 Inhibition of pyroptosis through targeting the activation or pore-formation of GSDMD.
Inhibition of pyroptosis through targeting the activation or pore-formation of GSDMD.
Abstract
The discovery of a previously unknown protein, gasdermin D (GSDMD), as the key effector that leads to pyroptosis and NETosis has created much excitement. Since its initial report in Oct. 2015, more than 200 papers have been published on studies of the structure and mechanism of GSDMD and its homologues. The clear connection between infection and inflammasome activation made GSDMD a promising target for the development of anti-infection treatment. In this mini review, we discuss first the current understanding of the structure and mechanism of GSDMD, focusing on its potential as a druggable target, and then recent efforts in the development of inhibitors to interfere with the pore-forming function of GSDMD and thus alleviate the detrimental effects due to pyroptotic cell death.
1. The role of GSDMD during microbial infection
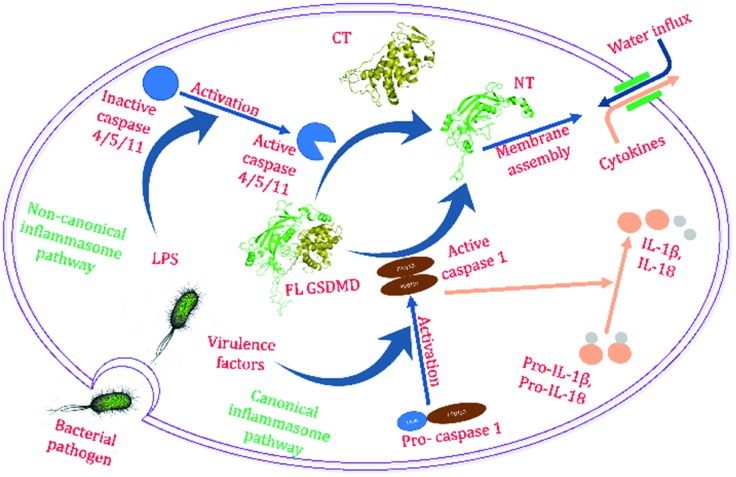
Infection kills millions of patients worldwide every year.1–4 With the spread of superbugs and dwindling of the developmental pipeline of new antibiotics, this number will only increase for years to come.5–7 Many infection-related deaths and prolonged hospitalization are due to sepsis, a common complication of infection.8–10 Sepsis is caused by the host inflammatory response triggered by an infection. Excessive inflammatory responses in the host induce cell death throughout the body and damage its own tissues and organs.11,12 As part of the responses, pyroptosis is an important form of programed cell death that plays a critical role in septic shock (Fig. 1).13–22
Fig. 1. Bacterial cellular components activate inflammatory caspases, which cleave GSDMD and lead to pyroptosis.
The detection of microbe invasion triggers activation of the inflammatory caspases (including 1, 4/5, and 11), which leads to the cleavage of the linker sequence between the N-terminal (NT) and C-terminal (CT) domains of the effector protein gasdermin D (GSDMD). GSDMD has recently been identified as the key effector in pyroptosis.23–25 In addition, an exciting new discovery was reported by Sollberger et al. a few months ago, which elucidated the key role played by GSDMD in the formation of neutrophil extracellular traps (NETs).26 Simultaneously, Chen et al. also independently established the role of GSDMD in NET formation.27 In neutrophils, GSDMD was cleaved by serine proteases including NE, rather than caspases.
The cleavage of the inter-domain linker lifts the auto-inhibition of the GSDMD CT domain on the NT domain, and the latter subsequently binds to predominantly the acidic lipid cardiolipins that are present in the mitochondria membrane and the inner leaflet of the plasma membrane, oligomerizes, and forms pores in the cell membranes.28,29 Pore formation by GSDMD elicits cell death (pyroptosis) and the release of the inflammatory cytokines such as IL-1β to trigger downstream responses.28–35 In NETosis, cleaved GSDMD forms pores in the plasma membrane to allow NET release. Deletion of the GSDMD gene has been shown to improve the survival rate in a murine sepsis model,31 demonstrating an essential role of GSDMD in sepsis. This observation suggests that inhibitors of GSDMD could serve as potential therapeutics to septic patients.36,37 Therefore, there is increasing interest in pursuing GSDMD as a new and alternative target for the development of anti-infection treatment, since disruption of GSDMD function may effectively alleviate the negative effects of pyroptosis on septic patients, and thus improve the outcome of sepsis treatment. As a proof-of-concept example, the IL-1 antagonist, anakinra, has been shown to effectively reduce mortality in a subset of sepsis patients, which demonstrates the therapeutic efficacy of inhibiting the inflammasome-pyroptosis pathway in the treatment of sepsis.38,39 Several groups have pioneered the search for inhibitors of GSDMD function, which are discussed later in this mini review.
