Abstract
The genome of recombinant adeno-associated virus 2 (rAAV2) remains a promising candidate for gene therapy for cystic fibrosis (CF) lung disease, but due to limitations in the packaging capacity and the tropism of this virus with respect to the airways, strategies have evolved for packaging an rAAV2 genome (up to 5.8 kb) into the capsid of human bocavirus 1 (HBoV1) to produce a chimeric rAAV2/HBoV1 vector. Although a replication-incompetent HBoV1 genome has been established as a trans helper for capsid complementation, this system remains suboptimal with respect to virion yield. Here, a streamlined production system is described based on knowledge of the involvement of HBoV1 nonstructural (NS) proteins NS1, NS2, NS3, NS4, and NP1 in the process of virion production. The analyses reveal that NS1 and NS2 negatively impact virion production, NP1 is required to prevent premature termination of transcription of the cap mRNA from the native genome, and silent mutations within the polyadenylation sites of the cap coding sequence can eliminate this requirement for NP1. It is further shown that preventing the expression of all NS proteins significantly increases virion yield. Whereas the expression of capsid proteins VP1, VP2, and VP3 from a codon-optimized cap mRNA was highly efficient, optimal virion assembly, and thus potency, required enhanced VP1 expression, entailing a separate VP1 expression cassette. The final NS protein-free production system uses three-plasmid co-transfection of HEK293 cells, with one trans helper plasmid encoding VP1 and the AAV2 Rep proteins, and another encoding VP2-3 and components from adenovirus. This system yielded >16-fold more virions than the prototypic system, without reducing transduction potency. This increase in virion production is expected to facilitate greatly both research on the biology of rAAV2/HBoV1 and preclinical studies testing the effectiveness of this vector for gene therapy of CF lung disease in large animal models.
Keywords: human bocavirus, adeno-associated virus, viral packaging, rAAV/HBoV vector production
Introduction
Cystic fibrosis (CF) is a lethal autosomal recessive disorder caused by defects in a single gene, cystic fibrosis transmembrane conductance regulator (CFTR).1,2 Malfunction or absence of the CFTR protein leads to deficiencies in chloride and bicarbonate transport in epithelial cells of the respiratory, digestive, and reproductive tracts. Although CF affects multiple organs, chronic bacterial infection and inflammation of the CF lung are the primary causes of morbidity and mortality.3,4 In spite of the fact that the genetic basis of CF is well understood, developing a gene therapy for the life-threatening lung disease that characterizes this disease has been more challenging than anticipated because the gene-transfer agents used in clinical trials to date have proved inefficient in restoring CFTR expression in the human lung.5–7 Specifically, these were recombinant viral vectors, including recombinant adenovirus8–10 and recombinant adeno-associated virus 2 (rAAV2).11–13 More recent efforts have involved plasmid-mediated CFTR delivery, with some beneficial effect in stabilizing CF lung function recorded when inhaled repeatedly on a monthly basis over the course of a year.14 However, due to inefficiencies in CFTR gene expression, this approach has also been abandoned.
The human airway has evolved highly effective barriers (both extra- and intracellular) to infection by foreign agents, but these can be overcome by certain viruses.15,16 It was reasoned that it should be possible to take advantage of the natural tropisms of such human respiratory viruses in developing novel airway transduction vectors. Human bocavirus 1 (HBoV1) is an autonomous parvovirus that was discovered in 200517 and naturally infects human airways, causing acute infection of the lower and upper respiratory tracts in infants and young children.18 HBoV1 infection occurs most frequently in young children, and the seroprevalence for HBoV1 capsid-specific immunoglobulin G is 13% and 59% in children and adults, respectively. However, seroconversion does not appear to prevent repeat infection by HBoV1,19–22 and thus the viral capsid can be considered for use in adults. To produce a safe and efficient airway transduction vector, a chimeric vector, rAAV2/HBoV1, was developed. The natural airway tropism of the HBoV1 capsid enables rAAV2/HBoV1 to transduce polarized human airway epithelia efficiently cultured at an air–liquid interface (HAE-ALI) in vitro,23 as well as intact airways of human bronchial xenografts grown ex vivo.24
Potential concerns about the pathogenicity of HBoV1 were circumvented by combining just its capsid with the rAAV2 genome that has been widely used in human gene therapy trials.25,26 Of note, rAAV2 is the first gene transfer agent to have been approved by the Food and Drug Administration for treating genetic diseases,27 and it has been tested in CF clinical trials. Although rAAV2-mediated gene therapy failed to produce a detectable level of CFTR mRNA in the human CF lung, its safety profile was favorable, and its genome was shown to persist in human airway epithelia for >60 days.11,28 The episomal persistence of the rAAV2 genome and its ability to support prolonged gene expression have been documented for many other tissues as well.26,29–31 More importantly, the genome of HBoV1 is 18% larger than that of AAV2 (5.54 kb vs. 4.68 kb).32 Indeed, the HBoV1 capsid comfortably accommodates an oversized rAAV2 genome and is capable of delivering a full-length CFTR coding sequence along with transcriptional elements necessary for efficient CFTR mRNA expression. In summary, exploiting the advantages of two parvoviruses, the rAAV2/HBoV1 vector overcomes the shortcomings of the low apical tropism and small cargo capacity of rAAV2 that prevented its success as a vector for CF gene therapy.
It was demonstrated that apical infection of HAE-ALI cultures derived from CF patients with the CFTR expression vector AV2/HBc.CBAhCFTR resulted in efficient expression of the CFTR protein, and that it partially restored Cl− channel function.23 It was also demonstrated that the rAAV2/HBoV1 vector is capable of efficiently transducing ferret lung in vivo.24 The next goal was to study the airway transduction biology of rAAV2/HBoV1, as well as to determine whether the CFTR expression delivered using this vector is sufficient to restore lung function in the context of a CF ferret model.33 However, it was found that the capacity to conduct such studies was limited by the low efficiency of the current system for producing this vector. HBoV1 and AAV2 are both human parvoviruses but belong to different genera (Bocaparvovirus and Dependoparvovirus, respectively).34 Although the efficiency of packaging of the rAAV2 genome is similar for the AAV2 capsid and those of other AAV serotypes,35 it was significantly lower for the HBoV1 capsid (yield was 5% of that for the rAAV2 vector) when the prototypic system was used. It was reasoned that limitations in parvovirus cross-genera pseudopackaging could be due to suboptimal complementation of the HBoV1 capsid during production using the prototypic system. Due to a lack of a detailed understanding of HBoV1 biology when the prototypic system was established and the fact that eukaryotic expression vectors encompassing the HBoV1 cap cDNA fail to express capsid proteins, the initial production systems used a HBoV1 replication-incompetent genome as trans helper plasmid, which also expressed all the nonstructural (NS) proteins.36 However, an enhanced understanding of the genome structure of HBoV1 and of the transcription profiles of HBoV1 genes involved in replication, both in HAE-ALI following infection with virus37–41 and in HEK293 cells following transfection with proviral plasmid,42–44 have now enabled more rational approaches to improving rAAV2/HBoV1 vector production.
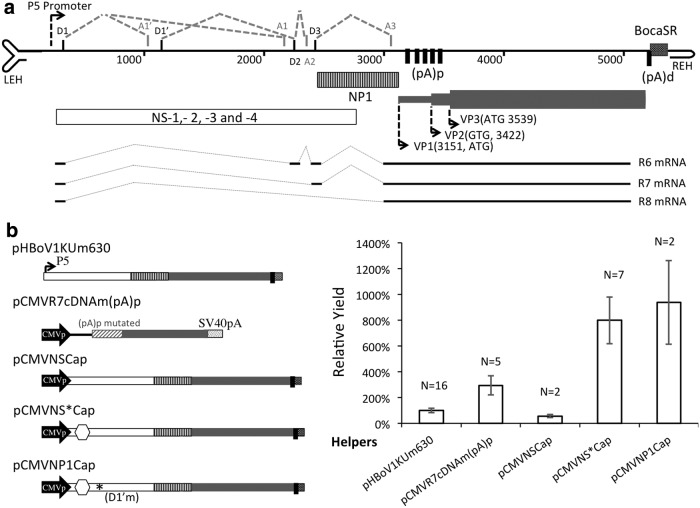
The HBoV1 genome (as depicted in Fig. 1a) consists of three sets of open reading frames (ORFs) and utilizes the sole promoter, P5, to express all of the viral proteins: five NS proteins (NS1, NS2, NS3, NS4, and NP1) and three capsid proteins (VP1, VP2, and VP3).43 One major ORF (at the left end of genome) encodes the NS proteins NS1–4, and a second (at the right end of genome) encodes the capsid proteins VP1–3. A third ORF (in the center of genome) encodes another NS protein, the nuclear protein NP1, which is required for efficient replication of viral DNA in all members of the Bocaparvovirus genus.45–47 As in AAV, the bocavirus cap gene contains overlapping ORFs that produce three capsid proteins VP1–3. The 74 kD VP1 and 64 kD VP2 ORFs encompass the 60 kD VP3 ORF (542 amino acids), containing 129 and 39 additional amino acids at their N-termini, respectively. Within the 3′ non-coding region of the positive strand (nucleotides [nt] 5,199–5,338), between the cap coding sequence and right-end hairpin, the HBoV1 genome encodes a 140 bp small non-coding RNA (BocaSR), which is transcribed by an intragenic promoter.41
Figure 1.
