Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) editing is being considered as a potential gene repair therapy to treat Duchenne muscular dystrophy, a dystrophin-deficient lethal muscle disease affecting all muscles in the body. A recent preliminary study from the Olson laboratory (Amoasii et al. Science 2018;362:89–91) showed robust dystrophin restoration in a canine Duchenne muscular dystrophy model following intramuscular or intravenous delivery of the CRISPR editing machinery by adeno-associated virus serotype 9. Despite the limitation of the small sample size, short study duration, and the lack of muscle function data, the Olson lab findings have provided important proof of principle for scaling up CRISPR therapy from rodents to large mammals. Future large-scale, long-term, and comprehensive studies are warranted to establish the safety and efficacy of CRISPR editing therapy in large mammals.
Keywords: CRISPR, DMD, dog, AAV, dystrophin, editing
Clustered regularly interspaced short palindromic repeats (CRISPR) is a novel gene editing technology that became widely used about 5 years ago.1–3 Despite its recent debut, it is rapidly advancing from a research tool to a potential therapeutic modality. CRISPR technology is relatively easy to understand and administer. A small sequence-specific guide RNA (gRNA) is used to direct the CRISPR-associated (Cas) nuclease to an intended genomic location for site-specific modification. For therapeutic application, a vector, such as adeno-associated virus (AAV), is used to deliver the gRNA and Cas nuclease to the diseased cells. The disease-causing mutation can be either corrected or removed by CRISPR/Cas-mediated editing in vivo, holding the promise of providing one-time curative therapeutic outcomes for patients. Though highly promising, significant challenges remain in applying CRISPR technology in human gene therapy. Creative strategies are needed to deliver the CRISPR/Cas system to the target cells efficiently and selectively, to achieve a sufficient level of editing for disease amelioration, to avoid and/or attenuate immune responses induced by the bacterial Cas protein and/or delivery vectors, and to prevent and/or minimize untoward off-target genomic alterations.4
Duchenne muscular dystrophy (DMD) is a relatively common X-linked inherited muscle disease caused by reading frame-aborting mutations in the DMD gene. The DMD gene is one of the largest genes in the body, and it encodes the 427 kD dystrophin protein. Dystrophin is a sub-sarcolemmal cytoskeletal protein, and it protects muscle from contraction-induced injuries. Muscle is essential for body movement, posture maintenance, respiration, and blood pumping. The absence of dystrophin leads to muscle degeneration, necrosis, inflammation, and fibrosis. As a consequence, DMD patients become wheelchair-bound in their early teens and die prematurely in their 20s or 30s from respiratory and/or heart failure. The human body has >600 muscles. These muscles consist of ∼45% body mass and are distributed throughout the body. Given the large size of the gene, the presence of muscle throughout the body, and the highly inflamed nature of the dystrophic muscle, DMD is considered one of the most challenging monogenic diseases amenable to gene therapy. Over the last 40 years, numerous viral and non-viral vector-delivered gene therapy strategies have been tested in mouse models of DMD.5–7 Many of these strategies have resulted in remarkable levels of dystrophin restoration and function improvement in mouse muscles. Yet, none of these “mouse therapies” has directly led to equivalent results in human patients. Preclinical study in the symptomatic canine model may help bridge the gap between mice and humans.8,9
CRISPR technology was first introduced for editing DMD gene mutations in 2014 to correct the germline DNA in mdx mice, a mouse DMD model (Table 1).10 Subsequently, a series of studies showed restoration of dystrophin expression in patient cells in vitro by co-transfection of the gRNA and Streptococcus pyogenes Cas9 (SpCas9) or Lachnospiraceae Cpf1 (a homolog of Cas9; Table 1).11–21 In vivo editing in postnatal animals was first achieved in 2016 in mdx mice by co-delivering the gRNA and Staphylococcus aureus Cas9 (SaCas9) or SpCas9 using adenovirus or AAV (Table 1).22–25 These studies revealed a moderate level of DNA editing (≤10%). However, robust dystrophin expression and muscle function improvement were observed in newborn and young adult mdx mice following local or systemic AAV injection. These findings were subsequently validated by other laboratories in different mouse models using SaCas9, SpCas9, Campylobacter jejuni Cas9, and adenine-based editors (Table 1).20,26–32 The next logical step was to see if this success can be reproduced in the canine model.33
Table 1.