2. Mechanism of pore formation by GSDMD
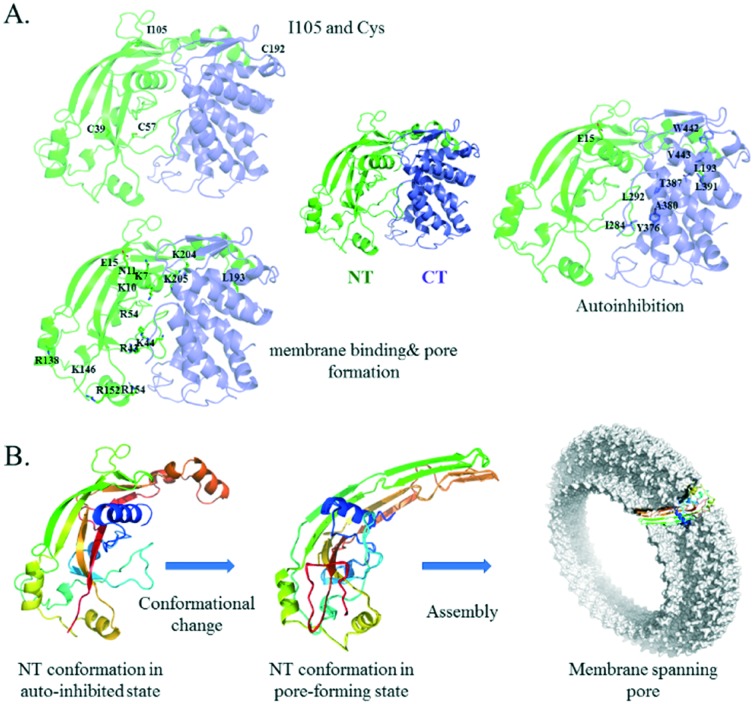
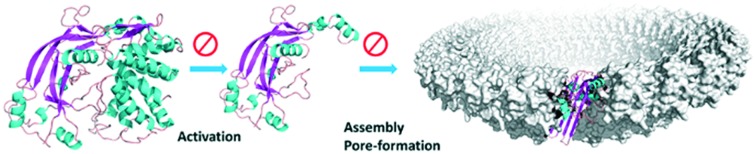
The central role of GSDMD activation and pore formation in inflammasome signalling has been established by many studies.26,27,40–44 GSDMD contains a well-defined NT domain of ∼30 kD and a CT domain of ∼26 kD. The crystal structure of the full length GSDMD is not yet available. The structure model was constructed using the online server of SWISS-MODEL and a homolog protein, GSDMA3, as the template (Fig. 2).29,45 Between the two domains is a long and flexible linker, and the structure of part of the linker (residues 249–263) is missing in the template. The mechanism of GSDMD-mediated pyroptosis and NETosis has been established by several recent studies.23–31,34,46,47 The CT domain (also called the p20 fragment) serves as an auto-inhibition module. Cleavage of the linker between the two domains upon activation frees the NT domain (also called the p30 fragment), which oligomerizes to form pores in the cell membrane. These pores increase the membrane permeability and facilitate the release of the NETs or inflammatory cytokines, which trigger a cascade of inflammatory response. Early analysis of the low-resolution electron microscopy images of the pores formed in liposomes revealed a 16-fold symmetry, suggesting that the pore contains 16 copies of the NT subunits.29,31,34 The opening of the pores is approximately 14–16 nm in diameter, large enough for the cytokines to leak out. More recently, Wu and coworkers reported the cryo-electron microscopy structures of the 27-fold and 28-fold single-ring pores formed by the N-terminal fragment of mouse GSDMA3 (GSDMA3-NT) at 3.8 and 4.2 Å resolutions, and of a double-ring pore at 4.6 Å resolution.48 In the 27-fold pore, a 108-stranded anti-parallel β-barrel is formed by two β-hairpins from each subunit capped by a globular domain. A positively charged helix α1, containing R9, R13, and R18, is positioned to interact with the acidic lipid cardiolipin and PI(4,5)P2. The NT domain undergoes significant conformational changes upon membrane insertion, in which stretches of sequences previously forming random coils in full length GSDMA3 transform into long, membrane-spanning β-strands (Fig. 2B). The diameter of the pore is ∼18 nm. Strategies that prevent the NT domain of GSDMD from forming pores in the cell membrane are expected to alleviate the inflammatory response due to pyroptosis.
Fig. 2. Structure of a full length and pore-forming NT mouse GSDMD structure model with mutation sites highlighted. A. Full length mouse GSDMD structure, with residues subjected to mutational studies highlighted. Residues are shown in three groups for clarity, based on their functional roles. The NT domain is coloured green and the CT domain blue. B. To assemble into a membrane spanning pore, the NT domain undergoes a large conformational change to convert from the auto-inhibited state to the pore-forming state upon activation. Secondary structures are coloured sequentially from blue to red. Structure model of full-length mouse GSDMD was generated from homology modelling using the structure of GSDMA3 (5b5r.pdb) as the template, and the NT structure model in the pore conformation was generated using the pore-formation structure of GSDMA3-NT (; 6cb8.pdb) as the template.
3. Structure/function studies of GSDMD by site-directed mutagenesis
Mutational studies provide valuable insights into the structure–function relationship of GSDMD and potentially help identify the hot spot for targeting GSDMD (Table 1). Initial studies were conducted in the absence of structural information. In June 2016, the crystal structure of a close homologue of GSDMD, GSDMA3, was reported.29 The structure model of GSDMD was then constructed based on the structure of GSDMA3, which provided structural insight and foundation to direct subsequent mutational studies.
Table 1. Summary of mutation studies of mouse and human GSDMD.
