Abstract
The oocytes, embryos, and cell-free lysates of the frog Xenopus laevis have emerged as powerful models for quantitative proteomic experiments. In the accompanying paper (Chapter 13) we describe how to prepare samples and acquire multiplexed proteomics spectra from those. As an illustrative example we use a 10-stage developmental time series from the egg to stage 35 (just before hatching). Here, we outline how to convert the ~700,000 acquired mass spectra from this time series into protein expression dynamics for ~9000 proteins. We first outline a preliminary quality-control analysis to discover any errors that occurred during sample preparation. We discuss how peptide and protein identification error rates are controlled, and how peptide and protein species are quantified. Our analysis relies on the freely available MaxQuant proteomics pipeline. Finally, we demonstrate how to start interpreting this large dataset by clustering and gene-set enrichment analysis.
Keywords: Quantitative multiplexed proteomics, TMT, Mass spectrum, MaxQuant, Xenopus laevis, Development, False discovery rate, Gene symbols, k-means clustering, Gene-set enrichment
1. Introduction
Xenopus laevis is a powerful amphibian model organism routinely used in numerous laboratories to investigate a wide range of problems in biology. The high synchrony of in vitro fertilization coupled with the large amounts of protein that are available (~25 μg/egg) [1] make the system ideal for proteomic analysis where a full experiment requires up to ~1 mg of protein. In a parallel publication we detail our sample preparation protocol for this workflow (Chapter 13) and how to acquire multiplexed proteomics spectra. Here, we describe how to identify proteins and extract their respective quantitative information from this data.
All of our protein identification and quantification is done with a mass spectrometer. Mass spectrometers measure the mass to charge ratio (m/z) of analytes. The goal of a proteomics experiment is to identify and quantify peptides via characteristic masses and their peak heights. However, for complex mixtures such as X. laevis lysates, simply measuring the m/z of a peptide is not sufficient for identification. Rather, a peptide is isolated inside the mass spectrometer, fragmented, and a second mass spectrum (MS/MS or MS2) is acquired. By comparing the obtained fragment ions with theoretical spectra from a reference database, peptides can be identified [2]. Matching peptide fragments to a sequence requires a protein reference database. An important step forward in performing sensitive X. laevis proteomics was the generation of a high-quality protein reference [3]. This matching process is imperfect and false positives (both on the peptide and protein level) occur. Below we will outline how to control these errors with a false discovery rate (FDR) so that only a user-specified percentage of peptides have been incorrectly assigned a sequence. Errors also arise when mapping peptide sequences to protein sequences which can also be controlled at a user-specified FDR [4]. Typically, 1% FDR is used on both the peptide and the protein level.
To understand many biological processes, identification of proteins present in a sample is often inadequate. Rather, quantification of the respective protein amounts in each sample is preferable. Many different forms of quantitative proteomics like label-free, SILAC, or DIA are available [5–7]. Our method of choice is multiplexed proteomics. For this approach up to 11 different conditions can be barcoded with tandem mass tags (TMT), mixed together, and ionized simultaneously onto a mass spectrometer. This approach allows the simultaneous quantification of peptides from all 11 conditions and the routine quantification of >8000 proteins in 2 days of instrument time (see Fig. 1). In its simplest implementation, TMT barcodes from each condition can be distinguished and quantified on the MS2 level [8, 9]. While the TMT-MS2 approach is advantageous in that it is implementable on a wide range of mass spectrometers, it has significant measurement distortion [10–12]. A recently introduced improvement is the TMT-MS3 approach where an additional gas purification step is performed in the mass spectrometer which drastically reduces measurement distortion [13]. Using this enhanced multiplexing approach, we have successfully extracted biological insight regarding nuclear composition in X. laevis oocytes [14], protein dynamics during early X. laevis development [15], and the proteome and phoshpho-protoeme changes during fertilization [16]. Below we detail how to quantify peptides and proteins from a TMT-MS3 experiment. In our experience, the most difficult part of quantitative proteomics experiments is to extract meaningful biological insight from the large datasets that are generated. This remains a massive challenge, and each study typically requires novel approaches. Nevertheless, we have found several common steps can be performed when a dataset is first acquired that can help to shed a preliminary understanding. Below we detail how we use k-means clustering to group similarly behaving proteins during early X. laevis development together and then use gene ontology term membership enrichment to determine which biological processes (if any) are over- or under-represented in a cluster.
Fig. 1.
Overview of quantitative multiplexed proteomics data analysis pipeline. (A) A preliminary quality control sample is made and analyzed on a mass spectrometer with a TMT-MS3 method. Peptide sequences are identified through a database search and matched to protein sequences. The protease digestion efficiency, tandem mass tagging efficiency, and cysteine alkylation efficiency are all estimated to determine if the sample preparation was satisfactory. (B) Full developmental time course X. laevis TMT-MS3 experiment. Samples at ten different stages in development were collected and processed as in Gupta et al. Analysis of these samples leads to the quantification of 9345 X. laevis proteins across ten stages in development. Illustrations of developmental stages courtesy of Garland Science [[21]]
2. Materials
2.1. MaxQuant
Version 1.6.0.16 at the time of publication. See below for download and installation instructions.