CRISPR editing of DMD gene mutations from 2014 to 2018
| Publication | Experiment details | In vivo editing outcome | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | Author | Ref. no. | Editing enzyme | Approach | Repair mechanism | Model | Age at injection | Study duration | Vector | Route | Dystrophin level (staining) | Dystrophin level (Western) | Histology | Muscle function | Heart function | |
| In vitro editing | 2015 | Li et al. | 12 | SphcCas9 | 1 cut with 1 gRNA | HDR and NHEJ | Patient iPSCs | Plasmid | ||||||||
| 2015 | Ousterout et al. | 11 | SpCas9 and hSpCas9 | 1 cut with 1 gRNA and 2 cuts with 2 gRNAs | NHEJ | Patient myoblasts | Plasmid | |||||||||
| 2016 | Young et al. | 13 | SpCas9 | 2 cuts with 2 gRNAs | NHEJ | Patient iPSCs | Plasmid | |||||||||
| 2016 | Iyombe-Engembe et al. | 19 | SaCas9 | 2 cuts with 2 gRNAs | NHEJ | Patient myoblasts | Plasmid | |||||||||
| 2016 | Wojtal et al. | 18 | SpCas9 | 1 cut with 1 gRNA | NHEJ | Patient myoblasts | Lentivirus | |||||||||
| 2016 | Maggio et al. | 17 | SpCas9 | 2 cuts with 2 gRNAs | NHEJ | Patient myoblasts | Adenovirus | |||||||||
| 2017 | Kyrychenko et al. | 15 | SpCas9 | 2 cuts with 2 gRNAs | NHEJ | Patient iPSCs | Plasmid | |||||||||
| 2017 | Lattanzi et al. | 16 | SpCas9 | 2 cuts with 2 gRNAs | NHEJ | Patient myoblasts | Lentivirus | |||||||||
| 2017 | Zhang et al. | 14 | LbCpf1 | 1 cut with 1 gRNA | HDR and NHEJ | Patient iPSCs | Plasmid | |||||||||
| 2018 | Long et al. | 21 | SpCas9 | 1 cut with 1 gRNA | NHEJ | Patient iPSCs and EHM | Plasmid | |||||||||
| 2018 | Duchêne et al. | 20 | SaCas9 | 2 cuts with 2 gRNAs | NHEJ | Patient myoblasts | Lentivirus | |||||||||
| Embryo editing | 2014 | Long et al. | 10 | SpCas9 | 1 cut with 1 gRNA | HDR and NHEJ | Mdx embryo | mRNA | ||||||||
| 2017 | Zhang et al. | 14 | LbCpf1 | 1 cut with 1 gRNA | HDR and NHEJ | Mdx embryo | mRNA | |||||||||
| In vivo editing in mice | 2015 | Xu et al. | 25 | SpCas9 | 2 cuts with 2 gRNAs | NHEJ | Mdx | P1–3 and 2 months old | 3 weeks | Adenovirus | i.m. | ND | ∼50% | ND | ND | ND |
| 2016 | Long et al. | 23 | SpCas9 | 2 cuts with 2 gRNAs | NHEJ | Mdx | P1–18 | 3–12 weeks | AAV | i.m., i.p., r.o. | ∼20–70% | ND | Improved | Grip force increased | ND | |
| 2016 | Nelson et al. | 22 | SaCas9 | 2 cuts with 2 gRNAs | NHEJ | Mdx | P2 and 1.5–2 months old | 7 weeks to 6 months | AAV | i.m., i.p., i.v. | ∼67% | ∼8% | Improved | TA force increased | ND | |
| 2016 | Tabebordbar et al. | 24 | SaCas9 | 2 cuts with 2 gRNAs | NHEJ | Mdx | P3 and 1.5 months old | 3–14 weeks | AAV | i.m., i.p., i.v. | ND | ∼0.03–12% | Improved | TA force increased | ND | |
| 2017 | Amoasii et al. | 28 | SpCas9 | 1 cut with 1 gRNA | NHEJ | ΔEx50 mice | P4–12 | 3–8 weeks | AAV | i.m., i.p. | ≤90% | ∼85% | Improved | Grip force increased | ND | |
| 2017 | Bengtsson et al. | 27 | SaCas9 and SpCas9 | 2 cuts with 2 gRNAs | HDR and NHEJ | Mdx4cv | 2–12 weeks old | 4–18 weeks | AAV | i.m., r.o. | 41–45% | ∼0.8–23% | Improved | TA force increased | ND | |
| 2017 | El Rafaey et al. | 26 | SaCas9 and SpCas9 | 2 cuts with 2 gRNAs | NHEJ | Mdx and Mdx/Utr+/– | P1–3 | 4–10 weeks | AAV and adenovirus | i.p., i.v., r.o. | 40% | ∼23% | Improved | ND | Muscle strip force increased | |
| 2017 | Young et al. | 30 | SpCas9 | 2 cuts with 2 gRNAs | NHEJ | ΔEx45 mice | 12–18 weeks old | 3–5 weeks | Plasmid | i.m.e. | ND | ND | ND | ND | ND | |
| 2018 | Ryu et al. | 32 | ABE | 1 cut with 1 gRNA | NHEJ | Exon20 disrupted | 7 weeks old | 8 weeks | AAV | i.m. | 17% | ND | ND | ND | ND | |
| 2018 | Duchêne et al. | 20 | SaCas9 | 2 cuts with 2 gRNAs | NHEJ | Δ52hDMD/mdx | 24–5 weeks old | 6 weeks | AAV | i.v. | ND | ND | ND | ND | ND | |
