Abstract
α-Synuclein fibrils are considered a hallmark of Parkinson’s disease and other synucleinopathies. However, small oligomers that formed during the early stages of α-synuclein aggregation are thought to be the main toxic species causing disease. The formation of α-synuclein oligomers has proven difficult to follow, because of the heterogeneity and transient nature of the species formed. Here, a novel bead-based aggregation assay for monitoring the earliest stages of α-synuclein oligomerization, α-Synuclein–Confocal Nanoscanning (ASYN-CONA), is presented. The α-synuclein A91C single cysteine mutant is modified with a trifunctional chemical tag, which allows simultaneous fluorescent labeling with a green dye (tetramethylrhodamine, TMR) and attachment to microbeads. Beads with bound TMR-labeled α-synuclein are then incubated with a red dye (Cy5)-labeled variant of α-synuclein A91C, and EtOH (20%) to induce aggregation. Aggregation is detected by confocal scanning imaging, below the equatorial plane of the beads, which is known as the CONA technique. On-bead TMR-labeled α-synuclein and aggregated Cy5-labeled α-synuclein from the solution are quantitatively monitored in parallel by detection of fluorescent halos or “rings”. α-Synuclein on-bead oligomerization results in a linear increase of red bead ring fluorescence intensity over a period of 5 h. Total internal reflection fluorescence microscopy was performed on oligomers cleaved from the beads, and it revealed that (i) oligomers are sufficiently stable in solution to investigate their composition, consisting of 6 ± 1 monomer units, and (ii) oligomers containing a mean of 15 monomers bind Thioflavin-T. Various known inhibitors of α-synuclein aggregation were used to validate the ASYN-CONA assay for drug screening. Baicalein, curcumin, and rifampicin showed concentration-dependent inhibition of the α-synuclein aggregation and the IC50 (the concentration of the compound at which the maxiumum intensity was reduced by one-half) were calculated.
Parkinson’s disease and other synucleinopathies are characterized by the misfolding and aggregation of α-synuclein. α-Synuclein is a small presynaptic protein whose main function is believed to occur at the presynaptic terminals and may play a role in regulating synaptic transmission.1 Its amino-acid sequence consists of three distinct regions with different properties. The N-terminus, which is defined by residues 1–60, contains an imperfect repeated sequence (KTKEGV) involved in the amphiphilic α-helical structure adopted when bound to lipids.2 The nonamyloid component (NAC), region, which is defined by residues 61–95, is highly hydrophobic and forms the core of the highly organized fibril structures.3 The C-terminus, which is defined by residues 96–140, is negatively charged and contains several proline residues, making it very flexible. This region is found to be unstructured in α-synuclein fibrils.4
α-Synuclein exists primarily as an unfolded monomer in equilibrium alongside some partially folded monomers and multimers, depending on the local environments of the protein.5−8 In disease, α-synuclein is believed to misfold, acquiring a conformation prone to aggregation that leads to the formation of highly organized fibrils.9 The structure of mature fibrils have recently been established in detail by solid-state NMR4 and cryo-EM10 to be Greek key-like. Fibrillation of α-synuclein is described as occurring in three phases, similar to the formation of other amyloid fibrils. The first stage of fibrillation is called the nucleation or lag phase, where the protein undergoes a change in conformation that allows the formation of small oligomers. The second phase is the elongation, in which the small oligomers rearrange into a conformation with a greater β-sheet content, forming protofibrils that subsequently elongate, forming the fibrils. The final phase is the stationary phase, in which fibrils reach equilibrium with the other α-synuclein species in solution.11 The kinetics of fibril formation are classically detected by fluorescence emission intensity changes of solvatochromic dyes such as thioflavinT, ThT, which exhibit enhanced fluorescence upon binding to β-sheet-rich fibrils. α-Synuclein fibrils are the species found to be the main components of Lewy bodies and Lewy neurites in the brains of patients.11,12 However, early oligomers, formed during the lag phase, are thought to be the main toxic species in disease.13,14
To date, the formation and structure of α-synuclein oligomers have proven difficult to investigate, because of their transient state and heterogeneous characteristics.15 The shape, size, and conformation of various different oligomers, and their kinetics of formation, have been described in the literature, but a consensus on the most disease relevant species has not yet been reached.15 Several groups have described the existence of two different types of oligomers on the pathway to fibril formation.16−19 The earliest oligomers detectable in the pathway have been shown to be smaller and less compact than the subsequent ones. Some studies have suggested that both types of oligomers are able to form fibrils,16,19 in disagreement with other studies that suggested that just one of the oligomers is able to continue through to fibril formation.20 It is not known if these studies were describing the same or different types of oligomer. Evidence of heterogeneity of oligomers has been reported many times. The different nature of these oligomers could explain the different fibrillation propensities. To our knowledge, the kinetics of the full oligomerization process has been quantified twice, both by single-molecule techniques. One by direct measuring of the aggregation of labeled α-synuclein,20 and the other one by measuring ThT anisotropy.21 Both studies focus on the conformational changes of the oligomers observed, describing the interconversion between two different types of oligomers and not on the development of an assay for HTS.
The variability of the oligomers formed could potentially be explained by the different experimental conditions used to induce the oligomerization process. To date, it appears that no universal standard detection method for oligomer formation has been established.15
This work describes a novel assay for monitoring early α-synuclein oligomerization on-bead, α-synuclein–confocal nanoscanning (ASYN-CONA). On-bead screening is a well-validated screening technique for the detection of the binding of small molecular entities synthesized on bead to target proteins in solution.22−25 Confocal nanoscanning (CONA) relies on the imaging of a monolayer of beads, using a scanning confocal microscope focused below the equatorial plane of the beads and scanned through the entire well, generating a cross section of the beads.26−28 When fluorescently labeled target protein is bound to the bead, a “halo” or “ring” is observed. Bead-based screening offers a series of advantages, including sensitivity, versatility, miniaturization, statistical significance, and multiplexing. The advantages of the bead-based screening platform have been exploited to develop ASYN-CONA, which focuses on studying the early oligomerization process of intrinsically unfolded proteins, exemplified with α-synuclein.
Materials and Methods
All commercially available reagents were purchased from Sigma–Aldrich or VWR Scientific, unless otherwise stated, and used as received taking the determined concentration after quality control by HPLC and LCMS.
Confocal Imaging
Bead images were taken on an Opera High Content screening system (PerkinElmer), using a 20× air objective, NA 0.45, with a 384 well plate (SWISSCI, Code PS384B-G175) and detection was performed by Peltier-cooled CCD cameras with 1.3 megapixel. The focal height was set to 30 μm above the well plate surface. A three-exposure setup was used, all in camera 2 with primary dichroic 445/561/640: exposure one, cw laser 561 nm, detection dichroic 568sp for camera 2, filter 585/40; exposure two (bright-field), top illumination, 50% LED, 160 ms exposure time, detection dichroic 650sp for camera 2, filter 690/70; and exposure three, cw laser 640 nm, detection dichroic 650sp for camera 2, filter 690/70. The specific laser powers and exposures times are specified in each experiment. The wells were imaged as 77 overlapping (20%) images over the entire well area, avoiding the edges.
