 The crystal structures of mithramycin and its analogue reveal how the chemistry of mithramycin shapes it for DNA binding.
The crystal structures of mithramycin and its analogue reveal how the chemistry of mithramycin shapes it for DNA binding.
Abstract
An aureolic acid natural product mithramycin (MTM) has been known for its potent antineoplastic properties. MTM inhibits cell growth by binding in the minor groove of double-stranded DNA as a dimer, in which the two molecules of MTM are coordinated to each other through a divalent metal ion. A crystal structure of an MTM analogue, MTM SA-Phe, in the active metal ion-coordinated dimeric form demonstrates how the stereochemical features of MTM define the helicity of the dimeric scaffold for its binding to a right-handed DNA double helix. We also show crystallographically and biochemically that MTM, but not MTM SA-Phe, can be inactivated by boric acid through formation of a large macrocyclic species, in which two molecules of MTM are crosslinked to each other through 3-side chain–boron–sugar intermolecular bonds. We discuss these structural and biochemical properties in the context of MTM biosynthesis and the design of MTM analogues as anticancer therapeutics.
Introduction
Mithramycin (MTM; Fig. 1) is a polyketide natural product generated by some Streptomyces bacteria. MTM is a potent antibiotic and anticancer agent, which functions by non-intercalative DNA binding resulting in inhibition of transcriptional activity of the target cell. Owing to these properties, MTM was previously used for the treatment of testicular carcinoma, Paget's disease and clinically tested against a potentially devastating bone cancer, Ewing sarcoma.1–6 The interest in MTM was recently renewed by the discovery of its potent antagonism of an oncogenic transcription factor EWS-FLI1,7 a causative agent of Ewing sarcoma. Likewise, MTM exhibited potent cytotoxicity against prostate cancer cells expressing an abnormal fusion TMPRSS2-ERG.8 Because EWS-FLI1 and TMPRSS2-ERG contain a nearly identical ETS-family DNA binding domain, MTM is thought to function by interfering with the function of this domain in these cancer cells. EWS-FLI1 and TMPRSS2-ERG are genetic aberrations present only in tumor cells; therefore, they are highly attractive targets for selective anticancer therapy. Even though preclinical studies of MTM in Ewing sarcoma xenografts in mice showed promise,7 a clinical phase I/II trial of MTM therapy demonstrated that an efficacious MTM concentration could not be achieved safely due to poor pharmacokinetic properties of MTM in human.9 Therefore, improved MTM analogues targeting Ewing sarcoma are highly desirable, and the effort to produce improved MTM analogues is underway in our and other groups.8,10 In order to develop such analogues in a rational way, we need to improve our understanding of the structure and function of this complex natural product.
Fig. 1. Chemical structures of MTM and MTM SA-Phe.
MTM and other aureolic acid natural products contain a tricyclic chromophore core with a specific dihydroxy-methoxy-oxo-pentyl side chain at C-3 and two oligosaccharide side chains appended at C-2 and C-6 (Fig. 1). The stereochemistry at C-2 and C-3 is shared by all aureolic acid molecules, whereas the identity of the sugars and their modifications differ greatly among the natural products of this group.11–14 Previous solution structures of MTM–DNA complexes determined by NMR15,16 as well as crystal structures of another aureolic acid, chromomycin A3 (ref. 17 and 18) in complex with DNA and our crystal structures of MTM analogue-DNA complexes19 revealed details of aureolic acid–DNA interactions. MTM binds DNA in the minor groove as a dimer, in which two chromophores are coordinated to each other through a divalent metal ion, likely Mg2+in vivo. The divalent metal is additionally coordinated to two water molecules, which are engaged in hydrogen bonds with the DNA recognition sequence (G/C)(G/C).20 In addition, the two chromophores are engaged in direct hydrogen bonds with the guanine bases of this sequence. The two MTM molecules in the DNA-bound dimer are oriented in an approximately antiparallel fashion, extending in the minor groove where they form an extensive network of hydrophobic interactions and hydrogen bonds with the DNA, dominated by contacts with the phosphodiester backbone. The 3-side chain of MTM protrudes nearly orthogonally outward into the solvent, which allowed us to design and synthesize 3-side chain analogues containing amino acid residue functionalities (MTM SA-Trp, MTM SA-Phe and others).8,21 We showed that these analogues exhibited similar dimerization and DNA binding properties to MTM.19,20 Not only did these analogues serve as critical molecules for obtaining MTM analogue-DNA crystals, but also some of them exhibited much greater selectivity against cancer cells expressing ETS-family oncogenic transcription factors.8 Most recently, by determining the crystal structures of unbound DNA of the same sequences, we showed that DNA undergoes structural adaptation in order to bind MTM.22 Binding of chromomycin A3 to DNA was also shown to induce DNA conformational changes,17 which differ from those occurring upon MTM binding, likely due to differences in sugars of the two natural products. In turn, comparison of the crystal structure of unbound chromomycin A3 with that of its complex with DNA showed that chromomycin A3 also undergoes conformational perturbations upon DNA binding.17 On the other hand, the conformational adaptation of MTM to binding DNA has been unclear due to the lack of the MTM structure in the unbound form. In this study, we aimed to fill this knowledge gap and establish how structural features of MTM encoded in its biosynthetic pathway enabled its efficient binding to DNA of the target cell.