| 2018 | Hakim and Wasala et al. | 29 | SaCas9 | 2 cuts with 2 gRNAs | NHEJ | Mdx | 6 weeks old | 6 weeks to 17 months | AAV | i.m., i.v. | ND | 2–20% | Improved | EDL force increased | ECG improved, hemodynamics enhanced | |
| 2018 | Koo et al. | 31 | CjCas9 | 1 cut with 1 gRNA | NHEJ | Exon23 disrupted | 8 weeks old | 7 weeks | AAV | i.m. | ∼26% | ND | ND | TA force increased | ND | |
| In vivo in dogs | 2018 | Amoasii et al. | 34 | SpCas9 | 1 cut with 1 gRNA | NHEJ | ΔEx50 dogs | 1 month old | 6–8 weeks | AAV | i.m., i.v. | ND | 5–92% | Improved | ND | ND |
AAV, adeno-associated virus; ABE, adenine base editors (engineered adenine deaminase and the Streptococcus pyogenes Cas9); Cas9, CRISPR associated protein 9; Cpf1, CRISPR from Prevotella and Francisella 1; CjCas9, Campylobacter jejuni Cas9; DMD, Duchenne muscular dystrophy; EDL, extensor digitorum longus; EHM, engineered heart muscle; gRNA, guide RNA; HDR, homologous recombination; i.m., intramuscular injection; i.m.e., intramuscular electroporation; iPSCs, induced pluripotent stem cells; i.p., intraperitonial injection; i.v., intravenous injection; LbCpf1, Lachnospiraceae bacterium Cpf1; ND, not done; NHEJ, non-homologous end joining; P1–3, postnatal days 1–3; P1–18, postnatal days 1–18; P2, postnatal day 2; P3, postnatal day 3; P4–12, postnatal days 4–12; r.o., retro-orbital injection; SaCas9, Streptococcus aureus Cas9; SpCas9, Streptococcus pyogenes Cas9; SphcCas9/hSpCas9, human codon-optimized SpCas9; TA, tibialis anterior.
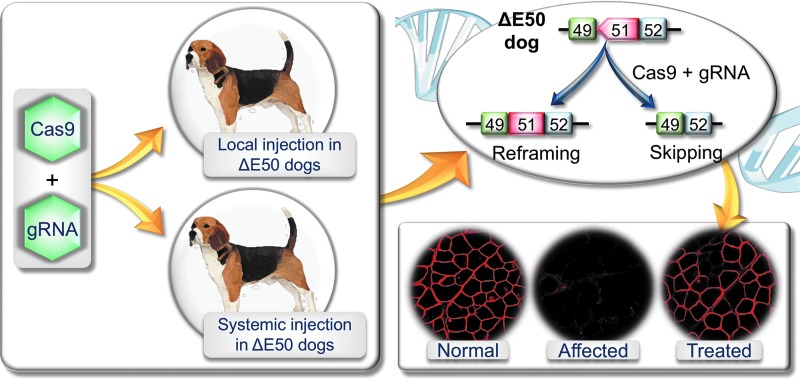
This milestone has now been accomplished according to a recent study by Amoasii et al. (Fig. 1).34 The authors packaged the gRNA and SpCas9 in two AAV serotype 9 (AAV9) vectors and tested them in the ΔEx50 canine DMD model (Fig. 1). In this model, a point mutation at the beginning of intron 50 of the DMD gene results in exon 50 skipping in the RNA transcript, reading-frame shift, and loss of dystrophin expression.35 Dystrophin expression can be recovered by reframing or removing exon 51. In the Cas9 vector, the authors used the muscle specific CKe promoter. In the gRNA vector, three copies of the same gRNA were expressed using three different pol III promoters. This gRNA targets the SpCas9 to the splicing acceptor site of exon 51. This genomic modification results in the skipping of exon 51 or reframing of the disrupted reading frame. The authors co-injected these two vectors into 1-month-old ΔEx50 puppies either intramuscularly in the absence of immune suppression (n = 2; 1.2 × 1013 vg/vector into the cranial tibialis muscle) or intravenously under immune suppression (n = 2; 2 × 1013 vg/kg/vector in one puppy and 1 × 1014 vg/kg/vector in another puppy; Fig. 1).34 The outcome of CRISPR editing was studied 6–8 weeks after injection. Specifically, the authors examined (1) gene editing on genomic DNA and cDNA by tracking indels by decomposition analysis and deep sequencing of the polymerase chain reaction product amplified from the target site; (2) dystrophin expression by immunostaining and Western blot; (3) dystrophin-associated glycoprotein complex restoration by β-dystroglycan immunostaining; (4) disease amelioration by qualitative hematoxylin and eosin (H&E) staining of muscle sections, developmental myosin heavy chain (a regeneration marker) expression, and serum creatine kinase levels; (5) safety by hematology, blood biochemistry, target site indel formation, and Cas9 expression in the testes, and deep sequencing of the predicted top off-target sites in muscle; (6) immune response by CD4+ and CD8+ T-cell immunostaining in the locally injected muscle.
