Abstract
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) loci and their associated (cas) genes encode an adaptive immune system that protects prokaryotes from viral1 and plasmid2 invaders. Upon viral (phage) infection, a small fraction of the prokaryotic cells are able to integrate a small sequence of the invader’s genome into the CRISPR array1. These sequences, known as spacers, are transcribed and processed into small CRISPR RNA (crRNA) guides3–5 that associate with Cas nucleases to specify a viral target for destruction6–9. Although, CRISPR-cas loci are widely distributed throughout microbial genomes and often display hallmarks of horizontal gene transfer10–12, the drivers of CRISPR dissemination remain unclear. Here we show that spacers can recombine with phage target sequences to mediate a form of specialized transduction of CRISPR elements. Phage targets in phage 85, ΦNM1, ΦNM4, and Φ12 can recombine with spacers in either chromosomal or plasmid-borne CRISPR loci in Staphylococcus, leading to either the transfer of CRISPR-adjacent genes or the propagation of acquired immunity to other bacteria in the population, respectively. Our data demonstrate that spacer sequences not only specify the targets of Cas nucleases, but also can promote horizontal gene transfer.
Bioinformatic analysis of CRISPR-cas loci have uncovered hallmarks of horizontal gene transfer (HGT)10–12 and CRISPR-Cas modularity13–15. Phylogenies based on either CRISPR repeats or the universal Cas1 protein revealed poor correlations between bacterial species trees, suggesting evidence of HGT of CRISPR-cas loci between distantly related bacterial species16–18. Furthermore, genomic studies have suggested that CRISPR systems evolved from a common ancestor and co-opted a diverse set of effector modules, potentially via HGT13–15. Plasmid conjugation16,18,19 and bacteriophage transduction20, fundamental routes for HGT21, have been implicated in the dissemination of CRISPR-cas loci22. Transduction occurs during viral infection, and can be divided into generalized or specialized23. Generalized transduction is a rare event that occurs when the phage machinery packages any DNA from the infected (donor) cell and subsequently delivers this DNA into a recipient cell. It is mediated by pac but not cos phages. In contrast, specialized transduction is a more specific event mediated by prophages (both pac and cos) that package genes located in the vicinity of their integration site. Recently it was shown that the type I-F CRISPR-cas locus of Pectobacterium atrosepticum can be mobilized through generalized transduction at rates ~10−8 transductants per virion22. Here we sought to determine if specific mechanisms are in place to mediate a more efficient horizontal gene transfer of CRISPR-Cas systems, their components, or their flanking sequences.
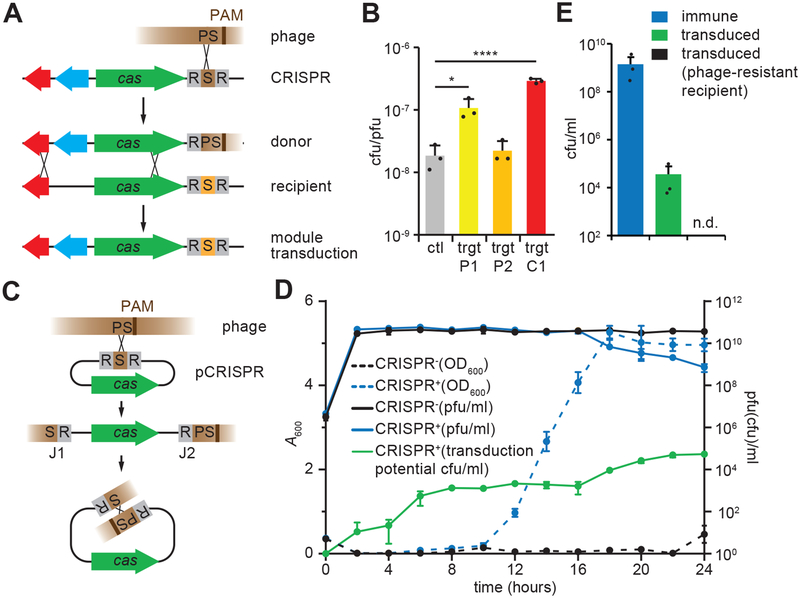
We explored whether recombination between newly acquired spacers and their targeted phage could mediate transduction. It is well established that even short sequences with homology to a phage directs recombination events that integrate the homologous DNA into the viral genome, leading to a drastic enhancement of the transduction rates of plasmids24–27, for example. The acquisition of a 30–40 base pair spacer sequence from the infecting virus during the CRISPR-Cas adaptive immune response would introduce homology between the phage genome and the CRISPR locus, and could facilitate recombination and elevated rates of transduction. Such a mechanism would lead to the transduction of genes adjacent to the CRISPR locus (Fig. 1A). To test this, we added an erythromycin resistance gene adjacent to the chromosomal type III-A system within the methicillin resistance cassette (SCCmec) of Staphylococcus aureus 08BA0217628, and inserted a target site for the first spacer of this CRISPR locus at two locations (P1 and P2) of the staphylococcal phage 85 genome29, or a new spacer (C1) matching orf28 (Fig S1A). This erythromycin-resistant strain was infected with each of the three phages or a non-targeted, wild-type phage as a control, and the lysates were used to transduce the marker into the wild-type strain. We observed that spacers P1 and C1 enhanced transduction of the antibiotic resistance cassette by one order of magnitude over the non-targeting control (Fig. 1B). Since there are no differences in the viability of the recipient cells (Fig. S1B), these results suggest that recombination between spacers P1 and C1 and the phages harboring their targets can direct the transfer of genomic locations adjacent to the CRISPR locus at rates that exceed those observed for generalized transduction (mediated by the non-targeted control phage). We also investigated the possibility of spacer-mediated transduction of entire chromosomal CRISPR-cas loci to CRISPR-lacking strains. Using two different empirical systems, the type I-F CRISPR-Cas system of Pseudomonas aeruginosa and the DMS3vir phage30 and the type II-A CRISPR-Cas system of Streptococcus thermophilus and the 2972 phage1, with adjacent chromosomal markers to track transduction (Fig. S1C), we observed generalized but not spacer-mediated transduction of the entire CRISPR-Cas system (Fig. S1D,E). Most likely this is a result of the presence of only one region of homology for integration (Fig. S1C).
Figure 1. Transfer of CRISPR-Cas elements through spacer-mediated transduction.