Experimental
Crystallization, X-ray diffraction data collection and crystal structure determination of MTM SA-Phe and the MTM–boron complex
MTM was produced, purified and characterized as previously described.11 The analogue MTM SA-Phe was generated semi-synthetically by our recently published procedure.19 MTM and MTM SA-Phe were dissolved in DMSO at 50 mM and 20 mM, respectively. Crystallization of these compounds was carried out by vapor diffusion in hanging drops. For MTM SA-Phe, the drops were prepared by mixing 1 μL of the aqueous sample solution (2.6 mM MTM SA-Phe in 1.4 mM zinc acetate and 25 mM Tris–HCl pH 8.0) with 1 μL of the reservoir solution (0.2 M MgCl2, 0.1 M Hepes pH 7.5 and 30% PEG 400). The drops were incubated against 1 mL of the reservoir solutions at 21 °C. Crystals of the MTM–boron complex were grown in an analogous manner. In this case, the sample solution was 1 mM MTM, 4 mM H3BO3, 1 mM zinc acetate, 25 mM Tris–HCl, pH 8.0, and the reservoir solution was 2 M ammonium sulfate, 2% PEG 400, 100 mM Hepes, pH 7.5. In both cases, the crystals were transferred from their mother liquor to the solution that had the same composition as the respective reservoir solution and included 20% glycerol. The crystals were then rapidly frozen in liquid nitrogen. X-ray diffraction data to ∼1.20 Å resolution for (MTM SA-Phe)2-Mg2+ were collected at beamline 22-ID of the Advanced Photon Source at the Argonne National Laboratory (Argonne, IL), at 100 K, with the X-ray wavelength set at 0.979 Å. The data were indexed, integrated and scaled with HKL-2000.23 Some apparent scan-truncation errors were treated using XABS2.24 X-ray diffraction data for the Zn2+ bound MTM–boron complex were collected using a Bruker-Nonius rotating anode diffractometer (CuKα X-rays) and processed using the APEX2 suite.25
Crystal structures of the (MTM SA-Phe)2-Mg2+ and (MTM)2-Zn2+-B2 complexes were determined by dual-space direct methods, using SHELXD26 for (MTM SA-Phe)2-Mg2+ and SHELXT27 for (MTM)2-Zn2+-B2, and refined by full-matrix least-squares using SHELXL.28 The P21 structure of (MTM SA-Phe)2-Mg2+ was a pseudo-C-centered orthorhombic twin, with major : minor twin ratio of 9 : 1, and it was handled using standard SHELX methods (commands TWIN and BASF). In each case, some of the solvent was too disordered to model with certainty; therefore it was accounted for using the SQUEEZE routine in PLATON.29 Data collection and structure related details are summarized in Table 1. The crystal structure coordinates and structure factor amplitudes for (MTM SA-Phe)2-Mg2+ and (MTM)2-Zn2+-B2 were deposited into the Cambridge Structural Database with accession numbers CCDC ; 1896888 and CCDC ; 1896889, respectively. The structural images were generated using Pymol (Schrödinger, Cambridge, MA).
Table 1. Crystallographic details for the crystal structures of (MTM SA-Phe)2-Mg2+ and (MTM)2-Zn2+-B2.
| Data and refinement statistics | (MTM SA-Phe)2-Mg2+ | (MTM)2-Zn2+-B2 |
| Temperature (K) | 100 (2) a | 90 (2) |
| Crystal size (mm) | 0.10 × 0.10 × 0.05 | 0.16 × 0.15 × 0.10 |
| Crystal system | Monoclinic | Orthorhombic |
| Space group | P21 | I222 |
| a (Å) | 28.073(5) | 18.7382(5) |
| b (Å) | 21.531(4) | 28.3032(8) |
| c (Å) | 28.536(5) | 31.3664(7) |
| β (°) | 104.17(3) | |
| Unit cell volume (Å3) | 16 723(6) | 16 635.2(7) |
| Source, λ (Å) | APS, 0.97872 | CuKα, 1.54178 |
| Total number of reflections | 51 177 | 167 676 |
| Number of unique reflections | 19 688 b | 15 202 b |
| Resolution (Å) | 1.20 | 0.83 |
| R int | 0.060 | 0.0783 |
| R 1 [I > 2σ(I)] | 0.0870 | 0.0547 |
| wR2(F2) (all data) | 0.2385 | 0.1727 |
| Goodness-of-fit | 1.662 c | 1.049 |
| Δρ min/max (e Å–3) | 0.464/–0.279 | 0.445/–0.460 |
aThe values in parentheses are standard uncertainties.
bAs per standard small-molecule protocol, Friedel pairs were not merged.
cIt was not possible to optimize the SHELXL weighting scheme in the usual way.