Figure 1.
Schematic illustration of clustered regularly interspaced short palindromic repeats (CRISPR) editing in the ΔE50 canine Duchenne muscular dystrophy model. An adeno-associated virus (AAV) CRISPR-associated protein 9 vector and an AAV guide RNA vector were co-delivered to ΔE50 dogs by intramuscular or intravenous injection. In ΔE50 dogs, exon 50 is skipped in the RNA transcript by a naturally existing point mutation. This leads to frame-shift and dystrophin deficiency. CRISPR therapy restores dystrophin expression by reframing or skipping exon 51. Please note: immunostaining images are for illustration purposes. They are not from the Amoasii et al. publication.
DNA-level editing was similar to that seen in mouse studies (1–14%).22–24 Immunostaining revealed widespread dystrophin expression. Western blots showed dystrophin restoration to 3–92% of wild-type levels. Remarkably robust dystrophin expression was detected in the heart and diaphragm (∼90% and 50% of normal, respectively, on Western blot) in the puppy that was systemically injected at the dose of 1 × 1014 vg/kg/vector. In the puppy that received the lower dose (2 × 1013 vg/kg/vector), dystrophin expression was consistently lower and more sparse. The authors also found some evidence suggesting a reduction of muscle damage in treated puppies. Importantly, in this short-term study, the authors did not detect T-cell response and germ cell editing. No safety concern was raised either.
The Amoasii et al. study is significant to the development of therapeutic CRISPR editing for DMD. First, this study demonstrated feasibility of systemic CRISPR editing in a diseased large mammal. Second, the level of dystrophin restoration, especially systemic delivery at the dose of 1 × 1014 vg/kg/vector, has reached the range considered to benefit patients according to mouse studies and clinical case reports.36 This dose is well tolerated in newborns with spinal muscular atrophy type 1 and is in line with the dose used in ongoing systemic AAV micro-dystrophin trials in DMD patients.37,38 Third, the strategy described by the authors may be directly applicable to ∼13% of DMD patients.39
As a proof of principle, the findings reported by Amoasii et al. are encouraging. However, the implication of the study is considerably limited due to the sample size, age of injection, duration of the treatment, and methods used to characterize the treatment outcome. A sample size of two for local injection and one for each dose of systemic injection is too small for statistical analysis. Additional studies in more dogs are needed in order to validate the findings.
In the Amoasii et al. study, the affected dogs were treated at 1 month of age. This is analogous to ∼1.5 years in humans. DMD patients are usually diagnosed between 3 and 5 years of age. Hence, the strategy described by Amoasii et al. might be suitable for treating neonatal patients identified through newborn screening. However, to meet the need of existing patients, the protocol has to be optimized in older affected dogs.
DMD is a chronic disease. DMD treatment requires continuous dystrophin expression.40 It was recently shown that persistent (up to 18 months) dystrophin restoration and muscle and cardiac function improvement can be achieved in young adult mdx mice from a single systemic AAV CRISPR injection (Table 1).29 Amoasii et al. terminated the study before treated puppies were weaned (2.5–3 months of age). At this age, affected puppies usually show mild clinical symptoms and signs.41 The short study duration makes it difficult to monitor clinical improvement, durability of gene editing, and potential untoward adverse events that arise later.