Structure of the human bocavirus 1 (HBoV1) genome and trans complementation of viral proteins for recombinant adeno-associated virus 2 (rAAV2)/HBoV1 production. (a) Schematic illustration of structures of the HBoV1 genome and the cap mRNAs. The HBoV1 genome (GenBank accession no. JQ92342232) harbors three sets of open reading frames (ORFs) that encode: the nonstructural (NS) proteins NS1–4 and NP1, and the capsid proteins VP1–3. Also, an intragenic promoter is used to transcribe the non-coding RNA BocaSR. The ORFs of capsid proteins VP1–3 overlap. All of the viral mRNA transcripts are generated from a single precursor transcribed from the P5 promoter, through alternative splicing at the marked donor (D) and acceptor (A) sites. Also, alterative polyadenylation occurs at both proximal polyadenylation (pA)p sites within the unique sequence of VP1/VP2 and distal polyadenylation (pA)d sites. mRNA R6, R7, and R8 behave similarly in terms of VP1–3 expression.43 NP1 is crucial to the generation of these cap mRNAs and to overcoming premature transcription caused by (pA)p. (b) Comparison of the effectiveness of HBoV1 helper plasmids. Left: Schematic illustration of helper plasmids. pHBoV1KUm630 is the replication-incompetent HBoV1 genome used in the prototype production system. pCMVR7cDNAm(pA)p encodes a modified HBoV1 R7 mRNA in which the (pA)p sites were eliminated by incorporating silent mutations. pCMVNSCap, pCMVNS*Cap, and pCMVNP1Cap are derivatives of pHBoV1KUm630 in which the P5 promoter is replaced with the CMV promoter. pCMVNScap expresses all of the structural and NS proteins of HBoV1, whereas pCMVNS*Cap lacks NS1/NS2 and pCMVNP1Cap lacks NS1-4. Right: Efficiencies of helper plasmids at left in producing rAAV2/HBoV1. Bars represent mean yield (N as indicated; error bars represent ±standard error of the mean [SEM] or difference [in the tests of N = 2]) relative to that of the prototype system, which typically yields 2.9 ± 0.5 × 1011 DNase I-resistant particles (DRP) of purified rAAV2/HBoV1 from forty 150 mm dishes of transfected HEK293 cells.
Elucidation of a HBoV1 transcription profile of genes involved in replication led to the investigation of the involvement of NS proteins in cap expression. It was found that polyadenylation signals in a region unique to VP1 and VP2 led to premature termination of transcription of cap mRNAs when NS proteins were not expressed. Yet, the expression of unneeded NS proteins from the prototypic helper hindered vector production. These polyadenylation sites (located in the center of the viral genome) were named as proximal polyadenylation sites, (pA)p, to distinguish them from the distal polyadenylation site (pA)d at the 3′ end of the genome. It was also found that NP1 is the only NS protein required for cap expression, and that it prevents such premature transcription termination, enabling production of the full-length cap mRNAs whose transcription terminates at the (pA)d site. Importantly, silent mutation of the (pA)p sites circumvented the requirement for NP1 in producing full-length 5′ modified cap mRNAs. These findings enabled capsid expression to be optimized in the absence of all five NS proteins, and a new rAAV2/HBoV1 NS-free production system to be produced that affords 16-fold higher yields than the prototype.
Methods
Chemicals and reagents
The proteasome inhibitor doxorubicin was purchased from Sigma–Aldrich (St. Louis, MO), and N-acetyl-L-leucine-L-leucine-L-norleucine (LLnL) was purchased from Boston Biochem (Cambridge, MA). Proteinase K was purchased from Roche (Indianapolis, IN). DNase I was purchased from MP Biochemicals (Santa Ana, CA). Ultroser G (USG) was purchased from Pall Corp. (Port Washington, NY). The firefly luciferase detection kit, Luciferase Assay System, was purchased from Promega (Madison, WI). The gaussia luciferase detection kit, BioLux® Gaussia Luciferase Assay Kit, was purchased from New England Biolabs (Ipswich, MA).
Generation of cis and trans helper plasmids for rAAV2/HBoV1 production
The cis rAAV transfer proviral plasmids for rAAV2/HBoV1 vectors were pAAV2.CMVmCherry-F5tg83fLuc and pAAV2.CBAmCherry-F5tg83gLuc.24 Adenovirus helper pAd4.1, AAV2 helper pAV-Rep2, and HBoV1 helper pHBoV1KUm630 are the previously described trans helper plasmids that were used in the prototype production system.23 Three previously reported HBoV1 capsid-expression plasmids—pCMVNSCap, pCMVNS*Cap, and pCMVR7cDNAm(pA)p—were also used in this study.43 In brief, pCMVNSCap is the derivative of pHBoV1KUm630. It was constructed by replacing the endogenous P5 with the sequence of the CMV promoter/enhancer. pCMVNS*Cap had been derived from pCMVNSCap by knocking out the expression of both NS1 and NS2 via a nonsense mutation at L65 of NS1/NS2. pCMVR7cDNAm(pA)p is a pcDNA3.0-based expression plasmid that harbors a modified HBoV1 R7 cDNA in which the (pA)p sites in the unique region to VP1 and VP2 were eliminated by introducing silent mutations.
In total, 10 new HBoV1 capsid-expression plasmids were constructed. Among these, three are intermediates, and seven (underlined below) were tested for effectiveness as helper plasmids for rAAV2/HBoV1 production. (1) pCMVNP1Cap expresses only NP1 and the capsid proteins; it was produced from pCMVNS*cap by introducing mutations that cause (a) early (E114) termination of translation of the NS3 ORF and (b) disruption of processing of the NS4 mRNA at splice-donor site D1. (2) pCMVint-NP1Cap also expresses only NP1 and the capsid proteins; it was constructed by replacing the 1.8 kb NS coding sequence of pCMVNSCap (up to splice-acceptor site A1) with a 134 bp sequence that encodes the intron IV of the human erythropoietin (Epo) gene. The two-in-one AAV-HBoV1 helper plasmids (3) pCMVNS*Cap-Rep2 and (4) pCMVint-NP1Cap-Rep2 were constructed by inserting the AAV2 rep expression cassette to pCMVNS*Cap and pCMVint-NP1Cap, respectively. (5) pcDNAoptVP, (6) pcDNAoptVP1(ATG), (7) pcDNAoptVP1, and (8) pcDNAoptVP2/3 were derived from a pcDNA3.0 variant that harbors a synthetic codon-optimized cap gene, and incorporate differences in the translation initiation sequences for VP1 and VP2, as detailed in the Results section. (9) pCMVoptVP1-Rep2 was produced from pAV-Rep2 by inserting the optVP1 expression cassette. (10) pAd4.1-CMVoptVP2/3 was produced from pAd4.1 by inserting the optVP2/3 expression cassette. The codon-optimized cap sequences for cloning into these helper plasmids were synthesized at GenScript (Piscataway, NJ). For ease of reference, the properties of all the HBoV1 capsid expression constructs tested in this study are summarized in Table 1.
Table 1.