2.2. Reference X. laevis Proteome
On our lab website (https://scholar.princeton.edu/wuehr/sample_prep) we provide a X. laevis proteome that can be used as a reference when matching mass spectra to peptide sequences. PHROG_09_26_2017. FASTA is a compilation of 79,215 X. laevis protein sequences that were generated by mRNA sequencing [3]. As of this publication PHROG_09_26_2017.FASTA is slightly superior in terms of number of peptides identified compared to JGI 9.1, the latest gene models release of the frog proteome available on Xenbase. However, we suspect that as newer versions are released newer versions of JGI will become superior. We encourage the reader to check for updated versions of JGI at http://www.xenbase.org/other/static/ftpDatafiles.jsp. Newer releases can be mapped to human gene symbols with the conveniently available web tool in Subheading 2.5.
2.3. X. laevis Developmental Time Series Mass Spectrometry Raw Data
The mass spectrometry proteomics data have been deposited to the ProteomeExchange Consortium via the PRIDE partner repository with the dataset identifier PXD007915 [20].
TGR_02788.raw is our quality control sample. The remaining raw files (TGR_02793.raw to TGR_02823.raw) correspond to the 24 reverse phase fractionated samples.
2.4. X. laevis Developmental Time Series Processed Data (Post MaxQuant)
On our website (https://scholar.princeton.edu/wuehr/sample_prep) we provide the MaxQuant output that should be obtained for both the sample quality control outlined below as well as for the entire dataset analysis using PHROG_09_26_2017.FASTA as a X. laevis reference proteome.
2.5. Mapping X. laevis Protein Sequences to Human Gene Symbols
We previously developed a convenient online resource for matching reference proteomes from X. laevis (or any other organism) to human gene symbols [3]. The tool is available at http://kirschner.med.harvard.edu/tools/genesym_assignment.html and can be used with future JGI releases. On our website (https://scholar.princeton.edu/wuehr/sample_prep) we provide spreadsheets with the human gene symbol mappings for PHROG_09_26_2017.FASTA.
2.6. Anaconda Python Distribution Package
Anaconda is a freely available compilation of python and many commonly used python libraries in scientific computing. The python scripts associated with this paper make use of several of these libraries: Pandas, NumPy, Matplotlib, and SciKitLearn. We encourage the reader to download the entire package, which should be able to run all scripts associated with this manuscript without any additional installation.
2.7. Python Scripts for Data Analysis of Data After Being Processed in MaxQuant
All of our Python scripts are available for download on our website in the folder maxquant_bioinformatics.zip.
tmt_labeling_efficiency.py estimates what fraction of peptides were labeled with TMT during the sample quality control search.
missed_cleavage_rate.py estimates what fraction of peptides were missed cleavages during the sample quality control search.
percent_cysteines.py estimates what fraction of peptides that were identified contain a cysteine during the sample quality control search.
map_gene_symbol.py takes a MaxQuant output file and maps the X. laevis proteins from the PHROG database to a gene symbol (if a good mapping exists) and generates a new .csv file containing this information.
k_means_clustering.py will perform k-means clustering on processed data that has been mapped to human gene symbols, generating a new .csv file containing this information. The script generates plots of the individual protein dynamics of each cluster in .png and .svg formats. Finally, separate .csv files of all of the gene symbols in each cluster are generated so that gene ontology term biological enrichment can be easily performed.
2.8. Reference MaxQuant Parameters and Screenshots of All Steps
Detailed screenshots of all operations within MaxQuant both for the sample quality control as well as the fully fractionated X. laevis developmental time series have been uploaded to our website (https://scholar.princeton.edu/wuehr/sample_prep). All MaxQuant parameters used in both searches are available in the parameters.txt file that is present in each output folder.
3. Methods
3.1. Downloading and Installing MaxQuant
3.1.1. Rationale
MaxQuant is a high-quality freely available proteomics software that allows users to analyze TMT-MS3 quantitative multiplexed proteomics experiments [17]. MaxQuant is fast and contains all features necessary to match mass spectra to peptide and protein sequences at controlled false discovery rates and faithfully extract their quantitative information.
3.1.2. Procedure
Go to http://www.coxdocs.org/doku.php?id=maxquant:common:download_and_installation
Register and accept licensing agreement. After receiving an email you can download the software.
Extract MaxQuant and place the MaxQuant_1.6.0.16 folder on your Desktop.
Open MaxQuant by opening the MaxQuant_1.6.0.16 folder, opening the MaxQuant folder and then running MaxQuant.exe.
3.2. Specifying Peptide Modifications in MaxQuant
3.2.1. Rationale
Our intention during the sample preparation procedure is to digest all proteins into peptides, modify all cysteines with N-ethyl maleimide (NEM) (see Note 1), and label all free n-termini and lysines with a tandem mass tag (TMT) reagent. Both of these reagents alter the mass of each peptide to which they are attached. MaxQuant must be told to add these mass modifications to all peptide sequences it is searching in the uploaded X. laevis protein database. Below we outline how to specify these modifications.
3.2.2. Procedure
- Adding the cysteine NEM modification:
- Navigate to the Configuration tab in MaxQuant.