As discussed above, Amoasii et al. performed a panel of assays to study editing effect, dystrophin restoration, and disease amelioration. There is no doubt that all these studies are important. However, a more thorough analysis should be included in future studies to assess the consequences of AAV CRISPR delivery fully in dystrophic large mammals. A significant safety concern of in vivo CRISPR editing is off-target editing. The authors only quantified indel formation in the predicted off-target sites in muscle. Unbiased approaches such as the recently reported verification of in vivo off-targets (VIVO) method should provide a more comprehensive view on genome-wide off-target editing.42 Besides indels, other important genetic changes such as AAV vector genome integration and large chromosomal alterations (e.g., large deletion, inversion, translocation, and duplications) should also be investigated. It is worth noting that a significant portion of systemically injected AAV vectors will end up in the liver. Hence, in addition to muscle, the liver should also be carefully examined for potential off-target genomic modification. To determine the safety, the authors also studied hematology and blood biochemistry. This can be further expanded to include the growth curve (body weight) and histology examination of major internal organs such as the liver, kidney, lung, and brain. Germline editing is an ethical/safety concern.43,44 Amoasii et al. did not find Cas9 expression and target-site indel formation in the testes. Similar analysis in germ cell forming tissues (the testes and ovaries) should be included in future large-scale, long-term studies.
Immune responses have been at the center stage of gene therapy for decades.45 These include innate, cellular, and humoral responses to the viral capsid, vector genome, and transgene product. The cellular immune response has been a major hurdle when intramuscular AAV delivery was initially tested in dog muscle. Robust cytotoxic immune rejection was evoked in normal adult dogs by AAV2, AAV6, and AAV9 that express the transgene from a ubiquitous promoter.46–48 In one study, cellular immunity was even detected in neonatal normal dogs.46 Interestingly, nominal CD4+ and CD8+ T-cell infiltration was found in locally injected dogs by Amoasii et al. This suggests that the use of the tissue-specific promoter is a powerful approach to prevent untoward cellular immune responses. While this finding is encouraging, a generalized conclusion that the CRISPR/Cas system does not induce severe innate and/or cellular immune response will require more thorough examination. For instance, (1) some muscle-specific promoters (such as the desmin and SPc5-12 promoter) have been shown to drive expression in antigen-presenting cells (APCs),49,50 (2) some AAV capsids may transduce APCs more effectively, and (3) age and breed influence the activity and profile of the dog immune system.51 There is a high likelihood that the T-cell response may still be a major hurdle for future AAV CRISPR local injection studies in the canine DMD model if a different promoter, different AAV capsid, different age, or different breed is used.
All three types of immune responses have been reported following high-dose systemic AAV delivery in neonatal large animals (reviewed by Duan36). Of note are two recent reports showing a fatal innate immune response in nonhuman primates and piglets.52,53 In the Amoasii et al. study, both systemically injected puppies survived to the scheduled experiment termination date, suggesting acute immunological death might not be a major concern if CRISPR AAV vectors are delivered to affected puppies using an experimental design and study protocol identical to that described by Amoasii et al.34 Several recent reports suggest that immune responses to the bacterial-originated Cas protein cannot be underestimated.54–56 Evaluation on anti-Cas9 T cells (by CD4, CD8, and Treg immunostaining, tetramer staining, INF-γ enzyme-linked immunosorbent spot, T-cell lineage, and functionality study and intracellular cytokine staining), anti-Cas9 antibodies (by in vitro neutralizing assay for the neutralizing antibody and by enzyme-linked immunosorbent assay [ELISA] for the binding antibody), and serum cytokines and complement (by bead-based multiplex cytokine analysis and complement pathway ELISA assay) should be included in future studies.57,58
The ultimate goal of CRISPR therapy is to mitigate dystrophic muscle disease. In this regard, the treatment effect should be quantified by morphometric analysis of muscle histology and physiological measurement of muscle function. Histopathology evaluation should include, but not be limited to, quantification of the percentage of centrally nucleated myofibers, analysis of myofiber size distribution, evaluation of macrophage and neutrophil infiltration, and qualitative (ideally quantitative) study of muscle fibrosis by Masson trichrome staining or picrosirius red staining (or by biochemical quantification of the collagen content in muscle). Similar assays have been routinely used to characterize morphological changes in dystrophic canine muscles in the literature.59–61
The mouse and dog DMD models were established at about the same time.62,63 While a quite comprehensive list of methods has been available for studying muscle function in mice, methods to study dog muscle function remain underdeveloped until recently. Over the past few years, a number of new assays have been developed, including in situ evaluation of a single dog muscle contractility,64 hind-limb muscle force measurement,65 method to study sympatholysis and functional ischemia in canine muscle,66 electrical impedance myography,67 noninvasive gait analysis,68–70 and noninvasive whole-body activity assay.69,71 The standard operating procedures have been published for many of these protocols and are freely accessible on the Treat-NMD Neuromuscular Network Web site (www.treat-nmd.eu/research/preclinical/overview/) and Parent Project Muscular Dystrophy Web site (http://join.parentprojectmd.org/site/PageServer?pagename=Advance_researchers_sops).72,73
Likely due to the limited sample size and the termination of the study at the pre-symptomatic age, the above-mentioned histological and physiological evaluations were not performed in the Amoasii et al. study. Inclusion of these assays in future studies will better inform the potential outcome when systemic CRISPR therapy moves to human patients.