HBoV1 capsid helper plasmids
| Helper name | Expression of HBoV1 proteins | Mutations of (pA)p sites | Cap codon-optimized | Other helper functions |
|---|---|---|---|---|
| pHBoV1KUm630 | NS1–4, NP1, VP1–3 | No | No | No |
| pCMVNSCap | NS1–4, NP1, VP1–3 | No | No | No |
| pCMVNS*Cap | NS3–4, NP1, VP1–3 | No | No | No |
| pCMVNP1Cap | NP1, VP1–3 | No | No | No |
| pCMVR7cDNAm(pA)p | VP1–3 | Yes | No | No |
| pCMVNS*Cap-Rep2 | NS3–4, NP1, VP1–3 | No | No | AAV2 Rep |
| pCMVint-NP1Cap-Rep2 | NP1, VP1–3 | No | No | AAV2 Rep |
| pCMVint-NP1Cap*-Rep2 | NP1 | No | No | AAV2 Rep |
| pcDNAoptVP | VP1–3 | Yes | Yes | No |
| pcDNAoptVP1(ATG) | VP1–3 | Yes | Yes | No |
| pCMVoptVP1-Rep2 | VP1 | Yes | Yes | AAV2 Rep |
| pAd4.1-CMVoptVP2/3 | VP2–3 | Yes | Yes | Adv VAI, E2a, E4 ORF6 |
HBoV1, human bocavirus 1; AAV2, adeno-associated virus 2; Adv, adenovirus.
Production of recombinant vector
rAAV2/HBoV1 vectors were generated in HEK293 cells by co-transfection with a rAAV proviral plasmid encoding gene of interest in cis (rAAV transfer vector) and helper plasmids at an equal molar ratio. In some cases, the cells were transfected with four plasmids, specifically the rAAV transfer vector, pAd4.1, pAV-rep2, and HBoV1 capsid-expression helper as in the previously described prototype system,23 and in others they were transfected with three plasmids either rAAV transfer vector, pAd4.1, and the two-in-one AAV-HBoV1 helper (pCMVint-NP1Cap-Rep2 or pCMVNS*Cap-Rep2) or rAAV transfer vector, pCMVoptVP1-Rep2, and pAd4.1-CMVoptVP2/3. In most of this study, virions were purified only from cell lysates. For such samples, cell pellets were collected at 72 h post transfection, at which time the cells were lysed in hypotonic buffer (10 mM Tris-HCl, pH 8.0) and treated with DNase I, and virions were subsequently purified through two rounds of CsCl ultracentrifugation and TaqMan polymerase chain reaction (PCR) used to determine titers as DNase I-resistant particles (DRP). The virion-containing fractions, whose densities ranged from 1.43 to 1.40 g/mL, were pooled and dialyzed against phosphate-buffered saline prior to use.
In the cases of samples for which virion secreted into the culture medium was also purified, virions were concentrated by PEG precipitation prior to further processing. In brief, after the cells were pelleted, the culture supernatants were mixed 4:1 with a 40% PEG-8000/2.5 M NaCl solution. After incubation on ice for 2 h, virions were precipitated by centrifugation at 3,000 g for 30 min and dissolved in 10 mM Tris-HCl, pH 8.0. Further purification steps and assessment of viral titers were as described for cell lysates.
TaqMan real-time PCR for quantification of the DRP in the virion preparations used sets of PCR primers and probes designed to identify the transgene, firefly luciferase cDNA, and gaussia luciferase cDNA.24 These primers were synthesized by IDT (Coralville, IA). The PCR reaction was performed, and results were analyzed using the Bio-Rad My IQ™ real-time PCR detection system and software (Bio-Rad, Hercules, CA), as described previously.24
Western blotting
rAAV2/HBoV1 (1.04 × 1010 DRP) or cell lysates from transfected HEK293 cells were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis using 10% gels. Following transfer to nitrocellulose membranes, Western blotting was performed, as previously described.43 The HBoV1 capsid proteins were probed with a rat anti-HBoV1 VP3 antiserum (recognizes the VP1, VP2, and VP3 proteins) at a dilution of 1:200 and horseradish peroxidase–conjugated anti-rat secondary antibody. After visualization of the HBoV1 capsid proteins, blots were re-probed with anti-β-actin antibody.
Cell culture and in vitro infection
HEK293 cells, which were used for virion production, were cultured as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin-streptomycin, and maintained in a 37°C incubator at 5% CO2. Human airway epithelial cultures were grown for 3 weeks at an air–liquid interface (HAE-ALI) by the In Vitro Models and Cell Culture Core of the University of Iowa, as previously described.48 Subsequent studies were performed using protocols that are compliant with University of Iowa policies and for which the Core has obtained Institutional Review Board approval. For apical infection of ALI cultures, 7.5 × 109 DRP of rAAV2/HBoV1 vector in a 50 μL inoculum was applied to the upper chamber of the transwell insert, and the epithelia were exposed to the virus for 16 h. With ∼7.5 × 105 cells present on each insert, the multiplicity of infection (MOI) was ∼10,000 DRP/cell. Infections of polarized HAE-ALI cultures were performed in the presence of proteasome inhibitors (5 μM doxorubicin and 40 μM LLnL), as described previously.23 The transduction assays for reporter expression of firefly luciferase or gaussia luciferase activity were conducted, as previously described,24 using the Luciferase Assay System from Promega or the BioLux® Gaussia Luciferase Assay Kit from New England Biolabs.
Results
Requirements for HBoV1 NS genes in the rAAV2/HBoV1 production system
It was hypothesized that the exclusion of any nonessential viral proteins from the rAAV2/HBoV1 production system would improve yield. The pCMVR7cDNAm(pA)p encodes a variant of the cap R7 mRNA in which the (pA)p sites are silently mutated. A previous study found that mutation of these (pA)p sites circumvented the requirement for NP1 in producing full-length 5′ modified cap mRNAs. pCMVR7cDNAm(pA)p was capable of expressing HBoV1 capsid proteins in the absence of all NS proteins (i.e., under NS-free conditions).43 In the first attempt to generate an NS-free vector production system, the effectiveness of pCMVR7cDNAm(pA)p in providing HBoV1 capsid complementation as a replacement for pHBoV1KUm630 was tested in the existing four-plasmid based prototypic system. HEK293 cells were transfected with the HBoV1 helper (pCMVR7cDNAm[pA]p or pHBoV1KUm630; Table 1) and the other three plasmids: the rAAV2 transfer vector, pAV-Rep2, and pAd4.1. In five independent preparations in which pCMVR7cDNAm(pA)p was used as helper, virion yield was 2.8 ± 0.7-fold higher than when pHBoV1KUm630 was used (Fig. 1b). Although this NS-free production system remains suboptimal, with the transfection of 40 plates (150 mm) yielding on average 8.7 ± 2.1 × 1011 DRPs, it represented a practical platform for further optimization.
In the prototypic system, the production of capsid from helper pHBoV1KUm630 is absolutely NP1 dependent. Given that the NS-free production system gave a higher yield and NS1–4 are not essential for the expression of VP1–3, it was hypothesized that the effectiveness of the prototypic NP1-dependent production system could be improved by exclusion of the expression of NS1–4 from the HBoV1 helper. Guided by the HBoV1 transcription profile, which clearly maps the transcription initiation and splice-donor and splice-acceptor sites for NS1–4 (Fig. 1a), new HBoV1 helper constructs pCMVNSCap, pCMVNS*Cap, and pCMVNP1Cap were used to probe the involvement of NS proteins in virion production. These and other helper constructs are summarized in Table 1 for their abilities to express HBoV1 viral proteins. While pCMVNP1Cap is a new construct, the expression profiles of pCMVNSCap and pCMVNS*Cap, as well as the pCMVR7cDNAm(pA)p and the prototype helper pHBoV1KUm630 (pHBoV1NSCap), were previously characterized at both the protein and RNA levels.43 In brief, the pCMVNSCap is a derivative of pHBoV1KUm630 in which the endogenous P5 promoter of the replication-incompetent genome is replaced with the strong CMV enhancer/promoter. Following transfection to HEK293 cells, the pCMVNSCap expressed higher levels of VP1–3, NP1, and NS1 than pHBoV1KUm630, but the expression levels of NS2–4 from these two plasmids were similar.43 First, it was hypothesized that enhancing expression of NS1 and NP1 would reduce virion production, although capsid expression could be elevated if pCMVNSCap was used as a trans helper. As anticipated, pCMVNSCap yielded only 56% of the virions produced by the prototypic system (Fig. 1b). Next, two helper derivatives of pCMVNSCap were evaluated that limit expression of specific NS proteins (pCMVNS*Cap and pCMVNP1Cap). pCMVNS*Cap expresses the three smaller NS proteins (NS3, NS4, and the essential NP1), as well as capsid proteins VP1–3. pCMVNP1Cap expresses only NP1 and the capsid proteins (Table 1). Elimination of either NS1–2 or NS1–4 increased the production of virion 7.7 ± 1.7- and 9.0 ± 3.1-fold, respectively (Fig. 1b). The similarity in efficiency of virion production from pCMVNS*Cap and pCMVNP1Cap suggested that although NS1–4 are not essential, the efficiency of rAAV2/HBoV1 production is negatively influenced by the expression of NS1 and NS2, but not by that of NS3 or NS4.