- Select Modifications in the Data tab.
- Select Add in the Table tab.
- Name the modification NEM.
- Leave description as New modification.
- On the Composition row select Change and enter H(7) C(6) N O(2) (see Note 2). The mass should be 125.0476784741.
- Position should be Anywhere.
- Type is Standard.
- New terminus is None.
- Under Specificities select the K and click (see Note 3).
- Select + under Specificities and select C.
- Under the Actions tab in Configuration section select Modify table.
- Under the Table actions tab in Configuration section select Save changes. The NEM modification is successfully added and is displayed at the bottom of the table on left.
- Adding the n-terminus TMT modification (see Note 4):
- Navigate to the Configuration tab in MaxQuant.
- Select Modifications.
- Select Add.
- Name the modification dynamicTMTnTerm.
- Leave description as New modification.
- On the Composition row select Change and enter H(20) C(8) Cx(4) N O(2) Nx (see Note 5). The mass should be 229.162932141.
- Position should be Any N-term.
- Type is Standard.
- New terminus is None.
- Under Specificities select the K and click −.
- Under Specificities select the + and add −.
- Under the Actions tab in Configuration section select Modify table.
- Under the Table actions tab in Configuration section select Save changes. The dynamicTMTnTerm modification is successfully added and is displayed at the bottom of the table on left.
- Adding the lysine TMT modification (see Note 4):
- Navigate to the Configuration tab in MaxQuant.
- Select Modifications.
- Select Add.
- Name the modification dynamicTMTonK.
- Leave description as New modification.
- On the Composition row select Change and enter H(20) C(8) Cx(4) N O(2) Nx. The mass should be 229.162932141.
- Position should be Anywhere.
- Type is Standard.
- New terminus is None.
- Under Specificities K should be on the only AA present (should be this way by default without doing anything).
- Under the Actions tab in Configuration section select Modify table.
- Under the Table actions tab in Configuration section select Save changes. The dynamicTMTonK modification is successfully added and is displayed at the bottom of the table on left.
- Specifying the Isotopic impurities of the TMT reagents used (see Note 6):
- Navigate to the Configuration tab in MaxQuant.
- Select Modifications.
- If you scroll down through the list you will find TMT10plex-Nter126C and further below it TMT10 plex-Lys126C. These both correspond to the TMT 126 tag from Thermo.
- Select one of them and scroll to the bottom on the right side. There will be rows denoted % − 2, % − 1, % + 1, % + 2. Here you can enter the isotopic impurity corrections from the manufacturer. Enter the same isotopic impurity corrections for both the “Nter” and Lys” version of each reagent (E.g., reagent 1 is 126C, reagent 2 is 127N, and so on).
- After each isotopic impurity has been entered, select Modify table and then Save changes to update the table.
- After all isotopic impurities have been entered MaxQuant must be restarted for this information to be correctly loaded.
After adding all desired modifications restart MaxQuant for them to be selectable in the drop down menus (see Note 7).
3.3. Providing a Protein Reference Database to MaxQuant
3.3.1. Rationale
Performing sensitive proteomics requires a reference database of protein sequences for MaxQuant to match acquired mass spectra to. We have provided a X. laevis protein reference database in the appropriate FASTA format that can be loaded directly into MaxQuant for use.
3.3.2. Procedure
Download PHROG.FASTA from (https://scholar.princeton.edu/wuehr/sample_prep), unzip and place PHROG_09_26_2017. FASTA in the MaxQuant directory (see Note 8).
Navigate to the Configuration tab in MaxQuant.
Select Sequence databases.
Select Add.
To the right of Fasta File name click Select and choose PHROG_09_26_2017.FASTA.
To the right of Identifier parse rule press Select and choose identifier parse rule: >(.*).
Under the Actions tab in Configuration section select Modify table.
Under the Table actions tab in Configuration section select Save changes. PHROG_09_26_2017.FASTA is successfully added and is displayed at the bottom of the table on left (see Note 9).
Close and restart MaxQuant for the changes to take effect (see Note 10).
Under the Global Parameters tab select Sequences.
To the right of the Fasta files header select Add File and then select PHROG_09_26_2017.FASTA and can be saved in any directory on your PC.
Ensure Include Contaminants is checked.
3.4. Specification of Various Mass Spectrometer Instrument Parameters in MaxQuant
3.4.1. Rationale
MaxQuant must be instructed how to generate all possible peptide sequences in silico that could be present in the sample given the protein sequences from the database. The type(s) of peptide sequences present are a consequence of the specificity of the enzyme(s) that were used during the digestion step in the sample preparation (Chapter 13). Typically, the specificity is either all peptides must have a c-terminal K (LysC only digest) or, all peptides must have a c-terminal K or R (LysC + Trypsin digest). The X. laevis time series was digested with only LysC. If the digestion was perfect every identified peptide sequence (excluding c-termini) would end in K.
To perform an initial sample quality control step (described in depth below) we also have to add TMT as a dynamic modification so that the database considers all peptide sequences with the TMT modification either present or absent. This will allow us to determine the fraction of peptides that were modified with TMT. We also need to specify any other mass modifications that were made. In this case, we need to add NEM as a fixed modification on cysteine.
