Abstract
Although the etiology of ALS remains poorly understood, impaired proteostasis is a common feature of different forms of ALS. Mutations in Ubiquilins, UBQLN2 and UBQLN4, cause familial ALS. The role of UBQLNs in proteasomal degradation is well established but their role in autophagy-lysosomal clearance is poorly defined. Here, we describe a crosstalk between ER stress, mTOR signaling, and autophagic flux in Drosophila and mammalian cells lacking Ubiquilins. We found that loss of Ubiquilins leads to ER stress, impairs mTORC1 activity, promotes autophagy, and causes the demise of neurons. We show that ubiquilin mutants display defective autophagic flux due to reduced lysosome acidification. Ubiquilins are required to maintain proper levels of V0a/V100 subunit of the v-ATPase and lysosomal pH. Feeding flies acidic nanoparticles alleviates defective autophagic flux in ubiquilin mutants. Hence, our studies reveal a conserved role for Ubiquilins as regulators of autophagy by controlling v-ATPase activity and mTOR signaling.
Keywords: Drosophila, proteostasis, ER stress, autophagy, lysosome acidification, ALS, v-ATPase
INTRODUCTION
Ubiquilin (UBQLN) proteins are highly conserved ubiquitin-binding proteins. Although flies have a single ubiquilin gene (ubqn), there are five human homologues: UBQLN1–4 and UBQLNL. Mutations in UBQLN2 and UBQLN4 cause amyotropic lateral sclerosis (ALS) with or without frontotemporal dementia (FTD)1,2.
Ubiquilins are characterized by a C-terminal ubiquitin-associated (UBA) domain and an N-terminal ubiquitin-like (UBL) domain that mediates interaction with the proteasome3,4. The middle region between the UBL and UBA domains contains a variable number of poorly characterized chaperone-binding motifs homologous to STI1. Ubiquilins are suggested to play a role in diverse biological processes such as cell signaling, cell cycle progression, endoplasmic reticulum (ER)-associated degradation, autophagosome maturation, and starvation induced autophagy5–8. Research in human cell lines suggests a role for UBQLNs in chaperoning mitochondrial membrane proteins and protein aggregate clearance via HSP70 and the proteasome9,10. Despite our understanding of Ubiquilin’s role in UPS-mediated degradation, its role in autophagy is ill-defined and controversial6,7.
Here we identified Drosophila Ubiquilin as a regulator of ER quality control, mTOR signaling, autophagy, and neuronal maintenance. We report a dual function of Ubiquilin that integrates the UPS and lysosomal degradation. We found that loss of Ubiquilin impairs mTORC1 activity and leads to increased autophagy induction in both flies and mammalian cells. Despite the promotion of autophagic vesicle formation, loss of Ubiquilin causes impaired autophagic flux. Ubiquilin interacts with subunits of the lysosomal proton pump, the vacuolar H+- ATPase (v-ATPase), and regulates v-ATPase function. Loss of Ubiquilins causes lysosome alkalization and affects lysosomal degradation due to impaired v-ATPase activity. Re-acidification of lysosomes with acidic nanoparticles ameliorates the impaired autophagic flux in ubiquilin mutants. Our data reveal a previously undocumented function for Ubiquilins in autophagy regulation by promoting v-ATPase activity and lysosomal acidification, which in turn may play a role in the demise of neurons.
RESULTS
Ubiquilin is broadly expressed and required in the developing nervous system
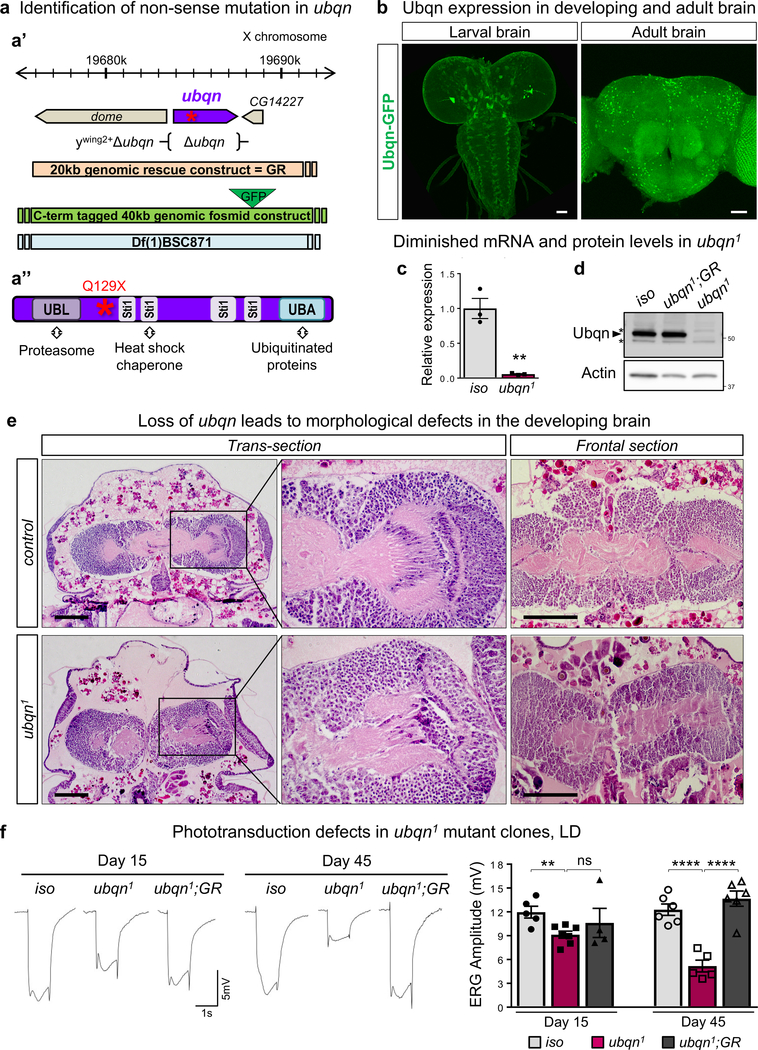
To isolate genes required for neuronal maintenance in Drosophila, we performed an unbiased forward genetic screen11. We identified a single nonsense mutation (Q129X) in ubiquilin, ubqn1, fly homologue of human UBQLNs12 (Fig. 1a and Supplementary Table 1). Ubqn is ubiquitously expressed in developing larvae but is most abundantly expressed in the larval and adult nervous systems as gauged by a C-terminal GFP-tagged ubqn genomic construct13 (Fig. 1b and Supplementary Fig. 1a).
Fig. 1. Ubiquilin Is Broadly Expressed and Required in the Developing and Adult Nervous System.
(a) (a’) Schematic representation of the molecular lesion in ubqn gene, ubqn deletion (ywing2+Δubqn), genomic rescue (GR) constructs and deficiency spanning the genomic region indicated (a”) Schematic representation of the nonsense mutation in Ubqn protein which contains an N-terminal UBiquitin-Like (UBL) domain, four Sti1 motifs, and a C-terminal UBiquitin-Associated (UBA) domain, capable of binding to proteasome, heat shock chaperones, and ubiquitinated proteins, respectively.
(b) Immunofluorescence staining with GFP antibody of larval brain, ventral nerve cord (VNC), and adult brain of flies expressing GFP-tagged genomic Ubqn transgene (Ubqn-GFP construct). Scale bars, 40 μm.
(c) qRT-PCR quantification showing ubqn transcript expression in ubqn1 wandering third instar larvae compared to iso (y w FRT19A) larvae normalized against housekeeping gene (GAPDH). n= 3 independent biological samples, and 3 PCR replicates for each biological sample. Mean ± s.e.m. **p= 0.0028.
(d) Western blot for Ubqn with protein lysates from third instar larvae of iso, ubqn1; GR, and ubqn1. Asterisks indicate non-specific bands.
(e) H&E staining in trans- and frontal- head sections of control (iso) and ubqn1 pre-pupae (P4 stage= 20h APF grown at room temperature). Neuropil is severely reduced in mutants (shown as light pink with H&E staining). Scale bars, 100 μm.
(f) ERG traces from 15 and 45 day-old ey-FLP clones of iso (control), ubqn1, and ubqn1; GR raised in 12h light/12h dark cycle (LD) with quantification of ERG amplitudes. n= 5 (iso 15d and ubqn1 45d), n= 6 (iso 45d and ubqn1; GR 45d) n= 7 (ubqn1 15d), n= 4 (ubqn1; GR 15d) flies. Mean ± s.e.m. ns, not significant; **p= 0.0057, ****p< 0.0001.
For all panels except 1e, three independent experiments were performed with similar results. For panel 1e, two independent experiments were performed with similar results. All statistics were determined by two-sided Student’s t-test. Statistics source data for Fig. 1c,f can be found in Table S9.
Both ubqn mRNA levels and Ubqn protein levels are significantly reduced in ubqn1 mutants (Fig. 1c,d). Ubqn loss causes developmental arrest as early pre-pupae (Supplementary Table 2). The lethality is rescued with a 20kb genomic rescue construct (GR)14 and with ubiquitous overexpression of ubqn cDNA (Supplementary Table 2 and Supplementary Fig. 1b). A null allele of ubqn (ywing2+Δubqn) generated by CRISPR/Cas9-mediated gene deletion15 does not complement ubqn1 lethality and shows pre-pupal lethality (Fig. 1a’, Supplementary Fig. 1c, and Supplementary Table 2). These data demonstrate that the ubqn1 is a severe loss-of-function or null allele of ubqn.
To determine if Ubqn plays a role during early development, we removed it in the female germline using the ovoD1 method16. Females with homozygous ubqn1 germline clones rarely laid eggs and these eggs do not develop (Supplementary Fig. 1d), showing that Ubqn is essential in oogenesis. Hence, development of ubqn1 mutants to the pre-pupal stage likely relies on maternally contributed Ubqn protein. We also noticed developmental defects in the pre-pupae prior to death. Analysis of H&E stained pre-pupae sections shows severely reduced neuropil in ubqn1 mutant nervous system (Fig. 1e). Mutants also exhibit incomplete limb eversion and loss of head fat body (Supplementary Fig. 1e,f). These data show that Ubqn is required for proper development of the nervous system as well as other tissues.