(A) Transduction of CRISPR-adjacent loci (blue arrow) after phage recombination with a chromosomal CRISPR-cas locus; “S” CRISPR spacer, “PS” protospacer. (B) Transducing particle production from S. aureus strain 08BA02176 tagged with an erythromycin resistance cassette. Liquid cultures were infected at a MOI (multiplicity of infection) of 50. Wild-type phage (ctl) or phage with the target site of the first spacer of the CRISPR array inserted at 15 kb (P1) or 20 kb (P2) positions on the phage genome were used. Wild-type phage was also used to infect a strain with a chromosomally inserted phage-targeting spacer (C1). Mean + STD of 3 biological replicates are reported. Two-tailed unpaired t-test was used to calculate P value, *p = 0.023, ****p = 0.00058 (C) Transduction of a plasmid containing a CRISPR-cas locus after recombination with the phage genome. J1 and J2, recombination junctions. (D) Growth and phage titers of infected cultures of bacteria containing plasmids with either the type II-A CRISPR-Cas system (CRISPR+) or the empty vector control (CRISPR−). Liquid cultures were infected at a MOI of 1 with ΦNM4γ4. The growth of cultures was determined by measurement of absorbance at 600 nm (A600). Titers, plaque forming units/ml (pfu/ml), and levels of transducing-immune phage particles, colony forming units/ml (cfu/ml), were determined every time point. No transducing-immune particles were detected using a vector control. Mean ± STD of 3 biological replicates are reported. (E) Levels of transduction during CRISPR adaption were determined by mixing cells at a 1 CRISPR+ (naïve, without a targeting spacer) to 5 CRISPR− ratio and infecting the mix with ΦNM4γ4 at an MOI of 1. Cultures were collected 20 hours post infection and survivors resulting from the acquisition of new spacers or the transduction of the adapted pCRISPR were measured by enumerating colonies on plates containing different antibiotic combinations. As a control, the experiment was repeated after mixing non-CRISPR resistant cells with naïve CRISPR+ staphylococci. Mean ± STD of 3 biological replicates are reported.
Although most CRISPR-Cas systems reside on chromosomes, an important fraction of CRISPR loci has been reported to be carried on plasmids31. Plasmid-borne CRISPR loci offer unique advantages for their lateral transfer via spacer recombination: the increased copy number elevates the probability of recombination27,32 and the circular nature allows the insertion of the entire CRISPR-cas locus into the phage genome, facilitating its packaging and re-circularization into the recipient host (Fig. 1C). To explore this, we first tested if the transduction of CRISPR-carrying plasmids can be mediated by the newly acquired spacers. We infected S. aureus RN422033 cells (which lack endogenous CRISPR-cas loci) carrying the type II-A locus of Streptococcus pyogenes5 (Fig. S2A) into the 2.9 kb staphylococcal plasmid pC194 (pCRISPR, conferring chloramphenicol resistance), with a staphylococcal pac phage carrying a virulent mutation, ΦNM4γ433. Staphylococci harboring pCRISPR, but not an empty vector control, recovered at 12 hours (Fig. 1D) through the acquisition of new spacers (Fig. S2B), at the same time as the phage titers began to decline (Fig. 1D). We then checked for the presence of pCRISPR-transducing particles in phage filtrates (which also contain infective phages) by infecting S. aureus RN4220 recipients and selecting for chloramphenicol-resistant colonies. We detected an increase of transduction starting at 16 hours post-infection, with a peak frequency of of 1 transduced colony per 104 plaque forming units in the filtrates (Figs. 1D and S2C–F). To determine if transduction could transfer expanded pCRISPR loci (and thus immunity) to CRISPR-Cas negative cells when both coexist in a mixed population, we combined naïve pCRISPR-harboring cells (chloramphenicol-resistant) and phage-sensitive recipients (kanamycin- and erythromycin-resistant) at a 1:5 ratio and infected with ΦNM4γ4. We were able to recover transduced colonies (resistant to all three antibiotics) at a frequency of ~ 10−5 with respect to CRISPR-adapted colonies (chloramphenicol-resistant) (Figs. 1E and S2G). Altogether, the data presented in Figs. 1 and S2 demonstrate that CRISPR-Cas plasmids can spread to naïve cells through transduction during the course of the CRISPR-Cas immune response.
Next, we investigated whether this transduction requires the presence of acquired spacers, as it seems to be the case for the transduction of chromosomal CRISPR-adjacent loci (Figs. 1AB). First, we compared the spacer repertoires of CRISPR-resistant and CRISPR-transduced cells, obtained 20 hours post-infection, using next-generation sequencing. Spacer sequences from four biological replicates were mapped onto the ΦNM4γ4 genome and plotted against their average number of reads (Fig. 2A, Supplementary Data File 1). The relative frequency of transduced spacers was consistent in each experiment (Fig. S3A) and we did not detect a correlation between the frequency of spacer acquisition and transduction (Fig. S3B and S3C). Instead, we found that spacers present in CRISPR-transduced cells were enriched in the 20–30 kb region of the phage, and depleted from the immediately preceding 5–20 kb region (Fig. 2A). These results suggest that there are CRISPR-cas loci containing specific spacer sequences that favor or limit their transduction. To test this we selected four spacer sequences, two with high (H1, H2), one with intermediate (I), and one with low (L) transduced/resistant ratio (Fig. 2A). These spacers were cloned into pCas9, a pCRISPR derivative unable to support new spacer acquisition due to the absence of cas1, cas2 and csn233. After infection with ΦNM4γ4, the populations carrying the high transducer spacers (H1 and H2) produced approximately three orders of magnitude more transducers than populations carrying the intermediate (I) or low (L)-transducing spacers (Fig. 2B). These results indicate that the sequence of the acquired spacer determines the frequency of transduction of the CRISPR-cas locus which harbors it. Similar results were obtained with a plasmid-borne type III-A system, which displayed enhanced levels of transduction when the system contains a spacer matching the infecting phage genome (Figs. S4AB).
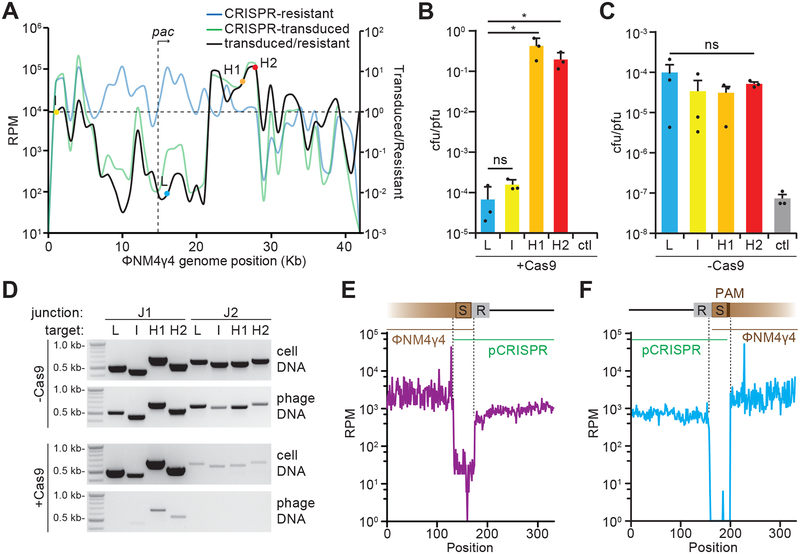
Figure 2. Spacers sequences determine frequency of pCRISPR transduction.