DNA binding assay
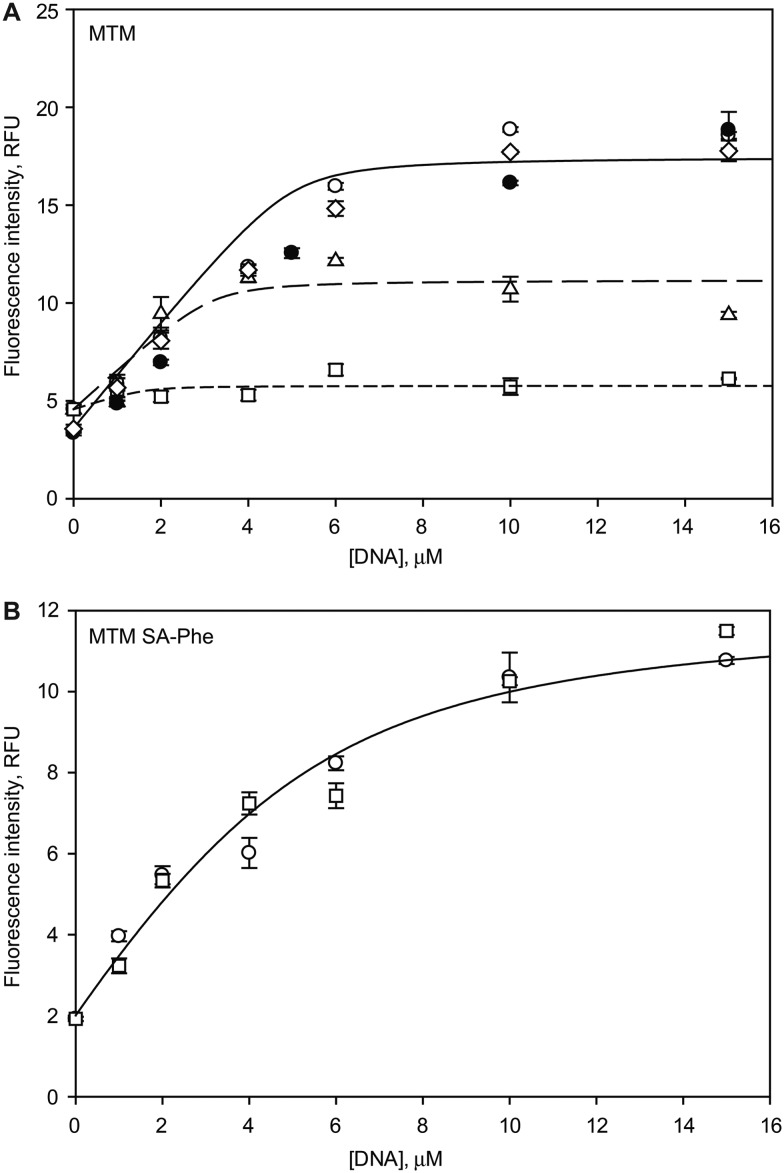
The DNA annealing and the binding assays were carried out by using the method analogous to that described previously by monitoring increase of intrinsic fluorescence of MTM and MTM SA-Phe upon DNA binding,20 with the following modifications. MTM or MTM SA-Phe were used at the concentration of 10 μM (5 μM dimers), and the double-stranded DNA oligomer GAGGCCTC containing one high-affinity MTM binding site (GGCC) was titrated in at specified concentrations. The assays were carried out in the binding buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2 or 1 mM Zn acetate) for 20 min at 21 °C, in triplicate. To perform a boron treatment, MTM, MTM SA-Phe (each at 2 mM) or DNA (at 0.4 mM) were preincubated with 4 mM or 20 mM of H3BO3 (as specified) in the binding buffer for 4 or 16 hours (as specified) at 21 °C prior to the DNA binding assay. Fluorescence was measured for the excitation and emission wavelengths of 470 nm and 540 nm, respectively, on a SpectraMax M5 microplate reader. The data were fit to a 1 : 1 MTM dimer : DNA binding isotherm using nonlinear regression with SigmaPlot 12.0 (Systat Software, San Jose, CA), as described previously. The binding data for MTM preincubated with boric acid were fit well with the fraction of MTM active in DNA binding as a fitting parameter.