In summary, understanding the genome structure of HBoV1 and the transcription profile of its genes has enabled us to generate a rAAV2/HBoV1 production system that is dependent on NP1 and free of NS1–4 proteins. Utilizing a cap cDNA whose (pA)p sites were silently mutated, helper pCMVR7cDNAm(pA)p made it possible to generate rAAV2/HBoV1 under conditions that were completely free of all five HBoV1 NS proteins. The yields from the NP1-dependent and the NS-free systems were around eight- and threefold higher, respectively, than those for the prototypic system.
NP1-dependent production of rAAV2/HBoV1
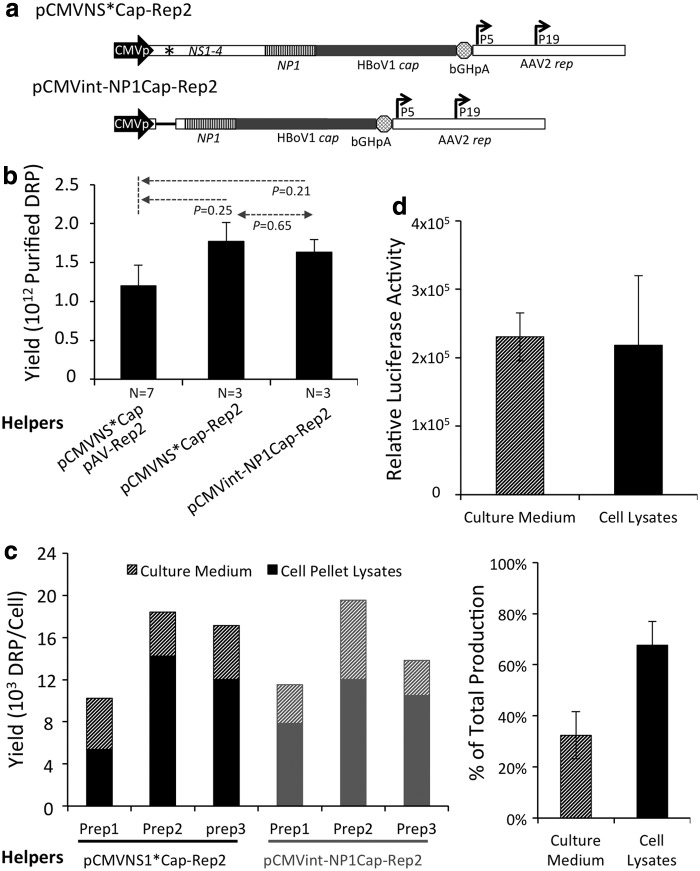
Given that most of the left end of the HBoV1 sequence, which encodes NS1–4, is not essential in the HBoV1 helper, an attempt was made to simplify the NP1-dependent production system further, moving from the four-plasmid co-transfection system in the prototypic version to a three-plasmid co-transfection system. To this end, two types of two-in-one helper plasmid were produced from the HBoV1 and AAV2 helper plasmids. In the case of one of these helper plasmids, a 1.8 kb HBoV1 sequence starting upstream of the NS1 start codon and ending at splice-acceptor site A1 in the helper pCMVNSCap was replaced with a 134 bp sequence from small intron IV of the human Erythropoietin (Epo) gene. Next, an AAV rep expression cassette was incorporated, producing a two-in-one HBoV1-AAV2 helper, pCMVint-NP1Cap-Rep2, which expresses NP1, the HBoV1 capsid proteins, and AAV rep proteins (Rep78/68 and Rep52/40). For the second helper plasmid, the same AAV rep expression cassette was incorporated into pCMVNS*Cap, generating another two-in-one helper, pCMVNS*Cap-Rep2 (Fig. 2a and Table 1). Notably, the efficiencies of these two helper plasmids in facilitating production of rAAV2/HBoV1 were similar in three side-by-side preparations, suggesting that the deleted 1.8 kb segment of the HBoV1 genome does not contain any cis function in regulating the cap expression. When 20 dishes (150 mm, ∼2 × 107 cells per plate) of HEK293 cells were transfected, ∼1.6–1.7 × 1012 DRP of purified virions were obtained from lysates of the cell pellet after two rounds of CsCl ultracentrifugation (Fig. 2b). This was not significantly higher than the result for the four-plasmid system. Of note, the total mass of DNA used in the four-plasmid system was larger than in the three-plasmid system, which could somewhat impact the transfection efficiency and lead to the observed small differences in yield. Notwithstanding the lack of improvement in the efficacy of production, the need for fewer plasmids in the transfection makes the three-plasmid system more cost-effective.
Figure 2.
Effectiveness of NP1-dependent rAAV2/HBoV1 production system. (a) Schematic illustration of structures of rAAV rep and HBoV1 cap two-in-one helper plasmids. bGHpA, bovine growth hormone polyadenylation signal; P5 and P19, the endogenous promoters in AAV2 genome for expressions of Rep78/68 and Rep52/40, respectively. (b) Comparison of yield of rAAV2/HBoV1 for four- versus three-plasmid co-transfection strategy. Bars and error bars represent mean ± SEM rAAV2/HBoV1 yield from cell pellets of twenty 150 mm dishes of transfected HEK293 cells (N as indicated). p-Values were determined by Student's t-test. (c) Comparison of yields of rAAV2/HBoV1 from culture medium and cell lysates. Left: Side-by-side comparison of yield for the two two-in-one helper plasmids in three independent preparations (preps) per plasmid. Yield includes virions collected from both the cell pellets and the culture medium. Right: Comparison of yield from culture medium versus cell lysates. Average for all preparations from both helper constructs is shown. (d) Comparison of transduction activities between the rAAV2/HBoV1 preparations isolated from culture medium and cell lysate. Human airway epithelia efficiently cultured at an air–liquid interface (HAE-ALI) were apically infected at a multiplicity of infection (MOI) of 10,000 DRP/cell with AV2/HBc-gLuc, and transgene expression was measured at 3 days post infection. Values represent mean ± SEM luciferase activity from three infections.
During the production of rAAV, depending on serotypes, substantial quantities of virion can be purified from the culture medium.49 It was observed that productive infection of HAE-ALI with HBoV1 results in a release of progeny virions.37 So, it was suspected that rAAV2/HBoV1 might be secreted into the culture medium at a late time point after transfection of HEK293 cells. Thus, for six independent transfections, the number of DRPs in both the culture medium and cell lysates was assessed. The estimated total yield of virions ranged from 1.02 × 104 to 1.96 × 104 DRP/cell prior to purification (Fig. 2c). This includes virions collected from the culture medium, which represented about 30% of the total yield. Notably, application of rAAV2/HBoV1 virions purified from the culture medium or cell lysates to the apical membrane of HAE-ALI cultures revealed that their potencies were similar (Fig. 2d).
rAAV2/HBoV1 production using the NS-free system
The potential of NP1-dependent rAAV2/HBoV1 production appeared to have been maximized under the test conditions, given that the simplicity of the three-plasmid system did not lead to better efficacy. Since the enhanced capsid expression from the NP1-dependent helper pCMVNS*Cap was accompanied by an elevation in NP1 level in the transfection,43 it was hypothesized that production by that system was limited by the overexpression of NP1. Although NP1 is indispensable in the context of the (pA)p sites in HBoV1, its expression at higher concentration than expected might negatively impact cellular machineries employed in virion production. Inducible NP1 expression could be one solution for further improvement. However, the possibility of developing an optimized NS-free system in which NP1 expression is not required was considered.