Stitching
Images were stitched using Fiji software29 with a batch stitching macro utilizing the ImageJ plugin Grid/Mosaic stitching.30
Image Analysis
Bead Ring Evaluation and Analysis of Data software (BREAD), developed in Auer lab, was used for the analysis of all image data (a manuscript describing BREAD in more detail is in preparation). In brief, beads are individually detected in each channel within a radius range defined by the number of pixels, here between 60 and 110. Where images are dark, or beads nonfluorescent, the bright-field channel is used to specify the location of beads in the fluorescent image, so that they can still be included for analysis to ensure correct statistical evaluation. Bead ring intensity is calculated from n profiles taken from each bead. In the analysis of the images in this project, n was always 10. The maxima of the intensity profiles at the two edges of the beads were averaged, and the 20th–80th percentiles of the profile intensity between these two points was subtracted to account for the intensity of the center of the bead. This number was referred to as the bead ring intensity. Bead ring intensity from each bead is calculated as the average of all the profiles of each bead. Mean bead ring intensity of each well in each channel is calculated as the average ring intensity of all the beads in the well. Ratiometric quantification was calculated by dividing the ring intensity in two different channels on an individual bead basis. Outlying beads (<5%) were deleted and not considered for the quantification. Beads were considered to be outlying if a bead was badly stitched or broken, when it was out of focus, when no ring was observed under exposure one, or when the bead brightness was at least 1 order of magnitude larger than average.
α-Synuclein Aggregation Assay
Ni-NTA agarose beads (Qiagen, No. 30250) were sieved manually using 120 μm (Merck Millipore, No. NY2H04700) and 100 μm (Falcon cell strainer, No. 352360) filters and beads with sizes between 100 μm to 120 μm were used for all of the on-bead experiments. Beads were stored in 20% EtOH in water and washed prior to any experiment with buffer (20 mM Tris, 100 mM NaCl, pH 7.1) three times by removing supernatant and adding more buffer and kept in a 50% slurry solution. Aliquots of protein stored at −80 °C were defrosted and used on the day. For the incubation of the α-synuclein-A91C-HTM with the beads, 275 μL of protein solutions for final concentrations of 0, 500, 200, 100, 50, and 25 nM were prepared and 25 μL of 50% slurry beads were added. The solutions were shaken at 1000 rpm and 22 °C for 20 min and washed 3 times to remove any unbound protein. Aliquots of 10 μL were taken and imaged at this stage. For the aggregation assays, α-synuclein-A91C-Cy5 (500 nM final concentration) and EtOH (0 for the control or 20% final concentration) were added. The reaction was shaken at 1000 rpm for a maximum of 6 h at 22 °C. Aliquots of 20 μL were taken every hour and imaged stitched and analyzed as described above: exposure one, 1500 μW and 120 ms; exposure two, LED 50% and 160 ms; exposure three, 1000 μW and 120 ms.
Elution of α-Synuclein Oligomers from Beads
After 5 h of incubation of the beads (1 mL of 500 nM aS-A91C-HTM, 2.5 μM aS-Cy5 and 20% EtOH) shaking at 22 °C, the supernatant was removed and the beads washed 3 times with 20 mM Tris, 100 mM NaCl, pH 7.1. 300 mM imidazole, 20 mM Tris, 100 mM NaCl, pH 7.1 (100 μL) was added to elute the oligomers from the beads. The solution was collected, flash frozen, and stored at −20 °C until required.
Sample Preparation for Single-Molecule TIRF
Microscope coverslips were prepared as described in the Supporting Information. Eluted α-synuclein was diluted to a concentration of 10 nM in 20 nm filtered buffer (20 mM Tris, 100 mM NaCl, pH 7.1) with 50 μM Thioflavin-T (Sigma–Aldrich. Product No. T3516) (for single aggregate visualization by enhancement (SAVE) imaging),31 before being added to the poly-l-lysine coated coverslip. Following 10 min of incubation, the coverslips were rinsed three times with 20 nm filtered buffer (20 mM Tris, 100 mM NaCl, pH 7.1), and imaged on the single-molecule total internal reflection fluorescence (TIRF) microscope.
Single-Molecule Imaging
Single-molecule imaging was performed using a custom-built, bespoke single-molecule TIRF microscope, which restricts the illumination to within 200 nm of the sample slide. The fluorophores were excited at either 405 nm (Thioflavin-T), 561 nm (TMR), or 638 nm (Cy5). Collimated laser light at wavelengths of 405 nm (Cobolt MLD Series 405-250 Diode Laser System, Cobolt AB, Solna, Sweden), 561 nm (Cobolt DPL Series 561-100 DPSS Laser System, Cobolt AB, Solna, Sweden), and 636 nm (Cobolt MLD Series 638-140 Diode Laser System, Cobolt AB, Solna, Sweden) were aligned and directed parallel to the optical axis at the edge of a 1.49 NA TIRF objective (CFI Apochromat TIRF 60XC Oil, Nikon, Japan), mounted on an inverted Nikon TI2 microscope (Nikon, Japan). The microscope was fitted with a perfect focus system, which autocorrects the z-stage drift during imaging. Fluorescence collected by the same objective was separated from the returning TIR beam by a dichroic mirror (Di01-R405/488/561/635 (Semrock, Rochester, NY, USA), and was passed through appropriate filters (405 nm: BLP01-488R-25 (Semrock, Rochester, NY, USA), 561 nm: LP02-568-RS, FF01-587/35 (Semrock, Rochester, NY, USA), 638 nm: FF01-692/40–25 (Semrock, Rochester, NY, USA). The fluorescence was then passed through a 2.5× beam expander and recorded on an EMCCD camera (Delta Evolve 512, Photometrics, Tucson, AZ, USA) operating in frame transfer mode (EMGain = 11.5 e–/ADU and 250 ADU/photon). Each pixel was 103 nm in length. Images were recorded with an exposure time of 50 ms with 638 nm (∼50 W cm–1) illumination, 561 nm illumination (∼50 W cm–1), followed by 405 nm excitation (∼100 W cm–1). The microscope was automated using the open source microscopy platform Micromanager.
Testing Inhibitors Using the ASYN-CONA Assay
Selegiline, curcumin, rifampicin, dopamine, baicalein, and nicotine were analyzed by HPLC and LCMS prior to use. The experiments were performed as described above. In more detail, the following conditions were applied. α-Synuclein-A91C-HTM was used at 100 nM and α-synuclein-A91C-Cy5 was used at 500 nM. The compounds were dissolved in EtOH and added to a final concentration of compound of 50 μM and 20% EtOH. Control samples with no compound, no compound, and no α-synuclein-A91C-HTM or no EtOH were included in the experiment. Imaging conditions were the same as described above.
For measurement of the concentration-dependent inhibition of baicalein, curcumin, and rifampicin, the experiments were performed exactly the same but with the addition of different concentrations of the inhibitors dissolved in EtOH.
With regard to IC50 fitting, where IC50 represents the concentration of the compound at which the maxiumum intensity (Imax) was reduced by one-half, the mean bead ring intensity data were plotted using GraFit v7.0.3 (Erithacus Software Limited).50 The standard IC50 equation in GraFit,
was used to perform a four-parameter fit of the data to obtain IC50 values for individual compounds, where y is the observed mean bead ring intensity; Imax and Imin are the maximum and minimum mean bead ring intensities, respectively; IC50 is the concentration of the compound at which Imax was reduced by half; n is the slope of the fit; and x is the compound concentration.
Results and Discussion
Attachment of α-Synuclein to Beads
A critical requirement for the development of the bead-based α-synuclein aggregation assay was the attachment of fluorescently labeled α-synuclein to microbeads. For this purpose, a trifunctional chemical tag was synthesized, enabling the simultaneous fluorescent labeling of the protein and specific functionalization with a reactive group to attach the protein to beads. The tag required three different functionalities: a reactive group to covalently bind to the protein, a fluorophore, and a specific group to bind to the beads (Figure 1). The functionalization was achieved by a synthetic trifunctional tag (HTM, 1). The three functionalities of the tag include a maleimide for protein labeling;32 a His6-tag for bead attachment, and tetramethylrhodamine (TMR)33 dye for fluorescent properties (Figure 1A). In brief, HTM (1) was successfully synthesized using standard Fmoc solid-phase peptide synthesis (see the Materials and Methods section).
Figure 1.