(A) Cultures containing pCRISPR were infected with ΦNM4γ4 at a MOI of 1. Expanded CRISPR arrays were analyzed by next generation sequencing. The reads per million (RPM) values of the acquired spacers were plotted against the ΦNM4γ4 genome (blue line, CRISPR-resistant). Lysates containing ΦNM4γ4 as well as pCRISPR transducing particles were collected at 20 hours post infection and were used to infect cells without CRISPR-cas at an MOI of 1. Cells were plated to collect pCRISPR transductants and their spacer content was analyzed by next-generation sequencing (green line, CRISPR-transduced). The ratio of transduced spacers over resistant spacers was also plotted for each acquired spacer (black line). Select spacers with low-(L), intermediate-(I), and high (H1, H2) ratios, along with phage pac site, are indicated. Mean of 4 replicates was reported. (B) Transduction efficiency, measured as the ratio of transductant colony forming units (cfu) to the total number of plaque forming units (pfu) of phage in the lysates obtained after infection of staphylococci harboring pCas9 carrying the L, I, H1 and H2 spacers, or no targeting spacer as a control (ctl). Mean + STD of 3 biological replicates are reported. Two-tailed unpaired t-test was used to calculate P values; ns, not significant (p = 0.1554), *p (H1) = 0.024, *p (H2) = 0.024 (C). Same as (B) but measuring the transduction of pSpacer plasmids; i.e. not carrying cas9. Mean + STD of 3 biological replicates are reported. One-way ANOVA was used to calculate P values; ns, not significant (p = 0.59). (D) PCR products after amplification of J1 and J2 junctions (Fig. 1C) either from lysates (phage DNA) or infected cells (cell DNA) obtained after infection of cells harboring pCas9 (+Cas9) or pSpacer (-Cas9) plasmids with L, I, H1 or H2 spacers. These results were representative of three independent experiments. (E) Next generation sequencing of phage DNA harvested after infection of cells pCas9(H1). Reads were aligned to the putative J1 junction. Reads per million (RPM) for each nucleotide within this region are shown. Dotted lines indicate a 75-nucleotide sequence that is unique to the junction. Results from a single experiment are shown. (F) Same as (E) but for the J2 junction. Results from a single experiment are shown.
To test if the spacer sequence itself could impact transduction we generated pSpacer plasmids containing only a repeat-spacer (H1, H2, I or L)-repeat unit, without cas9. After infection with ΦNM4γ4, we found that all four spacers equally increased the rate of pSpacer transfer when compared to a CRISPR array containing a control spacer that does not match the phage genome (Fig. 2C), suggesting that Cas9 targeting impacts the rate of transduction. Indeed, if pCRISPR transduction occurs through the formation of recombinants between the acquired spacer and the phage protospacer sequence, these recombinants will maintain a full target in one of the recombination junctions which could be cleaved by Cas9. To investigate this, staphylococci carrying the pSpacer or pCas9 plasmids were infected with ΦNM4γ4 and DNA was isolated from bacterial pellets (containing infected cells) or culture supernatants (containing virions) for PCR amplification of both recombination junctions (Fig. 2D) as well as chromosomal and viral genes as controls for the fractionation (Fig. S4C). pSpacer/phage recombinants were detected at both junctions, for all spacer sequences, both in infected cells and virions, a result that explains the equal transduction levels of these plasmids (Fig. 2C). In contrast, pCas9/phage recombinants were also detected but PCR products were much less abundant for the targeted junction (J2) in infected cells (Fig. 2D). In virions, we only detected the non-targeted junction (J1) for the constructs containing H1 and H2 spacers, along with a faint PCR product for the I-spacer construct; but we were unable to detect the PAM-flanked (J2) junction (Fig. 2D). These results were corroborated by next-generation sequencing of DNA extracted from the virion fraction after infection of cells containing pCas9(H1). We found abundant reads spanning the non-targeted phage-CRISPR junction (J1) (Fig. 2E), while the targeted junction (J2) had relatively fewer reads (Fig. 2F). Altogether, these experiments demonstrate two important aspects of spacer-mediated recombination. First, recombination between the spacer sequence in pCRISPR and the protospacer sequence in ΦNM4γ4 results in the formation of hybrid DNA molecules which can be encapsidated into virions during infection. This does not depend on the host RecA (Fig. S5A) and can also occur via the staphylococcal cos phage Φ12 (Fig. S5B). Second, Cas9 targeting of the PAM-flanked spacer/protospacer junction within these molecules reduces the efficacy of their packaging into viral capsids and therefore the efficiency of transduction.
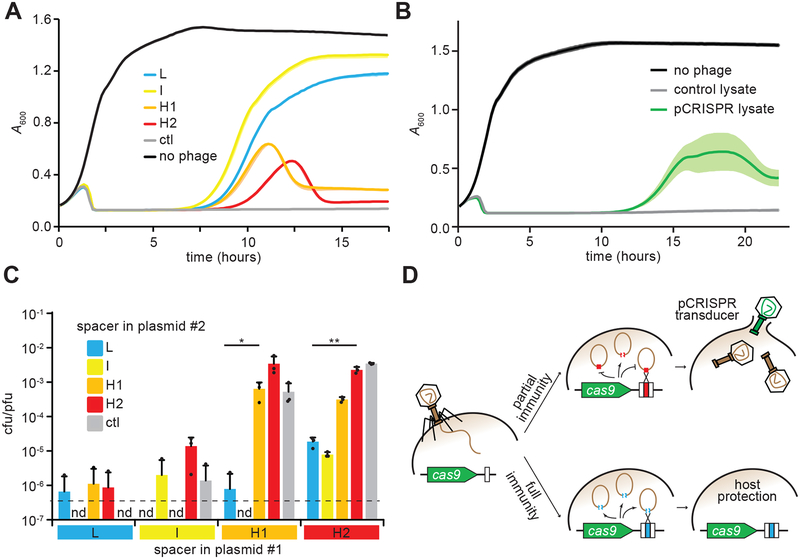
The presence of phage particles containing spacer/phage recombinants from infected CRISPR-immune cells suggests that incomplete protection of the host allows for the formation and release of the CRISPR-containing virions. Inefficient CRISPR immunity can result from at least two scenarios. One possibility is that the immunity provided by the acquired spacer can be bypassed by phages containing target mutations, known as “escapers”; in this case the spacers that target regions with high rate of mutation in the phage genome will be more prone to transduction. However the experiments described in Figure S6 ruled out this scenario. A second possibility is that the acquired spacer provides only partial immunity, i.e. a proportion of the adapted cells can be lysed by the phage and produce CRISPR-transducing particles. To test this, we measured immunity using an assay that reproduces the high multiplicity of infection faced by cells that acquire new spacers33. In these conditions, CRISPR-Cas systems programmed with the L and I spacers enabled the complete recovery of the host, and cells containing H1 and H2 spacers showed only a partial recovery of the infected staphylococci (Fig. 3A). To determine the strength of the immunity mediated by all the spacers present in the CRISPR-transducing particles (not just H1 and H2), we followed the survival of naïve staphylococci upon infection with phages collected during the CRISPR-Cas immune response (Fig. 1D, 22 hour time-point), which contain both ΦNM4γ4 as well as transducing particles that can provide immunity against the phage. We obtained similar partial survival curves to those provided by the H1- and H2-containing CRISPR-Cas systems (Fig. 3B). These results suggest that the complete destruction of the virus by the intermediate- and low-transducing spacers limits the formation of CRISPR-transducing particles. If true, these spacers should have a dominant effect on the H1 and H2 spacers, i.e. they will reduce their frequency of transduction. To test this, we combined different pairs of pCRISPR plasmids (with different antibiotic-resistance markers) in the same cell, infected them with ΦNM4γ4 and counted the number of transductants (Fig. 3C). We found that the combination of CRISPR-Cas systems harboring I or L spacers with either H1 or H2 spacers resulted in a low transduction frequency, i.e. the effect of I and L spacer predominate over H1 and H2. Similarly, a reduction in the transduction of a second plasmid (pE194, 2.9 kb) is observed in fully protected cells containing I and L spacers when compared to partially immune cells with H1 and H2 spacers (Fig. S7A). Finally, we looked at the transduction of pCRISPR plasmids harboring an inactivating mutation in Cas9 (dCas98). Corroborating our hypothesis, the reduction in immunity caused by this mutation enhanced the transduction rate for spacers I, H1, and H2 (Figs. S7BC). Altogether, these results demonstrate that spacer/phage recombination is the primary driver of spacer-enhanced transduction and that spacer sequences mediating highly efficient CRISPR immunity prevent the transduction of the CRISPR-cas locus.