3.4.2. Procedure
Navigate to the Global parameters tab.
Under Fixed Modifications select the Carbamidomethyl (C) fixed modification on the right and select < to remove it.
On the left hand side find NEM (sorted alphabetically) and select > to add it as a fixed modification.
Min. peptide length should be 7.
Max peptide mass [Da] should be 4600.
Min peptide length for unspecific search should be 8.
Max peptide length for unspecific search should be 25.
Navigate to the Group-specific parameters tab select Digestion.
Digestion mode should be Specific.
Under Enzyme select Trypsin/P and select < to remove it.
Under Enzyme select LysC and select > to add it (see Note 11).
Max. missed cleavages should be 2.
Select the Modifications panel.
Under Variable Modifications find dynamicTMTnTerm and select >.
Under Variable Modifications find dynamicTMTonK and select >.
On the right, remove Acetyl(Protein N-term) by selecting it and then pressing < (see Note 12).
Ensure that the max. Number of modifications per peptide is 5.
Navigate to the Global parameters tab. Select the Protein quantification panel.
To the right of Modifications used in protein quantification, select Acetyl (N-term) on the right and press <.
Select NEM and press >.
Select dynamicTMTnTerm and press >.
Select dynamicTMTonK and press >.
Select the Tables panel and deselect all boxes.
All other parameters can be left as the default.
3.5. Control Peptide and Protein Sequence Matching Error Rates
3.5.1. Rationale
Matching acquired mass spectra of peptides to the correct sequence is an imperfect process that has an unknown error rate. To estimate this error rate, the gold-standard in the field is to take the true protein sequences from the reference database and construct a second set of “decoy” sequences—sequences that do not exist in nature but have the same amino acid properties as the corresponding actual sequences [18]. This is accomplished by reversing all protein sequences (e.g., MYPEPTIDE becomes EDITPEPYM) and then generating a second set of in silico peptides which are also considered when each mass spectrum is matched to a peptide sequence from the database. Previous work has shown that peptides from these reverse sequences that are at least 7 amino acids in length almost never exist in the actual proteome [18]. Thus, one can assume that for every peptide that maps to a “decoy” peptide there is an equally likely chance of a peptide mapping to another peptide sequence that is from the actual proteome. Therefore, one can count the total number of “decoy” peptides that were matched and then double this number to estimate the number of incorrect peptide sequences from the actual proteome. Typically, a large fraction of spectra are incorrectly matched with the wrong sequence if no postfiltering is done. To overcome this, MaxQuant uses a machine learning approach to determine how various parameters from each mass spectrum (e.g., mass error of the match, number of matched peptide fragments observed, etc.) relates to the probability of whether a peptide is from the correct sequence or not. After predicting these probabilities, a user-defined false discovery rate (FDR) is specified to filter the matched peptide sequences down to say where only 1% of all peptide sequences are identified incorrectly. For large datasets, even if only 1% of all peptide sequences have been incorrectly assigned, large false discovery rates on the protein level (e.g., >20%) can be observed [19]. Thus, additionally a 1% FDR is also set when matching peptide sequences to their protein sequences [17] . Below we outline the parameters in MaxQuant that are used to set the identified peptide and protein false discovery rates.
3.5.2. Procedure
Under Global parameters select the Identification panel.
The PSM FDR stands for Peptide Spectral Match False Discovery Rate. We recommend using the default of 0.01 which corresponds to a 1% False Discovery Rate. This means 1% of all peptide sequences in the final peptide dataset from MaxQuant will be incorrect.
Similarly, we recommend using a Protein FDR of 0.01 (the default setting). This means that approximately 1% of all protein groups in the final protein dataset from MaxQuant will be incorrect.
3.6. Determining Sample Quality
3.6.1. Rationale
During the sample preparation protocol (Chapter 13), a prepared sample is subjected to an initial quality control step before it is fully processed and analyzed. This initial quality control step analyzes the sample with a TMT-MS3 method on a mass spectrometer for two hours. We will analyze a single unfractionated TMT-MS3 sample of a X. laevis developmental time series in MaxQuant to determine whether the sample preparation is of high quality. Specifically, we wish to determine (1) What was the labeling efficiency of n-terminal and lysine groups of peptides with TMT? Ideally >98% of all peptides detected are labeled with TMT. (2) How efficient was our protease digestion of each sample? Ideally, the number of missed cleavages (number of peptides that have a non-c-terminal Lysine is <10% for LysC only digests and a non-c-terminal Lysine or Arginine <20% for Trypsin/LysC digests). (3) Did the cysteine alkylation work? We expect to see least 5% of peptides containing a cysteine (see Note 1). First we need to load our raw data of a single unfractionated TMT-MS3 experiment into MaxQuant and adjust various parameters.
3.6.2. Procedure
- Loading the data.
- Open MaxQuant.
- In MaxQuant select the Raw data tab, select Load, and select the TGR_02788.raw file and click Open.
- If not already done, configure and upload the PHROG_09_26_2017.FASTA reference database as described above and select the correct protease specificity.
- All modifications should be as specified above.