Results and discussion
Crystal structure of an MTM analogue, MTM SA-Phe
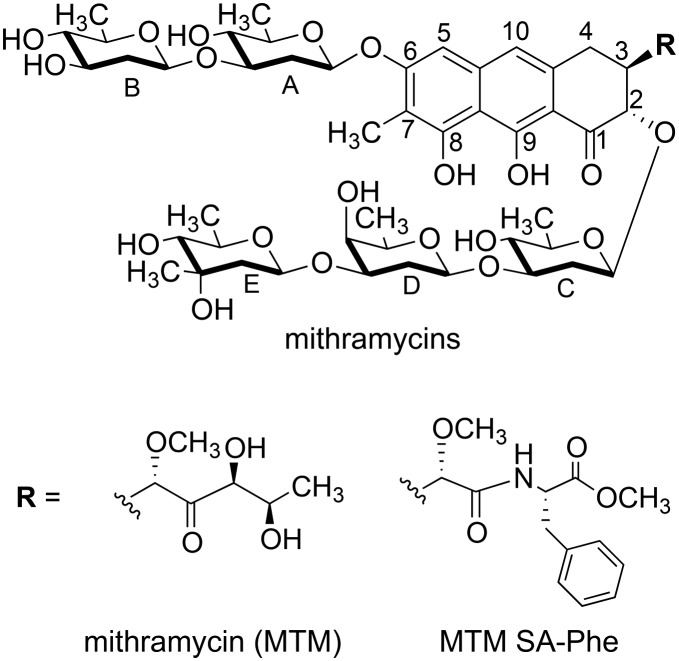
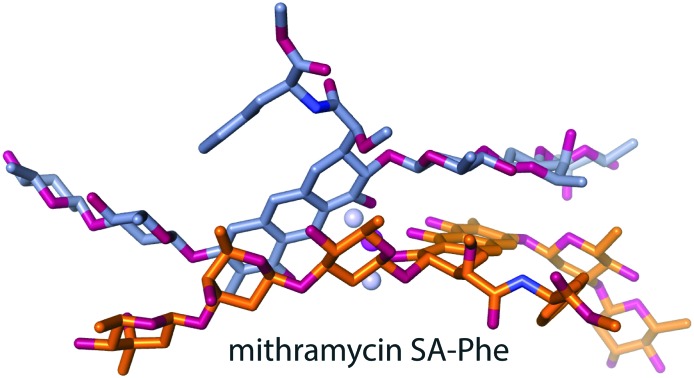
A crystal structure of MTM has been elusive, as our extensive efforts aimed at crystallizing MTM have not yielded crystals. Similarly, MTM–DNA complexes were refractory to crystallization, but 3-side chain analogues of MTM, MTM SA-Trp and MTM SA-Phe in complexes with Zn2+ and DNA were crystallized, and high-quality crystal structures were determined and analyzed.19 A similar strategy, crystallization of (MTM SA-Phe)2-Mg2+, yielded pseudo-merohedrally twinned crystals of this complex, which diffracted to the resolution of 1.2 Å. We determined the crystal structure of (MTM SA-Phe)2-Mg2+ by dual-space direct methods using SHELXD,26 albeit with some difficulty owing to the somewhat restrictive resolution (for a crystal of a small molecule) and twinning. The structure shows the chemical scaffold of an MTM molecule obtained for the first time at atomic resolution (Fig. 2A). The structure of MTM SA-Phe, except for the synthetic 3-side chain modification, is consistent in terms of chemical identity, connectivity and stereochemistry with the MTM structure derived two decades ago from the NMR analysis11 and that used previously in NMR and crystallographic structure determination of MTM and its analogues in complexes with DNA, at lower resolutions.15,16,19 The structure contains two dimers of MTM SA-Phe in the asymmetric unit in nearly identical conformations. We previously demonstrated that Mg2+-mediated dimerization of MTM and its analogues in solution occurs in the absence of DNA at physiological salt concentrations,20 in concordance with the dimeric assembly state in the crystal. The Mg2+ is coordinated octahedrally to the two molecules of MTM SA-Phe, where oxygens at positions 1 and 9 from each molecule and two water molecules form a full coordination shell, consistent with the metal coordination observed in our crystal structures of MTM SA Trp-DNA and MTM SA Phe-DNA complexes and in the crystal structures of chromomycin A3 alone and in complexes with DNA.17,18 The geometry of coordination is nearly ideal: the coordination bond angle deviations from the ideal geometry do not exceed approximately 1° and the bond lengths are all in the range 2.02–2.13 Å.
Fig. 2. Structures of MTM SA-Phe alone and in complex with DNA. A. The crystal structure of an MTM SA-Phe dimer (orange and dark grey sticks) coordinated by a Mg2+ ion (a purple ball). Two water molecules also coordinated by the Mg2+ are shown as light grey balls. B. Superimposition of the structure of unbound MTM SA-Phe dimer (as in panel A; orange and dark grey sticks) with our previously reported crystal structure of MTM SA-Phe dimer bound to a DNA oligomer (PDB ID: ; 5JW0; pale yellow and green sticks). The DNA is shown in light-grey. Oxygen and nitrogen atoms of MTM SA-Phe are shown in red and blue, respectively.
The structural connection between the chemistry of MTM and its DNA binding function
Without the structure of unbound MTM, its critical chemical features that have apparently evolved for the DNA binding function of this natural product have not been completely understood. Since the interactions of this molecule with DNA were described previously at a high level of detail,19 we will focus only on differences between the structures of unbound and DNA-bound MTM SA-Phe. In addition to the similar metal coordination, the conformation of the (MTM SA-Phe)2-Mg2+ complex (Fig. 2A) has a remarkable resemblance to the structure of this dimer in complex DNA (Fig. 2B), despite the presence of many rotatable bonds. Minor rotation of one chromophore relative to the other is observed, likely induced by interactions with the minor groove of the DNA. Other, also fairly minor conformational changes include rotations of the sugar moieties in the di- and the tri-saccharide side chains around the glycosidic bonds, which allow MTM to fit into the minor groove. The largest differences were observed for the terminal sugar moieties B and E, which are the least constrained. This induced fit-like conformational transition apparently maximizes interactions of these sugars with the DNA, whereby sugar B establishes several interactions with the DNA backbone and sugar E forms interactions both with the backbone and the bases, as previously described. Likewise, the phenyl rings of the 3-side chain in the two structures are related by an approximately 90° rotation. This side chain protrudes into the solvent and does not extensively interact with DNA.