Figure 3. Spacers that mediate high pCRISPR transduction provide poor immunity to the host.
(A) Cell survival of cultures containing pCas9 with spacers L, I, H1 or H2, monitored as growth after infection with ΦNM4γ4 at a MOI of 1. No spacer (ctl) and no phage experiments were used as controls. Growth was recorded by following the absorbance at 600 nm (A600) of the cultures. Mean - STD of 3 biological replicates are reported. (B) Cell survival of cultures lacking CRISPR-Cas immunity, monitored as growth after infection with a cell lysate obtained after infection of staphylococci harboring pCRISPR (or an empty vector as a control) with ΦNM4γ4 at a MOI of 1. This lysate contains both phage as well as plasmid transducing plasmids. Growth was recorded by following the absorbance at 600 nm (A600) of the cultures. Mean ± STD of 5 biological replicates are reported. (C) Transduction efficiency of plasmid #2, measured as transductant cfu / total pfu present in lysates of staphylococci harboring two pCas9 plasmids with different spacers, collected 90 minutes after infection with ΦNM4γ4 at a MOI of 50. Limit of detection is 1.5 cfu/ml (dotted line). Mean + STD of 3 biological replicates are reported. Two-tailed unpaired t-test was used to calculate P values, *p = 0.034, **p = 0.0012 (D) Model for spacer-mediated p CRISPR transduction. During the type II-A CRISPR-Cas immune response, efficient spacers fully protect the host and prevent the generation of both phage progeny and transducing particles. In contrast, spacers that mediate poor immunity allow the lysis of some of the infected hosts, enabling the release of pCRISPR transducing particles generated after the recombination of the plasmid and the phage genome at the spacer/protospacer sequences.
Here we show that spacers acquired by CRISPR-Cas systems can perform a form of specialized transduction that requires their recombination with the phage target as well as incomplete CRISPR immunity (Fig. 3D). The recombination between the spacer and its viral target connects the locus with the packaging sequences on the phage genome, enabling foreign DNA uptake at much higher rates than observed during typical generalized transduction in our experimental system. If the CRISPR locus resides in the host chromosome, this recombination can mediate the transfer of genes adjacent to the CRISPR locus and thus facilitate the dissemination and exchange of cas modules. If the CRISPR-Cas system resides in a circular genetic element, spacer-mediated recombination leads to the spread of CRISPR immunity among naïve CRISPR-negative hosts. Interestingly, the host RecA is not required for protospacer-spacer recombination, a result suggesting that this phenomenon is mediated by the phage’s own machinery, which can significantly elevate recombination rates34,35. Our analysis of four different spacers showed that transduction rates are higher for spacers that mediate poor cleavage of the pCRISPR/ΦNM4γ4 recombinants. We believe that the next-generation sequencing experiment shown in Fig. 2A, which includes data for all transduced spacers, supports this correlation: the sequences of the spacers mediating the lowest transduction rates (5 to 20 kb of the ΦNM4γ4 genome) are located around the pac site, which a previous study showed to be one of the regions of this phage best targeted by Cas936.
Our data shows that both types of spacer-enhanced transduction events we describe here occur at low frequencies, and in nature could be happening at even lower frequencies. However, as it is the case with most situations involving horizontal transfer of genetic material, the importance of these events relies not so much in their rate of occurrence, but in their capacity to increase the genetic pool of the recipients37; given the appropriate environmental conditions, the genes and plasmids transferred though spacer-mediated transductions could provide a crucial selective advantage to the population. For example, the exchange of CRISPR-adjacent modules could expand the repertoire of cas genes of a CRISPR locus and generate the genetic diversity13–15 required to stay ahead in the arms race with different phages and their anti-CRISPR inhibitors38. Plasmids and potentially excisable (circular) genomic islands harboring CRISPR-Cas loci are relatively common31,39, and their spread though spacer-mediated transduction could provide critical spacers and/or full defense cassettes for phage defense. Even if the transduced CRISPR-Cas locus does not harbor the most efficient spacers, as our data indicates, their spread could help increase the spacer diversity necessary to prevent the raise of phage escapers40 and/or provide partial defense to enable the acquisition of more potent sequences. Finally, it is worth noting that CRISPR-Cas systems have been identified within phage genomes41–43. Although we do not know their origin, it is possible that these arose by the type of spacer-mediated recombination we demonstrated in this study.
It is interesting to consider that acquired spacers could have a dual role during CRISPR immunity: a major one in the generation of crRNA guides and a minor one in mediating HGT. In support of this idea, recent work has found Tn7-like transposons that harbor CRISPR arrays without effector cas genes, in which dissemination is likely facilitated by spacers44. Circumstantial evidence that this second role may be important comes from the mechanism of crRNA biogenesis. In the S. pyogenes type II-A CRISPR-Cas pathway, the 10 nucleotides at the 5’ end of the spacer sequence on the crRNA are degraded and eliminated from the Cas9 ribonucleoprotein complex, making this region of the spacer dispensable for targeting8. As recombination increases with the extent of homology, it is possible that this additional 10 nucleotides in the spacer DNA could facilitate transduction (Fig. S8). Type III-A spacers also have 3’ end sequences that eliminated from the spacer RNA sequence during CRISPR-RNA maturation45, and while not shortened, crRNA-target homology at the 3’ end of type I-E spacers is not necessary for targeting46. Therefore, it is tempting to speculate that the acquisition of spacers has evolved not only to incorporate foreign sequences for defense against predation, but also as a means of hijacking the mobilization machinery of these elements to spread through prokaryotic populations.