(A) Structure of the HTM trifunctional tag (1). The tag consists of a His6-tag, for binding to Ni-NTA beads; a fluorophore (TMR); and a protein linkage point (maleimide). (B) The different steps required to attach the protein to the microbeads. First, the protein is covalently modified with the trifunctional tag. Subsequently, the functionalized protein is attached to microbeads, which will be used in the ASYN-CONA assay.
Since α-synuclein lacks cysteine residues in its sequence, a single cysteine mutation was generated to provide a specific reactive point for protein functionalization via a maleimide. Single-cysteine mutants of α-synuclein have been extensively used for its labeling and have shown no effect on its behavior when the mutation is located at the end of the NAC region or on the C-terminus of the protein (residues 90–140).17,18,20,34−37 Thirunavukkuaras et al. have used solvatochromic dyes at different positions in α-synuclein to study conformational changes during aggregation.34 The conclusions provided valuable information about regions of the protein in proximity to each other at different stages of the aggregation process, allowing the selection of labeling position that would have low impact on the aggregation.
For ASYN-CONA assay development, a cysteine was introduced in position A91. Briefly, α-synuclein-A91C was expressed in E. coli and purified by acid precipitation, followed by anion exchange chromatography (see the Materials and Methods section).
TMR was used as a dye, because of its optical stability and great brightness. It is of critical importance for the assay quality that the intensity of the TMR emission defines single bead loading by α-synuclein.
α-Synuclein-A91C was labeled with the trifunctional tag (1) and incubated with Ni-NTA agarose beads presieved to homogeneous sizes (100–120 μm) to allow optimized ring detection and quantification. Imaging of the TMR tagged on-bead α-synuclein (α-synuclein-A91C-HTM) was performed on a PerkinElmer Opera high-content screening system. The presence of protein on-bead was observed through a fluorescent ring around each bead (Figure 2). The attachment of TMR-labeled α-synuclein was shown to be concentration dependent (Figure 2). When the beads were incubated with increasing protein concentrations the bead ring intensities increased linearly as shown on Figure 2.
Figure 2.

Ring intensity is linearly proportional to the amount of α-synuclein-A91C-HTM incubated with the beads. Mean bead ring intensity in the TMR channel (λex = 561 nm, λem = 585/40 nm) is represented as a function of the concentration of α-synuclein-A91C-HTM (0, 25, 50, 100, 250, and 500 nM) used for attachment to the Ni-NTA agarose beads. Fitted line (red) represents linear regression of the data (R2 = 0.9984). The two different data points for each concentration originates from beads functionalized on the same day.
On-Bead Aggregation Assay
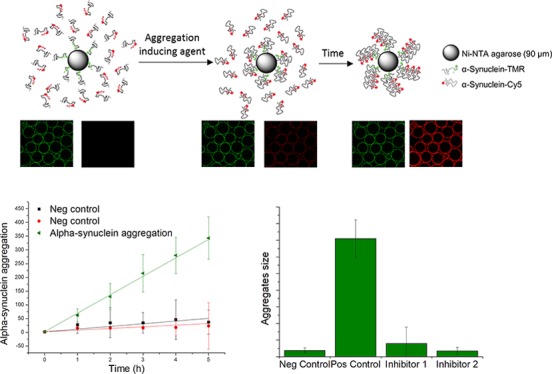
ASYN-CONA is performed on microbeads to which TMR-labeled α-synuclein is attached as described above. Aggregation is induced in the presence of Cy5-labeled α-synuclein in solution (Figure 3). On-bead and aggregated α-synuclein are observed via the fluorescence properties of the different dyes that form a fluorescent halo on the bead surface. The oligomerization is quantified via the increase of the fluorescent intensity of the bead rings.
Figure 3.

ASYN-CONA, a bead-based assay for α-synuclein aggregation. Upper panel shows a schematic representation of the assay. Lower panel shows example images acquired at different stages of aggregation. In the image on the left, α-synuclein labeled with TMR (green) is attached to the beads, which is observed as rings in fluorescence emission channel for TMR. Cy5-labeled α-synuclein is added to the solution but no bead rings are observed in Cy5 fluorescence emission channel (red), because aggregation has not yet started. In the middle image, upon treatment with an aggregation inducing agent, red rings of Cy5-labeled α-synuclein begin to form around the bead. In the image on the right, over time, α-synuclein aggregates increase in size, leading to an increase in the fluorescence emission ring intensity in the Cy5 channel.
α-Synuclein aggregation has often been described as a very heterogeneous process.15 Many aggregating agents have been reported, such as metal ions,38,39 organic solvents,40,41 acidity,42,43 temperature,43 sodium dodecyl sulfate (SDS),44 and liposomes.45 For the ASYN-CONA on-bead aggregation assay, 20% EtOH proved to be the most reliable aggregation inducing agent.41 Various other experimental conditions were tried, but none showed equally consistent results as EtOH. In the presence of DMSO, inhomogeneous bead rings were observed but did not increase in intensity over time (data not shown).
Following the attachment of the TMR-labeled α-synuclein to the microbeads, the aggregation of orthogonally labeled α-synuclein with Cy5 can be monitored.
Beads were conjugated with α-synuclein-A91C-HTM at different concentrations (25, 50, 100, 250, and 500 nM) and examined by confocal scanning microscopy. After this first step, α-synuclein-A91C-Cy5 (500 nM) and EtOH (20%) were added to the bead solutions to induce aggregation of the Cy5-labeled α-synuclein on the TMR-labeled α-synuclein on bead. The aggregation process was followed for 5 h while shaking. Aliquots of the beads were taken every hour and imaged by confocal scanning. The images were taken in two different fluorescent emission channels adapted to the fluorescent properties of Cy5 (ex. 640 nm, em. 690/70 nm) and TMR (ex. 561 nm, em. 586/40 nm); and a bright-field channel (see the Materials and Methods section). The bead images were then analyzed with a custom-developed software known as Bead Ring Evaluation and Analysis of Data (BREAD), for quantification of the bead ring intensities in the different detection channels. BREAD detects the presence and locations of beads in all fluorescent and bright-field channels, and then correlates and combines them. This reduces the risk that insufficiently fluorescent beads are excluded from the analysis. Intensity profiles (in this study, 10) are defined from each bead through the bead diameter, and the peak maxima of the profiles quantified and the bead interior fluorescence subtracted. The average of the profiles is calculated to reveal the individual bead ring intensity. The discrete bead ring intensities of all beads in a well are then averaged to produce the mean bead ring intensity in the well under study. Variable protein loading between beads is corrected via calculation of the ratios between the two fluorescence emission channels on a single-bead basis.
Two control experiments were included. In the first control experiment, the beads lacking preconjugated α-synuclein-A91C-HTM were exposed to the aggregation mix described above, to observe any unspecific interactions of the protein in solution with the beads in the presence of EtOH (Figure 4, black squares, 0 nM). The second control experiment was performed following an identical protocol to the aggregation experiment described above but in the absence of EtOH, to observe any noninduced aggregation processes that might occur (Figure 4, red circles, no EtOH). Both control experiments showed no increase of the mean bead ring intensity of Cy5, compared to the experiments in the presence of both α-synuclein-A91C-HTM and EtOH.
Figure 4.