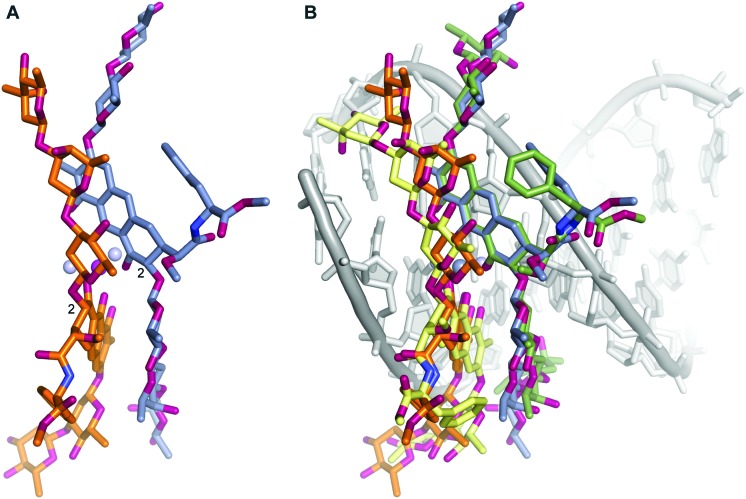
Clearly, numerous chemical features of MTM were encoded by evolution into the biosynthesis of MTM to result in the conformation appropriate for binding in the minor groove of the DNA. Based on the intermolecular interactions in the (MTM SA-Phe)2-Mg2+ complex (Fig. 2A), the core of the dimer is defined by the structure of the chromophore ring and the C–D rings of one molecule, which are stacked in an antiparallel fashion upon the C–D rings and the chromophore ring of its counterpart, respectively, in the dimer. This stacking conforms to the coordination of the two chromophores by the Mg2+ ion. This core is shaped even without the DNA to adopt a right-handed twist to fit snugly into the minor groove (Fig. 1B). A key chemical feature that enables the formation of this right-handed twisted core is the stereochemistry at C2 of the chromophore (Fig. 2A), which directs the trisaccharide side chain for the intermolecular stacking with the chromophore of the other molecule in the dimer. The opposite stereochemistry would direct the trisaccharide side chain away from the chromophore of the other MTM molecule. Even though it is yet to be demonstrated at which step in the MTM biosynthetic pathway the C2 stereocenter is formed, the tricyclic chromophore of MTM is formed in the penultimate step of MTM biosynthesis, as a result of the catalytic activity of Baeyer–Villiger monooxygenase MtmOIV.30–33 MtmOIV was shown to convert the tetracyclic premithramycin B, which we recently demonstrated to lack DNA binding activity,20 to an unstable intermediate MTM DK, which is then converted to MTM through the action of ketoreductase MtmW.34 This makes biological sense, as the formation of a bioactive species is postponed in the biosynthetic pathway by the evolution until the last steps, when the final product is ready to be secreted, likely to protect the producing organism from the potential toxicity of the intermediates.
