnight, allowed to warm to room temperature, and then quenched with methanol and washed with brine (×2). The combined organic layers were dried over anhydrous sodium sulfate, concentrated, and purified via flash silica gel chromatography (4 : 1, Hex : EtOAc) to afford 10a in 53% yield (406 mg). 1H NMR (400 MHz, chloroform-d) δ 13.70 (s, 1H), 6.24 (1H), 6.22 (s, 1H), 5.24 (s, 2H), 5.15 (s, 2H), 3.50 (s, 3H), 3.45 (s, 3H), 2.63 (s, 3H). 13C NMR (101 MHz, chloroform-d) δ 203.40, 167.02, 163.66, 160.57, 107.12, 97.32, 94.69, 94.21, 56.91, 56.64, 33.21. HRMS calcd. for C12H16ONa+m/z [M + Na]+ 279.0845; found 279.0842.
4.3.3. 1-(2,4,6-Tris(methoxymethoxy)phenyl)ethan-1-one (10b)
To a RBF under an argon atmosphere was added 9 (250 mg, 1.49 mmol, 1 eq.) and 8 mL of CH2Cl2, which was then cooled to 0 °C and then diisopropylethylamine (1.3 mL, 7.95 mmol, 5 eq.) and MOMCl (0.37 mL, 4.91 mmol, 3.3 eq.) was added. The solution was then stirred overnight, allowed to warm to room temperature, and then quenched with methanol and washed with brine (×2). The combined organic layers were dried over anhydrous sodium sulfate, concentrated, and purified via flash silica gel chromatography (4 : 1, Hex : EtOAc) to afford 10b in 39% yield (173 mg). 1H NMR (400 MHz, chloroform-d) δ 6.50 (s, 2H), 5.13 (s, 2H), 5.13 (s, 4H), 3.46 (s, 3H), 3.45 (s, 6H), 2.48 (s, 3H). 13C NMR (101 MHz, chloroform-d) δ 201.65, 159.68, 155.41, 117.16, 97.34, 94.99, 94.69, 56.55, 56.44, 56.34, 32.76. HRMS calcd. for C14H20O7Na+m/z [M + Na]+ 323.1107; found 323.1112.
4.3.4. 1-(2-Methoxy-4,6-bis(methoxymethoxy)phenyl)ethan-1-one (10c)
To a dried RBF under an argon atmosphere was added dry acetone (15 mL) to which was added 10a (400 mg, 1.56 mmol, 1 eq.) under vigorous stirring. Once a homogenous solution was obtained, potassium carbonate (1.07 g, 7.8 mmol, 5 eq.) and dimethyl sulfate (0.74 mL, 7.8 mmol, 5 eq.) were added. The reaction mixture was refluxed for 4 h, quenched with saturated ammonium chloride, and extracted with EtOAc (×2). The organic layers were combined, dried over sodium sulfate, concentrated, and purified via flash silica gel chromatography (7 : 3 Hex : EtOAc) to obtain 10c in 85% yield (358 mg). 1H NMR (400 MHz, chloroform-d) δ 6.45 (d, J = 2.0 Hz, 1H), 6.30 (d, J = 2.0 Hz, 1H), 5.15 (s, 2H), 5.13 (s, 2H), 3.78 (s, 3H), 3.47 (s, 3H), 3.45 (s, 3H), 2.47 (s, 3H). 13C NMR (101 MHz, chloroform-d) δ 201.84, 159.92, 158.11, 155.59, 116.07, 96.14, 95.01, 94.70, 94.08, 56.55, 56.41, 56.05, 32.72. HRMS calcd. for C13H19O6+m/z [M + H]+ 271.1182; found 271.1192.
4.3.5. (E)-1-(2-Hydroxy-4,6-bis(methoxymethoxy)phenyl)-3-phenylprop-2-en-1-one (11a)
To a RBF containing ethanol (10 mL) was added benzaldehyde (0.14 mL, 1.4 mmol, 1 eq.) and 10a (360 mg, 1.4 mmol, 1 eq.) and cooled to 0 °C. Once at temperature, KOH (1.34 g, 24 mmol, 17 eq.) dissolved in water was added to the reaction mixture and left for 16 h. The reaction was quenched with the addition of a 10% HCl solution (one volume eq.) and extracted with EtOAc (×3). The organic layers were combined, dried with sodium sulfate, and concentrated. The crude material was purified via flash silica gel chromatography (4 : 1 Hex : EtOAc) affording 11a in 93% yield (449 mg). 1H NMR (400 MHz, chloroform-d) δ 13.81 (s, 1H), 7.93 (d, J = 15.6 Hz, 1H), 7.79 (d, J = 15.7 Hz, 1H), 7.68–7.57 (m, 3H), 7.46–7.35 (m, 4H), 6.26 (d, J = 2.4 Hz, 1H), 5.29 (s, 2H), 5.19 (s, 2H), 3.51 (s, 3H), 3.50 (s, 3H). 13C NMR (101 MHz, chloroform-d) δ 193.15, 167.57, 163.74, 160.12, 142.72, 135.69, 130.40, 129.16, 128.54, 127.61, 107.76, 97.73, 95.38, 94.99, 94.30, 57.11, 56.70. HRMS calcd. for C19H21O6Na+m/z [M + Na]+ 345.1338; found 345.1354.