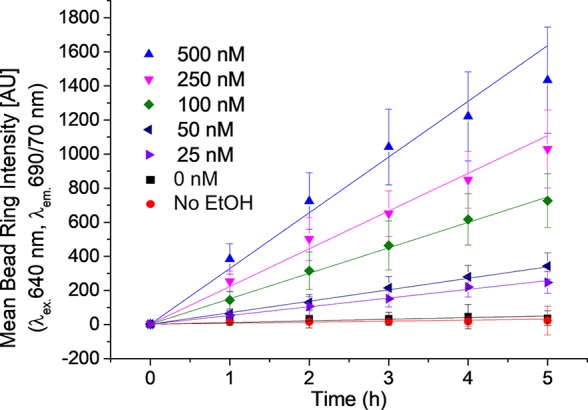
α-Synuclein-A91C-Cy5 aggregates in a linear manner. The plot shows the mean bead ring intensity detected in the Cy5 emission channel, representing the aggregated α-synuclein-A91C-Cy5 on the α-synuclein-HTM conjugated beads. Different colors and symbols represent the different concentrations of α-synuclein-A91C-HTM used to incubate with the beads. The rate of increase of the mean bead ring intensity is dependent on the concentration of the conjugated protein to the bead. Lines represent a linear regression of the data (data shown in Table 1). Six independent replicates of the experiment were performed in total, all of them showing the reproducibility of the assay; for the sake of clarity, only a single repetition of the experiment is shown, with the standard deviation corresponding to the individual experiment. After incubation of the beads with α-synuclein-A91C-HTM (500, 250, 100, 50, and 25 nM) and washing, α-synuclein-A91C-Cy5 (500 nM) and EtOH (20%) were added to the beads and left shaking. Aliquots were taken at time intervals for imaging and quantification.
Quantified bead ring intensities in the different detection channels correlate with the amount of the protein on the bead surface. As expected, the bead ring intensity measured via the fluorescence emission intensity detected in the TMR channel corresponding α-synuclein-A91C-HTM conjugated to beads remains stable during the course of the experiment (Figure S1 in the Supporting Information). Over time, the bead ring intensity measured via the fluorescence emission intensity detected in the Cy5 channel increases linearly, as shown in Figure 4. This increase of bead ring intensity represents the rate of α-synuclein-A91C-Cy5 aggregation onto bead-conjugated α-synuclein-A91C-HTM under the experimental conditions. The apparent rates of α-synuclein-Cy5 aggregation were dependent on the concentration of α-synuclein-HTM conjugated to the beads (see Figure 4, as well as Figure S2 in the Supporting Information) in a concentration range tested between 25 nM and 500 nM of α-synuclein-A91C-HTM. When the ratio between aggregated α-synuclein-A91C-Cy5 and on-bead α-synuclein-A91C-HTM was ≥5:1 (500 nM and 100 nM, respectively), the aggregation reaction followed a linear trend. At ratios of 1:1 or 2:1, the linear dependency began to deviate from linearity, most likely due to the depletion of available α-synuclein-Cy5 in solution. The mean bead intensity of α-synuclein-Cy5 over time was fitted by linear regression, as shown in Table 1.
Table 1. Observed Aggregation Rates of α-Synuclein-A91C-Cy5 (500 nM), as a Function of the Amount of α-Synuclein-A91C-HTM on Beada.
| Intercept
(AU) |
Slope (h–1) |
Statistics | |||
|---|---|---|---|---|---|
| αS-A91C-HTM | value | std er | value | std er | adj. R2 |
| 0 nM | 1.82 | 0.15 | 9.78 | 2.09 | 0.81 |
| 500 nM (no EtOH) | 1.85 | 0.34 | 6.01 | 1.11 | 0.85 |
| 500 nM | 2.19 | 0.36 | 326.94 | 17.56 | 0.99 |
| 250 nM | 2.06 | 0.18 | 221.31 | 8.74 | 0.99 |
| 100 nM | 2.45 | 0.07 | 149.22 | 2.74 | 1.00 |
| 50 nM | 2.31 | 0.08 | 67.24 | 1.68 | 1.00 |
| 25 nM | 2.34 | 0.07 | 51.07 | 1.16 | 1.00 |
Results from the fitting of the mean bead ring intensity in the Cy5 emission channel of data from Figure 4 for the different concentrations of α-synuclein-A91C-HTM incubated with the beads, using y = a + bx as a linear equation. At higher concentrations of α-synuclein-A91C-HTM, the slope of the fit increases, and the adjusted R2 decreases.
When the experiments were performed at 1:1 and 1:2 ratios of protein on bead to protein in solution, the observed aggregation rate is dependent on both the α-synuclein aggregation itself and the depletion of the protein in solution. On the other hand, in systems working at higher ratios, 1:5 to 1:20, the effect of protein depletion on the rate of α-synuclein aggregation rate is less significant, and the aggregation kinetics appear to be linear.
Under the experimental conditions examined here, the simplest model to explain the linear on-bead aggregation requires that each α-synuclein can only bind two other α-synuclein proteins (Figure 5A). In this model, the two potential mechanisms include one in which the interaction site is hampered by the bead on the α-synuclein-A91C-HTM and an alternative in which it is not (Figure 5A(i) and (ii). The characteristic kinetics of formation would differ only by changes in the gradient of the curve (Figure 5B). Any larger number of binding surfaces on the α-synuclein would lead to a characteristic pattern for aggregation corresponding to a geometric series (Figure 5B), which does not match the observed results. A one-to-one binding event cannot be discarded , because it would also match the observed results. Another possible explanation for the linear aggregation would involve unspecific binding of α-synuclein-A91C-Cy5 to the microbeads. However, this model does not correspond with the lack of increase in intensity as a function of time when no α-synuclein-A91C-HTM was attached to the beads (Figure 4, black squares, 0 nM).
Figure 5.

Models of on-bead α-synuclein aggregation: (A) α-synuclein is modeled to have two binding sites and the aggregation is shown as a linear increase of the fluorescence of the Cy5 labeled α-synuclein (bottom left) (i) one of the binding surfaces on α-synuclein-HTM is hampered by the microbead, and (ii) both binding surfaces on α-synuclein-HTM are exposed for aggregation); (B) α-synuclein is assumed to have three binding sites and the aggregation is shown as an increase in fluorescence intensity, following a geometric series (bottom right). The graph shown as an inset in panel (B) is set to the same scale as panel (A), for the sake of comparison. [Red features represent Cy-5-labeled α-synuclein, and green features represent HTM-labeled α-synuclein.]
In summary, the observed linear increase of the aggregated protein, ratios of ≥5:1 of in solution to on-bead α-synuclein, indicates that the mechanism of early aggregation under these experimental conditions might be more homogeneous and controlled than often assumed. Further characterization of the species formed on-bead is required to better understand the oligomerization mechanism detected under these experimental conditions.
The comparison with previously published results such as Munishkina et al.41 represents a challenge due to the different experimental conditions of the two systems. The results described by Munishkina are based on the increase of fluorescence of ThT upon binding to α-synuclein fibrils. It shows that the aggregation follows a sigmodal curve with a short lag phase in the presence of 20% EtOH. The 5 h time frame of the assay described here encompasses the lag phase and the beginning of the increase of fluorescence in the ThT, which correspond to the early aggregation of α-synuclein. However, care must be taken when comparing the two results, since different experimental conditions, such as concentration and temperature, among others, change the aggregation properties substantially.15
Two other α-synuclein mutants, V3C and I112C, were tested using the same assay as for A91C. Both α-synuclein mutants showed a linear aggregation process (data not shown), but with slightly different rates. The changes in the rate could be due to different aggregation propensities of the mutants, the relative position of the dyes, or other factors. However, the linearity observed is unaffected by the position of the mutation. The effect of position of the mutations is under further study and will be part of future research.