There are hundreds of muscles in the body (∼700 in a dog). Muscle disease in DMD patients is shaped by the level and extent of dystrophic changes in all body muscles, in particular the heart, diaphragm, and major pelvic limb muscles (e.g., quadriceps) because dystrophy in these muscles leads directly to death or ambulation loss. Following systemic injection, Amoasii et al. performed H&E staining on three skeletal muscles, dystrophin immunostaining on the heart and six skeletal muscles, dystrophin Western blot on the heart and five skeletal muscles, and Cas9 Western blot on the heart (left ventricle, right ventricle, and septum) and 10 skeletal muscles. The authors also quantified level of developmental myosin heavy chain in two skeletal muscles and on-target indel formation on the heart (left ventricle, right ventricle, and septum) and four skeletal muscles. Although Amoasii et al. included the heart and diaphragm in all these assays, the total number of skeletal muscles examined remains low compared to how many muscles a dog has. It will be worthwhile to include more muscles in future studies to appreciate bodywide dystrophin restoration and disease amelioration better.
In summary, the findings reported by Amoasii et al. are encouraging because they provide the critical proof of principle for continuing to explore systemic CRISPR editing as a potential therapy for DMD.33 The results of Amoasii et al. also suggest that the immune response to AAV9-mediated Cas9 expression may, under certain circumstances (as in the Amoasii et al. study), not be a severe concern as has been speculated. Validating these observations with additional studies, as suggested in this article, should prove to be extremely advantageous in the development of CRISPR technology as a therapeutic modality. The Amoasii et al. study has provided a good starting point for experimentally testing AAV-mediated CRISPR therapy in the canine DMD model (or other large mammalian models of human diseases). Large-scale, comprehensive, long-term studies are needed to determine the risk–benefit ratio of systemic AAV CRISPR therapy for DMD before considering testing this promising therapy in patients.
Acknowledgments
The authors thank the support from the National Institutes of Health (NIH; AR-69085 to D.D., GM-063732 and GM-117059 to S.C.), Department of Defense (MD150133 to D.D.), Hope for Javier (to D.D.), Jackson Freel DMD Research Fund (to D.D.), and Intramural Research Program of the NIH, NCATS (to N.N.Y. and C.H.H.).
Author Disclosure
D.D. is a member of the scientific advisory board for Solid Biosciences and an equity holder of Solid Biosciences. The Duan lab has received research supports unrelated to CRISPR editing from Solid Biosciences. C.H.H., D.D., N.B.W. have filed a patent disclosure on systemic CRISPR therapy.