| Mutation | Effect | Reference |
| Ile105 to Asn (mouse) | Loss of IL-1β release and pyroptotic cell death in BMDM | Kayagaki et al., 2015 (ref. 24) |
| Arg138, Lys146, Arg152, and Arg154 mutated to Ala (mouse) | When all four residues were mutated to Ala, pyroptosis and cell death were completely blocked; when only two or three residues were mutated to Ala, cell death was partially blocked | Liu et al., 2016 (ref. 33) |
| Arg138 to Ser, Arg152 to Ala (mouse) | When Arg138 was mutated to Ser and Arg152 was mutated to Ala simultaneously, cell death was almost completely blocked | Liu et al., 2016 (ref. 33) |
| Lys204, Lys205, Lys237, and Arg239 to Ala (mouse) | No effect on pyroptosis | Liu et al., 2016 (ref. 33) |
| Arg248 and Lys249 to Ala (mouse) | No effect on pyroptosis | Liu et al., 2016 (ref. 33) |
| Cys39 to Ala, Cys192 to Ala (mouse) | Impaired oligomerization | Liu et al., 2016 (ref. 33) |
| Ile104 to Asn (human) | Less effective in pore formation and inducing cell death | Sborgi et al., 2016 (ref. 34) Aglietti et al., 2016 (ref. 30) |
| Leu192 to Asp, Glu15 to Lys (human) | Decrease in pyroptotic activity. Effects were additive | Ding et al., 2016 (ref. 29) |
| Leu290, Tyr373, or Ala377 to Asp (human) | Disruption of auto-inhibition and leading to constitutive activation of GSDMD and subsequently pyroptosis | Ding et al., 2016 (ref. 29) |
| Leu292, Tyr376 or Ala380 to Asp (mouse) | Disruption of auto-inhibition and leading to spontaneous oligomerization of full length GSDMD | Rathkey et al., 2017 (ref. 49) |
| Phe283 to Ala, Asp or Tyr (human) | F283A and F283R disrupted auto-inhibition, whereas F283Y did not exhibit such an effect | Kuang et al., 2017 (ref. 47) |
| Cys191 to Ala (human) | Inhibited pyroptotic activity | Hu et al., 2018 (ref. 51) |
| Cys192 to Ala (mouse) | Inhibited pyroptotic activity | Hu et al., 2018 (ref. 51) |
| Cys191, Cys38, and Cys56 to Ala (human) | C191A reduced oligomerization and cell death. C38A and C56A mutations led to a similar effect to a lesser extent | Rathkey et al., 2018 (ref. 53) |
| Leu292, Tyr376 and Ala380 to Asp (mouse) | Compromised auto-inhibition and demonstrated spontaneous pore forming activity | Liu et al., 2018 (ref. 50) |
| Thr387, Leu391, Trp442 and Val443 to Asp (mouse) | No detectable detrimental effect on function | Liu et al., 2018 (ref. 50) |
| Arg7, Arg10 and Arg11 to Ala (human) | Reduced liposome leakage or pore forming activity | Ruan et al., 2018 (ref. 48) |
| Arg42, Lys43 and Arg53 to Ala (human) | No detectable detrimental effect on function | Ruan et al., 2018 (ref. 48) |
3.1. I104/105N (human/mouse)
The role of GSDMD in pyroptosis was established by two independent groups either through a CRISPR-Cas9 based genome-wide genetic screen or through an unbiased forward genetic screen using ethyl-N-nitrosourea (ENU)-mutagenized mice.23,24 Bone marrow derived macrophages (BMDMs) from mice carrying I105N mutation in GSDMD (GsdmdI105N/I105N) were found to be defective in IL-1β secretion and failed to undergo pyroptosis in response to cytoplasmic LPS.24GsdmdI105N/I105N BMDMs express a comparable amount of GSDMD as BMDMs from wild type mice, suggesting that the mutation impairs protein function rather than expression. Using a HEK293T transient overexpression system and membrane permeabilization assay in vitro, a follow up study by the same research team showed that the mouse I105N (corresponding to human I104N) mutation attenuated cell killing by GSDMD p30.30 Recombinant wild type human GSDMD and the I104N mutant were cleaved at a similar rate in vitro by a constitutively active form of caspase 11. Both resultant p30 fragments could bind to membranes and form pores in liposomes when examined under the electron microscope. However, the liposome leakage induced by the I104N p30 mutant fragment was slower compared to that by the wild type p30.
These observations were confirmed by another group independently, who also investigated the mechanism of human GSDMD and its mutant I104N.34 The human GSDMD I104N mutant was shown to behave similarly to wild type GSDMD by caspase cleavage. At high concentrations (i.e. 260 nM GSDMD incubated with 5 nM caspase-1), the mutant p30 fragment was as effective as the wild type p30 in forming functional pores in liposomes. However, at lower concentrations (130 nM GSDMD or less), the I104N mutant was less efficient. Overexpression of the mutant in HEK293T cells was less efficient in killing cells than that of the wild type protein. In addition, unlike wild type GSDMD, overexpression of this mutant GSDMD only partially restored pyroptotic cell death in the gsdmd–/– macrophages upon Salmonella infection.
Collectively, these studies indicate that I104N mutation affects human GSDMD itself rather than interactions with another protein in the signaling cascade. It was proposed to either hamper p30 membrane insertion and pore formation, or p30 oligomerization. With the recent determination of the membrane-spanning pore structure of GSDMA3-NT, the role of I104/105 emerged.48 I104/105 corresponds to V101 in GSDMA3, which is located close to the tip of the first β-hairpin in the pore structure, and thus is more likely to be involved in membrane insertion.
3.2. Auto-inhibition
Based on the GSDMD structure model, L192 and E15 from the NT domain are located at the domain interface and interact with residues from the CT domain, including L290, Y373, and A377. As expected, L290D, Y373D and A377D mutations led to disruption of auto-inhibition and constitutive activation of GSDMD.29 Pyroptotic cell death was observed when these mutated GSDMDs were expressed in HEK293T cells. The CT domain mutations presumably affect function via exposing L192/E15, thus these residues are likely to be important for pyroptosis. To test this hypothesis, direct mutations were also introduced to create L192D and E15K. These mutations will be discussed later.
Similar results were obtained with mouse GSDMD by Rathkey et al.49 Since these mutations in human GSDMD disrupted auto-inhibition, the corresponding mutations in mouse GSDMD are expected to behave similarly. Conserved residues L292, Y376 and A380 of mouse GSDMD were mutated into Asp. Mouse GSDMD was tagged with fluorescent protein mNeon or mRuby. These fluorescently tagged GSDMD mutants could be cleaved properly by caspase-11. Fluorescently tagged wild type GSDMD and mutants were expressed in HEK293T cells. Live cell imaging was performed using confocal microscopy to monitor the expression and localization of the fusion proteins. All GSDMD mutants were shown to form aggregate in cells, indicating that the disruption of auto-inhibition resulted in spontaneous GSDMD oligomerization. In addition, GSDMD mutants were found to be expressed at much lower levels, suggesting their cytotoxicity. In contrast, fusion proteins containing wild type or D276A GSDMD mutation (to disrupt the caspase cutting site) did not form any aggregates, suggesting that the fluorescent protein tag did not cause aggregation.