Loss of Ubqn leads to an age-dependent neuronal and glial degeneration
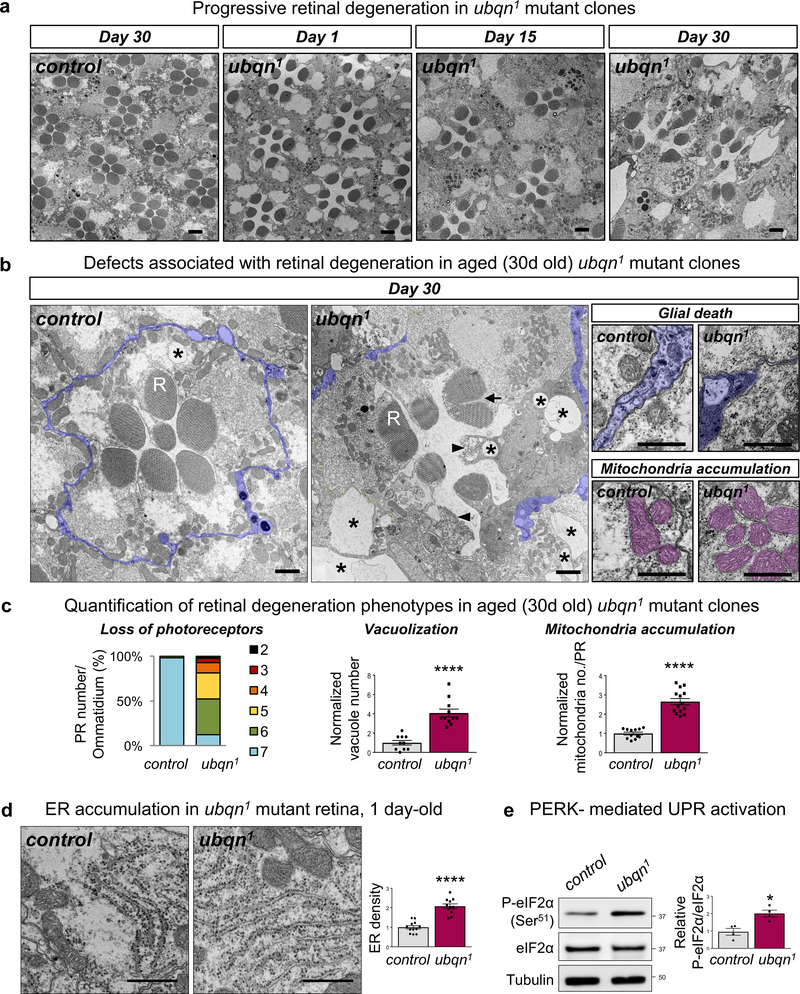
To assess the requirement of Ubqn in aging fly nervous system, we generated mosaic eye clones using the FLP-FRT system and recorded electroretinograms (ERGs) in mutant photoreceptors (PRs). ERG amplitudes are slightly reduced in 15 day-old flies raised in 12h Light/12h Dark (LD) cycle. However, flies aged 45 days exhibited a severe loss of ERG amplitude (Fig. 1f). To assess whether the phototransduction defect exhibited by ubqn1 clones is accompanied with structural defects, we used transmission electron microscopy (TEM) of 1, 15, and 30 day-old mutant eye clones raised in LD cycle. At day 1, mutant retinae display subtle defects, however, PR numbers are comparable to controls (Fig. 2a and Supplementary Fig. 2a). At day 15, the mutant retinae display vacuolization, electron dense aggregates, and PR loss (Fig. 2a and Supplementary Fig. 2a,b). Finally, at day 30, the mutant retinae exhibit numerous degenerative features, including PR loss, vacuolization, rhabdomere loss, split rhabdomeres, glial death, electron dense aggregate accumulation, and mitochondria accumulation (Fig. 2a–c and Supplementary Fig. 2c). Consistent with findings in the retina, 30 day-old laminae revealed that mutant synaptic terminals are severely affected as they are enlarged five-fold, which is accompanied by PR terminal loss (Supplementary Fig. 2d). Mutant flies aged under constant darkness (DD) also displayed decreased ERG amplitude and neurodegeneration (Supplementary Fig. 2e,f), indicating that neurodegeneration caused by Ubqn loss is activity-independent. These data show that Ubqn loss causes a slow and progressive demise of neurons, synaptic terminals, and glia.
Fig. 2. Ubqn Is Required for Neuronal Function and Maintenance.
(a) TEM images of the retinae of ey-FLP clones of ubqn1; GR (control) and ubqn1 raised in 12h light/12h dark cycle, at 1 day-old, 15 days-old, and 30 days-old. Scale bars, 2 μm.
(b) Higher magnification TEM images of the retinae of ey-FLP clones of ubqn1; GR (control) and ubqn1 raised in 12h light/12h dark cycle, at 30 days old. R stands for rhabdomere. ubqn1 retina displays degenerating photoreceptors, vacuolization (asterisks), rhabdomere loss (arrowheads), splitting rhabdomeres (arrow), glial death, and mitochondria accumulation. Glial cells are highlighted in purple and mitochondria are highlighted in magenta. Scale bars, left panels 1 μm, right panels 0.5 μm.
(c) Quantification of photoreceptor number per ommatidium (n= 53 (control), n= 42 (ubqn1) ommatidia from 3 biologically independent animals), vacuole number (n= 10 (control), n= 11 (ubqn1) images (7 ommatidia/image) from 3 biologically independent animals), and mitochondria number (n= 10 (control), n= 11 (ubqn1) ommatidia from 3 biologically independent animals) of TEM images from (b). Mean ± s.e.m. ****p< 0.0001.
(d) TEM images of the ER in the retinae of 1 day-old ey-FLP clones of ubqn1; GR (control) and ubqn1 with quantification of normalized ER length (μm)/area (μm2). Scale bars, 0.5 μm. n= 12 (control), n= 11 (ubqn1) images from 3 biologically independent animals. Mean ± s.e.m for vacuole and mitochondria number, and mean for quantification of % PR number/ommatidium. ****p< 0.0001.
(e) Western blot for Phospho-eIF2α (S51) and total eIF2α with protein lysates from third instar larvae of ubqn1; GR (control) and ubqn1 with quantification of Phospho-eIF2α (S51) to total eIF2α ratio in ubqn1; GR (control) and ubqn1. n= 4 biologically independent samples. Mean ± s.e.m. ns, not significant; ***p= 0.0009.
For all panels, three independent experiments were performed with similar results. All statistics were determined by two-sided Student’s t-test. Statistics source data for Fig. 2c-e can be found in Table S9.
Loss of Ubqn leads to ER expansion and proteostasis defects
Ubiquilins are implicated in regulation of ER-associated degradation (ERAD) in mammalian cells5. To test the involvement of Ubqn in ER proteostasis in vivo, we examined ER abundance and morphology in ubqn1 clones in larval salivary gland. ubqn1 clones display significantly expanded ER as indicated with a strong increase in the levels of the ubiquitously expressed ER marker, sqh-EYFP-ER17 (Supplementary Fig. 3a). Similarly, TEM reveals approximately two-fold increase in the rough ER density in 1 day-old ubqn1 retina (Fig. 2d).
An ER expansion phenotype was previously shown in flies lacking the fly homolog of vesicle-associated membrane protein-associated protein B (VAPB/ALS8). These flies also accumulated membrane proteins such as Chaoptin in the ER of a few adult neurons18. Similarly, neuron-specific knockdown of Ubqn, caused cytoplasmic accumulation of Chaoptin (Supplementary Fig. 3b), suggesting defective trafficking and clearance of Chaoptin and likely other membrane-bound proteins upon Ubqn loss.
ER expansion, coupled with defective ER protein trafficking in ubqn1 mutants could also promote ER stress and unfolded protein response (UPR). ubqn1 mutants show a significant increase in levels of an ER chaperone, BiP/Hsc3 (Supplementary Fig. 3c). We examined the activation of two UPR pathways: IRE1α (Inositol-requiring protein 1α) and PERK (Protein kinase RNA-like ER kinase)19. Ubqn does not affect IRE1α pathway (Supplementary Fig. 3d,e). However, ubqn1 mutants display an increase in phosphorylation of eIF2α, a component of PERK pathway that is phosphorylated during ER stress (Fig. 2e and Supplementary Fig. 3f). These data indicate that Ubqn is required for proper ER morphology and function and that its loss causes ER stress and PERK-mediated UPR activation.
Loss of Ubqn affects mTOR signaling and promotes autophagy induction
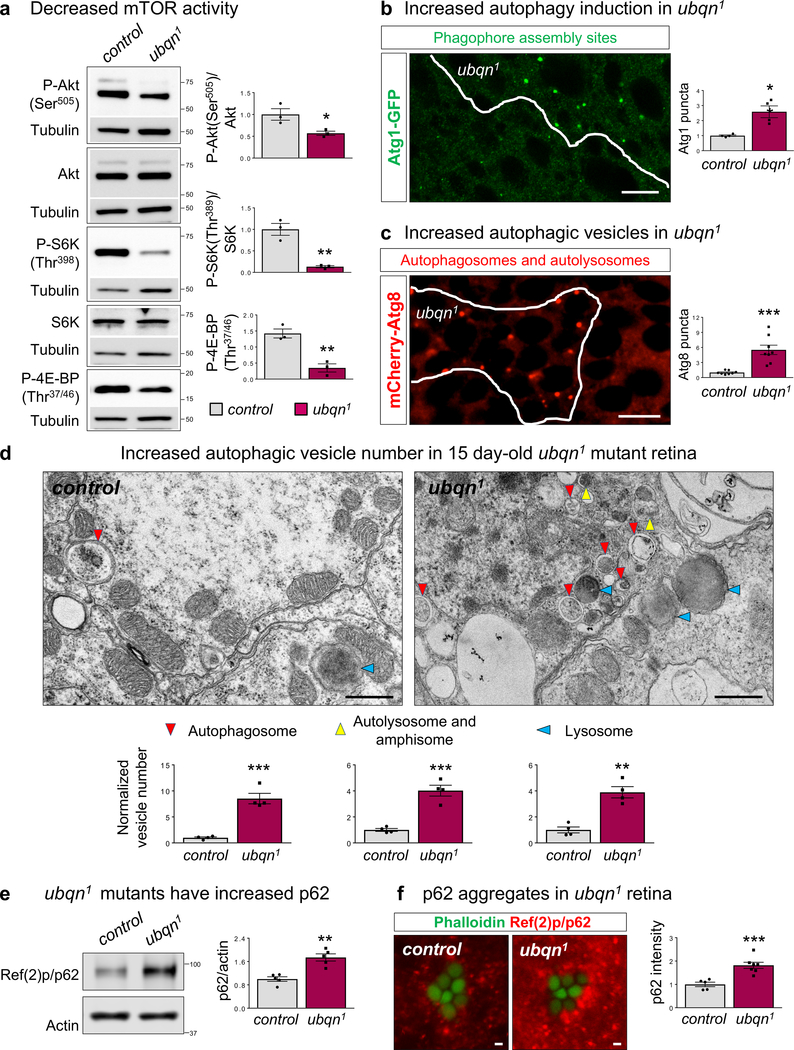
ER stress/UPR may mitigate proteostatic stress by enhancing autophagy through mechanistic target of rapamycin (mTOR) signaling20–24, a major autophagy regulator. To determine if Ubqn loss alters mTOR activity, we examined the phosphorylation status of direct targets of mTOR complexes, mTORC2 and mTORC1. Ubqn loss leads to diminished phosphorylation of the mTORC2 target, Akt, as well as mTORC1 targets, S6K and 4E-BP (Fig. 3a). Hence, mTOR signaling is decreased upon Ubqn loss.
Fig. 3. Loss of Ubqn Affects mTOR Signaling and Promotes Autophagy Induction.
(a) Western blot for mTOR-dependent phosphorylation of Akt, S6K, and 4E-BP with fat body protein lysates from third instar larvae of ubqn1; GR (control) and ubqn1. Quantification of relative P-Akt/Akt, P-S6K/S6K, and P-4EBP/Tubulin ratios. n= 3 biologically independent samples. Mean ± s.e.m. ns, not significant; *p= 0.0372, **p= 0.0035 (P-S6K/S6K), **p= 0.0047 (P-4EBP/Tubulin).
(b) ubqn1 clone in fat body of early third instar larvae expressing Atg1-GFP protein trap with quantification of normalized Atg1 punctae number/μm2. Scale bar, 10 μm. n= 4 biologically independent samples. Mean ± s.e.m. **p= 0.007.
(c) ubqn1 clone in fat body of early third instar larvae expressing UAS-mCherry-Atg8a with Cg-GAL4 driver and quantification of normalized Atg8 punctae number/μm2. Scale bar, 10 μm. n= 8 biologically independent samples. Mean ± s.e.m. ***p= 0.0003.
(d) TEM images of the retinae of 15 day-old ey-FLP clones of ubqn1; GR (control) and ubqn1 and quantification of autophagic vesicle numbers. Scale bars, 0.5 μm. n= 4 flies. **p= 0.001, ***p= 0.0003 (autophagosome), ***p= 0.0004 (autolysosome and amphisome).
(e) Western blot for p62 with fat body protein lysates from third instar larvae of ubqn1; GR (control) and ubqn1 and quantification of relative p62/actin levels. n= 5 biologically independent samples. Mean ± s.e.m. ***p = 0.0009.
(f) Immunofluorescence staining of p62 and phalloidin (labeling rhabdomeres in photoreceptors) in whole eye clones of 2 day-old control (iso) and ubqn1 and quantification of p62 fluorescence intensity. Scale bars, 1 μm. n= 5 (control), n= 7 (ubqn1) biologically independent animals. Mean ± s.e.m. ***p = 0.0009.
For all panels except 3b, three independent experiments were performed with similar results. For panel 3b, two independent experiments were performed with similar results. All statistics were determined by two-sided Student’s t-test. Statistics source data for Fig. 3 can be found in Table S9.