Imaging of Oligomers by Single-Molecule Total Internal Reflection Fluorescence (TIRF) Microscopy
Single-molecule total internal reflection fluorescence (TIRF) microscopy has previously been used to study the aggregation of α-synuclein.31,46,47 Utilizing the different fluorescent wavelengths of the fluorophores used in the assay (TMR and Cy5), it is possible to measure their oligomerization by observing the coincidence of the two α-synuclein species. In this study, aggregated on-bead α-synuclein (both HTM and Cy5 labeled) was eluted and imaged (Figures 6A–C). The coincidence between TMR (λex = 561 nm) and Cy5 (λex = 638 nm) indicates the presence of an oligomer, which are visible as yellow spots in Figure 6B and are highlighted in Figures 6D–F, in which only the coincident spots are shown. The association quotient, Q (see data analysis in the Supporting Information) is a measure of the level of coincidence of the two species. For these samples, Q = 0.090 ± 0.007 (mean ± S.D., n = 3), indicating that 9% of the Cy5 species detected were within a dual-labeled oligomer. By measuring the total brightness in each channel and comparing this with the mean brightness of the individual TMR or Cy5 fluorophores, the number of monomer units of each α-synuclein species in the oligomers can be calculated. The result from this calculation is a 6 ± 1 ratio of Cy5 to TMR monomer units, shown in the contour plot in Figure 6G. The sizes and natural logarithms of the stoichiometries of the detected oligomers are represented in the histograms in Figure 6H and Figure S3 in the Supporting Information, respectively.
Figure 6.
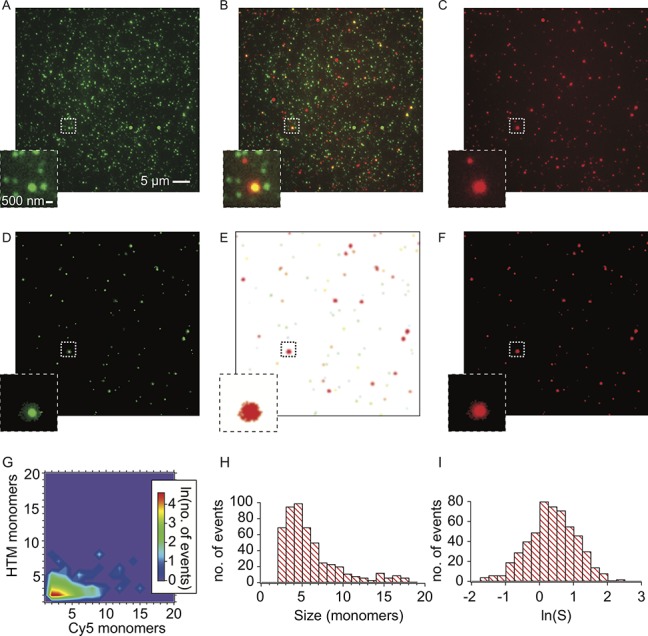
Single-molecule TIRF microscopy images of α-synuclein released from the beads. Following incubation with imidazole, the α-synuclein attached to the surface of the beads was eluted and imaged at the single-molecule level. The HTM labeled α-synuclein and Cy5 labeled α-synuclein in the same field of view are shown in panels (A) and (C), respectively. The two channels are merged to give panel (B), in which coincident spots corresponding to oligomers are clearly visible (shown in yellow). Panels (D) and (E) show the same fields of view in which only the coincident spots are shown in the HTM and Cy5 channels, respectively. Panel (E) shows a map of the oligomers detected. (G) Two-dimensional (2D) contour plot showing the number of Cy5 monomers and HTM monomers present in each detected oligomer. (H) Size histogram of all the detected oligomers. (I) Histogram of the natural logarithms of the stoichiometries of the oligomers S = size(Cy5)/size(TMR).
As described previously, ThT is able to bind to amyloid structures, leading to an increase in its fluorescence intensity by several orders of magnitude, making it an unusually sensitive and efficient reporter of extended β-sheet structure. It has previously been used with TIRF microscopy to detect individual aggregates within biofluids (single aggregate visualization by enhancement (SAVE) imaging).31 In order to gain some insights into the secondary structures of the on-bead oligomers, ThT was used to determine whether any of the oligomers contained extended β-sheet structure. The majority of oligomers detected were not ThT-active, indicating that they are early aggregates in the fibril-formation pathway, as expected from the assay conditions. In 1.8 ± 0.9% (mean ± S.D., n = 3) of the cases, however, there was enough extended β-sheet structure for the oligomers to be detected using ThT (see Figure S3 in the Supporting Information), highlighting that some of the oligomers have become more fibril-like.17,31
α-Synuclein Aggregation Inhibitors
The objective of this work was to develop an assay that (a) would allow monitoring of the earliest stages of oligomerization of α-synuclein, rather than later stage aggregation, and (b) is suitable for drug screening. The ASYN-CONA assay has demonstrated novel mechanistic insights into a so far undescribed oligomerization event. Given that the assay is reasonably miniaturized and fast, it was concluded that, in a next step, it could be investigated as a screening platform to find novel α-synuclein aggregation inhibitors. In order to validate ASYN-CONA as a screening method, several known inhibitors of α-synuclein aggregation were tested. Selegiline,48 curcumin,49,50 rifampicin,49,51 dopamine,52,53 baicalein,54,55 and nicotine56,57 were the natural products chosen to investigate their inhibitory properties under the assay conditions described above. The on-bead aggregation experiment was performed in the same way as previously described with the compounds dissolved in EtOH and added to the solution containing the beads with conjugated α-synuclein-A91C-HTM and in solution α-synuclein-A91C-Cy5. The aggregation of α-synuclein in the presence of the inhibitors was followed as described above.
Figure 7 shows the mean bead ring intensity and the ratio, representing the amount of aggregated α-synuclein-A91C-Cy5 onto on-bead α-synuclein-A91C-HTM after 5 h of incubation with inhibitors. At a compound concentration of 50 μM, only curcumin, baicalein, and rifampicin showed significant inhibition of α-synuclein aggregation. Most of the natural products used in the assay have been described in the literature to induce the formation of small nontoxic α-synuclein oligomers. α-Synuclein aggregation has also been described as being very dependent on the experimental conditions. The lack of an inhibitory effect of selegiline, dopamine, and nicotine, compared to other published results, could be due to the detection of early small oligomers with the ASYN-CONA assay, rather than the detection of larger oligomers common in other assays. Dopamine, selegiline, and nicotine may have more inhibitory properties on an alternative aggregation pathway than that observed on bead.
Figure 7.

Activity of α-synuclein aggregation inhibitors assessed using ASYN-CONA: (A) mean bead ring intensity quantified from the Cy5 fluorescence detection channel and (B) ratio of the Cy5 fluorescence intensity to the TMR fluorescence intensity. First, Ni-NTA agarose beads were incubated with α-synuclein-A91C-HTM (100 nM), shaken for 20 min at 22 °C, and washed. α-Synuclein-A91C-Cy5 (500 nM) and the inhibitor (50 μM) in EtOH (20%) were added to the solution and the beads incubated for 5 h. One aliquot was taken after 5 h for imaging and fluorescence intensity quantification. Three controls with, respectively, no on-bead α-synuclein-A91C-HTM, no EtOH, or no inhibitor present in the reaction sample were also included in the assay. All experiments were performed in duplicate, and the full experiment was repeated twice. The bars in graphs represent the average weighted by the number of beads of the four repetitions.
As a next step, the concentration dependence of the inhibitory activity of the three active compounds (baicalein, curcumin, and rifampicin) was assessed. The experiments were performed as described above. In brief, beads with conjugated α-synuclein-A91C-HTM were incubated with α-synuclein-A91C-Cy5 to which either baicalein, curcumin, or rifampicin dissolved in EtOH was added at different concentrations. All three compounds showed concentration dependent inhibition of aggregation (Figure 8). Using the equation
for curve fitting, where y is the observed mean bead ring intensity, Imax and Imin are the maximum and minimum mean bead ring intensities, respectively, IC50 is the concentration of the compound at which Imax was reduced by half, n is the slope of the fit, and x is the compound concentration, the IC50 for curcumin was determined to be 2.76 ± 0.89 μM, the IC50 for baicalein was 4.32 ± 0.60 μM, and the IC50 for rifampicin was 502.32 ± 360.36 μM.
Figure 8.