As discussed above, the NT and CT domains are connected by a long flexible loop, which also contains the caspase cutting site. To probe the role of the loop in regulating auto-inhibition, Kuang et al. mutated an aromatic residue in the CT domain, Phe283, which appears to interact with the loop according to the structure model.47 Three mutants were created, F283A, F283R, and F283Y. While wild type GSDMD largely exists as a monomer, these mutants tend to form higher order oligomers. Upon transfection into HEK293T cells, F283A and F283R mutants were found to lead to pyroptotic cell death, whereas F283Y did not.
Liu et al. determined the crystal structure of the CT domain of both human and murine GSDMD.50 They also used the structure of GSDMA3 by Ding et al. as a template to construct a structure model of the remaining NT region. They hypothesized that two regions at the interdomain interface, which they named site I and site II, are critical for the binding between the two domains. Site-directed mutagenesis was conducted at these two sites. The overexpression of the GSDMD mutants at site I, including L292D, Y376D, A380D, resulted in significant levels of cell death, although the level of cell death was much lower than the overexpression of the NT domain alone, suggesting compromised auto-inhibition. Mutation of residues at site II including T387D, L391D, W442D, and V443D did not have such an effect. The pyroptotic activity was also examined through propidium iodide (PI) uptake using immortalized mouse BMDMs (iBMDMs) upon Salmonella infection. Upon transfection of gsdmd deficient iBMDM, cells expressing Y376D showed comparable PI uptake to cells expressing WT protein when infected by Salmonella, although the expression level of Y376D was found to be much lower than that of wild type GSDMD. As controls, a low level of uptake was observed in the I105N mutant and no uptake was found in gsdmd deficient cells.
3.3. Membrane binding and pore formation
Since positively charged residues are often involved in membrane binding and pore formation in toxic proteins, Liu et al. examined the effects of the conserved positively charged residues in the NT domain of GSDMD on pyroptosis.33 By aligning the sequences of six mammalian GSDMDs, a group of conserved basic residues that might be involved in the assembly of GSDMD-NT to form membrane pores was identified in mouse GSDMD. Several mutants were created, in which Arg138, Lys146, Arg152, or Arg154 was replaced with Ala. Double, triple, and quadruple mutants containing Ala mutations of either two, three or all four sites were also constructed, as well as a mutant in which Arg138 was mutated into Ser and the other three residues into Ala. None of these mutations affected the overall structure and stability of GSDMD as verified using the melting temperature analysis. Ala mutation in all four residues, as well as R138S plus Ala mutation in the other three sites completely inhibited pyroptosis, whereas double or triple Ala mutation caused partial inhibition. R152A/R154A and K146A/R152A/R154A reduced cell death by more than 50%, whereas R138A/K146A and K146A/R152A reduced cell death by less than 50%. In gsdmd deficient iBMDM cells, transfection of WT gsdmd allowed the cell to undergo LPS-induced pyroptotic death, whereas transfection of the mutated gsdmd with quadruple Ala mutation did not. These observations strongly suggest that the conserved basic residues play important roles in pore formation, possibly in membrane binding. Several additional conserved basic residues tested, including Lys204, Lys205, Lys237 and Arg239, along with non-conserved basic residues Arg248 and Lys249, did not affect pyroptosis.
According to the pore structure, the first α-helix of GSDMD NT is important in membrane pore formation.48 Conserved basic residues of this region were individually mutated into Ala to create R7A, R10A and R11A. In addition, basic residues in the β1-2 regions including R42, K43 and R53 were also examined. After protein expression and purification, the recombinant GSDMD mutants were subjected to the liposome leakage assay. Caspase-11 was added to cleave the full-length protein. Mutants in the α1 region showed reduced liposome leakage, whereas mutation in the β1-2 region did not affect pore formation or liposome leakage. Hence, the basic residues in the α1 region are likely involved in lipid binding along with the basic residues in the α3 region (Arg138, Lys146, Arg152, or Arg154), which had been reported in an earlier study.33 However, since the α3 region is further away from the membrane than α1 according to the pore structure, basic residues in α1 appear to be the major sites of acidic lipid interaction.
As discussed above, E15K and L192D mutations in GSDMD NT were speculated to affect membrane insertion and pore formation. GSDMD p30 containing E15K or L192D mutation showed markedly decreased pyroptosis when transiently expressed in HEK293T cells and demonstrated impaired liposome binding and membrane pore formation. The detrimental effects of the two mutations were additive, as a double mutant containing both changes led to a further decrease in liposome binding, leakage, and pyroptosis when transfected into HEK293T cells.29 E15 and L192 in human GSDMD correspond to E14 and L184 in GSDMA3. According to the GSDMA3-NT pore structure, E14 and L184 are located at the inter-subunit interface between the globular domain and β-barrel domain respectively, and thus are likely to be involved in oligomerization. L184 is also involved in membrane insertion and its mutation to an acidic residue could therefore affect this process.
3.4. Cys reactivity and disulfide bond
The GSDMD p30 fragment was found to migrate as oligomers on SDS-PAGE when analyzed under non-reducing conditions, and the oligomer band disappeared in the presence of a reducing agent. Thus, Liu et al. proposed that formation of disulfide bonds might be important during oligomerization.33 Site-directed mutagenesis studies of all cysteines present in mouse GSDMD p30 revealed that Cys39 and Cys192 mutations impaired oligomerization. This is the first study suggesting a critical role of Cys192 in mouse GSDMD function (corresponding to human Cys 191). This discovery was confirmed by several additional studies as discussed below.