To determine if reduced mTORC1 activity in ubqn1 mutants affects autophagy induction, we assessed the levels of two autophagic markers, Atg1 and Atg8. Atg1/ULK1 kinase initiates autophagy and is inhibited by mTORC1-mediated phosphorylation25,26. ubqn1 clones of larval fat body show a significant increase in Atg1-GFP punctae (Fig. 3b), indicating increased autophagy induction, consistent with reduced mTORC1 activity. We monitored autophagic vesicles using mCherry-Atg8 or GFP-Atg8 reporters and observed a marked increase in both mCherry and GFP punctae in ubqn1 clones in larval fat body (Fig. 3c and Supplementary Fig. 4a). TEM data of 15 day-old photoreceptors confirms that Ubqn loss causes a robust increase in the number of autophagosomes, amphisomes, autolysosomes, and lysosomes (Fig. 3d and Supplementary Fig. 4b). Therefore, Ubqn loss leads to reduced mTOR signaling and enhanced autophagy induction.
Ubiquilin is required to maintain autophagic flux
To determine if autophagic flux is affected in ubqn1 mutants, we assessed the levels of Ref(2)P/p62 (also known as SQSTM1), an ALS and FTD-associated autophagy cargo receptor27. Impairment of autophagic degradation causes p62 accumulation, followed by formation of aggregates containing p62 and ubiquitinated proteins28. Indeed, loss of Ubqn caused a significant accumulation of p62 protein under both fed and starved conditions although it did not affect p62 transcript levels (Fig. 3e,f and Supplementary Fig. 4c–e). Midgut histolysis, a developmental process that requires autophagy29, was also impaired in ubqn1 mutants (Supplementary Fig. 4f).
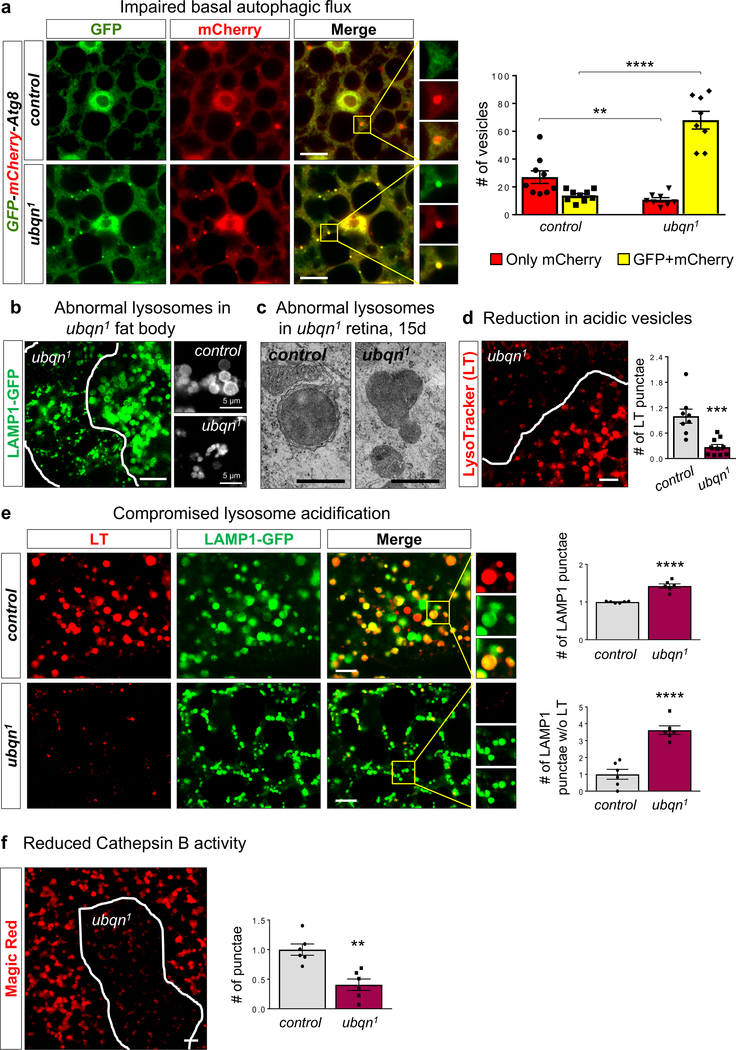
To more directly measure autophagic flux, we used a tandem-tagged mCherry-GFP-Atg8a construct30,31. Due to differential pH sensitivity of GFP and mCherry, non-acidic autophagic compartments (phagophores and autophagosomes) are labeled with both GFP and mCherry, however after fusing with a lysosome, autolysosomes become acidic and GFP fluorescence is quenched. Ubqn loss causes a prominent increase in non-acidic punctae (mCherry+GFP) number (Fig. 4a), indicating an overall increase in autophagosomes and non-acidic autolysosomes. However, the number of acidic punctae (only mCherry) was severely decreased in ubqn1 mutants, under both fed and starved conditions (Fig. 4a and Supplementary Fig. 4g). These data show that autophagic vesicle acidification and autophagic flux are compromised in ubqn1 mutants.
Fig. 4. Ubiquilin Facilitates Lysosomal Acidification and Function.
(a) Confocal images of fed early third instar larval fat body of y w (control) and ubqn1 expressing UAS-GFP-mCherry-Atg8a driven by Cg-GAL4 (drives expression in the fat body, hemocytes, and lymph gland) and quantification of only mCherry expressing and GFP+mCherry expressing punctae/section (135μm2). Scale bars, 10 μm. n= 9 (control), n= 8 (ubqn1) biologically independent samples. Mean ± s.e.m. **p= 0.006, ****p< 0.0001.
(b) ubqn1 clone in fat body of third instar larvae expressing UAS-LAMP1-GFP with Cg-GAL4 driver and higher magnification of LAMP1-GFP punctae to the right. Scale bar, 20 μm.
(c) TEM images of lysosomes in 15 day-old retinae of ey-FLP clones of ubqn1; GR (control) and ubqn1. Scale bar, 0.5 μm.
(d) Live imaging for LysoTracker (LT) dye in third instar larval fat body with ubqn1 clones and quantification of normalized LT punctae number/μm2 in ubqn1 clones compared to the surrounding wild-type cells. Scale bar, 10 μm. n= 8 (control), n= 10 (ubqn1) biologically independent samples. Mean ± s.e.m. ***p = 0.0004.
(e) Live imaging for LysoTracker (LT) in third instar larval fat bodies of iso (control) and ubqn1 expressing UAS-LAMP1-GFP with Cg-GAL4 driver and quantification of the normalized number of lysosomes (LAMP1-GFP punctae)/μm2 and the number of lysosomes without LysoTracker/μm2. Scale bars, 10 μm. n= 6 biologically independent samples. Mean ± s.e.m. ****p< 0.0001.
(f) Live imaging for Magic Red dye that detects active Cathepsin B in third instar larval fat body with ubqn1 clones and quantification of the number of punctae. Scale bar, 10 μm. n= 6 biologically independent samples. Mean ± s.e.m. **p = 0.0014.
For all panels, three independent experiments were performed with similar results. All statistics were determined by two-sided Student’s t-test. Statistics source data for Fig. 4a, d, e, and f can be found in Table S9.
Loss of Ubqn causes aberrant and dysfunctional lysosomes
To assess the role of lysosomes in ubqn1 phenotypes, we examined the expression of a lysosomal membrane marker, LAMP1-GFP. In second instar larvae, cells undergo minimal autophagy and contain a small number of lysosomes whereas in wandering third instar larvae, autophagic vesicles are readily observable due to induction of developmental autophagy32. ubqn1 clones in third instar larval fat body displayed aberrant and clustered LAMP1-GFP positive compartments (Fig. 4b). In second instar larval fat body, ubqn1 clones displayed a prominent increase in number and intensity of LAMP1-GFP positive compartments (Supplementary Fig. 4h). These punctae were dramatically enlarged upon starvation, suggesting non-degraded material accumulation in lysosomes. Lysosomal expansion with aberrant morphology was supported by TEM in photoreceptors of ubqn1 eye clones (Fig. 4c).
Lysosomes must maintain an acidic pH for the degradative activity of hydrolases such as Cathepsins. We assessed acidification of the vesicles in the endo-lysosomal pathway using LysoTracker, a dye that labels acidic vesicles33. Mutant clones in wandering third instar larval fat body displayed a strong reduction in LysoTracker staining (Fig. 4d). However, mutant clones in second instar larval fat body displayed LysoTracker staining similar to control cells despite a robust increase the total number of lysosomes as measured by LAMP1-GFP (Supplementary Fig. 4h,i). To confirm reduced lysosomal acidification, we examined the colocalization of LAMP1-GFP with LysoTracker. The vast majority of LAMP1 positive punctae in ubqn1 mutants did not contain LysoTracker (Fig. 4e), therefore most lysosomes are not acidified. Furthermore, ubqn1 clones displayed a severe reduction in activity of the lysosomal hydrolase Cathepsin B (Fig. 4f).
To determine if the lysosomal phenotypes are due to alterations in TFEB/MiTF pathway, we examined the expression levels of several TFEB/MiTF target genes. We did not observe an overall change in TFEB activity although we observed an increase in VhaM8.9 expression (Supplementary Fig. 4j). These findings provide compelling evidence that Ubqn loss, while increasing overall autophagosome and lysosome number, interferes with lysosomal acidification.
Ubiquilin interacts and colocalizes with cytoplasmic v-ATPase subunits
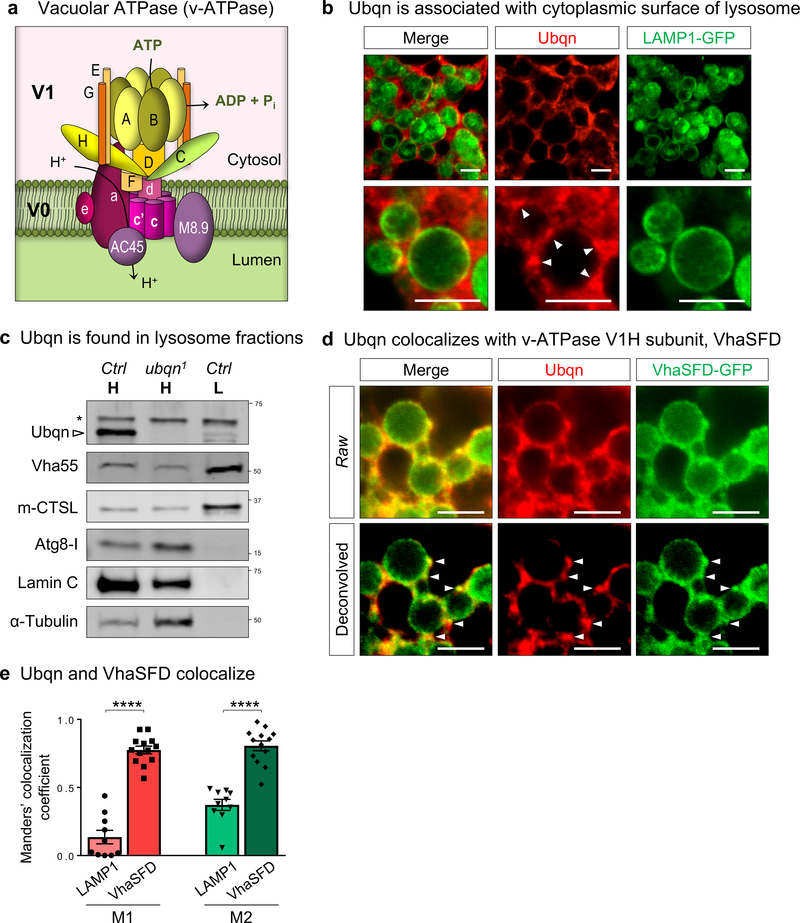
To determine Ubqn’s binding partners, we performed Immunoprecipitation (IP) and Mass Spectrometry (MS) using transgenic flies that express genomically tagged Ubqn-GFP. Intriguingly, eight v-ATPase subunits were identified (Supplementary Tables 3–4). The v-ATPase is an ATP-dependent lysosomal proton pump that generates and maintains the low intralysosomal pH. It consists of a peripheral domain (V1), an integral membrane domain (V0), and two accessory components (Fig. 5a). The IP-MS data identified almost all V1 subunits and one V0 subunit, V100/V0a, whose hydrophilic N-terminus is oriented towards the cytoplasm, suggesting that Ubqn may function at the cytoplasmic area of the v-ATPase. We examined Ubqn’s subcellular localization in larval fat body and found that Ubqn localizes to the cytoplasm and is enriched in punctae at the lysosomal surface (Fig. 5b). Subcellular fractionation showed that Ubqn is present at low levels in lysosome-enriched fractions (Fig. 5c). In addition, Ubqn and VhaSFD, an orthologue of V1H, colocalize on the lysosomal surface (Fig. 5d,e). Our findings indicate that Ubqn physically interacts with the cytoplasmic v-ATPase subunits.