As is the case for most parvoviruses, the HBoV1 capsid is a T = 1 icosahedron assembled from 60 copies of various combinations of VP1, VP2, and VP3, at a stoichiometry of 1:1:10. In AAV, VP1–3 with overlapping OFRs are expressed from two mRNAs produced by alternative splicing of the cap pre-mRNA transcribed from the P40 promoter. The minor spliced form produces VP1. The major splice form produces both VP2 and VP3, but a non-canonical start codon (ACG) is used for VP2 expression, and this causes expression of that protein to be limited.50 Current knowledge of the HBoV1 transcription profile did not suggest that a similar RNA splicing strategy is utilized to regulate the expression of VP1 and VP2/VP3, but the translation of HBoV1 VP2 is initiated from a non-canonical start codon, GTG, which lies 117 bp (39 amino acids) upstream of the VP3 ORF.43 Given that NP1 is the only NS protein required for cap expression, it was hypothesized that it contributes to the regulation of VP1, VP2, and VP3 expression at a ratio optimal for virion production (protein expression and capsid assembly). Due to the lack of such regulation in the NS-free production system in which pCMVR7cDNAm(pA)p serves as helper plasmid, the translation of capsid proteins from the (pA)p-mutated R7 mRNA may not be efficient in the absence of NP1, or translation may be hindered by structural changes in the mRNA caused by introduced mutations. Indeed, a previous study reported that pCMVR7cDNAm(pA)p expressed similar levels of VP1–3 proteins as pCMVNS*Cap following transfection of HEK293 cells, although the abundance of the full length (pA)d cap mRNA transcribed from pCMVR7cDNAm(pA)p was six- to sevenfold more than that from pCMVNS*Cap.43 It was hypothesized that the NS-free production system could be further improved through increasing the cap mRNA translation efficiency.
Next, two approaches were taken to increasing VP3 expression that involve modification of the cap mRNA. The first was to enhance cap expression by codon optimization globally, a strategy commonly used to enhance transgene expression. Essentially, silent mutations are used to adjust bias in codon usage, reduce secondary structure and unstable motifs in mRNAs, and silence cryptic splice sites and premature polyadenylation signals.51,52 Starting with the original R7 cDNA, codon optimization was applied to nearly the entire VP1 coding sequence, the exception being the codons thought to be crucial to initiation of the translation of VP2 and VP3. The second approach was to reduce relative level of VP1 expression, with the goal of generating an expression plasmid that expresses enhanced levels of VP1, VP2, and VP3 at a similar ratio to that of the prototype helper pHBoV1KUm630. To this end, the 5′ untranslated region (5′ UTR) and the VP1 start codon (ATG) of the R7 cDNA were replaced with a short sequence from the AAV2 genome (nt: 2,595–2,622 in the AAV2 genome; accession no. NC_001401.2) that includes the Kozak consensus sequence and the non-canonical start codon ACG. In effect, this causes the translation of HBoV1 VP1 to initiate through the same mechanism that AAV2 uses for VP2. These resulted in an artificial HBoV1 cap cDNA with 5′ UTR engineered and codon optimized. This sequence was designated as optVP, and it was used to substitute the (pA)p-mutated R7 cDNA in the helper pCMVR7cDNAm(pA)p, producing a new NP1-independent helper, pcDNAoptVP (Table 1).
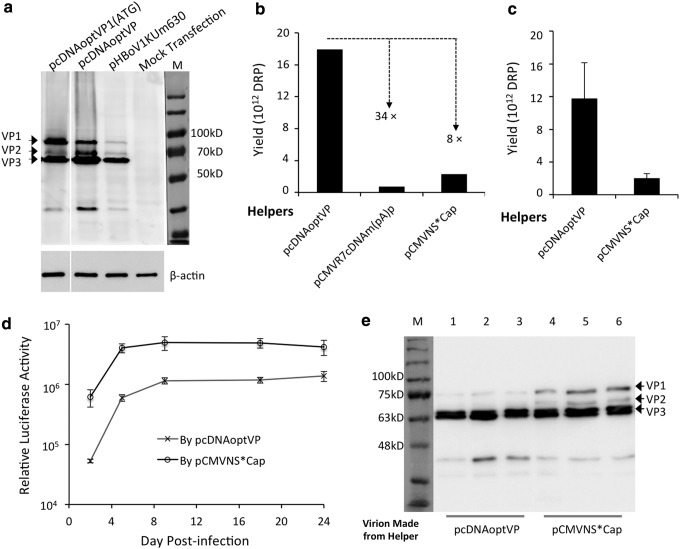
A comparison of cap expression patterns between pcDNAoptVP and pHBoV1KUm630 following transfection of HEK293 cells showed that pcDNAoptVP produced more cap globally, but that relative levels of VP1, VP2, and VP3 were similar to those produced by pHBoV1KUm630, the replication-incompetent HBoV1 genome clone (Fig. 3a). In the context of the four-plasmid transfection system for rAAV2/HBoV1 production, a side-by-side comparison demonstrated that pcDNAoptVP yielded 34- and 8-fold more virion than pCMVR7cDNAm(pA)p and pCMVNS*Cap, respectively (Fig. 3b). Further comparison between pcDNAoptVP and pCMVNS*Cap from four side-by-side preparations revealed that on average, the NS-free system in which pcDNAoptVP serves as helper was 7.3 ± 2.1-fold more efficient than its NP1-dependent counterpart, yielding >1013 DRP virions from 40 dishes (150 mm) after two rounds of CsCl ultracentrifugation (Fig. 3c). Of note, pCMVNS*Cap also expressed NS3 and NS4. Similar yields from pCMVNP1Cap and pCMVNS*Cap were previously observed (Fig. 1b), and the additional expression of NS3/4 from pCMVNS*Cap did not impact the virion production from the NP1-dependent system. However, the high production was accompanied by unexpected decreases in the potency of transduction and the kinetics of transgene expression (Fig. 3d). At 2 days following the infection of HAE-ALI cultures, the levels of transgene expression from the virions produced from pcDNAoptVP were one order of magnitude lower than those from pCMVNS*Cap. However, once the transduction plateaus were reached at later time points, these differences narrowed to around fourfold. Western blot analysis for the content of the capsid subunits VP1–3 using anti-HBoV1 capsid antibody revealed that all three batches of vector produced by pcDNAoptVP contained much less VP1 than those produced from pCMVNS*Cap (Fig. 3e).
Figure 3.
Effectiveness of NS-free rAAV2/HBoV1 production system. (a) Comparison of the expression of HBoV1 capsid proteins in transfected HEK293 cells. Expression of VP1, VP2, and VP3 was assessed by Western blot analysis following transfection with codon-optimized HBoV1 cap in helper pcDNAoptVP1(ATG), pcDNAoptVP, or replication-incompetent HBoV1 genome clone pHBoV1KUm630. Cell lysates from the transfections were resolved in same polyacrylamide gel, but several irrelevant lanes were removed. After probing for HBoV1 capsid proteins, the blot was re-probed for β-actin expression. (b and c) Quantitation of vector production. (b) Representative example of a side-by-side comparison of production yields from indicated HBoV1 helper plasmids for forty 150 mm plates of transfected HEK293 cells. (c) Comparison of average yields for NS-free helper pcDNAoptVP and NP1-depedent helper pCMVNS*Cap. Values represent mean ± SEM yield from four side-by-side preparations. (d) Comparison of transduction activities of rAAV2/HBoV1 preparations produced using pcDNAoptVP and pCMVNS*Cap. HAE-ALI cultures apically infected with AV2/HBc-gLuc at an MOI of 10,000 DRP/cell. Values represent mean ± SEM (n = 4) accumulated gaussia luciferase activity for the previous 24 h period at each time point. (e) Comparison of the content of HBoV1 capsid subunits in virions, as assessed by Western blot analysis. 1.3 × 1010 DRP of virions were loaded per lane. Lanes 1–3: virions generated using helper pcDNAoptVP. Lanes 4–6: virions generated using helper pCMVNS*Cap. VP1, VP2, and VP3 bands were detected using anti-HBoV1 VP3 antibody and visualized by horseradish peroxidase–conjugated anti-rat immunoglobulin G antibody. Note: Anti-HBoV1 VP3 visualized a low-molecular weight band on the blots from the transfection of capsid expression plasmid (a) and also from purified virions (e). It is unclear if this band is a degradative product or an unknown small VP component incorporated in the virions.
Optimization of the NS-free rAAV2/HBoV1 production system
In all parvoviruses except Amdoparvoviruses (e.g., Aleutian mink disease parvovirus), the N-terminal unique sequence of the VP1 ORF contains a phospholipase A2 activity domain and a nuclear-localization sequence. Both are critical for efficient transfer of the virus from late endosomal/lysosome compartments to the nucleus.53,54 Thus, in spite of the fact that VP1 subunits comprise <10% of all capsid subunits at normal stoichiometry, they are crucial to establishing a productive infection. It was reasoned that the reduction in VP1 level was the major cause of the observed reduction in transduction potency and transduction kinetics of virions produced with the helper pcDNAoptVP. Although HEK293 cells transfected with pcDNAoptVP express VP1, VP2, and VP3 at ratios similar to those produced using pHBoV1KUm630, the reduced potency in this context suggested that this stoichiometry is not sufficient for optimal assembly of the capsid. Of note, in some AAV serotypes, capsid assembly requires a small protein called assembly-activating protein (AAP), which is encoded by a sequence within the cap gene but a frameshifted ORF and requires a non-canonical initiation codon for translation. It remains unclear whether a similar AAP mechanism is involved in the assembly of HBoV1 virions. However, the codon optimization might be expected to interfere with the expression of any potent components that were encoded within the wild-type HBoV1 cap gene.