Hu et al. confirmed the important role of Cys191/192 (human/mouse) by mutating them into Ala.51 The Cys191/192A mutation completely abolished pyroptotic cell death, and thus they proposed that this Cys could be a potential target to prevent unwanted pyroptotic cell death. PROPKA was used to predict the reactivity of all Cys residues in GSDMD, and Cys191 (human)/Cys192 (mouse) was identified as the most reactive cysteine.52
Rathkey et al. also confirmed the importance of Cys191.49 Disulfide bond formation has been shown to be critical for oligomerization of the p30 fragment of GSDMD, thus the three conserved cysteines in the p30 fragment, C191, C38 and C56, were mutated into Ala. The C191A mutant, and to a lesser extent the other two mutants, had a decreased ability to oligomerize when analysed under non-reducing conditions. When expressed in HEK293T cells, the p30 fragment containing C191A mutation led to reduced cell death, whereas Cys38A and Cys56A mutations did not.53
The purified p30 fragment forms disulfide bond-linked oligomers. Thus, it has been implied that Cys191/192 should be important for oligomerization during pore formation, potentially through the formation of inter-subunit disulfide bonds. Interestingly, based on the recent GSDMA3-NT membrane pore structure, no disulfide bond was observed in the pore structure, and no two Cys residues were close enough to form inter- or intra-subunit disulfide bonds.48 The location of Cys191/192 is close to the tip of the second β-hairpin and the interface with the neighboring subunit, thus it is more likely to be involved in both membrane insertion and oligomerization, although not via formation of disulfide bonds. Then, why is the thiol reactivity so important? Did the disulfide bond involving Cys191/192 ever form transiently during the process of activation and pore formation? More research is needed to address these questions.
4. Recent progress in the identification and development of GSDMD inhibitors
Although the role of GSDMD in pyroptosis has only emerged in the last couple of years, researchers have already begun to search for small molecule inhibitors that block the function of GSDMD. While mutational studies have revealed several potential sites for targeting, so far, the most fruitful site has been Cys191/192 (human/mouse). Two research teams have reported their discovery of small molecule compounds that interrupt GSDMD function.51,53 Interestingly, the inhibitors found by both groups disrupt GSDMD function via a similar mechanism, namely, reacting with the free thiol group at Cys191/192 in GSDMD.
4.1. Inhibitors function through reacting with Cys191/192
There are 12 cysteine residues in the sequence of GSDMD. None of them is involved in the formation of intramolecular disulfide bonds according to the structure model of full length GSDMD (Fig. 2). However, the reactivity of Cys191/192(human/mouse) has been shown to be critical for pore formation. Due to the importance of this residue in pore-formation, and since the residue is well exposed and highly reactive in the protein structure, Abbott and coworkers have tested the idea of exploiting Cys-modifying compounds as inhibitors of GSDMD function.53 In parallel, Wu and coworkers also identified Cys-modifying inhibitors through high throughput screening.51
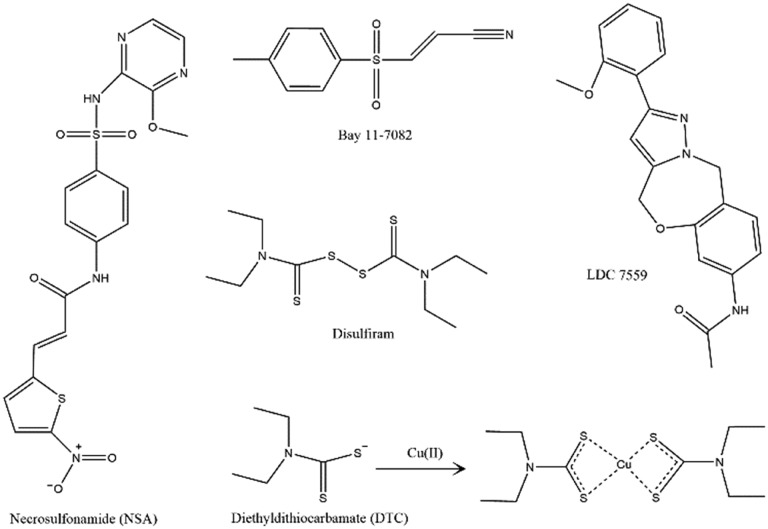
Abbott and coworkers found that necrosulfonamide (NSA) directly binds to GSDMD and inhibits p30-GSDMD oligomerization (Fig. 3). NSA was initially discovered though a screening of small-molecule inhibitors of necroptotic cell death in HT29 cells. It potentially inhibits necroptosis through binding to and disruption of disulfide bond formation of human mixed lineage kinase domain-like pseudokinase (MLKL).54 The authors speculated that NSA might also inhibit GSDMD oligomerization and pyroptotic cell death, since the reactivity of Cys191/192 has been shown to be critical for P30-GSDMD pore formation. The effect of NSA in inhibiting pyroptosis was examined using both primary and immortalized macrophages. To trigger the cellular responses leading to pyroptosis, macrophages were treated using LPS/nigericin or Salmonella typhimurium. For all conditions studied, 20 μM NSA had been shown to significantly reduce pyroptosis, as revealed by cell morphology change, aggregation of P30-GSDMD, PI uptake, release of cytokines, and cell death. NSA did not inhibit GSDMD cleavage, but effectively inhibited cell death induced by transient expression of the p30 fragment in HEK239T cells. According to the western blot analysis, GSDMD in the cell lysate mainly migrated as a dimer band and a distinctly higher order oligomer band. In the presence of 20 μM NSA, the dimer band remained but the higher order oligomer band disappeared. Direct binding of NSA to GSDMD was then confirmed using purified recombinant human GSDMD (expressed in E. coli) and NSA–biotin. After incubation, biotin–NSA could be immunoprecipitated using an anti-GSDMD antibody, and vice versa, GSDMD could be pulled down using streptavidin beads. Administration of NSA to mice significantly increased the survival rate of LPS-induced sepsis.