Fig. 5. Ubqn Localizes to the Periphery of Lysosomes and Interacts with v-ATPase.
(a) Schematic representation of v-ATPase with cytosolic V1 domain and integral membrane V0 domain.
(b) Immunofluorescence staining with Ubqn antibody in fat body of third instar larvae expressing UAS-LAMP1-GFP with Cg-GAL4 driver. Arrowheads indicate Ubqn punctae at the periphery of lysosomes. Scale bars, 5 μm.
(c) Western blot for Ubqn in homogenate (H) and lysosome (L) fractions from third instar larvae of iso (control) and ubqn1 homogenate. Western blots for Vha55 (a V1 subunit), m-CTSL (mature Cathepsin L), Atg8-I, Lamin C, and Tubulin are shown to indicate lysosome enrichment upon subcellular fractionation.
(d) Immunofluorescence staining with Ubqn antibody in fat body of third instar larvae expressing VhaSFD-GFP protein trap. Arrowheads indicate Ubqn punctae co-localizing with VhaSFD. Scale bars, 5 μm.
(e) Manders’ colocalization coefficients, M1 and M2 for Ubqn and VhaSFD-GFP fluorescence from (e) compared to M1 and M2 for Ubqn and LAMP1-GFP fluorescence from (c). n= 10 (LAMP1), n= 13 (VhaSFD) biologically independent samples. Mean ± s.e.m. ****p< 0.0001. For all panels except 5c, three independent experiments were performed with similar results. For panel 5c, two independent experiments were performed with similar results. Statistics were determined by two-sided Student’s t-test. Statistics source data for Fig. 5e can be found in Table S9.
Ubiquilin genetically interacts with v-ATPase and mediates vesicle acidification
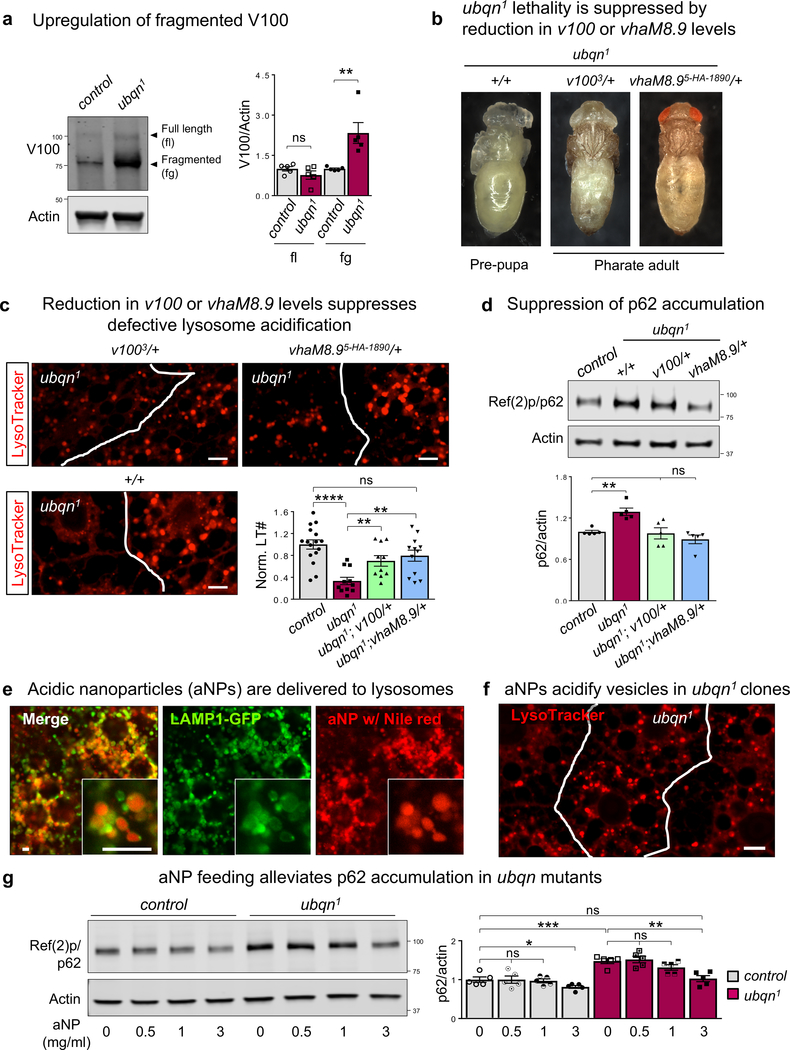
To assess v-ATPase levels in ubqn1 mutants, we tested V100–1 (V100) expression. We found that Ubqn loss causes a marked accumulation of fragmented V100 with no alteration in V100 transcript levels (Fig. 6a and Supplementary Fig. 4j). Loss of Ubqn did not alter levels of other v-ATPase subunits, Vha55 or VhaSFD (Supplementary Fig. 5a). To assess the functional link between the v-ATPase and Ubqn, we performed genetic interaction assays by selecting four subunits with different functions: V100/V0a (proton transport), Vha68/V1A (ATP hydrolysis), VhaSFD/V1H (regulatory), and VhaM8.9 (accessory). We examined a possible enhancement or suppression of ubqn1 pre-pupal lethality in by introducing one copy of a loss-of-function allele of these v-ATPase subunits. ubqn1 mutants with reduced levels of VhaSFD or Vha68 die earlier than ubqn1 as third instar larvae suggesting an enhancement of pre-pupal lethality (Supplementary Table 5). In contrast, ubqn1 mutants with reduced levels of VhaM8.9 or V100 develop beyond the pre-pupal stage and die as pharate adults (Fig. 6b). Based on these data, we hypothesized that a reduction in V100 or VhaM8.9 subunits of v-ATPase is sufficient to re-establish lysosomal acidification, leading to delayed developmental arrest. We examined LysoTracker staining in ubqn1 larval fat body clones that carry one copy of a v100 or vhaM8.9 loss-of-function allele. Indeed, reduced levels of V100 or VhaM8.9 caused a marked increase in LysoTracker punctae in ubqn1 mutant cells (Fig. 6c), showing a significant improvement in vesicle acidification. Reduced levels of V100 or VhaM8.9 also suppressed p62 accumulation (Fig. 6d). Hence, a reduction of two functionally related subunits of the V0 domain, VhaM8.9 and V10034, suppress the lethality and impaired autophagic flux associated with an increase in aberrant V100 in ubqn1 mutants (Supplementary Fig. 5b).
Fig. 6. Ubiquilin Genetically Interacts with v-ATPase and Mediates Vesicle Acidification.
(a) Western blot for V100 with protein lysates from third instar larvae of ubqn1; GR (control) and ubqn1 and quantification of full length (fl) and fragmented (fg) V100 protein levels. n= 5 biologically independent samples. Mean ± s.e.m. ns, not significant; **p = 0.0091.
(b) Images of ubqn1 pre-pupa, ubqn1; v1003/+ and ubqn1; vhaM8.9EY03616/+ pharate adults.
(c) Live imaging for LysoTracker dye in third instar larval fat body with ubqn1 clones of v1003/+ and vhaM8.95-HA−1890/+ and quantitative analysis of normalized LysoTracker positive punctae number/μm2. n= 16 (WT cell), n= 11 (ubqn1 cell), n= 10 (ubqn1; v1003/+ cell), and n= 12 (ubqn1; vhaM8.95HA/+ cell) independent samples. Mean ± s.e.m. ns, not significant; **p= 0.0053 (ubqn1 vs ubqn1; v1003/+), **p= 0.0013 (ubqn1 vs ubqn1; vhaM8.95HA/+), ****p< 0.0001. Scale bars, 10 μm.
(d) Western blot for p62 with fat body protein lysates from third instar larvae of ubqn1; GR (control), ubqn1, ubqn1; v1003/+, and ubqn1; vhaM8.9EY03616/+ and quantification of normalized p62/actin levels. n= 5 biologically independent samples. Mean ± s.e.m. ns, not significant; **p= 0.001.
(e) Live imaging of Nile red in fat body of third instar larvae expressing UAS-LAMP1-GFP driven by Cg-GAL4 driver after feeding the larvae with 1 mg/ml acidic nanoparticles (aNPs) loaded with Nile red for 3 hours. Scale bars, 10 μm.
(f) Live imaging for LysoTracker in third instar larval fat body with ubqn1 clones after feeding the larvae with 3 mg/ml aNPs for 3 hours. Scale bar, 10 μm.
(g) Western blot for p62 with fat body protein lysates from third instar larvae of ubqn1; GR (control) and ubqn1 and quantification of normalized p62/actin levels. Larvae are fed with 0, 0.5, 1, or 3 mg/ml aNPs (3 hours) and transferred to standard fly food (3 hours) before tissue dissection. n= 5 biologically independent samples. Mean ± s.e.m. ns, not significant; *p= 0.0461, **p= 0.002, ***p= 0.0009.
For all panels except 6e-g, three independent experiments were performed with similar results. For panels 6e-g, two independent experiments were performed with similar results. All statistics were determined by two-sided Student’s t-test. Statistics source data for Fig. 6a,c,d, and g can be found in Table S9.
To test whether fragmented V100 is inserted in the v-ATPase complex, we performed subcellular fractionation and found that it accumulates in lysosome fractions of ubqn1 mutants (Supplementary Fig. 5c). Next, we tested whether V100 overexpression phenocopies lysosomal degradation defects in ubqn1. Similar to Ubqn loss, V100 overexpression leads to fragmented V100 accumulation (Supplementary Fig. 5d). Moreover, it causes p62 accumulation and lysosome alkalization, but does not alter eIF2α phosphorylation (Supplementary Fig. 5d,e). Indeed, reduced V100 levels in ubqn1 mutants do not significantly restore ER expansion and ER stress (Supplementary Fig. 5f,g), indicating that Ubqn regulates ER function and lysosome activity independently. Importantly, mTORC1 activity was restored when V100 levels are reduced (Supplementary Fig. 5h), indicating that v-ATPase activity regulates mTORC1 in ubqn1 mutants. These findings indicate that Ubqn regulates v-ATPase activity and lysosome function through regulating V0 subunit, V100 levels.
Re-acidification of lysosomes suppresses defects in autophagic flux in ubqn1 mutants
The above data suggest that restoration of the lysosomal pH alone may ameliorate the diminished autophagic flux observed in ubqn1 mutants. Poly(DL-lactic-co-glycolic acid) (PLGA) nanoparticles can target and re-acidify late endosomes/lysosomes35–39. We used acidic nanoparticles (aNPs) to explore the effect of re-acidifying lysosomes. We fed ubqn1 mutants with aNPs composed of PLGA (Supplementary Fig. 6a,b). Nile red loaded aNPs were successfully delivered to lysosomes in fat body upon feeding of larvae with aNPs (Fig. 6e). We then examined whether aNP feeding can ameliorate compromised vesicle acidification in ubqn1 clones in fat body. LysoTracker staining showed that ubqn1 mutant cells display acidic vesicles comparable to surrounding wild-type cells upon aNP feeding (Fig. 6f). To assess the impact of lysosomal re-acidification by aNPs, we examined the autophagic flux in ubqn1 mutants. We tested p62 levels in third instar larvae fed with varying concentrations of aNPs and observed that adding 3mg/ml aNPs to the food significantly suppresses p62 accumulation in ubqn1 larvae (Fig. 6g), showing improved autophagic flux. We did not observe a suppression of UPR activation or reduced mTORC1 activity through aNP feeding (Supplementary Fig. 6c).