MTM–boron complex formation and its perturbing effects on the MTM structure and function
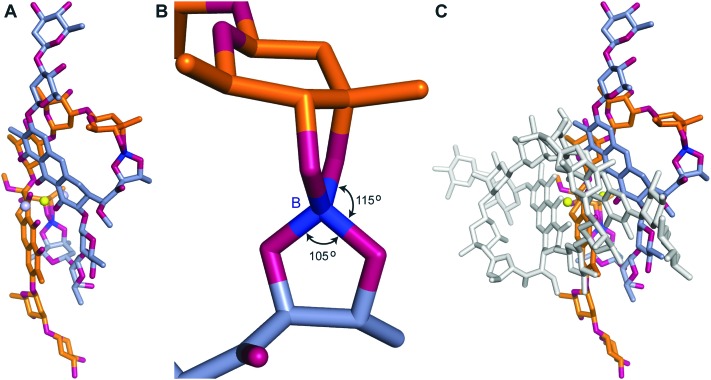
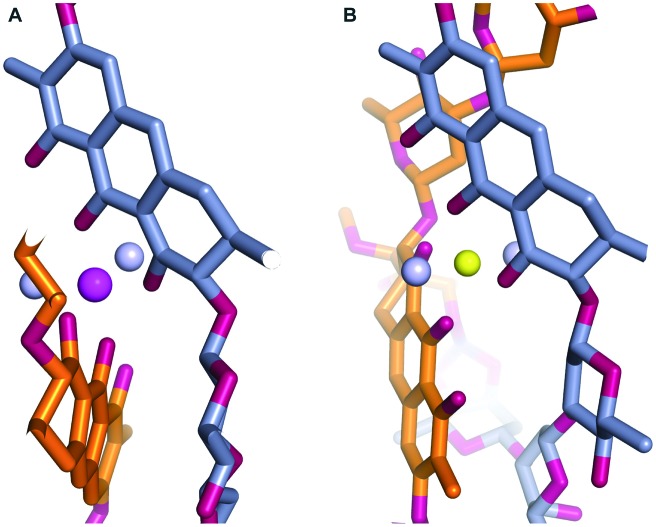
During our pursuit of crystals of MTM we observed that one of the MTM preparations formed well-diffracting crystals in the presence of Zn2+, with the data extending to 0.83 Å. The crystal structure, determined by direct methods, unexpectedly revealed two MTM molecules that were covalently bound to each other through two boron atoms. This preparation of MTM had a boron-containing contaminant, likely originating from chemical glassware. Subsequently, we have reproduced the same crystals with the MTM preparations that did not contain boron (and did not crystallize), after adding boric acid, in a slight stoichiometric excess of MTM, as described in Experimental. In the structure, a boron atom formed four covalent bonds: two with two oxygen atoms derived from the two hydroxyl groups on the E sugar of one MTM molecule and the other two with two oxygen atoms derived from the two hydroxyl groups of the 3-side chain (Fig. 3, panels A and B) of the other MTM molecule of a Zn2+-coordinated dimer. The other boron atom formed the identical reciprocal covalent bonds between the same two MTM molecules. As a result, the final structure contains two boron-mediated macrocycles joining the two molecules of the Zn2+-coordinated MTM dimer into one. Apparently, a similar spatial disposition of the two hydroxyl groups of the E sugar and the two hydroxyl groups of the 3-side chain favored boron binding. The boron–oxygen bond length are 1.44–1.48 Å and the bond geometry is slightly distorted from tetrahedral, with the O–B–O angles of 105–115° (Fig. 3B). In the crystal structure, a crystal symmetry-related copy of this boron-conjugated MTM dimer interacts with the first dimer through chromophore–chromophore stacking (Fig. 3C), facilitated by the nearly coplanar disposition of the two chromophore rings in the MTM2-Zn2+-B2 complex. As a consequence of structural distortions imposed by boron binding, one of the chromophores is rotated by nearly 180° with respect to its orientation in all other structures of MTM and its analogues (Fig. 4). This changes the topology of the Zn2+ coordination, where the two ligand water molecules are now coordinated in a collinear manner with the Zn2+ ion (Fig. 4B). The Zn2+ coordination remains octahedral, but it is slightly distorted due to the interactions with the second pair of chromophores in the crystal, requiring that one of the Zn2+-coordinated water molecules is shared by the two interacting B2-MTM2 complexes.
Fig. 3. The crystal structure of MTM in complex with Zn2+ and boron. A. The structure of a (MTM)2-Zn2+-B2 complex. Two MTM portions are shown by orange and dark grey sticks, the Zn2+ ion is shown as a yellow ball, water as light grey balls. The boron atoms are shown in dark blue. B. A zoomed-in view of the boron bonds. C. The crystal packing interactions between two (MTM)2-Zn2+-B2 complexes. The second (MTM)2-Zn2+-B2 complex is shown by white sticks.
Fig. 4. Zoomed-in views of the divalent metal ion coordination in the structures of (MTM SA-Phe)2-Mg2+ (A) and (MTM)2-Zn2+-B2 (B). The color schemes are the same as in Fig. 2 and 3.
Are the dramatic structural changes caused by the boron reactivity relevant in solution and, if so, do they perturb the DNA binding function of MTM? In order to answer these questions, we tested DNA binding of MTM and MTM SA-Phe in the absence and in the presence of boric acid, by using a well-established fluorescence-based assay (Fig. 5). The assay takes advantage of a strong increase of intrinsic fluorescence intensity of MTM and its analogues upon DNA binding, as reported by us and others.20,35 As expected, both MTM and MTM SA-Phe exhibited significant fluorescence enhancements upon titrating in DNA containing one high-affinity MTM binding site. The best fit to 1 : 1 (MTM dimer : DNA) binding isotherms yielded equilibrium binding constants Kd < 1 μM and Kd = (1.6 ± 0.6) μM, respectively (Fig. 5, open circles and solid curves). These binding affinities are in agreement with our previous DNA binding measurements for MTM and its MTM SA analogues.20 Consistent with previous reports,36,37 DNA binding was unaffected by using Zn2+ instead of Mg2+ (Fig. 5A). We observed that, in agreement with the structural perturbations, the fraction of MTM active in DNA binding decreases over time upon preincubation with boric acid at mildly basic pH (Fig. 5A, open triangles and squares) within several hours of preincubation, whereas MTM SA-Phe, which does not contain hydroxyl groups in the 3-side chain (Fig. 1) maintains its DNA binding function even after a 16 hour preincubation with boric acid at high concentration (Fig. 5B, open squares). Preincubation of DNA, but not MTM, did not result in the reduction of DNA binding affinity (Fig. 5A, open diamonds). These results indicate that not only is boron reactivity of MTM (but not of its SA analogue) relevant in solution, but that it abolishes the DNA binding activity of this natural product. These findings may have implications for the administration of MTM or its analogues in human, where one can expect MTM to be inactivated by boron (present at ∼10 μM in human plasma)38 in basic compartments of the human organism. MTM SA-Phe and similar analogues would not suffer from this metabolic instability.