- In the bottom left hand corner change the Number of processors to the # of CPUs on your PC ignoring hyperthreading—1 (see Note 13).
- Select Start in the bottom left hand corner to start the search (see Note 14).
- Progress of the search can be monitored under the Performance table.
- Determining TMT labeling efficiency:
- Once the search is complete by default the files will be stored in the same directory that MaxQuant is located in. Within this directory should be a folder named combined. Open it and then open the txt folder.
- Open modificationSpecificPeptides.txt in a spreadsheet viewing program such as Microsoft Excel. modificationSpecificPeptides.txt is a list of all peptides identified, including those with a variable modification present (Oxidation (M), dynamicTMTnTerm and dynamicTMTonK).
- To obtain a quantitative estimate of TMT labeling efficiency either use a custom function in your spreadsheet program to count the number of peptides in modificationSpecificPeptides.txt that have the string dynamicTMTnTerm in the Modifications column or use the tmt_labeling_efficiency.py python script that we provide on our website.
- To use the python script, place the tmt_labeling_efficiency. py script in the txt folder.
- Open the terminal by simultaneously pressing the windows button + R and typing cmd.
- In the terminal type cd Desktop\MaxQuant_1.6.0.16\MaxQuant\combined\txt and press enter.
- Type python tmt_labeling_efficiency.py and press enter. Your labeling efficiency results will be printed (see Note 15).
- Determining missed cleavage rate: The goal is to determine how many peptides present in the sample contain an internal amino acid (K in the case of LysC only digest and K or R in the case of a trypsin/LysC digest). These peptides are missed cleavages, the internal amino acid should have been digested but was not. If this number is too high (>10% for LysC only or >20% for LysC/Trypsin) then the digestion was not efficient and will introduce error in the measurements.
- Open the modificationSpecificPeptides.txt file.
- The Sequence column contains the sequences of all peptides that were identified.
- If it is convenient, you can specify a custom function either in your spreadsheet program or with your programming language of choice to count how many peptides have an internal K (lysC only) or K/R (trypsin + LysC).
- Alternatively, we provide a python script that will do this for you.
- Download missed_cleavage_rate.py from our website and place this in the txt folder.
- Open the terminal by pressing Windows button + R, typing cmd and pressing enter.
- In the terminal type cd Desktop\MaxQuant_1.6.0.16\MaxQuant\combined\txt and press enter.
- In the terminal type python missed_cleavage_rate.py --protease <protease_used> where <protease_used> is either lysc or trypsin (e.g., python missed_cleavage_rate.py --protease lysc).
- Your missed cleavage rate will be printed.
- Determining cysteine alkylation efficiency: During sample preparation (Chapter 13) cysteines are reduced to free thiols and alkylated with NEM. While we cannot quantitatively evaluate how well this worked, we can get a semi-quantitative evaluation by asking what fraction of all peptide sequences contain a cysteine. Typically for X. laevis samples coming from a complex mixture (egg extract, developing embryos, etc.) this number should be ~5–10%. If less than 3% of all peptides contain cysteines this suggests that alkylation efficiency was poor and cysteine containing peptides should be discarded prior to quantification (see Note 16). If cysteine alkylation is poor but TMT labeling efficiency was high and missed cleavage rates are acceptable, a full experiment can quantify proteins accurately with ~5% less proteins being quantified if all cysteine peptides are ignored.
- If convenient, the number of peptides containing a cysteine can be determined with a custom script of your choice by counting how many peptides contain a C in the Sequence column of modificationSpecificPeptides.txt. Alternatively, we provide a python script to calculate this.
- Download percent_cysteines.py from our website and place this in the txt folder.
- In the terminal type cd Desktop\MaxQuant_1.6.0.16\MaxQuant\combined\txt and press enter.
- In the terminal type python percent_cysteines.py.
- The percent of peptides containing cysteine will be printed.
3.7. Searching a Full TMT–MS3 X. laevis Developmental Time Course Experiment with MaxQuant
3.7.1. Rationale
Once a high-quality sample has been produced and then fractionated by medium pH reverse phase HPLC into 24 fractions and each fraction ionized onto a mass spectrometer (Chapter 13) the full experiment can be analyzed.
3.7.2. Procedure
Download the 24 fractionated samples from our ProteomeX change deposit (see Subheadings 2 and 3).
Store all of the samples in a folder titled 24fracs in the MaxQuant directory.
Select the Raw data tab in MaxQuant and then select Load folder and select 24fracs.
Select the Group-specific parameters tab and then select the Type panel. Under Type click the drop down menu and select Reporter ion MS3.
Select the 10plex TMT button below all of the label options. A total of 20 different TMT10plex options (e.g. Lys-126C, Lys-127N etc.) will appear on the right hand side.
Select the Digestion panel and select Trypsin/P from the right and select <. Then select LysC (see Note 17) on the left and select >.
Select the Global parameters tab and then select the Sequences panel. Under Fasta files click Add file and then select PHROG_09_26_2017.FASTA. Under Fixed modifications select carbamidomethyl on the right and select <. Then select NEM on the left and select >. Select the Protein quantification panel and under Modifications used in protein quantification select NEM and select > (see Note 18). Under tables deselect all options.