Ubiquilin’s role in lysosome acidification is evolutionarily conserved
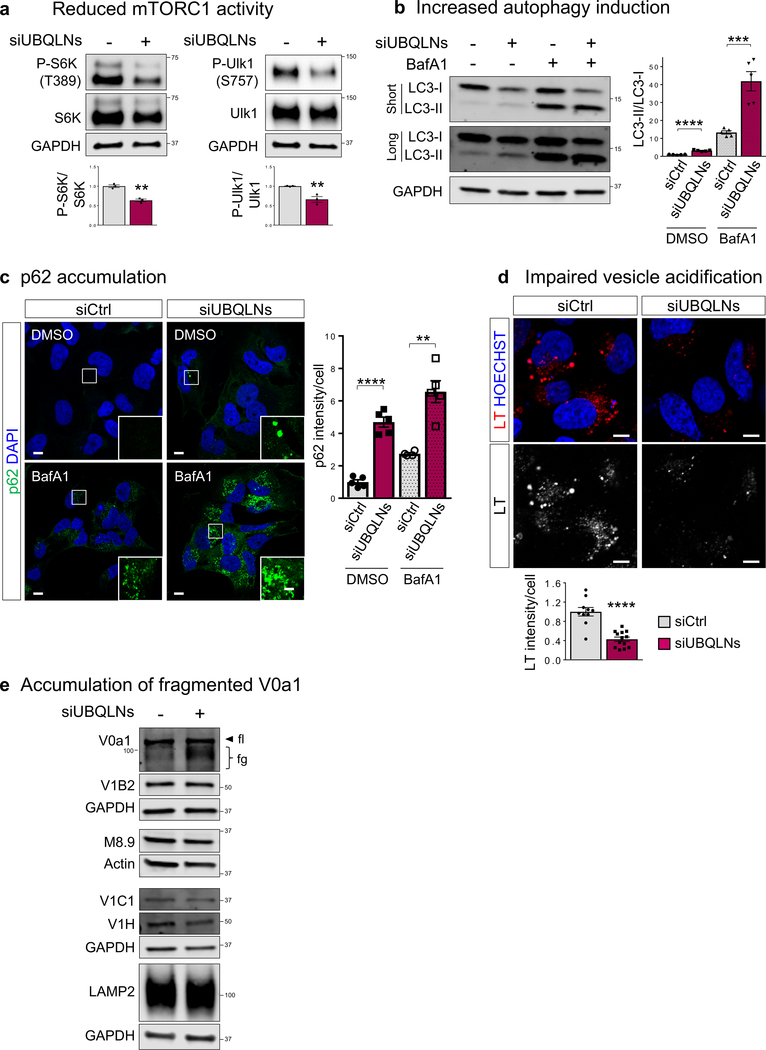
To assess the conservation of Ubqn function in mammalian cells, we performed a triple knockdown of UBQLN1, 2, and 4 (UBQLNs) in Daoy medullablastoma cells (Supplementary Fig. 7a). Consistent with the Drosophila data, depletion of UBQLNs caused reduced mTORC1 activity, revealed by a decrease in phosphorylation status of mTORC1 targets S6K and Ulk1, (Fig. 7a) similar to what we observe in ubqn1 mutant flies. UBQLNs depletion also caused increased eIF2α phosphorylation (Supplementary Fig. 7b). Next, we examined the effect of UBQLNs depletion in autophagy. We observed an increase in LC3 lipidation (LC3-II) and p62 punctae, suggesting increased induction of autophagy and/or impairment in degradation (Fig. 7b,c and Supplementary Fig. 7c). Treatment with the lysosomal inhibitor, bafilomycin A1, increases both p62 and LC3-II levels in UBQLNs depleted cells more strongly than the control (Fig. 7b,c). To test lysosomal degradation, we examined lysosomal acidification and found that loss of UBQLNs leads to reduced LysoTracker staining similar to ubqn1 mutants (Fig. 7d), suggesting impaired autophagic degradation. Consistent with the fly data, UBQLNs depletion in human cells also caused an accumulation of fragmented V0a1, the V100 homologue, but did not alter the levels of other v-ATPase subunits (Fig. 7e).
Fig. 7. Ubiquilin’s Role in Lysosomal Acidification Is Conserved in Human Neuronal Cells.
(a) Western blot for Phospho-S6K (T389), S6K, Phospho-Ulk1(S757), and Ulk1 with Daoy whole cell lysates of siCtrl and UBQLN1+2+4 knockdown (siUBQLNs) cells and quantification of phosphorylated/total protein ratios. n= 3 biologically independent samples. Mean ± s.e.m. **p= 0.0017 (P-S6K/S6K), **p= 0.0099 (P-Ulk1/Ulk1).
(b) Western blot for LC3-I and LC3-II with Daoy whole cell lysates of siCtrl and siUBQLNs cells with or without 4 hour Bafilomycin A1 treatment and quantification of LC3-II/LC3-I ratio. n= 5 biologically independent samples. Mean ± s.e.m. ***p= 0.0008, ****p< 0.0001.
(c) Immunofluorescence staining with p62 and DAPI in siCtrl and siUBQLNs Daoy cells with or without 4 hour Bafilomycin A1 treatment and quantification of normalized mean p62 fluorescence intensity per cell. n= 5 images (~40 cells). Mean ± s.e.m. **p= 0.0015, ****p< 0.0001. Scale bars, 10 μm.
(d) Live imaging for LysoTracker (LT) in siCtrl and siUBQLNs Daoy cells and quantification of normalized mean LT fluorescence intensity per cell. n= 10 (NC) and n= 13 (siUBQLNs) images (~100 cells). Mean ± s.e.m. ****p< 0.0001. Scale bars, 10 μm.
(e) Western blot for LAMP1 and v-ATPase subunits: V0a1, V1B2, M8.9, V1C1, and V1H with Daoy whole cell lysates of siCtrl and siUBQLNs cells. fl: full length, fg: fragmented.
For all panels, three independent experiments were performed with similar results. All statistics were determined by two-sided Student’s t-test. Statistics source data for Fig. 7a-d can be found in Table S9.
In addition, UBQLN1, 2, and 4 triple knockout HEK293 cells generated by CRISPR/Cas99 also showed increased autophagy induction, impaired autophagic flux, aberrant lysosomes, and fragmented V0a1 accumulation (Supplementary Fig. 7d–f). To determine whether the physical interaction between fly Ubqn and V100 identified through IP-MS is conserved, we performed co-immunoprecipitation (co-IP) experiments in HEK293T cells. Under basal conditions, we were unable to co-IP V0a1 using FLAG-tagged UBQLN1 or UBQLN2 unless a crosslinking reagent dithiobis (succinimidyl propionate) (DSP), was included, indicating a weak and/or transient physical association of Ubiquilins with V0a1 (Supplementary Fig. 7g).
In contrast to the role of the UBQLN1 and UBQLN2, a role of UBQLN4 in proteostasis has not been well-established. We examined the effect of UBQLN4 knockdown on UPR activation and autophagic degradation in Daoy cells. UBQLN4 depletion did not significantly alter eIF2α phosphorylation (Supplementary Fig. 8a–b). However, UBQLN4 depletion caused p62 accumulation and reduced LysoTracker staining similar to triple knockdown of UBQLN1+2+4 (Supplementary Fig. 8c,d), suggesting an important role for UBQLN4 in regulation of lysosomal degradation. These data show that Ubiquilin’s role in regulation of autophagic flux and v-ATPase mediated lysosome acidification is conserved between Drosophila and human.
Ubiquilin and ALS
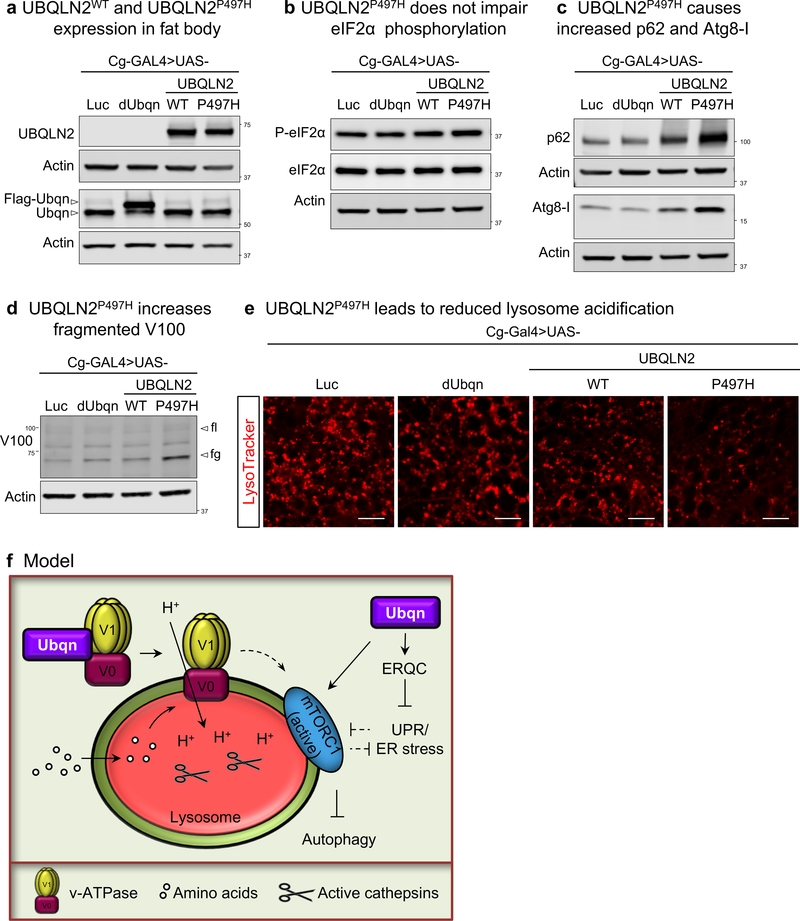
Previously, ALS-linked mutations in UBQLN2 were shown to be defective in chaperone binding and protein aggregate clearance, independent of autophagy10. The ALS-linked mutations in UBQLN2 are dominant and cluster in the PXXP domain2 that is not conserved in fly Ubqn even though the proteins are highly conserved. Hence, we cannot introduce ALS-linked mutations and replace the conserved residues in fly Ubqn. We therefore resorted to the ubiquitous overexpression of UBQLN2WT in wild-type flies and showed that it is toxic as it causes lethality. We overexpressed both UBQLN2WT and the ALS-variant UBQLN2P497H in fat body to assess their effects on ER stress and lysosomal degradation. UBQLN2P497H does not alter eIF2α phosphorylation but causes defects consistent with loss of Ubiquilins observed in flies and human cells: accumulation of p62, fragmented V100, and reduced LysoTracker staining (Fig. 8a–e). These findings suggest that UBQLN2P497H may act as a dominant negative mutation and that it affects the interaction of Ubiquilin with v-ATPase to impair lysosome activity.
Fig. 8. Expression of ALS-variant containing UBQLN2 (UBQLN2P497H) impairs lysosomal degradation.
(a-d) Western blots for UBQLN2, dUbqn, Phospho-eIF2α (S51), total eIF2α, p62, Atg8, and V100 with fat body protein lysates from third instar larvae of w; Cg-GAL4/UAS-Luciferase, w; Cg-GAL4/UAS-FLAG-dUbqn, w; Cg-GAL4/UAS-UBQLN2WT-HA, and w; Cg-GAL4/UAS-UBQLN2P497H-HA. Two independent experiments were performed with similar results.
(e) Live imaging for LysoTracker (LT) in third instar fat body of w; Cg-GAL4/UAS-Luciferase, w; Cg-GAL4/UAS-FLAG-dUbqn, w; Cg-GAL4/UAS-UBQLN2WT-HA, and w; Cg-GAL4/UAS-UBQLN2P497H-HA. Two independent experiments were performed by n=10 biologically independent samples with similar results. Scale bars, 20 μm.
(f) Ubiquilins regulate autophagic flux through mTOR signaling and lysosomal acidification
DISCUSSION
Here, we show that Ubqn interacts with the v-ATPase and is required for lysosomal acidification. Lysosomal re-acidification by acidic nanoparticles ameliorates the impaired autophagic flux in ubqn1 mutants. Ubqn’s role in regulation of autophagy and lysosomal activity is conserved in human cells. Our data provide compelling evidence that Ubqn affects two key players in proteostasis: ER quality control and autophagy by regulating lysosomal acidification (Fig. 8f).