4.3.6. (E)-3-Phenyl-1-(2,4,6-tris(methoxymethoxy)phenyl)prop-2-en-1-one (11b)
To a RBF containing ethanol (4 mL) was added benzaldehyde (0.05 mL, 0.5 mmol, 1 eq.) and 10b (170 mg, 0.5 mmol, 1 eq.) and cooled to 0 °C. Once at temperature, KOH (480 mg, 8.48 mmol, 17 eq.) dissolved in water was added to the reaction mixture and left for 16 h. The reaction was quenched with the addition of a 10% HCl solution (one volume eq.) and extracted with EtOAc (×3). The organic layers were combined, dried with sodium sulfate, and concentrated. The crude material was purified via flash silica gel chromatography (4 : 1 Hex : EtOAc) affording 11b in a quantitative yield (195 mg). 1H NMR (400 MHz, chloroform-d) δ 7.54–7.49 (m, 2H), 7.39–7.33 (m, 4H), 6.97 (d, J = 16.2 Hz, 1H), 6.57 (s, 2H), 5.18 (s, 3H), 5.11 (s, 4H), 3.50 (s, 3H), 3.38 (s, 6H). 13C NMR (101 MHz, chloroform-d) δ 194.50, 159.88, 156.06, 145.22, 135.02, 130.61, 129.22, 129.12, 128.55, 114.99, 97.36, 94.99, 94.81, 94.79, 56.51. HRMS calcd. for C21H24O7Na+m/z [M + Na]+ 411.1420; found 411.1423.
4.3.7. (E)-1-(2-Methoxy-4,6-bis(methoxymethoxy)phenyl)-3-phenylprop-2-en-1-one (11c)
To a RBF containing ethanol (15 mL) was added benzaldehyde (0.16 mL, 1.55 mmol, 1 eq.) and 10b (350 mg, 1.29 mmol, 1 eq.) and cooled to 0 °C. Once at temperature, KOH (1.23 g, 21.9 mmol, 17 eq.) dissolved in water was added to the reaction mixture and left for 16 h. The reaction was quenched with the addition of a 10% HCl solution (one volume eq.) and extracted with EtOAc (×3). The organic layers were combined, dried with sodium sulfate, and concentrated. The crude material was purified via flash silica gel chromatography (7 : 3 Hex : EtOAc) affording 11c in a 80% yield (370 mg). 1H NMR (400 MHz, chloroform-d) δ 7.52 (dq, J = 6.5, 4.0, 3.0 Hz, 2H), 7.38–7.34 (m, 4H), 6.96 (d, J = 16.1 Hz, 1H), 6.50 (dd, J = 6.2, 2.0 Hz, 1H), 6.37 (d, J = 2.0 Hz, 1H), 5.20 (s, 1H), 5.11 (s, 1H), 3.76 (s, 3H), 3.51 (s, 3H), 3.39 (s, 3H). 13C NMR (101 MHz, chloroform-d) δ 194.56, 160.10, 158.77, 156.16, 144.91, 135.13, 130.52, 129.19, 129.07, 128.58, 96.15, 94.84, 94.80, 94.21, 56.53, 56.48, 56.16. HRMS calcd. for C20H23O6+m/z [M + H]+ 359.1494; found 359.1495.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This work could not have been undertaken without the gracious financial support from the Johnson Cancer Research Center of Kansas State University and Startup Capital from Kansas State University. Funding for this research, and for funding for Johnathan Dallman, was provided by the NSF REU program under grant number CHE-1460898.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9md00010k
References
- Hitchings G. H., Elion G. B. Pharmacol. Rev. 1963;15:365–405. [PubMed] [Google Scholar]
- Torpy J. M., Lynm C., Glass R. M. JAMA, J. Am. Med. Assoc. 2009;301:452. doi: 10.1001/jama.301.4.452. [DOI] [PubMed] [Google Scholar]
- Moriyama T., Nishii R., Lin T.-N., Kihira K., Toyoda H., Jacob N., Kato M., Koh K., Inaba H., Manabe A. Pharmacogenet. Genomics. 2017;27:236–239. doi: 10.1097/FPC.0000000000000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrik C., Bergers J., De Jong W., Steerenberg P. Cancer Chemother. Pharmacol. 1992;29:413–429. doi: 10.1007/BF00684841. [DOI] [PubMed] [Google Scholar]
- Eden O. J. Clin. Pathol. 2000;53:55–59. doi: 10.1136/jcp.53.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuffari C., Seidman E., Latour S., Theoret Y. Can. J. Physiol. Pharmacol. 1996;74:580–585. [PubMed] [Google Scholar]