Methods
Bacterial strains and growth conditions
Culture of Staphylococcus aureus RN422047 was carried out in brain-heart infusion (BHI) medium at 37°C with agitation at 220 revolutions per minute. Liquid experiments were carried out in 3 milliliters of medium in 15 ml conical tubes unless otherwise noted. S. aureus media was supplemented with 10 μg/ml chloramphenicol, 10 μg/ml erythromycin, or 25 μg/ml kanamycin for plasmid maintenance and/or chromosomal marker selection.
Culture of Streptococcus thermophilus was carried out in M17 media supplemented with 10% lactose at 37°C without agitation, unless otherwise noted. Liquid experiments were carried out in 5 milliliters of media in 15 ml conical tubes. M17 media was supplemented with 5 μg/ml chloramphenicol, 200 μg/ml spectinomycin, or 2.5 μg/ml erythromycin for chromosomal marker selection.
Culture of Pseudomonas aeruginosa was carried out in LB media at 37°C with agitation at 180 revolutions per minute. LB media was supplemented with 100 μg/ml streptomycin or 30 μg/ml gentamycin for chromosomal marker selection
All strains are listed in Table S1.
Quantification of CRISPR-Cas transducing particles
In S. aureus, overnight cultures of pWJ4033 or pC19448 were diluted 1:100 in fresh BHI with appropriate antibiotics and 5 mM CaCl2. At OD600 = 0.4, cultures were infected with ΦNM4γ433 at a multiplicity of infection (MOI) 1. Phage was collected at indicated time points and filtered with 0.45-μm syringe filters (Acrodisc). Harvested phage were then used to infect lawns of S, aureus strain OS249 suspended in 50% BHI supplemented with 5 mM CaCl2 at an MOI of 1 on a BHI base supplemented with erythromycin and chloramphenicol to select for recipient cells and CRISPR-Cas transduction. For quantification of transducing particles produced from strains already containing CRISPR-immunity, overnight cultures were diluted 1:100 in fresh BHI with appropriate antibiotics and 5 mM CaCl2. At OD600 = 0.4, cultures were infected with ΦNM4γ4 or Φ12γ336 at a MOI of 50. 90 minutes post infection, phage were collected and filtered with 0.45-μm syringe filters (Acrodisc). Harvested phage were then used to infect lawns of OS2 suspended in 50% BHI supplemented with 5mM CaCl2 at an MOI of 1 on a BHI base supplemented with erythromycin and chloramphenicol to select for recipient cells and CRISPR-Cas transduction. Phages that were not of sufficient titers to infect at an MOI of 1 were supplemented with the appropriate phage prepared from RN4220.
Detection of spacer acquisition
To check for spacer acquisition in S. aureus, transduced colonies were resuspended in colony lysis buffer (250 mM KCl, 5 mM MgCl2 50 mM Tris-HCl at pH 9.0, 0.5% Triton X-100), treated with 200 ng/μl lysostaphin and incubated at 37°C for 20 minutes, then 98°C for 10 minutes. Samples were centrifuged and supernatant was used for PCR amplification with primers L400 and H50.
CRISPR adaptation and escaper phage generation
For P. aeruginosa, to monitor the effect of increased homology between the CRISPR system and the phage DMS3vir genome, we cultured PA14-Sm in the presence of DMS3vir and isolated a phage-resistant mutant that had acquired an additional spacer targeting the phage, following procedures previously described30. Next, we isolated DMS3vir ‘escape’ mutants by inoculating a 96 well plate with 200 μl of the CRISPR-resistant PA14-Sm strain and ~6*107 DMS3vir. After a 24-hour incubation at 37°C phages were isolated by chloroform extraction and spotted onto a lawn of the CRISPR-resistant PA14-Sm. Individual ‘escape’ phage clones were isolated, followed by sequencing of the amplicon containing the protospacer and PAM sequences. A single ‘PAM-escape’ mutant was used in the transduction assays (G>A, position 25926) along with the WT DMS3vir phage.
For S. thermophilus, we isolated bacterial colonies that had acquired spacers in the erythromycin-tagged CRISPR1 locus of JAV28 following infection by phage 2972 using procedures previously described50. Genomic DNA from strain JAV33 was amplified and sequenced with AV638-AV724 and found to have a spacer targeting the top strand beginning at position 26,553 of 2972. Phage 2972 was passaged on this strain for escapers on soft-agar. Single plaques were isolated and re-passaged to single plaques on JAV33. Phage DNA was extracted by boiling the phage and 2972α1 DNA was amplified and sequenced with AV868-AV869. 2972α1 contained a mutation in the PAM region (A>G, 26,588)
Quantification of transduction
For S. aureus, overnight donor cultures were diluted 1:100 in fresh BHI with appropriate antibiotics and 5 mM CaCl2. At OD600 = 0.4, cultures were infected with either ΦNM4γ4 at a MOI of 1 or 85α1–3 and ΦNM1γ6 at an MOI of 50. Following lysis of the culture at 2 hours, phages were collected and filtered with a 0.45-μm syringe filters (Acrodisc). Overnight recipient cultures were diluted 1:100 in fresh BHI with appropriate antibiotics and 5 mM CaCl2. At OD600 = 0.4, cultures were infected at an MOI of 1 with the transducing phage. 20 minutes post-infection, 40 mM of sodium citrate was added to the cultures. For erythromycin transduction, the cells were incubated for an additional 40 minutes then pelleted and washed twice with fresh BHI supplemented with 40 mM sodium citrate, while for chloramphenicol transduction cells were washed immediately. Cells were then plated on BHI plates supplemented with the antibiotics selecting for the recipient strain and transduction marker along with 20 mM sodium citrate for type II-A plasmids and no sodium citrate for type III-A plasmids.
For P. aeruginosa, bacterial lawns with near-confluent lysis were generated by mixing 200 μl of PA14-Sm on overnight cultures with 20μl of ~104 PFU DMS3vir and 10 mL soft LB agar. Phage only controls were included by applying the same protocol, but excluding the addition of bacteria. After 24-hour incubation at 37°C, phages were harvested by soaking the lawns in 3 mL of M9 salts buffer for 1 hour at room temperature followed by chloroform extraction and titration of the resulting phage stock. As recipients, we used P. aeruginosa PA14 ΔCRISPR-Cas51 transformed with pHERD30T (conferring gentamycin resistance). 10 mL LB overnight culture supplemented with 30 μg mL−1 gentamycin of each recipient was spun down (3000 rpm, 10 min) and re-suspended in 1 mL of LB. 100 μl of lysate was then added and statically incubated for 25 minutes. Each culture was then spun down and the whole culture was plated on LB agar supplemented with 100 μg mL−1 streptomycin and 30 μg mL−1 gentamycin (to prevent carry over of PA14-Sm cells). To estimate transduction frequency, 48 colonies were picked per replicate experiment, and screened by PCR using primers specific for the CRISPR 2 locus primers CR2_F-CR2_R.