Fig. 3. Chemical structures of all the compounds discussed.
In parallel, Wu and coworkers identified two compounds, disulfiram, a drug used to treat alcohol addiction,55 and Bay 11-7082, a previously identified NF-κB inhibitor56 that inhibited GSDMD pore formation. Disulfiram was identified through screening 3752 small molecules from the Harvard ICCB-Longwood collection using a fluorogenic liposome leakage assay. The efficacy of hit compounds was first examined using human and mouse macrophages in which inflammasome was activated by treatment of LPS electroporation or nigericin. Disulfiram was found to inhibit cytokine (IL-1β) release and pyroptotic cell death in both human and mouse monocyte/macrophage cell lines. Disulfiram is rapidly metabolized to diethyldithiocarbamate (DTC) in cells.57 In the presence of Cu2+, both disulfiram and DTC are potent inhibitors of pyroptosis with an IC50 of ∼0.4 μM. In an LPS-induced murine sepsis model, disulfiram pretreatment prevented the death of mice that were injected with a low dose of LPS (15 mg kg–1) for at least 96 h (while 3/8 of control mice died), and delayed death by approximately 24 hours among mice injected with a high-dose LPS (50 mg kg–1). Serum IL-1β concentrations were also reduced in the disulfiram-pretreated mice.
Since disulfiram is capable of inactivating reactive Cys residues by covalent modification,58 Wu et al. confirmed that disulfiram could covalently modify human GSDMD Cys191 using tryptic digestion coupled with mass spectrometry analysis and mutational studies. Pre-treatment of disulfiram with N-acetylcysteine abolished the activity of disulfiram, further confirming the importance of reactivity toward the thiol group. Furthermore, disulfiram has been shown to inhibit caspases (including caspases 1 and 11) by reacting with the active site Cys. Thus, it might work on multiple targets, including the upstream caspases and the substrate GSDMD. A set of analogues were synthesized, and their activity was evaluated. But none of them was more potent than disulfiram.
In an effort to identify additional inhibitors, an additional 86 050 compounds in the ICCB-Longwood collection were screened using the same fluorogenic liposome leakage assay. Only two hit compounds emerged from the screening effort that inhibited pyroptotic cell death by more than 50%, including a pan-caspase inhibitor z-VAD-fmk and a previously known NF-κB inhibitor Bay 11-7082. Surprisingly, Bay 11-7082 functioned with a similar mechanism to disulfiram, namely through covalently modifying the reactive Cys191. In addition, as a thiol-reactive compound it also inhibited caspases. Overall the potency of Bay 11-7082 was weaker than that of disulfiram.
Disulfiram and Bay 11-7082 have been shown to inhibit multiple steps leading to pyroptosis and inflammatory cytokine release, which is not surprising due to the non-specific nature of Cys-modification and the presence of reactive thiol groups in many molecules along the inflammatory pathway. Although not tested, NSA is likely to have similar effects to a certain degree. The repeated identification of cysteine-modifying drugs as inhibitors of GSDMD, especially through random screening, is intriguing. In the liposome leakage assay, purified human GSDMD was first incubated with liposome in the presence of testing compounds for 1 h, before caspase 11 was added to cleave and activate GSDMD. Thus thiol-reactive inhibitors, which can potentially modify and inactivate both caspase 11 and GSDMD, are clear winners in the campaign. Insights gained from these inhibitor hunting efforts echoed the mechanistic studies, which also established Cys191/192 as a key residue in the function of GSDMD.
As alkylating compounds likely targeting cysteine residues, GSDMD and MLKL are not likely to be the only proteins altered by NSA. Similarly, while disulfiram and Bay 11-7082 have been confirmed to modify and inactivate both caspases and GSDMD, their targets in vivo are not limited to these proteins. There is a long way to develop them into anti-infection medicines. But these studies provide the proof-of-principle that GSDMD pore-formation can be effectively inhibited and validate GSDMD as a viable pharmaceutical target. It is interesting to note that while Cys191 is crucial for binding and interaction with the thiol-modifiers, the C191A mutation affects death to a much lesser extent than exposure to the modifier. Since mutation is 100% conversion of the target residue, while chemical modification is rarely complete, this observation suggests that attachment of a large chemical moiety like NSA at the Cys191 site may disrupt oligomerization to a greater extent than the removal of the reactivity of the thiol group. It is also possible that the more significant inhibition is due to modification of other molecules in the pyroptosis pathway upstream of GSDMD that contains free cysteines.
4.2. Inhibitors targeting GSDMD NT at an unknown site
Similar to pyroptosis, the release of NETs is also a double-edged sword. While it captures microorganisms and is beneficial in infections, at the same time it also exposes autoantigens and promotes thrombosis.59 In an effort to identify inhibitors of mitogen-induced NETosis, a library of 182 710 small molecules was screened and a compound class with the pyrazolo-oxazepine scaffold was identified. A non-toxic member of this class, LDC7559 (Fig. 3), was found to bind to GSDMD. GSDMD was identified via a pull down assay, in which a derivative of LDC7559 was immobilized and used as the bait, and LDC7559 was used to confirm that the binding is specific through competitive elution. Subsequent mass spectrometry peptide fingerprinting analysis of the pulldowns revealed the identity of GSDMD as the most enriched protein.