In the bottom left hand corner change the number of processors to # of CPUs on your computer (ignoring threads)—1.
Select Start at the bottom and the progress of the searches can be monitored under the Performance tab. This will take at least several hours.
3.8. Obtaining X. laevis Protein Relative Abundance Quantitation and Mapping X. laevis Sequences to Human Gene Symbols
3.8.1. Rationale
MaxQuant will write a .tsv file that can be opened in excel to find the measured protein abundances for each X. laevis protein. We find it very convenient to map the X. laevis protein sequences to human gene symbols which have a lot of metadata associated with them from the literature [3] (see Fig. 2). Most, but not all X. laevis proteins map to a human gene symbol with the PHROG reference database. A total of 15,672 unique human gene symbols are present in the entire database.
Fig. 2.
Mapping X. laevis protein sequences to human gene symbols. (A) X. laevis protein sequences are mapped to human gene symbols by using a bi-directional blast approach [3]. (B) Of the 79,215 X. laevis sequences present in our database, 59,596 of these have enough homology with the human sequences to map to the human gene symbol. These 59,596 mapped sequences collapse to a total of 15,673 unique human gene symbols. Part of this redundancy is from protein isoforms (X. laevis is pseudotetraploid) and part of it may come from splice-isoforms or errors during the protein reference database construction, e.g., fragmented proteins
3.8.2. Procedure
After the search is finished, open the 24fracs folder, open the combined folder, and then open the txt folder.
The proteinGroups.txt file can be opened in the spreadsheet viewing program of your choice.
The Protein IDs column lists all of the X. laevis protein identifiers for a given row (see Note 19). The number of proteins in a given row is indicated by the Number of proteins column. The Peptides column indicates how many different peptides were identified and quantified from the protein(s) in that row.
The TMT-MS3 quantitative relative abundance metrics are stored in the columns titled Reporter intensity correct x where x ranges from 0 to 9. Reporter intensity correct 0 corresponds to the TMT 126C TMT tag, Reporter intensity correct 1 corresponds to the TMT 127N TMT tag, and so on. Reporter intensity correct 9 corresponds to the TMT 131N tag.
These values can be used for any analysis the user is interested in doing on their own.
To create a new spreadsheet that maps the X. laevis protein(s) in each row to a gene symbol, copy the map_gene_symbol.py script from our website into the txt folder.
Download PHROG_annotation.csv from the phrog_annotation.zip file on our website (Materials and Equipment section). Unzip the folder and place PHROG_annotation.csv into the txt folder.
In the terminal type
cd Desktop\MaxQuant_1.6.0.16\MaxQuant\24fracs\combined\txt and press enter.
Type python map_gene_symbol.py and press enter. A new file proteins_mapped_to_gene_symbols.csv will be created.
3.9. k-Means Clustering of Proteins During X. Laevis Development
3.9.1. Rationale
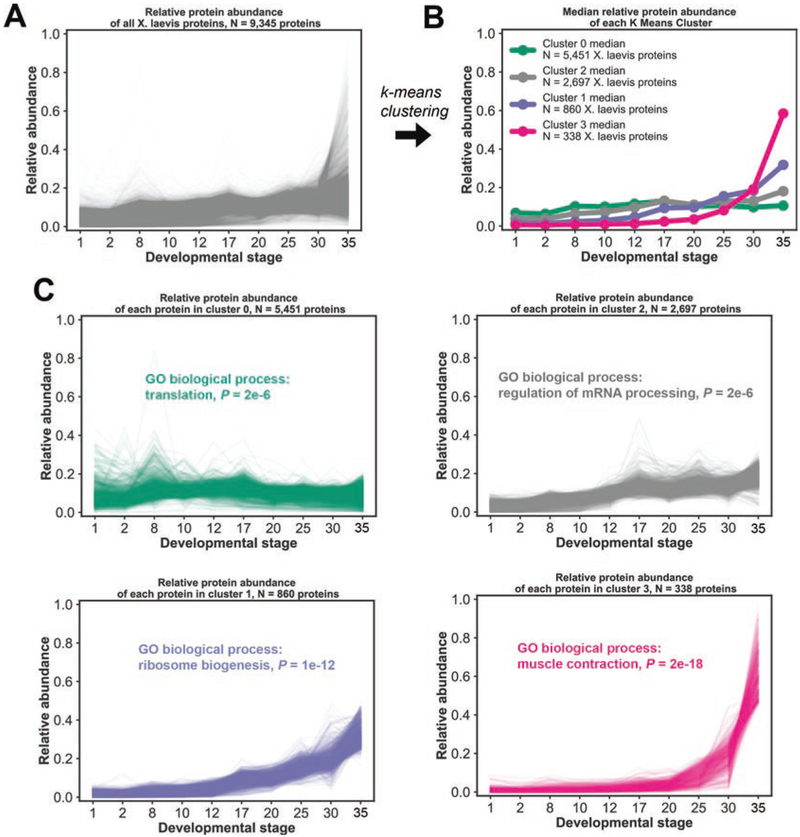
One useful way of analyzing large X. laevis proteomic datasets is to cluster proteins into distinct groups based on their relative abundance within the set of collected conditions. Similarly behaving proteins will be grouped within the same cluster (see Fig. 3a–c).