Role of Ubiquilin in ERAD and Lysosomal Degradation
Ubiquilins are largely studied in the context of protein quality control through proteasomal degradation. Previous findings have documented delayed degradation of membrane bound and luminal ERAD substrates upon UBQLN1 or UBQLN2 depletion in vitro5,40. We find that Ubqn loss in vivo causes membrane protein accumulation and a marked increase in ER volume coupled with ER stress and induction of the PERK branch of UPR pathway. Although it is plausible that chronic ER stress is detrimental, some data argue that induction of low-level ER stress has protective roles41–44. We propose that activation of the PERK pathway may play a role in increased autophagy induction and autophagosome formation in ubqn1 mutants as previous reports showed that ER stress can trigger autophagy to induce compensatory degradation21,45,46. Our findings show that Ubqn loss leads to lysosome alkalization and diminished autophagic flux. Hence, we argue that lysosomal dysfunction in ubqn1 mutants causes a blockage in autophagic flux despite increased autophagy induction.
Ubiquilin is a Regulator of mTOR Activity, Autophagy, and Lysosomal Acidification
UBQLN1 and UBQLN2 were previously shown to interact with the autophagosomal marker, LC3, and proposed to have a role in autophagy6,7. N’Diaye et al. argue that UBQLN1 and 2 knockdown in Hela cells increases autophagosome formation but reduces starvation induced autophagy by delaying the delivery of autophagosomes to lysosomes6. In contrast, Rothenberg et al. argue that UBQLN1 and 2 knockdown affects maturation of cytosolic LC3-I to membrane-bound LC3-II and leads to reduced autophagosome formation7. Although these studies suggest an involvement of Ubiquilin in autophagic degradation, the mechanisms by which Ubiquilin regulate autophagy in vivo were not defined. Furthermore, Ubiquilins were not previously implicated in lysosomal acidification.
Under nutrient-rich conditions, mTORC1 senses lysosomal amino acids through the v-ATPase and localizes to the lysosomal surface where it becomes activated47,48. Active mTORC1 inhibits autophagy through phosphorylation-dependent Atg1/ULK1 inhibition. Lysosomal mTORC2 has also been shown to inhibit chaperone-mediated autophagy49. Our data show that Ubqn loss compromises both mTORC2 and mTORC1 activity. Given that the v-ATPase is required for the lysosomal localization and activation of mTORC1, it is likely that mTORC1 activity is affected due to reduced v-ATPase activity in ubqn1 mutants. Indeed, restoration of v-ATPase activity by manipulating V100 subunit levels in ubqn1 mutants leads to a restoration of reduced mTORC1 activity. Interestingly, ER stress can induce autophagy by inhibition of Akt/mTOR pathway24,50. Hence, both ER stress and diminished v-ATPase activity in ubqn1 mutants may contribute to increased autophagy induction by converging on mTORC1 inhibition.
Ubqn’s localization at the periphery of the lysosome, its physical interaction with v-ATPase, and compromised lysosomal acidification in ubqn1 mutants indicate that Ubqn is involved in regulation of v-ATPase activity. We argue that loss of Ubqn impairs the levels of aberrant V100, disrupting the composition and function of the v-ATPase complex. Mammalian VhaM8.9 (ATP6AP2) affects the level of mammalian V100 (ATP6V0a1)34. Indeed, reduction of VhaM8.9 levels reduces fragmented V100 accumulation and suppresses the ubqn1-associated phenotypes. Consistent with these data, V100 overexpression phenocopies several lysosomal degradation defects observed in ubqn1 mutants. Moreover, loss of V100 or Ubqn reveal striking similarities51,52. First, eyFLP-mediated eye clones of both mutants show a slow and progressive neuronal degeneration and dysfunction. Second, similar to ubqn1 clones, mutant clones of v100 show autophagosome accumulation and reduced LysoTracker signal in the photoreceptor51. Additionally, vhaM8.9 mutant clones in the pre-pupal wings cause reduced LysoTracker-positive vesicle number53 and VhaM8.9 knockdown causes p62 accumulation in the adult brain54. We therefore propose that Ubqn acts as a chaperone for the V100 subunit and promotes stable v-ATPase formation by maintaining the stoichiometric balance between its subunits. In summary, we identify a previously undocumented Ubqn function in regulation of lysosomal activity.
METHODS
Live imaging of LysoTracker and Magic Red Staining
Second or third instar Drosophila fat bodies were dissected in PBS and then stained with 1X LysoTracker Red DND-99 (L7528, ThermoFisher) or 1X Magic Red cresyl violet-(RR)2 (Immunochemistry Technology) for 3 mins on a shaker. After quickly washing with PBS, samples were mounted in 80% glycerol and were imaged with Zeiss LSM880 confocal microscope (Carl Zeiss).
Acidic Nanoparticle (aNP) Preparation
Synthesis: Poly (lactic-co-glycolic acid) (PLGA) nanoparticles were synthesized based on previously published protocols36,38,55. PLGA (Resomer 503H, Sigma) was dissolved in dichloromethane (DCM) to form a solution of 10mg/mL. 10mL of this solution was added to 60mL of 2wt% PVA (87–90% hydrolyzed; MW 30,000–70,000, Sigma) and sonicated for 2.5 minutes. The resulting oil-in-water emulsion was allowed to stir for 4 hours to remove any residual DCM. The aNPs were recovered via centrifugation at 10,000×g for 25 minutes, following washing twice with refrigerated millipore water. The nanoparticles were flash frozen in liquid nitrogen and dried for 48 hours. Nile Red-loaded aNPs were prepared as above except Nile red (Sigma) was added at a concentration of 0.3 mg/mL to the DCM phase prior to sonication. Coating: aNPs were covalently coated in Bovine Serum Albumin (BSA) (Sigma) using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) (ThermoFisher) and N-hydroxysuccinimide (NHS) (ThermoFisher)56. Briefly, dried aNPs were resuspended at a concentration of 10mg/mL in 0.1 M 2-morpholinoethanesulfonic acid (MES) buffer (pH 5.5). 5x molar excess (in regards to the PLGA) of EDC was added and the solution was allowed to stir for 10 minutes after which 2x molar excess of NHS was added. After stirring for 2 hours, the pH of the solution was titrated to 7.2 using sodium bicarbonate (Sigma) and an equimolar amount of BSA (with respect to the PLGA) was added. The resulting solution was stirred for 24 hours prior to recovery of aNPs by centrifugation. After two subsequent wash steps the recovered aNPs were flash frozen and vacuum dried for 48 hours. Characteristics of aNPs can be found in Supplementary Table 6.
Acidic Nanoparticle (aNP) Feeding
aNPs were added in standard fly food in the following concentrations: 0.5 mg/ml, 1 mg/ml, and 3 mg/ml. Third instar larvae are fed with aNP containing food for 3 hours. Subsequently, the larvae are either dissected for confocal imaging or transferred to standard food for 3 hours for recovery before protein extraction.
Transmission Electron Microscopy (TEM)
Both retinae and laminae in adult flies were processed for TEM imaging as previously described57,58.
Drosophila Genetics and Strains
Fly stocks are maintained at room temperature (RT) (~22°C) and crosses for experiments are kept either at RT or at 25°C. For clonal analysis in fat body and salivary gland, Drosophila eggs are heat shocked (37°C) for 1 hour within 4–6 hours after egg laying to induce clones. Collected eggs are kept at RT until 3rd instar larvae stage for further experiments. Null allele of ubqn (ubqn1) was generated by X-chromosome chemical mutagenesis performed in the Bellen lab59. ywing2+Δubqn allele was generated by CRISPR-Cas9 mediated gene deletion as previously described15. Genomic rescue line (y w ubqn1 FRT19A; 20kb P[acman]) was generated by fly crosses using a 20kb genomic rescue construct. 20kb genomic rescue fragment (P[acman] BAC CH322–10F17) was injected into embryos with VK32 attP docking site to generate the rescue line. Transgenic flies with UAS-dUbqn and UAS-FLAG-dUbqn were generated as previously described60. Briefly, Ubqn cDNA clone (LD38919, DGRC) was amplified by PCR and used for construction of pUASTattB-dUbqn and pUASTattB-FLAG-Ubqn. PCR reactions were performed by using the following primers before subcloning into pUASTattB vector (linearized with Bgl2 and NotI): Ubqn_F BamHI: (5’ - ccattttGGATCCaaaATGGCGGAAGGCGGCAGCAAG- 3’) Ubqn_R NotI: (5’- agttaaGCGGCCGCTTAACTCAAGGACAACTGGTTGAG-3’) Ubqn-Nter-Flag_F BamHI: (5’- ccattttGGATCCaaaATGGACTACAAGGACCACGATATTGATTACAAGGACGATGACGACAAGGGAAGCGGCATGGCGGAAGGCGGCAGCAAG-3’). The final constructs were injected into embryos with VK37 attP docking site (BDSC #9752) to generate the transgenic flies.
P497H mutation in UBQLN2 DNA template (clone from Ultimate ORF LITE collection, ThermoFisher) was induced by megaprimer mutagenesis with M13F (5′-GTAAAACGACGGCC −3′) and M13R (5′-AACAGCTATGACC −3′) primers, and P497H mutation-specific primer (5′-cctgtaggcccagtcacccAcataggccccataggccctatagtcccttt −3′) using KOD Hot Start DNA Polymerase (Novagen). PCR reactions were then recombined into destination vector, pUASg-HA-attB, using LR Clonase (Life Technologies) and transgenic flies (y w;UAS-UBQLN2WT and y w;UAS-UBQLN2P497H) were generated by injecting the WT or mutation-containing vector into embryos with VK37 attP docking site (BDSC #9752). Transgenic flies expressing Ubqn-GFP were generated by injecting C-terminal GFP tagged ubqn genomic construct13 into embryos with VK33 attP docking site (BDSC #32542: y1 w* P{nos-phiC31\int.NLS}X; PBac{y+-attP-3B}VK00033). Df(1)BSC871, Cg-Gal4, Act-Gal4, da-GAL4, nSyb-Gal4, y1 w1118 P{neoFRT}19A; P{eyFLP}, P{Ubi-mRFP.nls}, w* P{hsFLP}, P{neoFRT}19A; P{UASp-mCherry-Atg8a}2, UAS-Luciferase RNAi, UAS-Luciferase, y1 v1; UAS-V100 RNAi, w*; P{w[+mC]=sqh-EYFP-ER}3, y1 w1118; P{UASp-GFP-mCherry-Atg8a}2, y1 w*; Mi{PT-GFSTF.0}Atg1MI07056-GFSTF.0, w1118; P{PTT-GA}VhaSFDG00259, y1w67c23; P{EPgy2}VhaSFDEY04644, w1118; P{EP}Vha68–2EP2364, and y1w67c23; P{EPgy2}VhaM8.9EY03616 were obtained from Bloomington Drosophila Stock Center. w1118; P{RS5}VhaM8.95-HA−1890 was obtained from Kyoto Stock Center. y w; FRT82B v100[3] and UAS-V100 were from the Bellen lab61. The following lines were obtained from Vienna Drosophila Research Center: P{KK107676}VIE-260B (RNAi against VhaM8.9), UAS-Ubqn RNAi was a gift from Dr. Ming Guo, UAS-LAMP1-GFP was a gift from Dr. Helmut Kramer, and cl(1), P{neoFRT}19A/Dp(1;Y) y+ v+; ey-FLP was a gift from Drs. John Olson and Utpal Banerjee. Xbp1-EGFP was a gift from Dr. Don Ryoo.
Pupal histology
Pupal histology processing was performed as previously described58. Briefly, P4 stage pupae grown at room temperature were fixed in 8% glutaraldehyde (EM grade) and embedded in paraffin. Coronal (5 μm) and sagittal sections (5 μm) were prepared on the microtome (Leica) and stained with hematoxylin and eosin. At least 3 animals were examined for each genotype and preparation.