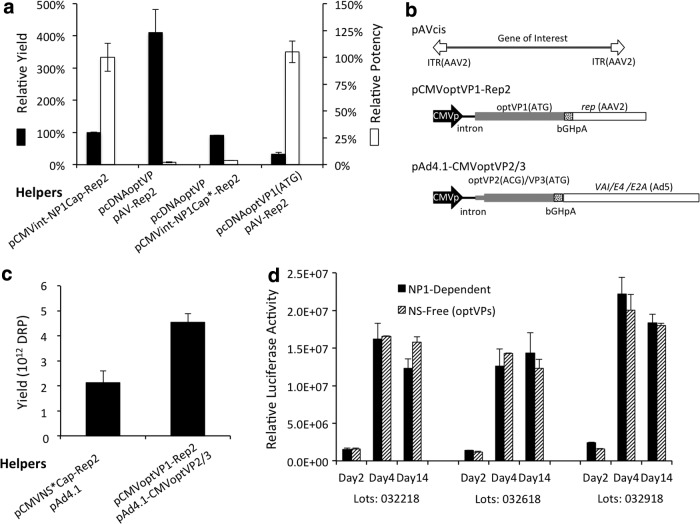
To test this hypothesis, cap expression from the NP1-dependent helper pCMVint-NP1Cap-Rep2 was disrupted by introducing two point mutations: one leading to early termination of VP1 translation at G28 (GAA to TAA), and the other leading to premature termination of the translation of VP2 at N47 and of VP3 at N7 (CAA to TAA). Neither introduces a nonsense mutation in the +1 or +2 reading frame. The resultant pCMVint-NP1Cap*-Rep2 could then be used to replace pAV-Rep2. It also transcribes the non-coding RNA BocaSR, expresses NP1, and preserves any potent trans functions that might be disrupted in pcDNAoptVP. In spite of these careful manipulations, the addition of pCMVint-NP1Cap*-Rep2 did not improve on the potency of virion produced using pcDNAoptVP as helper. Instead, it sharply reduced yield to a level similar to that for the best-performing NP1-dependent helper (Fig. 4a). This outcome suggests that even if pCMVint-NP1Cap*-Rep2 preserves functions that pcDNAoptVP lacks, such as the BocaSR, they are not necessary for virion assembly.
Figure 4.
Optimization of NS-free production system. (a) Comparison of production yield and transduction potency. Virions produced using the indicated helper plasmids demonstrated trade-off in production yield versus transduction potency. Values represent mean ± SEM yield (left y-axis) and potency (right y-axis) as percentages relative to those for virion produced using pCMEpoIVNP1Cap-Rep2 (values were set at 100% yield and potency). (b) Schematic illustration of three plasmids used in NS-free production system. HBoV1 capsid proteins VP1, VP2, and VP3 are expressed from the codon-optimized cap genes by using distinct expression plasmids. (c) Comparison of production yield from most efficient NP1-dependent and NS-free production systems. Values represent mean ± SEM yield in three side-by-side preparations using the indicated helper plasmids. (d) Comparison of transduction (luciferase) activities of rAAV2/HBoV1 preparations, as described, for HAE-ALI cultures apically transduced with three side-by-side paired lots of virions, as described in (c). Transgene expression was assessed on days 2, 4, and 14 post infection. Bars and error bars represent mean ± SEM (n = 4) gaussia luciferase activity in the culture medium accumulated in 24 h intervals.
The negative impact of imposed NP1 expression (using pCMVint-NP1Cap*-Rep2) on the yield of pcDNAoptVP was somewhat predictable. This outcome supports our assumption that NP1 expression might account for the relatively low yield of the NP1-dependent system in spite of being indispensable for cap expression. However, the failure to improve the potency of the virion by means of imposed NP1 expression suggests that the endogenous mechanism for regulating cap expression and virion assembly, which is retained in the NP1-dependent production system, is dependent on either DNA- or RNA-sequence, and thus that the absence of NP1 is not responsible for the reduced transduction potency. The latter was further supported by the testing of pcDNAoptVP1(ATG), a variant of pcDNAoptVP in which the endogenous HBoV1 core Kozak sequence and an ATG codon are used to initiate VP1 translation. The potency of virions generated from pcDNAoptVP1(ATG) was similar to those of virions produced by the NP1-dependent helper (pCMVint-NP1Cap-Rep2), in spite of the fact that their yield was dramatically reduced (Fig. 4a).
Although pcDNAoptVP1(ATG) transcribes a cap mRNA whose primary sequence is almost identical to that of the parental pcDNAoptVP, enhanced VP1 expression from pcDNAoptVP1(ATG) (Fig. 3a) led to optimal virion potency. Notably, when VP3 expression was predominant from pcDNAoptVP, production was high and potency was impaired, and when VP1 expression was elevated from pcDNAoptVP1(ATG), the effect was reversed. It was hypothesized that these opposite outcomes could be balanced by using separate plasmids to increase the expression of both VP1 and VP3 simultaneously.
To test this hypothesis, the VP1 expression cassette was incorporated into pAV-Rep2 and the VP2/VP3 cassette into pAd4.1, producing a new NS-free production system in the context of three-plasmid transfection (Fig. 4b). The following two-in-one trans helpers were used: pCMVoptVP1-Rep2 and pAd4.1-CMVoptVP2/3 (Table 1). pCMVoptVP1-Rep2 expresses HBoV1 VP1 from the codon-optimized cap gene, which incorporates (1) an optimal Kozak sequence and ATG to initiate VP1 expression, and (2) the mutation M130L (VP1) to eliminate VP3 expression from this construct. pAd4.1-CMVoptVP2/3 mimics AAV2 with respect to expressing VP2 and VP3 from a single mRNA transcript. The original GTG start codon for VP2 and the upstream sequence encoding the unique amino acids of VP1 in the synthetic optVP sequence were replaced with the short AAV2 sequence that had been used in pcDNAoptVP to provide VP1 expression. Thus, pAd4.1-CMVoptVP2/3 drove attenuated expression of VP2 from the non-canonical start codon ACG and higher levels of VP3 expression from the internal ATG that also encodes the M40 of VP2. In spite of the fact that this strategy resulted in lower virion production than when pcDNAoptVP was used, the complementation of HBoV1 capsid proteins from separate VP1 and VP2/VP3 helpers increased the yield to double that of the best-performing NP1-dependent system (Fig. 4c). The estimated yield per cell from the three-plasmid transfections of HEK293 cells was 3.2 ± 0.3 × 104 DRP. From 40 plates (150 mm) of transfected HEK293 cells, a total of 4.5 × 1012 DRP purified virions can be produced. Importantly, this high yield was not accompanied by loss of virion functionality. Side-by-side comparison of three independent preparations showed that the virions generated from the new NS-free system had transduction potency and kinetics similar to those of virions generated using the NP1-dependent system (Fig. 4d).
Discussion
The cross-genera pseudopackaging of the rAAV2 genome into the HBoV1 capsid to generate rAAV2/HBoV1 vectors requires transgene expression from both the AAV2 rep and the HBoV1 cap plasmids, in addition to helper function from adenovirus. However, when inserted into eukaryotic expression vectors, the HBoV1 cap cDNA on its own is not sufficient to drive production of capsid, suggesting that the functional expression of HBoV1 cap requires other NS component(s) from the HBoV1 genome. The prototype production system uses a cloned HBoV1 replication-incompetent genome (pHBoV1KUm630) for trans complementation of the HBoV1 capsid, but the rAAV2/HBoV1 vector produced from this genome in HEK293 cells is equivalent to only 5% of the rAAV2 produced at the same scale of transfection. pHBoV1KUm630 expresses three capsid proteins (VP1–3), the non-coding RNA BocaSR, and all five of the HBoV1 NS proteins (NS1–4 and NP1). It is now known that NP1 is the only one of the NS components that is required for efficient capsid production and that the low yield of the prototypic system using this helper was a consequence of overexpression of unneeded NS proteins. Indeed, this notion was supported by the fact that production from pHBoV1KUm630 dropped sharply when the expression of HBoV1 proteins was increased globally by replacing the endogenous P5 promoter with the CMV promoter (Fig. 1b).