Fig. 3.
k-Means clustering and gene ontology biological enrichment of developmental time series. (a) Time series profiles throughout developmental stages for all 9345 X. laevis proteins that were quantified. (b) Median relative abundances and their membership number using a k-means clustering approach based on Euclidean distance with the number of clusters set to 4. (c) Relative abundances of proteins in individual clusters and the top gene ontology (GO) biological enrichment term associated with each cluster
3.9.2. Procedure
Copy the k_means_clustering.py script from our website into your txt folder after you have made the proteins_mapped_to_gene_symbols.csv file as outlined in Subheading 3.8.
In the terminal type
Cd Desktop\MaxQuant_1.6.0.16\MaxQuant\24fracs\combined\txt and press enter.
Type python k_means_clustering.py --num_clusters <number> --line_opacity <opacity> --folder_name <name> where num_clusters is the number of clusters you wish to make. line_opacity is a tuning parameter that controls the opacity of each line in the cluster, decrease the number (a good starting point is 0.01) to make lines more opaque. folder_name is the name of the folder all of the generated files will be stored (e.g., python k_means_clustering.py --num_clusters 4 --line_opacity 0.01 --folder_name clustering_results). We recommend playing around with this number and inspecting the generated plots of each cluster to determine if increasing the number of clusters produces additional clusters with distinct behavior. In the case of the X. laevis developmental time series dataset the most informative number appears to be 4.
A new file proteins_mapped_to_gene_symbols_w_kmeans_clusters.csv is generated. This contains all previous information as well as column cluster_num which indicates the cluster number each protein belongs to.
For each cluster, a cluster_num_x.svg and cluster_num_x.png file (where x is a cluster number) will be generated that can be used to visually inspect the individual behaviors in each cluster.
A cluster_num_x.csv (where x is a cluster number) will be generated which contains a list of human gene symbols that can be used for gene ontology biological enrichment analysis (see Subheading 3.10).
A protein_background.csv file is generated which contains a list of the unique gene symbols identified in the entire dataset and will be used in the gene enrichment analysis below.
3.10. Gene Ontology Biological Function Enrichment Analysis of k-Means Clusters
3.10.1. Rationale
The human gene symbols of the proteins in each protein cluster can be analyzed with the bioinformatics tool Gene Ontology which determines if certain gene sets belonging to specific biological processes are either over- or under-enriched (see Fig. 3c). This can often give a helpful course grained picture of the underlying biological response. An important aspect of enrichment analysis is the notion of a background: the set of all proteins from which subsets will be analyzed for enrichment. By default the background is set to be the entire genome. However, while the genome contains ~20,000 gene symbols, typically we only map all X. laevis proteins quantified in a TMT-MS3 experiment to ~7000 gene symbols. Therefore, we typically only use the ~7000 gene symbols that were detected in the experiment as background.
3.10.2. Procedure
Open a web browser and go to http://pantherdb.org/webservices/go/overrep.jsp
Under Step 1. Press choose file and select one of the cluster_num_x.csv files located in the txt file from the MaxQuant searches.
Under Step 2. Select Homo sapiens.
Under Step 3. Select Statistical overrepresentation test and use the default settings.
Select submit.
To the right of Reference list (this is the background) select Change. Select the protein_background.csv file that was generated in Subheading 3.9.
To the right of Annotation Data Set you can select different types of analyses. We recommend starting with GO biological process complete.
The category, fold enrichment, and multiple hypothesis testing corrected P values will be displayed.
4. Notes
If cysteines are not modified with NEM (as in Chapter 13), free thiols are extremely reactive and will almost certainly be oxidized. There are several oxidation states that can be produced, which are difficult to reliably detect with current mass spectrometry technology and will not be identified. Thus, if alkylation was not carried out successfully, essentially no peptides identified will contain a cysteine.
To successfully enter a chemical formula the drop down menu for an element must be selected and then the correct number entered. The drop down menu for the next element must then be selected for the previous selection to be registered. On the last element select ok.
By default MaxQuant puts a Lysine (K) here. Remove it in this case because with the correct pH NEM will only modify cysteines.
MaxQuant already has the TMT modification incorporated if one wants to assume it is a static modification, which we will make use of later. However, we first use it as a dynamic modification, allowing MaxQuant to consider the full database for all possible peptides with or without TMT present so that we can determine how many peptides were modified with TMT.
Cx is C13 and Nx is N15.
Each condition in a multiplexed experiment is barcoded by being labeled with a different TMT reagent, which are distinguishable after fragmentation and have different numbers of heavy isotopes. However, these TMT reagents are commercially synthesized and there are isotopic impurities in the tags that will distort the resulting ratios unless MaxQuant normalizes them. The exact impurity ratios depend on the LOT# you have purchased from Thermo and can be obtained from them.
These modifications will be saved in the future and only need to be configured once until you install a new version of MaxQuant.
E.g., if the MaxQuant folder is on your desktop the directory would be C:\Users\YourUserName\Desktop\MaxQuant_1.6.0.16\MaxQuant where you replace YourUser Name with the name of your PC.