Fig. 5. Effects of boron on DNA binding by MTM and MTM SA-Phe. A. DNA binding by MTM (5 μM dimers) in the absence of boron in Mg2+ (open circles), in Zn2+ (filled circles), in Mg2+ after the DNA (but not MTM) was preincubated with 20 mM boric acid for 16 hours at 21 °C (open diamonds), after MTM was preincubated with 4 mM boric acid for 4 hours (open triangles) and for 16 hours (open squares). The curves correspond to the best fits of the 1 : 1 (MTM dimer : DNA) binding isotherms with Kd < 1 μM and the fractions of MTM active in DNA binding of 1 (solid curve), 0.51 ± 0.06 (long dashes) and 0.09 ± 0.05 (short dashes) in the absence of boric acid, for the 4 hour and for the 16 hour preincubation with boric acid, respectively. B. DNA binding by MTM SA-Phe in the absence of boric acid (open circles) and after MTM SA-Phe was preincubated with 20 mM boric acid for 16 hours at 21 °C (open squares). The curve is the best fit of the 1 : 1 (MTM SA-Phe) binding isotherm, Kd = (1.6 ± 0.6) μM.
Conclusions
In summary, in this study we presented the first crystal structures of MTM and its SA analogues and elucidated that, despite many degrees of conformational freedom of this large natural product, its chemical structure encodes both the divalent metal ion coordination and the preferred conformation of the dimer of MTM for proper DNA binding. As a result, only minor induced-fit adaptation is needed for the MTM dimer to bind in the minor groove. The inactivation of MTM by its reaction with boron highlights the importance of developing its more promising inert SA analogues as next generation anticancer agents targeting cancers caused by oncogenic transcription factors.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We thank sector SER-CAT of the Advanced Photon Source at the Argonne National Laboratory for the support during the remote collection of the X-ray diffraction data. This work was supported by US Department of Defense grant PC150300P1 to O. V. T. and J. R., by National Institutes of Health grant CA091901 to J. R. and by National Science Foundation MRI grants CHE-0319176 and CHE-1625732 to S. P. and O. V. T.
Footnotes
†CCDC 1896888 and 1896889. For crystallographic data in CIF or other electronic format see DOI: 10.1039/c9md00100j
References
- Ryan W. G., Schwartz T. B., Northrop G. JAMA, J. Am. Med. Assoc. 1970;213:1153–1157. [PubMed] [Google Scholar]
- Kennedy B. J. J. Urol. 1972;107:429–432. doi: 10.1016/s0022-5347(17)61046-2. [DOI] [PubMed] [Google Scholar]
- Ream N. W., Perlia C. P., Wolter J., Taylor, 3rd S. G. JAMA, J. Am. Med. Assoc. 1968;204:1030–1036. [PubMed] [Google Scholar]
- Elias E. G., Evans J. T. J. Bone Jt. Surg. 1972;54:1730–1736. [PubMed] [Google Scholar]
- Kennedy B. J., Torkelson J. L. Med. Pediatr. Oncol. 1995;24:327–328. doi: 10.1002/mpo.2950240511. [DOI] [PubMed] [Google Scholar]
- Kofman S., Perlia C. P., Economou S. G. Cancer. 1973;31:889–893. doi: 10.1002/1097-0142(197304)31:4<889::aid-cncr2820310419>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Grohar P. J., Woldemichael G. M., Griffin L. B., Mendoza A., Chen Q. R., Yeung C., Currier D. G., Davis S., Khanna C., Khan J., McMahon J. B., Helman L. J. J. Natl. Cancer Inst. 2011;103:962–978. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P., Eckenrode J. M., Mandal A., Jha A. K., Salem S. M., Leggas M., Rohr J. J. Med. Chem. 2018;61:8001–8016. doi: 10.1021/acs.jmedchem.8b01107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohar P. J., Glod J., Peer C. J., Sissung T. M., Arnaldez F. I., Long L., Figg W. D., Whitcomb P., Helman L. J., Widemann B. C. Cancer Chemother. Pharmacol. 2017;80:645–652. doi: 10.1007/s00280-017-3382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osgood C. L., Maloney N., Kidd C. G., Kitchen-Goosen S., Segars L., Gebregiorgis M., Woldemichael G. M., He M., Sankar S., Lessnick S. L., Kang M., Smith M., Turner L., Madaj Z. B., Winn M. E., Nunez L. E., Gonzalez-Sabin J., Helman L. J., Moris F., Grohar P. J. Clin. Cancer Res. 2016;22:4105–4118. doi: 10.1158/1078-0432.CCR-15-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlert S. E., Kunzel E., Machinek R., Mendez C., Salas J. A., Rohr J. J. Nat. Prod. 1999;62:119–121. doi: 10.1021/np980355k. [DOI] [PubMed] [Google Scholar]