In parvoviruses, expression from the cap gene usually requires the participation of NS proteins, either directly or indirectly. For example, in the case of AAV, Rep78 and Rep68 positively regulate transcription of cap mRNAs from the P40 promoter. Yet, expression occurs even in the absence of these NS proteins.55 In HBoV1, NP1 is indispensable to capsid expression from the native genome, but the mechanism whereby NP1 interacts with the transcription complex to prevent premature transcription termination remains unclear. However, it is notable that silent mutations eliminating the (pA)p sites circumvent the need for NP1. This enables HBoV1 capsid proteins to be expressed from a simple eukaryotic expression plasmid harboring a (pA)p-sites mutated cap cDNA or codon-optimized cap gene. Indeed, when pCMVR7cDNAm(pA)p or pcDNAoptVP was used as helper, rAAV2/HBoV1 could be produced under NS-free conditions.
In general, the production of recombinant vector benefits from the exclusion of any nonessential proteins in the transfected cells. Indeed, eliminating the nonessential NS1–2 genes, or even NS1–4, from the pHBoV1KUm630 greatly increased the efficiency of NP1-dependent rAAV2/HBoV1 production by around eightfold. Furthermore, using the pcDNAoptVP as helper, 34-fold more efficacy was achieved in the context of completely NS-free rAAV2/HBoV1 production. Featuring a fully codon-optimized synthetic cap coding sequence that lacks the (pA)p sites and attenuates VP1 expression by using a non-canonical start codon, pcDNAoptVP reproduces VP1–3 at a ratio similar to that seen from pHBoV1KUm630 but with higher efficiency due to the absence of all NS proteins (Fig. 3a). The higher efficiency of capsid protein expression from this NS-free system versus the NP1-dependent production counterpart (Fig. 3b) and the fourfold reduction in pcDNAoptVP-based production in the context of imposed NP1 expression (Fig. 4a) suggest that NP1 has a negative impact in spite of the fact that it is indispensable to cap expression in the replication-incompetent HBoV1 genome and its derivatives. Notwithstanding the improvement of pcDNAoptVP with respect to yield, it has a key limitation. The 34-fold increase in virion production with this helper was offset by a significant reduction in transduction potency of virions. The Western blot analysis of the proportion of VP1–3 of the virions suggested the suboptimal assembly. The reduced VP1 content in the virion produced from pcDNAoptVP likely accounts for the impaired potency.
Key questions that remain to be resolved with respect to parvovirus biology include how AAV and HBoV parvoviruses assemble icosahedral virions from 60 subunits of VP1, VP2, and VP3 at the 1:1:10 stoichiometry required for optimal infectivity. Previous thinking was that in AAV, optimal virion assembly results from predominant VP3 expression and attenuated expression of VP1 and VP2, under the control of alternative mRNA splicing and non-canonical translation initiation (VP2). In HBoV1, the expression of these proteins is most likely regulated by NP1, with VP2 expression also limited by non-canonical initiation of translation. However, the recent discovery of the requirement for AAP in AAV assembly has suggested that regulation of the formation of functional virion is more complex than producing the VP1, VP2, and VP3 capsid proteins at the correct stoichiometry. Moreover, it remains unclear why some but not all AAV serotypes require AAP for virion assembly.56,57 Whether the HBoV1 genome encodes an AAP-like protein remains an open question. The production of fully transduction-active rAAV2/HBoV1 from the codon-optimized cap gene suggests that rAAV2/HBoV1 packaging might not require such a function, at least in the context of the new NS-free system, in which VP1 expression was intentionally enhanced. However, when pcDNAoptVP alone was used as the NS-free helper, even though it expressed VP1, VP2, and VP3 at ratios similar to those produced in the context of NP1-dependent cap expression, this was not sufficient for assembly of rAAV2/HBoV1 virions with optimal VP1 content. The fact that imposed NP1 expression failed to improve the potency of the virion suggested that the mechanism whereby cap expression and virion assembly are retained during NP1-dependent production does not apply to the NS-free system using codon-optimized cap gene, and thus the mechanism appears to be dependent on the DNA or/and RNA sequence.
Interestingly, the assembly of rAAV vector might not strictly obey the rule of VP1, VP2, and VP3 at a stoichiometry of 1:1:10. While sufficient VP1 content in the virion is crucial to infectivity, the presence of VP2 appears not to be essential for the packaging and potency of rAAV,58 and a recent study reported that the transduction efficiency of rAAV correlates with the abundance of VP1, rAAV with higher abundance of VP1 demonstrated improved transduction activity.59 It was found that the NS-free rAAV2/HBoV1 production system required enhanced level of VP1 expression. The strategy is to increase the VP1 and VP3 expression simultaneously in an NS-free production system. Expression of VP1 and VP2–3 from distinct helper plasmids under the control of the same strong promoter/enhancer (CMV) enables efficient production of fully infectious rAAV2/HBoV1. The new NS-free production system is a three-plasmid base transfection using two helper plasmids for all the trans functions: the VP1 expression cassette and the VP2–3 expression cassette (with attenuated VP2 expression) are incorporated into the AAV rep helper plasmid and the adenovirus helper plasmid, respectively. Although this system does not match the efficacy of pcDNAoptVP with respect to virion production, it nevertheless exceeds the best-performing NP1-dependent method for producing transduction-competent virions by more than a factor of two. Moreover, it is 16- to 18-fold more efficient than the prototypic system.
In summary, an efficient rAAV2/HBoV1 production system free of the expression of all HBoV1 NS proteins was developed, with an estimated yield of 3–4 × 104 DRP/cell before purification. It was also found that ∼30% of the infectious virion can be recovered from the culture medium, suggesting that production could be further increased using a procedure that recovers virions from the culture medium as well as cell pellets. The current yield of rAAV2/HBoV1 remains slightly lower than that of rAAV produced by the routine manufacturing method, which involves three-plasmid transfection in adherent cultures of HEK293 cells. However, current capacity satisfies the demand for the preclinical studies, and it is expected that further optimization is possible. Importantly, the separate capsid expression cassettes generated for the production of rAAV2/HBoV1 could be applied in other approaches that increase rAAV yield. One involves the use of suspension cultures of HEK293 cells.60 Another such approach takes advantage of baculovirus expression system, which has been used successfully to produce HBoV1 virus-like particles comprised of VP3.61,62
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL051670 to J.F.E.; AI105543, and AI112803 and AI139572 to J.Q.), Cystic Fibrosis Foundation (YAN15XX0 to Z.Y.), the University of Iowa Center for Gene Therapy (DK54759), and the Roy J. Carver Chair in Molecular Medicine (to J.F.E.).
Author Disclosure
The authors have no conflicts of interest to declare.