Subcellular fractionation
Lysosomes were enriched by centrifugation from a large pool of L3 larvae in a discontinuous Nycodenz density gradient, as described previously62,63, with modifications. In brief, homogenate was prepared in 0.25 M sucrose (pH 7.2) and centrifuged in succession at 4,800 × g, 5 min, and 17,000 × g, 10 min. The sediment of the second centrifugation was washed at 17,000 × g, 10 min, resuspended 1:1 vol/vol in 85.6% Nycodenz, and placed on the bottom of an Ultraclear (Beckman) tube. On top, a discontinuous gradient of Nycodenz was constructed (from bottom to top: 32.8%, 26.3%, and 19.8% Nycodenz). Centrifugation was for 1 h in an SW 40 Ti rotor (Beckman) at 141,000 × g. Top layers containing lysosomes were collected and diluted in assay buffer and centrifuged at 37,000 × g, 15 min. Pellet was resuspended in assay buffer and standard western blotting protocol is followed.
Drosophila antibodies
Immunofluorescence
Primary antibodies were used at following concentration: Rabbit anti-p62/ref(2)p: 1:1000 (gift from Dr. Sheng Zhang); Chicken anti-GFP: 1:500 (Abcam, ab13970), Rat anti-ELAV: 1:500 (DSHB: 7E8A10); Mouse anti-Chaoptin 1:200 (DSHB: 24B10); Rabbit anti-Ubqn 1:200 (gift from Dr. Ming Guo). Detailed primary antibody information can be found in Supplementary Table 7. Secondary antibodies used: Alexa 488, and Cy3 or Cy5 conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were used at 1:500. Alexa 488-conjugated Phalloidin (Invitrogen) is used at 1:250.
Western blotting
Primary antibodies were used at the following concentrations: Rabbit anti-Ubqn: 1:2000 (gift from Dr. Ming Guo); Mouse anti-actin: 1:10,000 (ICN Biomedicals: C4); Mouse anti-α-Tubulin: 1:10,000 (Sigma T9026); Rabbit anti-P-eIF2α (S51): 1:2000 (CST #9721); Rabbit anti-eIF2α (eIF2S1): 1:1000 (Abcam ab26197); Rabbit anti-GFP: 1:7500 (Invitrogen #A-11122); Rabbit anti-DsRed: 1:2000 (Clontech #632496); Rabbit anti-Phospho-Drosophila-Akt (Ser505): 1:1000 (CST #4054); Rabbit Akt (pan): 1:1000 (CST #4691, C67E7); Rabbit anti-Phospho-Drosophila-S6K (Thr398): 1:1000 (CST #9209); Rabbit anti-S6K: 1:1000 (SCBT sc-9027); Rabbit anti-Phospho-4E-BP (Thr37/46): 1:1000 (CST #2855); Rabbit anti-p62/ref(2)p: 1:2000 (gift from Dr. Sheng Zhang); Guinea pig anti-Vha100: 1:10,00061; Mouse anti CTSL: 1:1000 (R&D Systems, MAB22591); Rabbit anti-ATP6V1B2/Vha55: 1:1000 (CST #14617); Guinea pig anti-Bip/Hsc3: 1:1000 (gift from Dr. Don Ryoo); Rabbit anti-Drosophila Atg8: 1:1000 (gift from Dr. Linda Partridge); Mouse anti-Lamin C: 1:1000 (DSHB, LC28.26). Detailed primary antibody information can be found in Supplementary Table 7. Secondary antibodies, IRdye 680RD and IRdye 800CW (Li-COR Biosciences), were used at 1:10,000 and HRP-conjugated secondary antibodies (Jackson ImmunoResearch) were used at 1:5000. Images have been cropped for presentation. Full size images are presented in Fig. S9.
Electroretinogram (ERG) Assay
For ERG recordings, y w ubqn1 FRT19A/FM7c, Kr-Gal4, UAS-GFP flies were crossed to y w P{w+} cl(1) FRT19A/Dp(1;Y)y+; eyFLP to generate flies with mutant clones in the eyes and flies were aged at 25°C in 12h light/12h dark cycle or in 24h darkness. ERG recordings were performed as previously described57. At least 5 flies were examined for each genotype and timepoint.
RNA extraction and Quantitative Real-Time PCR
Total RNA was isolated from 10 third instar larvae using Trizol (Life Technologies). Reverse transcription was performed using the iScript Reverse Transcription Supermix (Bio-Rad, Hercules, CA). RT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and CFX96 Touch Real-Time PCR Detection System. RT-PCR was done with 3–6 PCR replicates for each biological sample, 3–4 biological replicates. Primers used for the RT-PCR were listed in the Supplementary Table 8.
Immunoprecipitation and mass spectrometry
The larvae or adult head pellet was lysed in 3 volumes of NETN buffer (50 mM Tris pH 7.3, 170 mM NaCl, 1 mM EDTA, 0.5% NP-40) using beads-beater. The lysate was removed after spun at 12,000g for 5 min, then ultracentrifuged at 200,000g for 20min at 4°C. The lysate was incubated in 20 μl of protein A sepharose slurry (GE Healthcare Life Sciences, 17–0780-01) for 1 hr to reduce the non-specific binding. After pre-clearing, the lysate was incubated with GFP-Trap beads (ChromoTek GFP-Trap®, Germany) for 1 hr at 4°C. The beads were briefly washed with lysis buffer, boiled in 2X NUPAGE® LDS sample buffer (Invitrogen) and subjected to SDS-PAGE (NuPAGE 10% Bis-Tris Gel, Invitrogen). The eluted proteins were visualized with Coomassie Brilliant blue-stain and excised into gel pieces according to the molecular weight. The individual gel piece was destained and subject to in-gel digestion using trypsin (GenDepot T9600, Houston, TX). The tryptic peptides were resuspended in 10 μl of loading solution (5% methanol containing 0.1% formic acid) and subjected to nanoflow LC-MS/MS analysis with a nano-LC 1000 system (Thermo Scientific) coupled to Orbitrap Fusion™ Tribrid™ (Thermo Scientific) mass spectrometer. The peptides were loaded onto a Reprosil-Pur Basic C18 (1.9 μm, Dr. Maisch GmbH, Germany) pre-column of 2cm × 100μm size. The pre-column was switched in-line with an in-housed 5 cm × 150 μm analytical column packed with Reprosil-Pur Basic C18 equilibrated in 0.1% formic acid/water. The peptides were eluted using a 35 min discontinuous gradient of 4–26% acetonitrile/0.1% formic acid at a flow rate of 800nl/min. The eluted peptides were directly electro-sprayed into Orbitrap Fusion™ mass spectrometer. The instrument was operated in the data-dependent mode acquiring fragmentation under direct control of Xcalibur software (Thermo Scientific). Precursor MS spectrum was scanned at 375–1300 m/z with 120,000 resolution at 400 m/z, 5×105 AGC target (50 ms maximum injection time) by Orbitrap. Then, the top 35 strongest ions were fragmented by collison-induced dissociation (CID) with 35 normalized collision energy and 1.6 m/z isolation width and detected by Iontrap with 30 sec of dynamic exclusion time, 1×104 AGC target and 100 ms of maximum injection time.
Database Search and Data quantification for MS
Obtained MS/MS spectra were searched against target-decoy Drosophila Flybase in Proteome Discoverer 1.4 interface (Thermo Scientific) with Mascot algorithm (Mascot 2.4, Matrix Science). The precursor mass tolerance was confined within 20 ppm with fragment mass tolerance of 0.5 dalton and a maximum of two missed cleavage allowed. Dynamic modification of methionine oxidation, protein N-terminal acetylation, destreak on cysteine and ubiquitination on lysine was allowed. The peptides identified from mascot result file were validated with 5% false discover rate (FDR).
Daoy cell siRNA transfection and cell maintenance
The following siRNAs were used for transient interference of UBQLN1, UBQLN2, and UBQLN4:
UBQLN1 (ThermoFisher Scientific: HSS121163 and HSS121164)
GGAACCAAUGCUGAGUGCUGCACAA and CCUUGUUACAGAUUCAGCAGGGUUU
UBQLN2 (ThermoFisher Scientific: HSS121160) GCCCUGAGAUGAUGAUCCAAAUAAU
UBQLN4 (ThermoFisher Scientific: (HSS183586) GCAGACAAUGGAGCUUGCUCGGAAU
Daoy cells were maintained in DMEM medium supplemented with 10% FBS, 1X GlutaMAX, 1X pyruvate and 1X penicillin and streptomycin. All reagents were purchased from ThermoFisher.
For the inhibitor treatments: Daoy cells were treated with Bafilomycin A1 (200 nM for 4 hours) before harvesting or fixation.
UBQLN½/4 triple knockout HEK293 Flp-in TRex cells
UBQLN1+2+4 triple knockout cells (TKO) were a gift from Ramanujan S. Hedge. They were generated via CRISPR/Cas9 in HEK293 Flp-in TRex cells. Cells were grown and maintained as described previously9. For the inhibitor treatments: HEK293 cells were treated with Bafilomycin A1 (200 nM for 4 hours) before harvesting or fixation.
HEK293T cell cDNA Transfection
The UBQLN1 (p4458 FLAG-hPLIC-1; #8663, Addgene) or UBQLN2 (p4455 FLAG-hPLIC-2; #8661, Addgene) cDNA were transiently transfected to HEK293T cells using Lipofectamine 3000 (#L3000008, ThermoFisher) following the manufacturer’s instructions.
In Vivo Crosslinking
HEK293T cells transfected as indicated above were washed twice with cold PBS, and incubated with 2 mM DSP (#A35393, Life Technologies) freshly prepared in PBS for 2 hr on ice. Unreacted DSP was quenched by adding 1 M Tris (pH 7.5) to a final concentration of 20 mM for 15 min. Crosslinked cells were then washed with cold PBS and lysed with 0.3% CHAPS lysis buffer (150 mM NaCl, 20 mM HEPES pH7.5, 10% Glycerol, 0.3% CHAPS and supplemented with protease inhibitors (Roche)). EZview™ Red ANTI-FLAG® M2 Affinity Gel (#F2426, Sigma), was incubated with the pre-cleared cell lysates for overnight at 4°C. Co-immunoprecipitated proteins were uncrosslinked by adding beta-mercaptoethanol in sample buffer prior to western blotting analysis.
Mammalian antibodies
Immunofluorescence
Daoy cells seeded on coverslips were fixed with 4% paraformaldehyde in 1X PBS at room temperature for 10 minutes. The fixed cells were permeabilized in blocking buffer (0.25% Saponin and 10% non-immune goat serum in 1XPBS). After permeabilization, cells were used for immunofluorescence staining using the following antibodies diluted in blocking buffer: Rabbit anti-p62: 1:500 (MBL: PM045); Rabbit anti-LC3B: 1:250 (CST: D11); and Alexa 488 conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) 1:500. The cells were mounted in DAPI Fluoromount-G (0100–20, SouthernBiotech) and kept at 4 °C before imaging under a Zeiss LSM710 confocal microscope (Carl Zeiss). Detailed primary antibody information can be found in Supplementary Table 7.
Live imaging of LysoTracker staining
Daoy cells seeded on coverslips were treated with 50 nM of LysoTracker Red DND-99 (L7528, ThermoFisher) for 30 minutes at 37°C. Cell nuclei were stained by 1 μg/ml of Hoechst 33342 (H3570, ThermoFisher). Images were taken immediately after incubation by a Zeiss LSM710 confocal microscope (Carl Zeiss).