As an inhibitor of GSDMD is expected to interfere with GSDMD-dependent processes, the effect of LDC7559 on pyroptosis was evaluated in human primary monocytes THP-1 and murine iBMDMs. LDC7559 was found to inhibit IL-1β release upon inflammasome activation, and significantly blocked the lethal effect of both human and murine GSDMD NT domains transfected into HEK293T cells at concentrations of 1 and 5 μM, respectively. These studies confirmed that LDC7559 worked directly through blocking the activity of the GSDMD NT domain, rather than interfering with the activation and cleavage process. The exact molecular details of the mechanism of inhibition remain to be determined.
4.3. GSDMD-derived peptide
A rationally designed GSDMD-inhibitor is derived from its own caspase digestion site, peptide FLTD (D is the site of digestion).60 FLTD was found to specifically bind to inflammatory caspases, and thus it was fused to a halomethyl ketone group to generate N-acetyl-Phe-Leu-Thr-Asp-chloromethylketone (Ac-FLTD-CMK), aiming at covalently modifying the catalytic site Cys residue. Ac-FLTD-CMK specifically inhibited GSDMD cleavage by inflammatory caspases 1, 4, 5, and 11 in both biochemical and cellular assays. Ac-FLTD-CMK also effectively inhibited pyroptosis as revealed by decreased release of IL-1β and reduced cell death. It does not directly interact with GSDMD, but rather works through the prevention of its cleavage by the inflammatory caspases. During cleavage GSDMD forms a stable complex with its corresponding caspases, 1 and 11. The interaction is recapitulated by its cleavage site peptide, FLTD. The work leads to new insights into the design of bona fide GSDMD inhibitors targeting the activation step. The effectiveness of targeting the cleavage process has been demonstrated through blocking the active site of the corresponding caspases. A more desirable inhibitor should directly target GSDMD to reduce the potential side effect of targeting caspases. The crystal structure of the peptide FLTD in complex with human caspase-1 has been reported, which illustrated critical interactions between the substrate and the enzyme. A more complete picture showing the complex of GSDMD with caspases will be valuable in directing the design of inhibitors that target the interaction site through binding to GSDMD.
5. Concluding remarks
While the invasion of microbes leads to infection, in many cases patients die of excessive activation of their own immune system. Programmed cell death including pyroptosis is an important mechanism of host self-protection, yet excessive activation of the system leads to a dysregulated cytokine release (named cytokine storm), which causes multiple organ failure and eventually patient death. When the immune system escalates to eliminate the invading alien microbes, (patho)physiological responses to the cytokine storm and signal molecules cause complications which are the true culprit behind many patient deaths. A series of significant papers have established the critical role of GSDMD in pyroptosis and septic shock.31,33,34,46 Deficiency of GSDMD has been shown to increase the survival rate in murine sepsis models, posing it as a potential target for sepsis treatment. Many studies targeting the same pathway focused on the reduction of the levels of pro-inflammatory cytokines such as TNFα and interleukin using soluble cytokine receptors or neutralizing antibodies.61–63 Inhibiting pyroptosis by targeting GSDMD could effectively block the release of different cytokines, thereby reducing the overall excessive inflammation in sepsis. Thus, GSDMD represents a novel target and a new anti-infection approach. In addition, the function of pyroptosis in inflammation is not limited to sepsis. Inflammasome activation contributes to many other human diseases, ranging from cardiovascular disease, inflammatory bowel disease, type II diabetes, rheumatoid arthritis, cancer, to Alzheimer's disease.64–68 GSDMD inhibitors may also be useful in the treatment of these disorders.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
Y. W. is supported by AHA Great Rivers Affiliate Grant-in-Aid 17GRNT33410327, NSF CHE-1709381, and NIH, NIAID, R56AI137020 and R21AI142063. Z. L. is supported by NIH, NHLBI, HL123927.
References
- Jawad I., Luksic I., Rafnsson S. B. J. Glob. Health. 2012;2:010404. doi: 10.7189/jogh.02.010404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger R. P. Intensive Care Med. 2012;39:165. [Google Scholar]
- Davis R. P., Miller-Dorey S., Jenne C. N. Clin. Transl. Immunol. 2016;5:e89. doi: 10.1038/cti.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann C. Am. J. Respir. Crit. Care Med. 2016;193:259. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- Dombrovskiy V. Y., Martin A. A., Sunderram J., Paz H. L. Crit. Care Med. 2007;35:1244. doi: 10.1097/01.CCM.0000261890.41311.E9. [DOI] [PubMed] [Google Scholar]
- Laxminarayan R. Lancet. 2016;387:168. doi: 10.1016/S0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- Mayr F. B., Yende S., Angus D. C. Virulence. 2014;5:4. doi: 10.4161/viru.27372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone R. C. Ann. Intern. Med. 1991;115:457. doi: 10.7326/0003-4819-115-6-457. [DOI] [PubMed] [Google Scholar]
- van der Poll T., Opal S. M. Lancet Infect. Dis. 2008;8:32. doi: 10.1016/S1473-3099(07)70265-7. [DOI] [PubMed] [Google Scholar]
- Cinel I., Opal S. M. Crit. Care Med. 2009;37:291. doi: 10.1097/CCM.0b013e31819267fb. [DOI] [PubMed] [Google Scholar]
- Stearns-Kurosawa D. J., Osuchowski M. F., Valentine C., Kurosawa S. and Remick D. G., Annual Review of Pathology-Mechanisms of Disease, ed. A. K. Abbas, S. J. Galli and P. M. Howley, 2011, vol. 6, p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S., Tanaka M. Nat. Rev. Immunol. 2017;17:333. doi: 10.1038/nri.2016.153. [DOI] [PubMed] [Google Scholar]
- Cookson B. T., Brennan M. A. Trends Microbiol. 2001;9:113. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- Labbe K., Saleh M. Cell Death Differ. 2008;15:1339. doi: 10.1038/cdd.2008.91. [DOI] [PubMed] [Google Scholar]