For S. thermophilus, transducing phage stocks were made by infecting mid-log growth JAV33 at 42°C supplemented with 10 mM CaCl2 with either 2972 or 2972α1 at an MOI of 1. Phage stocks were harvested and filtered using 0.45-μm syringe filters (Acrodisc) after the culture had cleared. JAV27 were used as recipient cells and were grown to at 42°C supplemented with 10 mM CaCl2 and infected at an MOI of 0.5 when cultures reached OD600 = 0.4. 10 minutes following infection, 20 mM sodium citrate was added to the cultures. After 1 hour incubation at 42°C, the cultures were washed two times in M17 media supplemented with 20 mM sodium citrate and then plated on erythromycin M17 plates. Transductants were confirmed by streaking out colonies on M17 chloramphenicol plates to confirm antibiotic resistance engineered into the CRISPR3 locus.
Detection of phage-CRISPR junctions
Overnight cultures were diluted 1:100 in fresh BHI with appropriate antibiotics and 5 mM CaCl2. At OD600 = 0.4, cultures were infected with ΦNM4γ4 at a MOI of 50 for targeting strains or 1 for non-targeting strains. Phages were collected from indicated strains 60 minutes post-infection. Supernatants were filtered using a 0.45-μm filter and then concentrated with Ultra-4 100k centrifugal 50-ml spin columns (Amicon). Concentrates were resuspended with DNase I buffer, 20 mM Tris-HCl pH 8.0 and 2 mM MgCl2 and re-concentrated two times. The suspension was then treated with 25 units of DNase I (Sigma) for one hour. Following DNAse I treatment, the enzyme was inactivated by heating at 70°C for 10 minutes and the addition of 5 mM EDTA. Phages were then incubated with 8 units of proteinase K (NEB) and 0.5% SDS at 37°C for one hour. Phage DNA was isolated using a phenol/chloroform/isoamyl alcohol extraction (Fisher). Cellular DNA was collected 15 minutes and 60 minutes post infection for non-targeting strains and targeting strains respectively. Approximately 109 cells were pelleted and resuspended in 100 μl of 50 mM EDTA and 1 mg/ml lysostaphin (AMBI Products) and incubated at 37°C for one hour. DNA was then extracted with the Wizard genomic purification kit (Promega) according to the manufacturer’s instructions. For the non-PAM junction, primer JM117 was used with NP255, AV547, AV469, AV471 for L, I, H1, and H2, respectively. For the PAM junction, primer L400 was used with AV457, AV458, AV456, AV459 for L, I, H1, and H2, respectively. For loading controls oGG38-oGG40 were used to amplify gp14 and JW96-W964 for recA.
High-throughput sequencing
Overnight cultures of pWJ40 were diluted 1:100 in fresh BHI with appropriate antibiotics and 5 mM CaCl2. At OD600 = 0.4, cultures were infected with ΦNM4γ4 at a MOI of 1. 20 hours post-infection DNA was collected from recovered cells (CRISPR-resistant). Phages were also collected and filtered with 0.45-μm syringe filters (Acrodisc). Overnight cultures of OS2 were diluted 1:100 in fresh BHI with appropriate antibiotics and 5 mM CaCl2. At OD600 = 0.4, cultures were infected with ΦNM4γ4 collected from the pWJ40 culture at a MOI of 1. 20 hours post-infection DNA was collected from recovered cells (CRISPR-transduced). Spacers were amplified with RH50 and JW655-JW662 for sample barcoding. The sequences of the oligonucleotides used in this study are listed in Table S2. Adapted bands were gel-extracted and subjected to Illumina MiSeq sequencing. Data analysis was performed in Python. Spacer reads were extracted from the raw MiSeq FASTA files and aligned to the phage genome. Number of reads and PAM were designated for each spacer. Spacers were normalized as reads per million and plotted against the ΦNM4γ4 genome in 2000-base-pair bins.deep sequence phage-CRISPR DNA junctions, cultures containing spacer H1 were infected with ΦNM4γ4 at a MOI of 50 and phages were collected 90 minutes post infection. Phage DNA was isolated as described above. DNA was then prepped with the Illumina TruSeq Nano kit according to the manufacturer’s instructions. Prepped DNA was then subject to NextSeq sequencing. BWA-MEM (arXiv:1303.3997v1) was used to align sequenced DNA to the PAM junction, which contains 200 base pairs of the upstream CRISPR sequence (leader and direct repeat) and 205 base pairs of the downstream phage sequence (Spacer, PAM, and phage genome) or the repeat junction, which contains 205 base pairs of the upstream phage sequence (phage genome and spacer, and 200 base pair downstream CRISPR sequence (direct repeat and downstream plasmid sequence). A python script was then used to sort and bin reads spanned the full 75-nucleotide read length allowing for one mismatch.
Phage titer assay
Phage titer assays were performed as previously described52.
Efficiency of plaquing assays
Efficiency of plaquing assays were performed as previously described52.
Simulation of CRISPR immunization
Simulation of CRISPR immunization was performed as previously described53.
Strain construction
To make the recA knockout JAV9, the allelic replacement system developed by Wenyan Jiang using pWJ244 was applied as previously described36. Briefly, pAV44 was transformed into RN4220 and integrants were isolated. Double crossover events were selected for by a temperature sensitive cat targeting Cas9 phagemid, pWJ326. RecA deletion was confirmed by primers outside the homology arms, AV223 and AV224. To make JAV21, OS2 was infected with ΦNM1γ652 at an MOI of 1 to produce transducing particles carrying the genomic erythromycin cassette. These particles were used to infect JW26336 as described in quantification of transduction. Colonies that were resistant to kanamycin and erythromycin were struck out 2 times on plates supplemented with 20mM sodium citrate, kanamycin, and erythromycin. JAV29 and JAV32 were constructed by transforming suicide vectors pAV253 and pAV282. Integration was confirmed using primers AV594 and AV812 for pAV253 and AV648 and AV525. JAV33 was made by infecting RN4220 at MOI of 1 in soft agar with ΦNM4γ4. After a 24-hour incubation, a resistant colony was picked, restreaked two times, and confirmed to be insensitive to ΦNM4γ4 infection.
To create a P. aeruginosa PA14 strain carrying a streptomycin resistance cassette immediately adjacent to the Type I-F CRISPR-Cas system in the genome (PA14-Sm, with the Sm gene inserted at position 2937360), we used homologous recombination. The streptomycin (Sm) resistance gene and its promoter were PCR amplified from pBAM1-Sm54 using primers pB_Sm_F and pB_Sm_R, and inserted into the NheI restriction site of pHERD30T, flanked by amplicons FL1 (flank1, generated using primer pairs FL1_F and FL1_R) and FL2 (flank2, generated using primer pairs FL2_F and FL2_R). To select for recombinants, a crRNA targeting the junction between the flanking sequences was expressed from the same plasmid.