References
- 1. Collins FS. Cystic fibrosis: molecular biology and therapeutic implications. Science 1992;256:774–779 [DOI] [PubMed] [Google Scholar]
- 2. Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 1989;245:1066–1073 [DOI] [PubMed] [Google Scholar]
- 3. O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet 2009;373:1891–1904 [DOI] [PubMed] [Google Scholar]
- 4. Welsh MJ, Ramsey BW, Accurso F, et al. Cystic fibrosis. In: Scriver CR, Beaudet AL, Sly WS, et al., eds. The Metabolic and Molecular Basis of Inherited Disease. New York: McGraw–Hill, 2001:5121–5188 [Google Scholar]
- 5. Driskell RA, Engelhardt JF. Current status of gene therapy for inherited lung diseases. Annu Rev Physiol 2003;65:585–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prickett M, Jain M. Gene therapy in cystic fibrosis. Transl Res 2013;161:255–264 [DOI] [PubMed] [Google Scholar]
- 7. Griesenbach U, Pytel KM, Alton EW. Cystic fibrosis gene therapy in the UK and elsewhere. Hum Gene Ther 2015;26:266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zabner J, Couture LA, Gregory RJ, et al. Adenovirus-mediated gene transfer transiently corrects the chloride transport defect in nasal epithelia of patients with cystic fibrosis. Cell 1993;75:207–216 [DOI] [PubMed] [Google Scholar]
- 9. Boucher RC, Knowles MR, Johnson LG, et al. Gene therapy for cystic fibrosis using E1-deleted adenovirus: a Phase I trial in the nasal cavity. The University of North Carolina at Chapel Hill. Hum Gene Ther 1994;5:615–639 [DOI] [PubMed] [Google Scholar]
- 10. Perricone MA, Morris JE, Pavelka K, et al. Aerosol and lobar administration of a recombinant adenovirus to individuals with cystic fibrosis. II. Transfection efficiency in airway epithelium. Hum Gene Ther 2001;12:1383–1394 [DOI] [PubMed] [Google Scholar]
- 11. Moss RB, Milla C, Colombo J, et al. Repeated aerosolized AAV-CFTR for treatment of cystic fibrosis: a randomized placebo-controlled Phase 2B trial. Hum Gene Ther 2007;18:726–732 [DOI] [PubMed] [Google Scholar]
- 12. Flotte TR, Zeitlin PL, Reynolds TC, et al. Phase I trial of intranasal and endobronchial administration of a recombinant adeno-associated virus serotype 2 (rAAV2)-CFTR vector in adult cystic fibrosis patients: a two-part clinical study. Hum Gene Ther 2003;14:1079–1088 [DOI] [PubMed] [Google Scholar]
- 13. Aitken ML, Moss RB, Waltz DA, et al. A Phase I study of aerosolized administration of tgAAVCF to cystic fibrosis subjects with mild lung disease. Hum Gene Ther 2001;12:1907–1916 [DOI] [PubMed] [Google Scholar]
- 14. Alton E, Armstrong DK, Ashby D, et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: a randomised, double-blind, placebo-controlled, Phase 2b trial. Lancet Respir Med 2015;3:684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eisele NA, Anderson DM. Host Defense and the airway epithelium: frontline responses that protect against bacterial invasion and pneumonia. J Pathog 2011;2011:249802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanders N, Rudolph C, Braeckmans K, et al. Extracellular barriers in respiratory gene therapy. Adv Drug Deliv Rev 2009;61:115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Allander T, Tammi MT, Eriksson M, et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 2005;102:12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allander T, Jartti T, Gupta S, et al. Human bocavirus and acute wheezing in children. Clin Infect Dis 2007;44:904–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jula A, Waris M, Kantola K, et al. Primary and secondary human bocavirus 1 infections in a family, Finland. Emerg Infect Dis 2013;19:1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jartti T, Hedman K, Jartti L, et al. Human bocavirus—the first 5 years. Rev Med Virol 2012;22:46–64 [DOI] [PubMed] [Google Scholar]
- 21. Meriluoto M, Hedman L, Tanner L, et al. Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study, Finland. Emerg Infect Dis 2012;18:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wagner JC, Pyles RB, Miller AL, et al. Determining persistence of bocavirus DNA in the respiratory tract of children by pyrosequencing. Pediatr Infect Dis J 2016;35:471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan Z, Keiser NW, Song Y, et al. A novel chimeric adenoassociated virus 2/human bocavirus 1 parvovirus vector efficiently transduces human airway epithelia. Mol Ther 2013;21:2181–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan Z, Feng Z, Sun X, et al. Human bocavirus type-1 capsid facilitates the transduction of ferret airways by adeno-associated virus genomes. Hum Gene Ther 2017;28:612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carter BJ. Adeno-associated virus vectors in clinical trials. Hum Gene Ther 2005;16:541–550 [DOI] [PubMed] [Google Scholar]
- 26. Samulski RJ, Muzyczka N. AAV-mediated gene therapy for research and therapeutic purposes. Annu Rev Virol 2014;1:427–451 [DOI] [PubMed] [Google Scholar]
- 27. Smalley E. First AAV gene therapy poised for landmark approval. Nat Biotechnol 2017;35:998–999 [DOI] [PubMed] [Google Scholar]
- 28. Moss RB, Rodman D, Spencer LT, et al. Repeated adeno-associated virus serotype 2 aerosol-mediated cystic fibrosis transmembrane regulator gene transfer to the lungs of patients with cystic fibrosis: a multicenter, double-blind, placebo-controlled trial. Chest 2004;125:509–521 [DOI] [PubMed] [Google Scholar]
- 29. Duan D, Sharma P, Yang J, et al. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J Virol 1998;72:8568–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ehrhardt A, Xu H, Kay MA. Episomal persistence of recombinant adenoviral vector genomes during the cell cycle in vivo. J Virol 2003;77:7689–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ofri R, Averbukh E, Ezra-Elia R, et al. Six years and counting: restoration of photopic retinal function and visual behavior following gene augmentation therapy in a sheep model of CNGA3 achromatopsia. Hum Gene Ther 2018;29:1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Q, Deng X, Yan Z, et al. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog 2012;8:e1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun X, Olivier AK, Liang B, et al. Lung phenotype of juvenile and adult cystic fibrosis transmembrane conductance regulator-knockout ferrets. Am J Respir Cell Mol Biol 2014;50:502–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cotmore SF, Agbandje-McKenna M, Chiorini JA, et al. The family Parvoviridae. Arch Virol 2014;159:1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther 2006;14:316–327 [DOI] [PubMed] [Google Scholar]
- 36. Chen AY, Cheng F, Lou S, et al. Characterization of the gene expression profile of human bocavirus. Virology 2010;403:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deng X, Yan Z, Luo Y, et al. In vitro modeling of human bocavirus 1 infection of polarized primary human airway epithelia. J Virol 2013;87:4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen W, Deng X, Zou W, et al. Identification and functional analysis of novel nonstructural proteins of human bocavirus 1. J Virol 2015;89:10097–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deng X, Yan Z, Cheng F, et al. Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLoS Pathog 2016;12:e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng X, Zou W, Xiong M, et al. Human parvovirus infection of human airway epithelia induces pyroptotic cell death by inhibiting apoptosis. J Virol 2017;91:e01533–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Z, Shen W, Cheng F, et al. Parvovirus expresses a small noncoding RNA that plays an essential role in virus replication. J Virol 2017;91:e02375–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shen W, Deng X, Zou W, et al. Analysis of cis and trans requirements for DNA replication at the right-end hairpin of the human bocavirus 1 genome. J Virol 2016;90:7761–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zou W, Cheng F, Shen W, et al. Nonstructural protein NP1 of human bocavirus 1 plays a critical role in the expression of viral capsid proteins. J Virol 2016;90:4658–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deng X, Xu P, Zou W, et al. DNA Damage Signaling Is Required for Replication of Human Bocavirus 1 DNA in Dividing HEK293 Cells. J Virol 2017;91:e01831–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun Y, Chen AY, Cheng F, et al. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol 2009;83:3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lederman M, Patton JT, Stout ER, et al. Virally coded noncapsid protein associated with bovine parvovirus infection. J Virol 1984;49:315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cotmore SF, Tattersall P. Parvovirus diversity and DNA damage responses. Cold Spring Harb Perspect Biol 2013;5:a012989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karp PH, Moninger TO, Weber SP, et al. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol 2002;188:115–137 [DOI] [PubMed] [Google Scholar]
- 49. Lock M, Alvira M, Vandenberghe LH, et al. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther 2010;21:1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berns KI, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol 1996;218:1–23 [DOI] [PubMed] [Google Scholar]
- 51. Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet 2008;42:287–299 [DOI] [PubMed] [Google Scholar]
- 52. Quax TE, Claassens NJ, Soll D, et al. Codon bias as a means to fine-tune gene expression. Mol Cell 2015;59:149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zadori Z, Szelei J, Lacoste MC, et al. A viral phospholipase A2 is required for parvovirus infectivity. Dev Cell 2001;1:291–302 [DOI] [PubMed] [Google Scholar]
- 54. Cotmore SF, Tattersall P. Parvoviral host range and cell entry mechanisms. Adv Virus Res 2007;70:183–232 [DOI] [PubMed] [Google Scholar]
- 55. Weger S, Wistuba A, Grimm D, et al. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J Virol 1997;71:8437–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Earley LF, Powers JM, Adachi K, et al. Adeno-associated virus (AAV) assembly-activating protein is not an essential requirement for capsid assembly of AAV serotypes 4, 5, and 11. J Virol 2017;91:e01980–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sonntag F, Kother K, Schmidt K, et al. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol 2011;85:12686–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Warrington KH, Jr, Gorbatyuk OS, Harrison JK, et al. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J Virol 2004;78:6595–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Q, Wu Z, Zhang J, et al. A robust system for production of superabundant VP1 recombinant AAV vectors. Mol Ther Methods Clin Dev 2017;7:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grieger JC, Soltys SM, Samulski RJ. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol Ther 2016;24:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gurda BL, Parent KN, Bladek H, et al. Human bocavirus capsid structure: insights into the structural repertoire of the Parvoviridae. J Virol 2010;84:5880–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kantola K, Hedman L, Arthur J, et al. Seroepidemiology of human bocaviruses 1–4. J Infect Dis 2011;204:1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]