References
- 1. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas Systems. Science 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mali P, Yang LH, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med 2015;21:121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdul-Razak H, Malerba A, Dickson G. Advances in gene therapy for muscular dystrophies. F1000Res 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duan D. Dystrophin gene replacement and gene repair therapy for Duchenne muscular dystrophy in 2016. Hum Gene Ther Clin Dev 2016;27:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chamberlain JR, Chamberlain JS. Progress toward gene therapy for Duchenne muscular dystrophy. Mol Ther 2017;25:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duan D. Duchenne muscular dystrophy gene therapy in the canine model. Hum Gene Ther Clin Dev 2015;26:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGreevy JW, Hakim CH, McIntosh MA, et al. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis Model Mech 2015;8:195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Long C, McAnally JR, Shelton JM, et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 2014;345:1184–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ousterout DG, Kabadi AM, Thakore PI, et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 2015;6:6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li HL, Fujimoto N, Sasakawa N, et al. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 2015;4:143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Young CS, Hicks MR, Ermolova NV, et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell 2016;18:533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y, Long C, Li H, et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv 2017;3:e1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kyrychenko V, Kyrychenko S, Tiburcy M, et al. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight 2017;2:e95918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lattanzi A, Duguez S, Moiani A, et al. Correction of the exon 2 duplication in DMD myoblasts by a single CRISPR/Cas9 system. Mol Ther Nucl Acids 2017;7:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maggio I, Liu J, Janssen JM, et al. Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci Rep 2016;6:37051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wojtal D, Kemaladewi DU, Malam Z, et al. Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am J Hum Genet 2016;98:90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iyombe-Engembe JP, Ouellet DL, Barbeau X, et al. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol Ther Nucl Acids 2016;5:e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duchêne BL, Cherif K, Iyombe-Engembe JP, et al. CRISPR-induced deletion with SaCas9 restores dystrophin expression in dystrophic models in vitro and in vivo. Mol Ther 2018;26:2604–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Long C, Li H, Tiburcy M, et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 2018;4:eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016;351:403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016;351:400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tabebordbar M, Zhu K, Cheng JK, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016;351:407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu L, Park KH, Zhao L, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther 2016;24:564–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. El Refaey M, Xu L, Gao Y, et al. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res 2017;121:923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bengtsson NE, Hall JK, Odom GL, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 2017;8:14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amoasii L, Long C, Li H, et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med 2017;9: pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hakim CH, Wasala NB, Nelson CE, et al. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Young CS, Mokhonova E, Quinonez M, et al. Creation of a novel humanized dystrophic mouse model of Duchenne muscular dystrophy and application of a CRISPR/Cas9 gene editing therapy. J Neuromusc Dis 2017;4:139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koo T, Lu-Nguyen NB, Malerba A, et al. Functional rescue of dystrophin deficiency in mice caused by frameshift mutations using Campylobacter jejuni Cas9. Mol Ther 2018;26:1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ryu SM, Koo T, Kim K, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 2018;36:536–539 [DOI] [PubMed] [Google Scholar]
- 33. Duan D. CRISPR alleviates muscular dystrophy in dogs. Nat Biomed Eng 2018;2:795–796 [DOI] [PubMed] [Google Scholar]
- 34. Amoasii L, Hildyard JCW, Li H, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018;362:86–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, et al. A Duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient Cavalier King Charles Spaniels is amenable to exon 51 skipping. PLoS One 2010;5:e8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duan D. Systemic AAV micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol Ther 2018;26:2337–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mendell JR, Al-Zaidy S, Shell R, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 2017;377:1713–1722 [DOI] [PubMed] [Google Scholar]
- 38. Duan D. Micro-dystrophin gene therapy goes systemic in Duchenne muscular dystrophy patients. Hum Gene Ther 2018;29:733–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aartsma-Rus A, Fokkema I, Verschuuren J, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat 2009;30:293–299 [DOI] [PubMed] [Google Scholar]
- 40. Wasala NB, Lai Y, Shin J-H, et al. Genomic removal of a therapeutic mini-dystrophin gene from adult mice elicits a Duchenne muscular dystrophy-like phenotype. Hum Mol Genet 2016;25:2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kornegay JN. The golden retriever model of Duchenne muscular dystrophy. Skelet Muscle 2017;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akcakaya P, Bobbin ML, Guo JA, et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 2018;561:416–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cwik B. Designing ethical trials of germline gene editing. New Engl J Med 2017;377:1911–1913 [DOI] [PubMed] [Google Scholar]