- Lombo F., Menendez N., Salas J. A., Mendez C. Appl. Microbiol. Biotechnol. 2006;73:1–14. doi: 10.1007/s00253-006-0511-6. [DOI] [PubMed] [Google Scholar]
- Jayasuriya H., Lingham R. B., Graham P., Quamina D., Herranz L., Genilloud O., Gagliardi M., Danzeisen R., Tomassini J. E., Zink D. L., Guan Z., Singh S. B. J. Nat. Prod. 2002;65:1091–1095. doi: 10.1021/np010642f. [DOI] [PubMed] [Google Scholar]
- Katahira R., Uosaki Y., Ogawa H., Yamashita Y., Nakano H., Yoshida M. J. Antibiot. 1998;51:267–274. doi: 10.7164/antibiotics.51.267. [DOI] [PubMed] [Google Scholar]
- Sastry M., Fiala R., Patel D. J. J. Mol. Biol. 1995;251:674–689. doi: 10.1006/jmbi.1995.0464. [DOI] [PubMed] [Google Scholar]
- Sastry M., Patel D. J. Biochemistry. 1993;32:6588–6604. doi: 10.1021/bi00077a012. [DOI] [PubMed] [Google Scholar]
- Tseng W. H., Chang C. K., Wu P. C., Hu N. J., Lee G. H., Tzeng C. C., Neidle S., Hou M. H. Angew. Chem., Int. Ed. 2017;56:8761–8765. doi: 10.1002/anie.201703989. [DOI] [PubMed] [Google Scholar]
- Hou M. H., Robinson H., Gao Y. G., Wang A. H. Nucleic Acids Res. 2004;32:2214–2222. doi: 10.1093/nar/gkh549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Weidenbach S., Cano K. E., Wang Z., Mitra P., Ivanov D. N., Rohr J., Tsodikov O. V. Nucleic Acids Res. 2016;44:8990–9004. doi: 10.1093/nar/gkw761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenbach S., Hou C., Chen J. M., Tsodikov O. V., Rohr J. J. Inorg. Biochem. 2016;156:40–47. doi: 10.1016/j.jinorgbio.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D., Chen J. M., Bae Y., Rohr J. Chem. Biol. Drug Des. 2013;81:615–624. doi: 10.1111/cbdd.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Tsodikov O. V. Acta Crystallogr., Sect. D: Struct. Biol. 2019;75:32–40. doi: 10.1107/S205979831801553X. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parkin S., Moezzi B., Hope H. J. Appl. Crystallogr. 1995;28:53–56. [Google Scholar]
- Bruker, Bruker AXS Inc., Madison, Wisconsin, USA, 2012.
- Sheldrick G. M. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. Acta Crystallogr., Sect. A: Found. Adv. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. Acta Crystallogr., Sect. C: Struct. Chem. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spek A. L. Acta Crystallogr., Sect. C: Struct. Chem. 2015;71:9–18. doi: 10.1107/S2053229614024929. [DOI] [PubMed] [Google Scholar]
- Beam M. P., Bosserman M. A., Noinaj N., Wehenkel M., Rohr J. Biochemistry. 2009;48:4476–4487. doi: 10.1021/bi8023509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosserman M. A., Downey T., Noinaj N., Buchanan S. K., Rohr J. ACS Chem. Biol. 2013;8:2466–2477. doi: 10.1021/cb400399b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M., Nur-e-alam M., Lipata F., Oliveira M. A., Rohr J. J. Am. Chem. Soc. 2005;127:17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- Prado L., Fernandez E., Weissbach U., Blanco G., Quiros L. M., Brana A. F., Mendez C., Rohr J., Salas J. A. Chem. Biol. 1999;6:19–30. doi: 10.1016/s1074-5521(99)80017-9. [DOI] [PubMed] [Google Scholar]
- Remsing L. L., Gonzalez A. M., Nur-e-Alam M., Fernandez-Lozano M. J., Brana A. F., Rix U., Oliveira M. A., Mendez C., Salas J. A., Rohr J. J. Am. Chem. Soc. 2003;125:5745–5753. doi: 10.1021/ja034162h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majee S., Sen R., Guha S., Bhattacharyya D., Dasgupta D. Biochemistry. 1997;36:2291–2299. doi: 10.1021/bi9613281. [DOI] [PubMed] [Google Scholar]
- Reyzer M. L., Brodbelt J. S., Kerwin S. M., Kumar D. Nucleic Acids Res. 2001;29:E103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi P. G., Pal S., Banerjee R., Dasgupta D. J. Inorg. Biochem. 2007;101:127–137. doi: 10.1016/j.jinorgbio.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Usuda K., Kono K., Yoshida Y. Biol. Trace Elem. Res. 1997;56:167–178. doi: 10.1007/BF02785390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.