If this step is not done correctly when you initiate the search you will obtain an error saying “WARNING: fasta file <fasta_directory_here> is not configured.” If MaxQuant does not know how to correctly parse the header of the FASTA file it will error out when writing out the spreadsheets after the search is over and you will not be able to obtain your searched data.
The fasta file, isotopic impurity modifications as well as the NEM, dynamicTMTonK, and dynamicTMTnTerm will be saved in the future and only need to be configured once unless a new version of MaxQuant is installed.
Trypsin/P is appropriate when a trypsin digest alone is carried out, as Trypsin cannot cut lysines that are followed by a proline. However, when a tandem LysC/Trypsin digest is used as we outlined in our previous paper, LysC is able to digest lysines that are followed by proline and Trypsin should be used. If only LysC was used then select LysC.
Including acetylated n-termini is useful when analyzing a full dataset but during sample quality control analysis we want to estimate what fraction of n-termini on peptides were labeled with TMT so we will ignore any peptides with endogenous acetyl groups (this is typically a small fraction: <1% of all peptides).
E.g., if you have an intel i7-6700K with four cores and eight threads this number would be 3.
This should take anywhere from 30 min to several hours depending on how many cores your CPU has.
Ideally this number should be >98%. Lower labeling efficiency will typically result in noisier data.
Cysteine peptides can be ignored for quantification by not adding NEM under the protein quantification panel.
If samples were digested with Trypsin and LysC then choose Trypsin instead. Trypsin/P should be used if only Trypsin (and not LysC) was used during the digestion.
Only do this if cysteine alkylation worked.
More than one X. laevis protein is often assigned to a given row because a peptide mapped to both sequences and it cannot be determined which sequence the peptide came from (e.g., protein isoforms).
Acknowledgments
We thank Lillia Ryazanova for help with the sample preparation, and Felix Keber for comments and suggestions on the manuscript. MS was supported by a NIH F31 pre-doctoral fellowship 5F31GM116451. This work was supported by NIH grant 1R35GM128813 and by Princeton University startup funding.
References
- 1.Gurdon JB, & Wakefield L (1986). Microinjection of amphibian oocytes and eggs for the analysis of transcription. Microinjection and Organelle Transplantation Techniques, 269–299. [Google Scholar]
- 2.Eng JK, McCormack AL, Yates JR (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5(11):976–989. 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- 3.Wühr M, Freeman RM Jr, Presler M, Horb ME, Peshkin L, Gygi S, Kirschner MW (2014) Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr Biol 24(13):1467–1475. 10.1016/j.cub.2014.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M (2015) A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteomics 14(9):2394–2404. 10.1074/mcp.M114.046995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13(9):2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1(5):376–386 [DOI] [PubMed] [Google Scholar]
- 7.Selevsek N, Chang CY, Gillet LC, Navarro P, Bernhardt OM, Reiter L, Cheng Ly, Vitek O, Aebersold R (2015) Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Mol Cell Proteomics 14(3):739–749. 10.1074/mcp.M113.035550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75(8):1895–1904 [DOI] [PubMed] [Google Scholar]
- 9.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3(12):1154–1169. 10.1074/mcp.M400129-MCP200 [DOI] [PubMed] [Google Scholar]
- 10.Ting L, Rad R, Gygi SP, Haas W (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8(11):937–940. 10.1038/nmeth.1714 pii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wühr M, Haas W, McAlister GC, Peshkin L, Rad R, Kirschner MW, Gygi SP (2012) Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Anal Chem 84(21):9214–9221. 10.1021/ac301962s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hebert AS, Merrill AE, Bailey DJ, Still AJ, Westphall MS, Strieter ER, Pagliarini DJ, Coon JJ (2013) Neutron-encoded mass signatures for multiplexed proteome quantification. Nat Methods 10(4):332–334. 10.1038/nmeth.2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP (2014) MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86(14):7150–7158. 10.1021/ac502040v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wühr M, Guttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ, Kirschner MW, Gygi SP (2015) The nuclear proteome of a vertebrate. Curr Biol 25(20):2663–2671. 10.1016/j.cub.2015.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peshkin L, Wühr M, Pearl E, Haas W, Freeman RM Jr, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW (2015) On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev Cell 35(3):383–394. 10.1016/j.devcel.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Presler MS, Van Itallie E, Klein AM, Kunz R, Coughlin P, Peshkin L, Gygi S, Wühr M, Kirschner M (2017) Proteomics of phosphorylation and protein dynamics during fertilization and meiotic exit in the Xenopus egg. bioRxiv 2017:145086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyanova S, Temu T, Cox J (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11(12):2301–2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 18.Elias JE, Gygi SP (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4(3):207–214. 10.1038/nmeth1019 nmeth1019 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143(7):1174–1189. 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vizcaíno JL, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q-W, Wang R, Hermjakob H, (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Research 44 (D1):D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubrecht-Laboratorium (Embryologisch Instituut), Nieuwkoop PD, Faber J (1967). Normal Tables of Xenopus Laevis:(Daudin) a Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of the Metamorphosis. North-Holland. [Google Scholar]