To create S. thermophilus strains, PCR products were generated with homology arms approximately 2 kilobases long that flank antibiotic resistant cassettes and transformed into the wildtype strains. For JAV27, CRISPR1 was eliminated by amplifying homology arms with AV664-AV665 and AV666-AV667. The spectinomycin resistance cassette was amplified from pLZ12spec55 with AV672-AV673 and a three piece Gibson assembly was used to create the final product for transformation. Also in JAV27, CRISPR3 was eliminated by amplifying homology arms with AV668-AV669 and AV682-AV683. The chloramphenicol resistance cassette was amplified from pC19448 with W1055-W1056 and a three piece Gibson assembly was used to create the final product for transformation. JAV27 was made by first knocking out CRISPR1 and then repeating the procedure for CRISPR3. For JAV28, CRISPR1 was tagged with erythromycin resistance by amplifying homology arms with AV667-AV692 and AV693-AV694. The erythromycin cassette was amplified from pE19456 with AV177-AV695 and a three piece Gibson assembly was used to create the final product for transformation. To transform assembled DNA fragments into cells, an overnight culture was washed once in chemically-defined medium (CDM)57, then diluted 1:100 in one milliter of CDM. Following 1.5 hours of incubation at 37°C, 10 μl of the Gibson product along with 1 μM ComS17–24 peptide58 (LPYFAGCL, Genescript) were added. Following a 4-hour incubation, cells were plated with the appropriate antibiotic and incubated for 36 hours.
Phage construction
To create phages to study transduction in S. aureus 08BA0217628, phage 8529 was used to infect this strain at a high MOI on soft-agar. 85α1 was isolated for its ability to form plaques on 08BA02176. To make 85α2, the 08BA02176 type III-A target was inserted site early-genome. 85α1 was passaged on soft-agar on a strain containing pAV247, a plasmid containing ~1 kilobase phage-homology arms where a small, unessential portion of the phage genome was replaced with the type III-A spacer 1 target. This phage stock was then passaged on soft-agar on a strain containing pGG1252, a plasmid containing a CRISPR-Cas system that targets the portion of the phage replaced with the 08BA02176 spacer 1 target. Plaques were picked from this passage and re-passaged to single plaques on soft-agar a second time. Phages were then amplified and sequenced with oGG38-oGG40 to confirm target insertion. To make 85α3, the 08BA02176 type III-A target was inserted site mid-genome. 85α1 was passaged on soft-agar on a strain containing pAV282, a plasmid containing ~1 kilobase phage-homology arms with an insertion into the phage genome with the type III-A spacer 1 target. This phage stock was then passaged on soft-agar on a strain containing pAV284, a plasmid containing a CRISPR-Cas system that targets the portion of the phage interrupted with the 08BA02176 spacer 1 target. Plaques were picked from this passage and re-passaged to single plaques on soft-agar a second time. Phages were then amplified and sequenced with AV876-AV877 to confirm target insertion.
Plasmid construction
All plasmids were constructed using electro-competent cells as described elsewhere52. The sequences of the oligonucleotides used in this study are listed in Table S2. To create recA allelic exchange vector pAV44, a three-piece Gibson assembly was performed using W1005-W1055 to amplify pWJ24436, with AV206-AV208 and AV207-AV209 to amplify the homology arms from RN4220. AV149, pAV150, pAV153, pAV155, high- and low-transducing spacers targeting ΦNM4γ4, were assembled by using BsaI cloning described in detail elsewhere59. Primer pairs AV404-AV405, AV406-AV407, AV412-AV413, and AV416-AV417 were annealed and ligated into pDB11459, to construct the respective plasmids. To make Φ12γ336 targeting plasmids, pAV293, pAV294, pAV295, and pAV296, BsaI cloning was used to insert JW600-JW601, JW604-JW605, JW620-JW621, and JW695-JW696 into pDB114 respectively. To make pAV158, pAV159, pAV162, pAV164, and pAV165, one piece Gibson assembly was performed using H235-H236 to remove cas9 from pAV149, pAV150, pAV153, pAV155, and pDB114, respectively. To transfer high and low-transducing spacers to a pE19456 background, a two-piece Gibson assembly was used. AV176 and AV177 were used to amplify pE194 and AV423-AV424 were used to amplify the tracrRNA, cas9, and CRISPR array cassette. pAV149, pAV150, pAV153, and pAV155 were used as templates for pAV175, pAV173, pAV174, and pAV176, respectively. To make pAV185, the last 10-basepairs of H1 were complemented. BsaI cloning was used to insert annealed oligonucleotides AV485-AV486 into pDB114. pAV195 was made with a one-piece Gibson assembly, where pAV185 was amplified with H235-H236 to remove cas9. To create phage 8529 editing plasmid pAV247 a three-piece Gibson was performed where pC19448 was amplified with AV186-AV204, and phage homology arms were amplified AV607-AV611 and AV609-AV610. To create the construct to tag the type III-A locus with erythromycin a two-piece Gibson assembly was performed, where pTM40260 was amplified with AV590-AV591 and the homology arm was amplified with AV622-AV623 from 08BA02176 and grown in strain TM1760. To create phage 85 editing plasmid pAV281 a three-piece Gibson was performed where pC194 was amplified with AV186-AV204, and phage homology arms were amplified AV862-AV864 and AV863-AV865. To add a spacer that targets phage 85 (5’-TTTCAACATTCTTCAACATACGCTGTCCTTGTGAGT-3’) to 08BA02176, pAV282 was made with a 3-piece Gibson assembly, where pTM402 was amplified with AV590-AV591, and homology arms were amplified with AV879-AV880 and AV878-AV881. pAV282 was grown in TM17. To make phage 85 portal-targeting plasmid pAV284, BsaI cloning was used to insert AV866-AV867 into pDB114. To make dcas9 contstructs pAV305, pAV306, pAV307, pAV308, and pAV309 gibson assembly was performed where B338-B339 were used to amplify cas9 from pDB114 and B337-B340 were used to amplify the plasmid backbone and spacer from pAV149, pAV150, pAV153, pAV155, and pDB114, respectively.
Statistics and Reproducibility
All experiments were independently reproduced three times unless stated otherwise in the figure legend.
Data availability statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Raw sequencing data is available upon request.
Code availability statement
All code used in this study is available upon request.
Supplementary Material
Acknowledgements.
We would like to thank Joshua W. Modell, Jakob T. Rostol, and Alissa M. Pham for helpful discussion and critical reading of the manuscript. JWM also provided strains pAV293–296. AV is supported by the Arnold O. Beckman Postdoctoral Fellowship. LAM is supported by the Rita Allen Scholars Program and an NIH Director’s Pioneer Award (DP1GM128184–01). The work carried out by ERW and SM was supported by the Biotechnology and Biological Sciences Research Council (BB/N017412/1) and Natural Environment Research Council (NE/M018350/1).
Footnotes
Supplementary information
Supplementary information Figures S1–S6.
Competing Interests
L.A.M. is a cofounder and Scientific Advisory Board member of Intellia Therapeutics and a cofounder of Eligo Biosciences. R.B. is a cofounder and Scientific Advisory Board member of Intellia Therapeutics, a cofounder of Locus Biosciences, an advisor to Inari Ag, and a shareholder of DuPont and Caribou Biosciences. ERW and SM declare no conflict of interest.