- Miao E. A. Nat. Immunol. 2010;11:1136. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P. Nature. 2015;526:642. doi: 10.1038/nature15632. [DOI] [PubMed] [Google Scholar]
- Liu X., Lieberman J. A. Adv. Immunol. 2017;135:81. doi: 10.1016/bs.ai.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. J., Gao W. Q., Shao F. Trends Biochem. Sci. 2016;42:245. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Man S. M., Karki R., Kanneganti T. D. Immunol. Rev. 2017;277:61. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglietti R. A., Dueber E. C. Trends Immunol. 2017;38:261. doi: 10.1016/j.it.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Yatim N., Cullen S., Albert M. L. Nat. Rev. Immunol. 2017;17:262. doi: 10.1038/nri.2017.9. [DOI] [PubMed] [Google Scholar]
- Jorgensen I., Rayamajhi M., Miao E. A. Nat. Rev. Immunol. 2017;17:151. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. Nature. 2015;526:660. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Kayagaki N. Nature. 2015;526:666. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- He W. T. Cell Res. 2015;25:1285. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollberger G. Sci. Immunol. 2018;3:aar6689. [Google Scholar]
- Chen K. W. Sci. Immunol. 2018;3:aar6676. doi: 10.1126/sciimmunol.aar6676. [DOI] [PubMed] [Google Scholar]
- Chen X. Cell Res. 2016;26:1007. doi: 10.1038/cr.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. Nature. 2016;535:111. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- Aglietti R. A. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7858. doi: 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt M. M., Hornung V. EMBO J. 2016;35:2167. doi: 10.15252/embj.201695415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I., Zhang Y., Krantz B. A., Miao E. A. J. Exp. Med. 2016;213:2113. doi: 10.1084/jem.20151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Nature. 2016;535:153. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborgi L. EMBO J. 2016;35:1766. doi: 10.15252/embj.201694696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl S. Science. 2018;362:956. doi: 10.1126/science.aar7607. [DOI] [PubMed] [Google Scholar]
- Chen Y. L. Am. J. Transl. Res. 2016;8:5685. [PMC free article] [PubMed] [Google Scholar]
- Liu Z. J. J. Pineal Res. 2017;63:e12414. doi: 10.1111/jpi.12414. [DOI] [PubMed] [Google Scholar]
- Shakoory B. Crit. Care Med. 2016;44:275. doi: 10.1097/CCM.0000000000001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., Dinarello C. A. Front. Pharmacol. 2018;9:1157. doi: 10.3389/fphar.2018.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill E. EMBO J. 2018;37:e98321. doi: 10.15252/embj.201798321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig R. Eur. J. Immunol. 2018;48:584. doi: 10.1002/eji.201747404. [DOI] [PubMed] [Google Scholar]
- Evavold C. L. Immunity. 2018;48:35. doi: 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S. M., Kanneganti T. D. Cell Res. 2015;25:1183. doi: 10.1038/cr.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs S. B., Miao E. A. Trends Cell Biol. 2017;27:673. doi: 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. Nucleic Acids Res. 2018;46:W296. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Gao W., Shao F. Trends Biochem. Sci. 2017;42:245. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Kuang S. Y. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10642. doi: 10.1073/pnas.1708194114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J., Xia S., Liu X., Lieberman J., Wu H. Nature. 2018;557:62. doi: 10.1038/s41586-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathkey J. K. J. Biol. Chem. 2017;292:14649. doi: 10.1074/jbc.M117.797217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Structure. 2018;26:778. doi: 10.1016/j.str.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. J., Liu X., Zhao J., Xia S., Ruan J., Luo X., Kim J., Lieberman J. and Wu H., BioRxiv, 2018, 10.1101/365908. [DOI] [Google Scholar]
- Sanchez R., Riddle M., Woo J., Momand J. Protein Sci. 2008;17:473. doi: 10.1110/ps.073252408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathkey J. K. Sci. Immunol. 2018;3:aat2738. doi: 10.1126/sciimmunol.aat2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. Cell. 2012;148:213. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Wright C., Moore R. D. Am. J. Med. 1990;88:647. doi: 10.1016/0002-9343(90)90534-k. [DOI] [PubMed] [Google Scholar]
- Pierce J. W. J. Biol. Chem. 1997;272:21096. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- Shen M. L., Johnson K. L., Mays D. C., Lipsky J. J., Naylor S. Biochem. Pharmacol. 2001;61:537. doi: 10.1016/s0006-2952(00)00586-4. [DOI] [PubMed] [Google Scholar]
- Castillo-Villanueva A. Int. J. Parasitol.: Drugs Drug Resist. 2017;7:425. doi: 10.1016/j.ijpddr.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M. J., Radic M. J. Immunol. 2012;189:2689. doi: 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6792. doi: 10.1073/pnas.1800562115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. Biochim. Biophys. Acta. 2011;1813:878. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- Chaudhry H. In Vivo. 2013;27:669. [PMC free article] [PubMed] [Google Scholar]
- Schulte W., Bernhagen J., Bucala R. Mediators Inflammation. 2013:165974. doi: 10.1155/2013/165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. Oncol. Rep. 2018;40:1971. doi: 10.3892/or.2018.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I. Cancer Immunol. Res. 2013;1:145. doi: 10.1158/2326-6066.CIR-13-0102. [DOI] [PubMed] [Google Scholar]
- Man S. M. Nat. Rev. Gastroenterol. Hepatol. 2018;15:721. doi: 10.1038/s41575-018-0054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Transl. Stroke Res. 2018;9:555. doi: 10.1007/s12975-018-0666-3. [DOI] [PubMed] [Google Scholar]
- Cheng K. T. J. Clin. Invest. 2017;127:4124. doi: 10.1172/JCI94495. [DOI] [PMC free article] [PubMed] [Google Scholar]