- 44. Kohn DB, Porteus MH, Scharenberg AM. Ethical and regulatory aspects of genome editing. Blood 2016;127:2553–2560 [DOI] [PubMed] [Google Scholar]
- 45. Mingozzi F, High KA. Overcoming the host immune response to adeno-associated virus gene delivery vectors: the race between clearance, tolerance, neutralization, and escape. Annu Rev Virol 2017;4:511–534 [DOI] [PubMed] [Google Scholar]
- 46. Yuasa K, Yoshimura M, Urasawa N, et al. Injection of a recombinant AAV serotype 2 into canine skeletal muscles evokes strong immune responses against transgene products. Gene Ther 2007;14:1249–1260 [DOI] [PubMed] [Google Scholar]
- 47. Wang Z, Allen JM, Riddell SR, et al. Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of Duchenne muscular dystrophy. Hum Gene Ther 2007;18:18–26 [DOI] [PubMed] [Google Scholar]
- 48. Yue Y, Ghosh A, Long C, et al. A single intravenous injection of adeno-associated virus serotype-9 leads to whole body skeletal muscle transduction in dogs. Mol Ther 2008;16:1944–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boisgerault F, Gross DA, Ferrand M, et al. Prolonged gene expression in muscle is achieved without active immune tolerance using MicrorRNA 142.3p-regulated rAAV gene transfer. Hum Gene Ther 2013;24:393–405 [DOI] [PubMed] [Google Scholar]
- 50. Wang L, Dobrzynski E, Schlachterman A, et al. Systemic protein delivery by muscle-gene transfer is limited by a local immune response. Blood 2005;105:4226–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Day MJ. Immune system development in the dog and cat. J Comp Pathol 2007;137:S10–15 [DOI] [PubMed] [Google Scholar]
- 52. Hinderer C, Katz N, Buza EL, et al. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum Gene Ther 2018;29:285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hordeaux J, Wang Q, Katz N, et al. The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol Ther 2018;26:664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chew WL, Tabebordbar M, Cheng JK, et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods 2016;13:868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Simhadri VL, McGill J, McMahon S, et al. Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol Ther Methods Clin Dev 2018;10:105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Charlesworth CT, Deshpande PS, Dever DP, et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. bioRxiv 2018. Jan 5 [Epub ahead of print]; DOI: 10.1101/243345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Calcedo R, Chichester JA, Wilson JM. Assessment of humoral, innate, and T-cell immune responses to adeno-associated virus vectors. Hum Gene Ther Methods 2018;29:86–95 [DOI] [PubMed] [Google Scholar]
- 58. Martino AT, Herzog RW, Anegon I, et al. Measuring immune responses to recombinant AAV gene transfer. Methods Mol Biol 2011;807:259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shin J-H, Pan X, Hakim CH, et al. Microdystrophin ameliorates muscular dystrophy in the canine model of Duchenne muscular dystrophy. Mol Ther 2013;21:750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith BF, Yue Y, Woods PR, et al. An intronic LINE-1 element insertion in the dystrophin gene aborts dystrophin expression and results in Duchenne-like muscular dystrophy in the corgi breed. Lab Invest 2011;91:216–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kodippili K, Hakim CH, Pan X, et al. Dual AAV gene therapy for Duchenne muscular dystrophy with a 7-kb mini-dystrophin gene in the canine model. Hum Gene Ther 2018;29:299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sicinski P, Geng Y, Ryder-Cook AS, et al. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science 1989;244:1578–1580 [DOI] [PubMed] [Google Scholar]
- 63. Cooper BJ, Winand NJ, Stedman H, et al. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature 1988;334:154–156 [DOI] [PubMed] [Google Scholar]
- 64. Yang HT, Shin J-H, Hakim CH, et al. Dystrophin deficiency compromises force production of the extensor carpi ulnaris muscle in the canine model of Duchenne muscular dystrophy. PLoS One 2012;7:e44438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Childers MK, Grange RW, Kornegay JN. In vivo canine muscle function assay. J Vis Exp 2011; (50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kodippili K, Hakim CH, Yang HT, et al. Nitric oxide dependent attenuation of norepinephrine-induced vasoconstriction is impaired in the canine model of Duchenne muscular dystrophy. J Physiol 2018;596:5199–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hakim CH, Mijailovic A, Lessa TB, et al. Non-invasive evaluation of muscle disease in the canine model of Duchenne muscular dystrophy by electrical impedance myography. PLoS One 2017;12:e0173557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jenkins GJ, Hakim CH, Yang NN, et al. Automatic characterization of stride parameters in canines with a single wearable inertial sensor. PLoS One 2018;13:e0198893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shin J-H, Greer B, Hakim CH, et al. Quantitative phenotyping of Duchenne muscular dystrophy dogs by comprehensive gait analysis and overnight activity monitoring. PLoS One 2013;8:e59875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barthelemy I, Barrey E, Aguilar P, et al. Longitudinal ambulatory measurements of gait abnormality in dystrophin-deficient dogs. BMC Musculoskelet Disord 2011;12:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hakim CH, Peters AA, Feng F, et al. Night activity reduction is a signature physiological biomarker for Duchenne muscular dystroophy dogs. J Neuromuscul Dis 2015;2:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Duan D, Rafael-Fortney JA, Blain A, et al. Standard operating procedures (SOPs) for evaluating the heart in preclinical studies of Duchenne muscular dystrophy. J Cardiovasc Transl Res 2016;9:85–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nagaraju K, Willmann R, Network T-N, et al. Developing standard procedures for murine and canine efficacy studies of DMD therapeutics: report of two expert workshops on “Pre-clinical testing for Duchenne dystrophy”: Washington DC, October 27th–28th 2007 and Zurich, June 30th–July 1st 2008. Neuromuscul Disord 2009;19:502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]