The authors have no conflicting financial interests.
References
- 1.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712, (2007). [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA & Sontheimer EJ CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouns SJ et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carte J, Wang R, Li H, Terns RM & Terns MP Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22, 3489–3496, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jore MM et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol 18, 529–536, (2011). [DOI] [PubMed] [Google Scholar]
- 7.Samai P et al. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell 161, 1164–1174, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasiunas G, Barrangou R, Horvath P & Siksnys V Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A 109, E2579–2586, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Aravind L, Grishin NV, Rogozin IB & Koonin EV A DNA repair system specific for thermophilic Archaea and Bacteria predicted by genomic context analysis. Nucleic Acids Res 30, 482–496, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 13, 722–736, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen R, Embden JD, Gaastra W & Schouls LM Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol 43, 1565–1575, (2002). [DOI] [PubMed] [Google Scholar]
- 13.Shmakov SA, Makarova KS, Wolf YI, Severinov KV & Koonin EV Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci U S A 115, E5307–E5316, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin EV & Makarova KS Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back. Genome Biol Evol 9, 2812–2825, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westra ER, Dowling AJ, Broniewski JM & Houte S v. Evolution and Ecology of CRISPR. Annual Review of Ecology, Evolution, and Systematics 47, 307–331, (2016). [Google Scholar]
- 16.Haft DH, Selengut J, Mongodin EF & Nelson KE A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol 1, e60, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty S et al. Comparative network clustering of direct repeats (DRs) and cas genes confirms the possibility of the horizontal transfer of CRISPR locus among bacteria. Mol. Phylogenet. Evol 56, 878–887, (2010). [DOI] [PubMed] [Google Scholar]
- 18.Godde JS & Bickerton A The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol 62, 718–729, (2006). [DOI] [PubMed] [Google Scholar]
- 19.Millen AM, Horvath P, Boyaval P & Romero DA Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS One 7, e51663, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinder ND & Lederberg J Genetic exchange in Salmonella. J. Bacteriol 64, 679–699, (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas CM & Nielsen KM Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol 3, 711–721, (2005). [DOI] [PubMed] [Google Scholar]
- 22.Watson BNJ, Staals RHJ & Fineran PC CRISPR-Cas-Mediated Phage Resistance Enhances Horizontal Gene Transfer by Transduction. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Touchon M, Moura de Sousa JA & Rocha EP Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol 38, 66–73, (2017). [DOI] [PubMed] [Google Scholar]
- 24.Orbach MJ & Jackson EN Transfer of chimeric plasmids among Salmonella typhimurium strains by P22 transduction. J. Bacteriol 149, 985–994, (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deichelbohrer I, Alonso JC, Luder G & Trautner TA Plasmid transduction by Bacillus subtilis bacteriophage SPP1: effects of DNA homology between plasmid and bacteriophage. J. Bacteriol 162, 1238–1243, (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick RP, Edelman I & Lofdahl S Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. J. Mol. Biol 192, 209–220, (1986). [DOI] [PubMed] [Google Scholar]
- 27.Maniv I, Jiang W, Bikard D & Marraffini LA Impact of different target sequences on Type III CRISPR-Cas immunity. J. Bacteriol 198, 941–950, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golding GR et al. Whole-genome sequence of livestock-associated ST398 methicillin-resistant Staphylococcus aureus Isolated from humans in Canada. J. Bacteriol 194, 6627–6628, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwan T, Liu J, DuBow M, Gros P & Pelletier J The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A 102, 5174–5179, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westra ER et al. Parasite Exposure Drives Selective Evolution of Constitutive versus Inducible Defense. Curr. Biol 25, 1043–1049, (2015). [DOI] [PubMed] [Google Scholar]
- 31.Lange SJ, Alkhnbashi OS, Rose D, Will S & Backofen R CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res 41, 8034–8044, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann BA & Slauch JM Transduction of low-copy number plasmids by bacteriophage P22. Genetics 146, 447–456, (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heler R et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Paepe M et al. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-like recombinases. PLoS Genet 10, e1004181, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopes A, Amarir-Bouhram J, Faure G, Petit MA & Guerois R Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res 38, 3952–3962, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modell JW, Jiang W & Marraffini LA CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544, 101–104, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polz MF, Alm EJ & Hanage WP Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29, 170–175, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bondy-Denomy J, Pawluk A, Maxwell KL & Davidson AR Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho Sui SJ, Fedynak A, Hsiao WW, Langille MG & Brinkman FS The association of virulence factors with genomic islands. PLoS One 4, e8094, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Houte S et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minot S et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21, 1616–1625, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seed KD, Lazinski DW, Calderwood SB & Camilli A A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494, 489–491, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sebaihia M et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet 38, 779–786, (2006). [DOI] [PubMed] [Google Scholar]
- 44.Peters JE, Makarova KS, Shmakov S & Koonin EV Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci U S A 114, E7358–E7366, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatoum-Aslan A, Maniv I & Marraffini LA Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc. Natl. Acad. Sci. U.S.A 108, 21218–21222, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenova E et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A 108, 10098–10103, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kreiswirth BN et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712, (1983). [DOI] [PubMed] [Google Scholar]
- 48.Horinouchi S & Weisblum B Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol 150, 815–825, (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneewind O, Model P & Fischetti VA Sorting of protein A to the staphylococcal cell wall. Cell 70, 267–281, (1992). [DOI] [PubMed] [Google Scholar]
- 50.Hynes AP et al. Detecting natural adaptation of the Streptococcus thermophilus CRISPR-Cas systems in research and classroom settings. Nat Protoc 12, 547–565, (2017). [DOI] [PubMed] [Google Scholar]
- 51.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR & O’Toole GA The CRISPR/Cas Adaptive Immune System of Pseudomonas aeruginosa Mediates Resistance to Naturally Occurring and Engineered Phages. J. Bacteriol 194, 5728–5738, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldberg GW, Jiang W, Bikard D & Marraffini LA Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGinn J & Marraffini LA CRISPR-Cas systems optimize their immune response by specifying the site of spacer integration. Mol. Cell 64, 616–623, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Garcia E, Calles B, Arevalo-Rodriguez M & de Lorenzo V pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol 11, 38, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Husmann LK, Scott JR, Lindahl G & Stenberg L Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect. Immun 63, 345–348, (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horinouchi S & Weisblum B Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J. Bacteriol 150, 804–814, (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letort C & Juillard V Development of a minimal chemically-defined medium for the exponential growth of Streptococcus thermophilus. J. Appl. Microbiol 91, 1023–1029, (2001). [DOI] [PubMed] [Google Scholar]
- 58.Fontaine L et al. Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl. Environ. Microbiol 76, 7870–7877, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bikard D et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol 32, 1146–1150, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Claveau D, Vaillancourt JP, Roemer T & Meredith TC High-frequency transposition for determining antibacterial mode of action. Nat. Chem. Biol 7, 720–729, (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Raw sequencing data is available upon request.