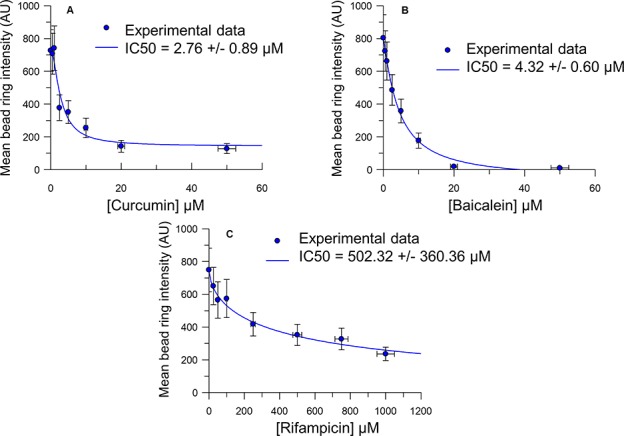
Baicalein, curcumin, and rifampicin inhibit aggregation of α-synuclein in a concentration-dependent manner, as determined using ASYN-CONA. Mean bead ring intensity in the Cy5 fluorescence emission channel detected after 5 h of incubation in the presence of different concentrations of curcumin, baicalein, and rifampicin. First, Ni-NTA agarose beads were incubated with α-synuclein-A91C-HTM (100 nM), shaken for 20 min at 22 °C, and washed. α-Synuclein-A91C-Cy5 (500 nM) and the inhibitor (baicalein, curcumin, or rifampicin), at different concentrations in EtOH (20%), were added to the solution and the beads were incubated for 5 h. One aliquot (20 μL) was taken after 5 h, for imaging and quantification. Three controls with, respectively, no on-bead α-synuclein-A91C-HTM, no EtOH, and no inhibitor were also included in the assay. The data show the average intensity values weighted by the number of beads of six repetitions. The blue lines represents the fit to the full four-parameter equation for IC50 determination, y = (Imax – Imin)/[1 + (x/IC50)n] + Imin.
Acknowledgments
The authors thank Prof. Chris Rochet for kindly providing the plasmid of α-synuclein-A91C. M.A. acknowledges financial support from the Scottish Universities Life Sciences Alliance ((SULSA), http://www.sulsa.ac.uk), the Medical Research Council ((MRC), www.mrc.ac.uk, No. J54359) Strategic Grant. T.K. acknowledges funding support from Parkinson’s UK (No. F-0902). I.P. acknowledges financial support from the MSD Scottish Life Sciences fund. M.H. acknowledges funding from UCB to build the total internal reflection fluorescence microscope (https://www.ucb.com), N.P acknowledges financial support from the Wellcome Trust (Grant 201531/Z/16/Z). The opinions expressed in this research are those of the authors and do not represent those of MSD, nor its affiliates.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b03842.
Supplementary figures covering the mean bead ring intensity of α-synuclein-A91C-HTM over time (Figure S1), and the derived apparent aggregation rate of α-synuclein-A91C-Cy5, as a function of α-synuclein-A91C-HTM on bead (Figure S2); more-detailed information covering experimental procedures for HPLC-MS, protein expression and purification, synthesis of HTM and protein labeling; details of the single-molecule data analysis, together with a figure showing ThT active oligomers (PDF)
Author Present Address
∇ Optos Plc, Carnegie Campus, Enterprise Way, Dunfermline, KY11 8GR, U.K. E-mail: d.a.evans@me.com.
Author Contributions
I.P. coinvented and codeveloped the assay, codesigned, performed and analyzed the experiments and wrote the manuscript; N.P. contributed to confocal microscopy experiments and data fitting; D.E. created the BREAD software for data analysis and supported analysis of the results; J.K. supported assay development and data analysis; K.D. contributed to development of the α-synuclein mutation used in this work; M.H. performed analysis of oligomers by TIRF; T.K. coinvented the assay, provided plasmids; M.A. developed the concept of the ASYN-CONA assay, coinvented the assay and wrote the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Villar-Piqué A.; Lopes da Fonseca T.; Outeiro T. F. Structure, function and toxicity of alpha-synuclein: the Bermuda triangle in synucleinopathies. J. Neurochem. 2016, 139, 240–255. 10.1111/jnc.13249. [DOI] [PubMed] [Google Scholar]
- Eliezer D.; Kutluay E.; Bussell R. Jr; Browne G. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001, 307 (4), 1061–1073. 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- Rodriguez J. A.; Ivanova M. I.; Sawaya M. R.; Cascio D.; Reyes F. E.; Shi D.; Sangwan S.; Guenther E. L.; Johnson L. M.; Zhang M.; Jiang L.; Arbing M. A.; Nannenga B. L.; Hattne J.; Whitelegge J.; Brewster A. S.; Messerschmidt M.; Boutet S.; Sauter N. K.; Gonen T.; Eisenberg D. S. Structure of the toxic core of α-synuclein from invisible crystals. Nature 2015, 525, 486. 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle M. D.; Comellas G.; Nieuwkoop A. J.; Covell D. J.; Berthold D. A.; Kloepper K. D.; Courtney J. M.; Kim J. K.; Barclay A. M.; Kendall A.; Wan W.; Stubbs G.; Schwieters C. D.; Lee V. M. Y.; George J. M.; Rienstra C. M. Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat. Struct. Mol. Biol. 2016, 23 (5), 409–415. 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb P. H.; Zhen W.; Poon A. W.; Conway K. A.; Lansbury P. T. NACP, A Protein Implicated in Alzheimer’s Disease and Learning, Is Natively Unfolded. Biochemistry 1996, 35 (43), 13709–13715. 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- Burré J.; Vivona S.; Diao J.; Sharma M.; Brunger A. T.; Südhof T. C. Properties of native brain α-synuclein. Nature 2013, 498 (7453), E4–E6. 10.1038/nature12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet F.-X.; Binolfi A.; Bekei B.; Martorana A.; Rose H. M.; Stuiver M.; Verzini S.; Lorenz D.; van Rossum M.; Goldfarb D.; Selenko P. Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature 2016, 530 (7588), 45–50. 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- Gould N.; Mor D.; Lightfoot R.; Malkus K.; Giasson B.; Ischiropoulos H. Evidence of Native α-Synuclein Conformers in the Human Brain. J. Biol. Chem. 2014, 289, 7929. 10.1074/jbc.C113.538249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breydo L.; Wu J. W.; Uversky V. N. α-Synuclein misfolding and Parkinson’s disease. Biochim. Biophys. Acta, Mol. Basis Dis. 2012, 1822 (2), 261–285. 10.1016/j.bbadis.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Guerrero-Ferreira R.; Taylor N. M. I.; Mona D.; Ringler P.; Lauer M. E.; Riek R.; Britschgi M.; Stahlberg H. Cryo-EM structure of alpha-synuclein fibrils. eLife 2018, 7, e36402 10.7554/eLife.36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D.; Mehra S.; Sahay S.; Singh P. K.; Maji S. K. α-synuclein aggregation and its modulation. Int. J. Biol. Macromol. 2017, 100, 37. 10.1016/j.ijbiomac.2016.10.021. [DOI] [PubMed] [Google Scholar]