Western blotting
Daoy cells were homogenized in Modified RIPA buffer (50 mM Tris-Cl, 150 mM NaCl, 1 % NP-40, 1 % Sodium deoxycholate, 0.1 % SDS, 50 mM NaF, 1 mM Na3VO4, 10 % Glycerol and Roche protease inhibitor mix) on ice. HEK293 cells were homogenized in SDS lysis buffer (2% SDS, 50mM Tris-HCl, pH 7.4, 2mM EDTA) on ice. Isolated lysates heated in an appropriate volume of Laemmli buffer were loaded into gels, separated by SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad). Primary antibodies used in this study were as follows: Rabbit anti-GAPDH: 1:5000 (CST #2118); Mouse anti-actin: 1:10,000 (ICN Biomedicals: C4); Rabbit anti-UBQLN1: 1:1000 (CST #14526); Rabbit anti-UBQLN2: 1:1000 (CST #85509); Rabbit anti-UBQLN4: 1:1000 (Abcam ab106443); Rabbit anti-Phospho S6K (T389): 1:1000 (CST #9205); Rabbit anti-S6K (SCBT sc-9027); Rabbit anti-Phospho ULK1 (S757): 1:1000 (CST #6888); Rabbit anti-ULK1: 1:1000 (CST #8054, D8H5); Rabbit anti-p62: 1:5000 (MBL: PM045); Rabbit anti-LAMP1: 1:1000 (CST #9091); Mouse anti-LAMP2: 1:2000 (SCBT: H4B4); Rabbit anti-LC3B: 1:1000 (CST: D11); Rabbit anti-ATP6V0A1: 1:1000 (Novus Biologicals: NBP1–89342); Rabbit anti-ATP6V1B2: 1:1000 (CST #14617); Mouse anti-ATP6V1C1: 1:500 (SCBT: sc-271077); Mouse anti-ATP6V1D1: 1:500 (SCBT: sc-390384); Mouse anti-ATP6V1H: 1:500 (SCBT: sc-166227); Rabbit anti-Renin R/M8.9: 1:2000 (Novus Biologicals: NBP1–90820); Mouse anti-FLAG M2: 1:1000 (Sigma: F1804). Detailed primary antibody information can be found in Supplementary Table 7. Secondary antibodies, IRdye 680RD and IRdye 800CW (Li-COR Biosciences), were used at 1:10,000 and HRP-conjugated secondary antibodies (Jackson ImmunoResearch) were used at 1:5000. Images have been cropped for presentation. Full size images are presented in Fig. S9.
Nanoparticle Characterization
Size and Zeta Potential Measurements: aNP size and zeta potential were measured using Zetasizer Nano: Malvern Zen 3600 Zetasizer. 0.1 mg of particles were added to 1 mL of filtered Millipore water, allowed to swell for 6 hours, and then measured. BSA loading: Amount of BSA per mg of aNP was measured using the Micro BSA Protein Assay Kit (ThermoFisher). Briefly, nanoparticles were suspended in Millipore water at a concentration of 0.1 mg/mL and added to an equal volume of working solution. After 2 hours of incubation at 37˚C, the suspension was centrifuged, and the supernatant removed for analysis. For the analysis, BSA-coated PLGA nanoparticles were compared to blank uncoated PLGA nanoparticles, and BSA-coated Nile Red-loaded PLGA nanoparticles were compared to Nile Red-loaded uncoated PLGA nanoparticles as some of the Nile Red was released from the nanoparticles during the incubation period. SEM: Vacuum dried nanoparticles were coated with 10 nm of gold using the Denton Desk V Sputter system. Particles were imaged with FEI Quanta 400 ESEM FEG using 10 kV.
Statistics and Reproducibility
All quantitative data were derived from multiple independent experiments. Each exact n value is indicated in the corresponding figure legend. Two-tailed Student’s t-test was used to analyze data and a P value < 0.05 was considered statistically significant. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments. Experiments in the manuscript were performed at least three times except for Figures 1e, 3b, 5c, 6e–g, S1c, S1f, S3d–f, S4d, S4e–f, S5f, S5h, S6c, S7g, S8d, and larval IP/MS experiments (in Supplementary Tables 3–4) which were performed twice and adult head IP/MS experiment (in Supplementary Tables 3–4) which was performed once.
Code Availability
There is no custom code used in this study.
Data Availability
The mass spectrometry proteomics data can be accessed from the ProteomeXchange Consortium via the MassIVE repository (MSV000083259) under accession code PXD012104. The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information files. Source data for Figures 1c, 1f, 2c–e, 3b–f, 4a, 4d–f, 5e, 6a, 6c–d, 6g, 7a–d, S1f, S2a, S2d–e, S3a–c, S4b–c, S4g, S4j, S5a, S5e–f, S7b–c, and S8b–d have been provided as Supplementary Table 9. All other data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank past and current members of the Bellen lab for technical assistance and discussions. We thank Thibault Orand, Wan Hee Yoon, Alberto Di Ronza, Hemanjani Venkata Bhavana, Hongling Pan, and Lita Duraine for technical support. We thank Sonal Nagarkar Jaiswal for helpful discussions and comments. We thank Karen Schulze, Megan Campbell, and Berrak Ugur for their comments on the manuscript. We thank the BDSC (NIH P40OD018537) for Drosophila stocks. We thank Sung Yun Jung, Antrix Jain and the BCM Pathway Discovery Proteomics Core (CPRIT RP120092 and P30CA125123). Microscopy was supported in part by the Baylor College of Medicine IDDRC Neurovisualization Core (U54HD083092) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. E.W. received support from a Ruth L. Kirschstein Fellowship from the National Institute of Dental and Craniofacial Research (F31 DE027586). H.J.B. is supported by the Robert A. and Renee E. Belfer Family Foundation, the Huffington Foundation, Target ALS and the NIH Office of the Director (R24OD022005). HJB is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests
The authors declare no competing financial interests.
REFERENCES
- 1.Edens BM et al. A novel ALS-associated variant in UBQLN4 regulates motor axon morphogenesis. Elife 6, 3797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng H-X et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko HS, Uehara T, Tsuruma K & Nomura Y Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 566, 110–114 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Kleijnen, MF. et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Molecular Cell 6, 409–419 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Lim PJ et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J. Cell Biol 187, 201–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N’Diaye E-N et al. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 10, 173–179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothenberg C et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet 19, 3219–3232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biggins S, Ivanovska I & Rose MD Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol 133, 1331–1346 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itakura E et al. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Molecular Cell 63, 21–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjerpe R et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 166, 935–949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto S et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159, 200–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haelterman NA et al. Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res. 24, 1707–1718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarov M et al. A genome-wide resource for the analysis of protein localisation in Drosophila. Elife 5, e12068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venken KJT et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li-Kroeger D et al. An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. Elife 7, e1002472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou TB, Noll E & Perrimon N Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119, 1359–1369 (1993). [DOI] [PubMed] [Google Scholar]
- 17.LaJeunesse DR et al. Three new Drosophila markers of intracellular membranes. BioTechniques 36, 784–8–790 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Moustaqim-Barrette A et al. The amyotrophic lateral sclerosis 8 protein, VAP, is required for ER protein quality control. Hum. Mol. Genet 23, 1975–1989 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hetz C & Saxena S ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol 13, 477–491 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Ogata M et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol 26, 9220–9231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid H-O, Yadav RK, Kim H-R & Chae H-J ER stress: Autophagy induction, inhibition and selection. Autophagy 11, 1956–1977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Høyer-Hansen M & Jäättelä M Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 14, 1576–1582 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Senft D & Ronai ZA UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 40, 141–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin L, Wang Z, Tao L & Wang Y ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6, 239–247 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Kamada Y et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol 150, 1507–1513 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuura A, Tsukada M, Wada Y & Ohsumi Y Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245–250 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Le Ber I et al. SQSTM1 mutations in French patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol 70, 1403–1410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Denton D et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol 19, 1741–1746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura S, Noda T & Yoshimori T Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Mauvezin C, Nagy P, Juhász G & Neufeld TP Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun 6, 7007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neufeld TP & Baehrecke EH Eating on the fly: function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy 4, 557–562 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott RC, Schuldiner O & Neufeld TP Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167–178 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Kinouchi K et al. The role of individual domains and the significance of shedding of ATP6AP2/(pro)renin receptor in vacuolar H(+)-ATPase biogenesis. PLoS ONE 8, e78603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bareford LM & Swaan PW Endocytic mechanisms for targeted drug delivery. Adv. Drug Deliv. Rev. 59, 748–758 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baltazar GC et al. Acidic nanoparticles are trafficked to lysosomes and restore an acidic lysosomal pH and degradative function to compromised ARPE-19 cells. PLoS ONE 7, e49635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Krieger V & Hensel M Application of fluorescent nanoparticles to study remodeling of the endo-lysosomal system by intracellular bacteria. J Vis Exp e52058–e52058 (2015). doi: 10.3791/52058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourdenx M et al. Nanoparticles restore lysosomal acidification defects: Implications for Parkinson and other lysosomal-related diseases. Autophagy 12, 472–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J-H et al. Presenilin 1 Maintains Lysosomal Ca2+ Homeostasis via TRPML1 by Regulating vATPase-Mediated Lysosome Acidification. Cell Reports 12, 1430–1444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim T-Y, Kim E, Yoon SK & Yoon J-B Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem. Biophys. Res. Commun 369, 741–746 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Mendes CS et al. ER stress protects from retinal degeneration. EMBO J. 28, 1296–1307 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Popko B, Tixier E & Roos RP Guanabenz, which enhances the unfolded protein response, ameliorates mutant SOD1-induced amyotrophic lateral sclerosis. Neurobiol. Dis 71, 317–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Popko B & Roos RP An enhanced integrated stress response ameliorates mutant SOD1-induced ALS. Hum. Mol. Genet 23, 2629–2638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao D et al. VAMP associated proteins are required for autophagic and lysosomal degradation by promoting a PtdIns4P mediated endosomal pathway. Autophagy [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandey UB et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447, 859–863 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Appenzeller-Herzog C & Hall MN Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 22, 274–282 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Sancak Y et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoncu R et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arias E et al. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Molecular Cell 59, 270–284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C-H et al. ER stress inhibits mTORC2 and Akt signaling through GSK-3β-mediated phosphorylation of rictor. Sci Signal 4, ra10–ra10 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Williamson WR, Wang D, Haberman AS & Hiesinger PR A dual function of V0-ATPase a1 provides an endolysosomal degradation mechanism in Drosophila melanogaster photoreceptors. J. Cell Biol 189, 885–899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haberman A et al. The synaptic vesicle SNARE neuronal Synaptobrevin promotes endolysosomal degradation and prevents neurodegeneration. J. Cell Biol 196, 261–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hermle T, Guida MC, Beck S, Helmstädter S & Simons M Drosophila ATP6AP2/VhaPRR functions both as a novel planar cell polarity core protein and a regulator of endosomal trafficking. EMBO J. 32, 245–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubos A et al. Conditional depletion of intellectual disability and Parkinsonism candidate gene ATP6AP2 in fly and mouse induces cognitive impairment and neurodegeneration. Hum. Mol. Genet 24, 6736–6755 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang S et al. Oral Administration and Selective Uptake of Polymeric Nanoparticles in DrosophilaLarvae as an in VivoModel. ACS Biomaterials Science & Engineering 1, 1077–1084 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Chung HJ, Kim HK, Yoon JJ & Park TG Heparin immobilized porous PLGA microspheres for angiogenic growth factor delivery. Pharm. Res 23, 1835–1841 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Verstreken P et al. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron 40, 733–748 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Chouhan AK et al. Uncoupling neuronal death and dysfunction in Drosophila models of neurodegenerative disease. Acta Neuropathol Commun 4, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto S et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159, 200–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon WH et al. Loss of Nardilysin, a Mitochondrial Co-chaperone for α-Ketoglutarate Dehydrogenase, Promotes mTORC1 Activation and Neurodegeneration. Neuron 93, 115–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hiesinger PR et al. The v-ATPase V0 Subunit a1 Is Required for a Late Step in Synaptic Vesicle Exocytosis in Drosophila. Cell 121, 607–620 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aniento F, Roche E, Cuervo AM & Knecht E Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem 268, 10463–10470 (1993). [PubMed] [Google Scholar]
- 63.di Ronza A et al. CLN8 is an endoplasmic reticulum cargo receptor that regulates lysosome biogenesis. Nat. Cell Biol 17, 701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data can be accessed from the ProteomeXchange Consortium via the MassIVE repository (MSV000083259) under accession code PXD012104. The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information files. Source data for Figures 1c, 1f, 2c–e, 3b–f, 4a, 4d–f, 5e, 6a, 6c–d, 6g, 7a–d, S1f, S2a, S2d–e, S3a–c, S4b–c, S4g, S4j, S5a, S5e–f, S7b–c, and S8b–d have been provided as Supplementary Table 9. All other data that support the findings of this study are available from the corresponding author upon reasonable request.