- Rampton D. S. BMJ [Br. Med. J.] 1999;319:1480. doi: 10.1136/bmj.319.7223.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinen M. L., van Asseldonk D. P., de Boer N. K., Losekoot N., Smid K., Mulder C. J., Bouma G., Peters G. J. and van Bodegraven A. A., Optimalisation of conventional therapies in inflammatory bowel disease, 2016, vol. 7, pp. 812–819. [Google Scholar]
- Roblin X., Biroulet L. P., Phelip J. M., Nancey S., Flourie B. Am. J. Gastroenterol. 2008;103:3115. doi: 10.1111/j.1572-0241.2008.01743.x. [DOI] [PubMed] [Google Scholar]
- Gearry R. B., Barclay M. L. J. Gastroenterol. Hepatol. 2005;20:1149–1157. doi: 10.1111/j.1440-1746.2005.03832.x. [DOI] [PubMed] [Google Scholar]
- Nelson J. A., Carpenter J. W., Rose L. M., Adamson D. J. Cancer Res. 1975;35:2872–2878. [PubMed] [Google Scholar]
- Haglund S., Taipalensuu J., Peterson C., Almer S. Br. J. Clin. Pharmacol. 2008;65:69–77. doi: 10.1111/j.1365-2125.2007.02985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland K., Lennard L., Lilleyman J. S. Xenobiotica. 1999;29:615–628. doi: 10.1080/004982599238434. [DOI] [PubMed] [Google Scholar]
- Tidd D. M., Paterson A. R. P. Cancer Res. 1974;34:738–746. [PubMed] [Google Scholar]
- Karran P. Br. Med. Bull. 2006;79–80:153–170. doi: 10.1093/bmb/ldl020. [DOI] [PubMed] [Google Scholar]
- Karran P., Attard N. Nat. Rev. Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- Chouchana L., Fernández-Ramos A. A., Dumont F., Marchetti C., Ceballos-Picot I., Beaune P., Gurwitz D., Loriot M.-A. Genome Med. 2015;7:37. doi: 10.1186/s13073-015-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervieux T., Brenner T. L., Hon Y. Y., Zhou Y., Hancock M. L., Sandlund J. T., Rivera G. K., Ribeiro R. C., Boyett J. M., Pui C.-H., Relling M. V., Evans W. E. Blood. 2002;100:1240–1247. doi: 10.1182/blood-2002-02-0495. [DOI] [PubMed] [Google Scholar]
- Carethers J. M., Hawn M. T., Chauhan D. P., Luce M. C., Marra G., Koi M., Boland C. R. J. Clin. Invest. 1996;98:199–206. doi: 10.1172/JCI118767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxler R. and Brown S., in Bilirubin, CRC Press, 2018, pp. 1–38. [Google Scholar]
- Hullah E. A., Blaker P. A., Marinaki A. M., Escudier M. P., Sanderson J. D. J. Oral Pathol. Med. 2015;44:761–768. doi: 10.1111/jop.12274. [DOI] [PubMed] [Google Scholar]
- Siva P. P., Murali K. M., Deepak A., Murali S., Michael S., Sandhya M. Drug Metab. Lett. 2016;10:264–269. [Google Scholar]
- Weeramange C. J., Binns C. M., Chen C., Rafferty R. J. J. Pharm. Biomed. Anal. 2018;151:106–115. doi: 10.1016/j.jpba.2017.12.058. [DOI] [PubMed] [Google Scholar]
- Torres Hernandez A. X., Weeramange C., Desman P., Fatino A., Haney O., Rafferty R. J. MedChemComm. 2019;10:169–179. doi: 10.1039/c8md00463c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows R. B., Cintron C. Anal. Biochem. 1983;130:376–378. doi: 10.1016/0003-2697(83)90602-4. [DOI] [PubMed] [Google Scholar]
- Gaffney P. C., Cooper W. M. Ann. N. Y. Acad. Sci. 1954;60:478–482. doi: 10.1111/j.1749-6632.1954.tb40044.x. [DOI] [PubMed] [Google Scholar]
- Singh J., Schwarz L., Wiebel F. Biochem. J. 1980;189:369–372. doi: 10.1042/bj1890369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde A. S., Thelen A. M., Barycki J. J., Simpson M. A. J. Biol. Chem. 2013;288:35049–35057. doi: 10.1074/jbc.M113.519090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B. J., Barycki J. J., Simpson M. A. J. Biol. Chem. 2004;279:23590–23596. doi: 10.1074/jbc.M401928200. [DOI] [PubMed] [Google Scholar]
- Chou T.-C., Talalay P. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.