- Spillantini M. G.; Schmidt M. L.; Lee V. M. Y.; Trojanowski J. Q.; Jakes R.; Goedert M. Alpha-Synuclein in Lewy bodies. Nature 1997, 388 (6645), 839–840. 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Conway K. A.; Lee S.-J.; Rochet J.-C.; Ding T. T.; Williamson R. E.; Lansbury P. T. Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early-onset Parkinson’s disease: Implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (2), 571–576. 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner B.; Jappelli R.; Maji S. K.; Desplats P. A.; Boyer L.; Aigner S.; Hetzer C.; Loher T.; Vilar M.; Campioni S.; Tzitzilonis C.; Soragni A.; Jessberger S.; Mira H.; Consiglio A.; Pham E.; Masliah E.; Gage F. H.; Riek R. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (10), 4194–4199. 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Diggelen F.; Tepper A. W. J. W.; Apetri M. M.; Otzen D. E. α-Synuclein Oligomers: A Study in Diversity. Isr. J. Chem. 2017, 57, 699. 10.1002/ijch.201600116. [DOI] [Google Scholar]
- Kaylor J.; Bodner N.; Edridge S.; Yamin G.; Hong D.-P.; Fink A. L. Characterization of Oligomeric Intermediates in α-Synuclein Fibrillation: FRET Studies of Y125W/Y133F/Y136F α-Synuclein. J. Mol. Biol. 2005, 353 (2), 357–372. 10.1016/j.jmb.2005.08.046. [DOI] [PubMed] [Google Scholar]
- Cremades N.; Cohen S. I. A.; Deas E.; Abramov A. Y.; Chen A. Y.; Orte A.; Sandal M.; Clarke R. W.; Dunne P.; Aprile F. A.; Bertoncini C. W.; Wood N. W.; Knowles T. P. J.; Dobson C. M.; Klenerman D. Direct Observation of the Interconversion of Normal and Toxic Forms of α-Synuclein. Cell 2012, 149 (5), 1048–1059. 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosatto L.; Horrocks M. H.; Dear A. J.; Knowles T. P. J.; Dalla Serra M.; Cremades N.; Dobson C. M.; Klenerman D. Single-molecule FRET studies on alpha-synuclein oligomerization of Parkinson’s disease genetically related mutants. Sci. Rep. 2015, 5, 16696. 10.1038/srep16696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese A.; Bader B.; Bieschke J.; Schaffar G.; Odoy S.; Kahle P. J.; Haass C.; Kretzschmar H. Single particle detection and characterization of synuclein co-aggregation. Biochem. Biophys. Res. Commun. 2005, 333 (4), 1202–1210. 10.1016/j.bbrc.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Iljina M.; Garcia G. A.; Horrocks M. H.; Tosatto L.; Choi M. L.; Ganzinger K. A.; Abramov A. Y.; Gandhi S.; Wood N. W.; Cremades N.; Dobson C. M.; Knowles T. P. J.; Klenerman D. Kinetic model of the aggregation of alpha-synuclein provides insights into prion-like spreading. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (9), E1206–E1215. 10.1073/pnas.1524128113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela J. A.; Rodrigues M.; De S.; Flagmeier P.; Gandhi S.; Dobson C. M.; Klenerman D.; Lee S. F. Optical Structural Analysis of Individual α-Synuclein Oligomers. Angew. Chem., Int. Ed. 2018, 57 (18), 4886–4890. 10.1002/anie.201710779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintersteiner M.; Ambrus G.; Bednenko J.; Schmied M.; Knox A. J. S.; Meisner N.-C.; Gstach H.; Seifert J.-M.; Singer E. L.; Gerace L.; Auer M. Identification of a Small Molecule Inhibitor of Importin β Mediated Nuclear Import by Confocal On-Bead Screening of Tagged One-Bead One-Compound Libraries. ACS Chem. Biol. 2010, 5 (10), 967–979. 10.1021/cb100094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintersteiner M.; Kallen J.; Schmied M.; Graf C.; Jung T.; Mudd G.; Shave S.; Gstach H.; Auer M. Identification and X-ray Co-crystal Structure of a Small-Molecule Activator of LFA-1-ICAM-1 Binding. Angew. Chem., Int. Ed. 2014, 53 (17), 4322–4326. 10.1002/anie.201310240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H.; Prodinger W. M.; Kopeček J. Two-Step Fluorescence Screening of CD21-Binding Peptides with One-Bead One-Compound Library and Investigation of Binding Properties of N-(2-Hydroxypropyl)methacrylamide Copolymer–Peptide Conjugates. Biomacromolecules 2006, 7 (11), 3037–3046. 10.1021/bm060508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivos H. J.; Bachhawat-Sikder K.; Kodadek T. Quantum Dots As A Visual Aid For Screening Bead-Bound Combinatorial Libraries. ChemBioChem 2003, 4 (11), 1242–1245. 10.1002/cbic.200300712. [DOI] [PubMed] [Google Scholar]
- Hintersteiner M.; Auer M. Single-bead, Single-molecule, Single-cell Fluorescence. Ann. N. Y. Acad. Sci. 2008, 1130 (1), 1–11. 10.1196/annals.1430.055. [DOI] [PubMed] [Google Scholar]
- Hintersteiner M.; Buehler C.; Uhl V.; Schmied M.; Müller J.; Kottig K.; Auer M. Confocal Nanoscanning, Bead Picking (CONA): PickoScreen Microscopes for Automated and Quantitative Screening of One-Bead One-Compound Libraries. J. Comb. Chem. 2009, 11 (5), 886–894. 10.1021/cc900059q. [DOI] [PubMed] [Google Scholar]
- Hintersteiner M.; Buehler C.; Auer M. On-Bead Screens Sample Narrower Affinity Ranges of Protein–Ligand Interactions Compared to Equivalent Solution Assays. ChemPhysChem 2012, 13 (15), 3472–3480. 10.1002/cphc.201200117. [DOI] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S.; Saalfeld S.; Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25 (11), 1463–1465. 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks M. H.; Lee S. F.; Gandhi S.; Magdalinou N. K.; Chen S. W.; Devine M. J.; Tosatto L.; Kjaergaard M.; Beckwith J. S.; Zetterberg H.; Iljina M.; Cremades N.; Dobson C. M.; Wood N. W.; Klenerman D. Single-Molecule Imaging of Individual Amyloid Protein Aggregates in Human Biofluids. ACS Chem. Neurosci. 2016, 7 (3), 399–406. 10.1021/acschemneuro.5b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C. M.; Wissner R. F.; Warner J. B.; Wang Y. J.; Ferrie J. J.; Covell D. J.; Karpowicz R. J.; Lee V. M. Y.; James Petersson E. Comparison of strategies for non-perturbing labeling of alpha-synuclein to study amyloidogenesis. Org. Biomol. Chem. 2016, 14 (5), 1584–1592. 10.1039/C5OB02329G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd G.; Pérez Pi I.; Fethers N.; Dodd P. G.; Barbeau O. R.; Auer M. A general synthetic route to isomerically pure functionalized rhodamine dyes. Methods Appl. Fluoresc. 2015, 3 (4), 045002. 10.1088/2050-6120/3/4/045002. [DOI] [PubMed] [Google Scholar]
- Thirunavukkuarasu S.; Jares-Erijman E. A.; Jovin T. M. Multiparametric Fluorescence Detection of Early Stages in the Amyloid Protein Aggregation of Pyrene-labeled α-Synuclein. J. Mol. Biol. 2008, 378 (5), 1064–1073. 10.1016/j.jmb.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Gallea J. I.; Celej M. S. Structural insights into amyloid oligomers of the Parkinson disease related protein α-synuclein. J. Biol. Chem. 2014, 289, 26733. 10.1074/jbc.M114.566695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinotsi D.; Buell A. K.; Galvagnion C.; Dobson C. M.; Kaminski Schierle G. S.; Kaminski C. F. Direct Observation of Heterogeneous Amyloid Fibril Growth Kinetics via Two-Color Super-Resolution Microscopy. Nano Lett. 2014, 14 (1), 339–345. 10.1021/nl4041093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z.; Krasnoslobodtsev A. V.; Zhang Y.; Ysselstein D.; Rochet J.-C.; Blanchard S. C.; Lyubchenko Y. L. Direct Detection of a-Synuclein Dimerization Dynamics: Single-Molecule Fluorescence Analysis. Biophys. J. 2015, 108, 2038–2047. 10.1016/j.bpj.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka M.; Högen T.; Danzer K. M.; Levin J.; Habeck M.; Wirth A.; Wagner R.; Glabe C. G.; Finger S.; Heinzelmann U.; Garidel P.; Duan W.; Ross C. A.; Kretzschmar H.; Giese A. Single Particle Characterization of Iron-induced Pore-forming α-Synuclein Oligomers. J. Biol. Chem. 2008, 283 (16), 10992–11003. 10.1074/jbc.M709634200. [DOI] [PubMed] [Google Scholar]
- Uversky V. N.; Li J.; Fink A. L. Metal-triggered Structural Transformations, Aggregation, and Fibrillation of Human α-Synuclein: A possible molecular link between Parkinson’s disease and heavy metal exposure. J. Biol. Chem. 2001, 276 (47), 44284–44296. 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- Wagner J.; Ryazanov S.; Leonov A.; Levin J.; Shi S.; Schmidt F.; Prix C.; Pan-Montojo F.; Bertsch U.; Mitteregger-Kretzschmar G.; Geissen M.; Eiden M.; Leidel F.; Hirschberger T.; Deeg A.; Krauth J.; Zinth W.; Tavan P.; Pilger J.; Zweckstetter M.; Frank T.; Bähr M.; Weishaupt J.; Uhr M.; Urlaub H.; Teichmann U.; Samwer M.; Bötzel K.; Groschup M.; Kretzschmar H.; Griesinger C.; Giese A. Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathol. 2013, 125 (6), 795–813. 10.1007/s00401-013-1114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munishkina L. A.; Phelan C.; Uversky V. N.; Fink A. L. Conformational Behavior and Aggregation of α-Synuclein in Organic Solvents: Modeling the Effects of Membranes. Biochemistry 2003, 42 (9), 2720–2730. 10.1021/bi027166s. [DOI] [PubMed] [Google Scholar]
- Saraiva M. A.; Jorge C. D.; Santos H.; Maçanita A. L. Earliest events in α-synuclein fibrillation probed with the fluorescence of intrinsic tyrosines. J. Photochem. Photobiol., B 2016, 154, 16–23. 10.1016/j.jphotobiol.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Uversky V. N.; Li J.; Fink A. L. Evidence for a Partially Folded Intermediate in α-Synuclein Fibril Formation. J. Biol. Chem. 2001, 276 (14), 10737–10744. 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- Giehm L.; Oliveira C. L. P.; Christiansen G.; Pedersen J. S.; Otzen D. E. SDS-Induced Fibrillation of α-Synuclein: An Alternative Fibrillation Pathway. J. Mol. Biol. 2010, 401 (1), 115–133. 10.1016/j.jmb.2010.05.060. [DOI] [PubMed] [Google Scholar]
- Wrasidlo W.; Tsigelny I. F.; Price D. L.; Dutta G.; Rockenstein E.; Schwarz T. C.; Ledolter K.; Bonhaus D.; Paulino A.; Eleuteri S.; Skjevik Å. A.; Kouznetsova V. L.; Spencer B.; Desplats P.; Gonzalez-Ruelas T.; Trejo-Morales M.; Overk C. R.; Winter S.; Zhu C.; Chesselet M.-F.; Meier D.; Moessler H.; Konrat R.; Masliah E. A de novo compound targeting alpha-synuclein improves deficits in models of Parkinson’s disease. Brain 2016, 139, 3217. 10.1093/brain/aww238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiten D. R.; Zuo Y.; Calo L.; Choi M.-L.; De S.; Flagmeier P.; Wirthensohn D. C.; Kundel F.; Ranasinghe R. T.; Sanchez S. E.; Athauda D.; Lee S. F.; Dobson C. M.; Gandhi S.; Spillantini M.-G.; Klenerman D.; Horrocks M. H. Nanoscopic Characterisation of Individual Endogenous Protein Aggregates in Human Neuronal Cells. ChemBioChem 2018, 19 (19), 2033–2038. 10.1002/cbic.201800209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-E.; Sang J. C.; Rodrigues M.; Carr A. R.; Horrocks M. H.; De S.; Bongiovanni M. N.; Flagmeier P.; Dobson C. M.; Wales D. J.; Lee S. F.; Klenerman D. Mapping Surface Hydrophobicity of α-Synuclein Oligomers at the Nanoscale. Nano Lett. 2018, 18 (12), 7494–7501. 10.1021/acs.nanolett.8b02916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga C. A.; Follmer C.; Palhano F. L.; Khattar E.; Freitas M. S.; Romão L.; Di Giovanni S.; Lashuel H. A.; Silva J. L.; Foguel D. The Anti-Parkinsonian Drug Selegiline Delays the Nucleation Phase of α-Synuclein Aggregation Leading to the Formation of Nontoxic Species. J. Mol. Biol. 2011, 405 (1), 254–273. 10.1016/j.jmb.2010.10.027. [DOI] [PubMed] [Google Scholar]
- Ono K.; Yamada M. Antioxidant compounds have potent anti-fibrillogenic and fibril-destabilizing effects for α-synuclein fibrils in vitro. J. Neurochem. 2006, 97 (1), 105–115. 10.1111/j.1471-4159.2006.03707.x. [DOI] [PubMed] [Google Scholar]
- Pandey N.; Strider J.; Nolan W. C.; Yan S. X.; Galvin J. E. Curcumin inhibits aggregation of α-synuclein. Acta Neuropathol. 2008, 115 (4), 479–489. 10.1007/s00401-007-0332-4. [DOI] [PubMed] [Google Scholar]
- Li J.; Zhu M.; Rajamani S.; Uversky V. N.; Fink A. L. Rifampicin Inhibits α-Synuclein Fibrillation and Disaggregates Fibrils. Chem. Biol. 2004, 11 (11), 1513–1521. 10.1016/j.chembiol.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Li H.-T.; Lin D.-H.; Luo X.-Y.; Zhang F.; Ji L.-N.; Du H.-N.; Song G.-Q.; Hu J.; Zhou J.-W.; Hu H.-Y. Inhibition of α-synuclein fibrillization by dopamine analogs via reaction with the amino groups of α-synuclein. FEBS J. 2005, 272 (14), 3661–3672. 10.1111/j.1742-4658.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- Conway K. A.; Rochet J.-C.; Bieganski R. M.; Lansbury P. T. Kinetic Stabilization of the α-Synuclein Protofibril by a Dopamine-α-Synuclein Adduct. Science 2001, 294 (5545), 1346–1349. 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- Zhu M.; Rajamani S.; Kaylor J.; Han S.; Zhou F.; Fink A. L. The Flavonoid Baicalein Inhibits Fibrillation of α-Synuclein and Disaggregates Existing Fibrils. J. Biol. Chem. 2004, 279 (26), 26846–26857. 10.1074/jbc.M403129200. [DOI] [PubMed] [Google Scholar]
- Lu J.-H.; Ardah M. T.; Durairajan S. S. K.; Liu L.-F.; Xie L.-X.; Fong W.-F. D.; Hasan M. Y.; Huang J.-D.; El-Agnaf O. M. A.; Li M. Baicalein Inhibits Formation of α-Synuclein Oligomers within Living Cells and Prevents Aβ Peptide Fibrillation and Oligomerisation. ChemBioChem 2011, 12 (4), 615–624. 10.1002/cbic.201000604. [DOI] [PubMed] [Google Scholar]
- Ono K.; Hirohata M.; Yamada M. Anti-fibrillogenic and fibril-destabilizing activity of nicotine in vitro: Implications for the prevention and therapeutics of Lewy body diseases. Exp. Neurol. 2007, 205 (2), 414–424. 10.1016/j.expneurol.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Hong D.-P.; Fink A. L.; Uversky V. N. Smoking and Parkinson’s disease: Does nicotine affect α-synuclein fibrillation?. Biochim. Biophys. Acta, Proteins Proteomics 2009, 1794 (2), 282–290. 10.1016/j.bbapap.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


