Abstract
Measuring and modeling the integrated behavior of biomolecular-cellular networks is central to systems biology. Over several decades, systems biology has been shaped by quantitative biologists, physicists, mathematicians, and engineers in different ways. However, the basic and applied versions of systems biology are not typically distinguished, which blurs the separate aspirations of the field and its potential for real-world impact. Here, we articulate an engineering approach to systems biology, which applies educational philosophy, engineering design, and predictive models to solve contemporary problems in an age of biomedical Big Data. A concerted effort to train systems bioengineers will provide a versatile workforce capable of tackling the diverse challenges faced by the biotechnological and pharmaceutical sectors in a modern, information-dense economy.
Introduction
The term “systems biology” is now ~15 years old,1 long enough for it to be projected onto biochemistry,2 pharmacology,3 genetics,4 and other areas of basic science, as well as federal funding agencies (Table 1). Systems biology has also influenced applied disciplines such as engineering,5 and an engineer’s view of systems biology is distinct from those who approach the field from chemistry, mathematics, or physics. The intersection of engineering research and systems biology has given rise to a type of “systems bioengineering” that tightly integrates predictive modeling, engineering design, and quantitative analysis with the molecular mechanisms of cellular pathways. Despite interesting similarities and key differences between basic systems biology and applied systems bioengineering, we are not aware of any prior literature that directly appraises one separately from the other. Conflation of basic and applied perspectives is perhaps why there remains confusion about what “systems biology” truly means to education and academic-industrial research.
Table 1.
Various definitions of systems biology.
| Source | Quoted definition | Reference |
|---|---|---|
| Ideker, Galitski, and Hood | Systems biology does not investigate individual genes or proteins one at a time, as has been the highly successful mode of biology for the past 30 years. Rather, it investigates the behavior and relationships of all of the elements in a particular biological system while it is functioning. | Ideker et al., 20011 |
| Wikipedia | A biology-based inter-disciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach (holism instead of the more traditional reductionism) to biological research. | https://en.wikipedia.org/wiki/Systems_biology |
| National Institute of General Medical Sciences | An integrated approach in biomedical research using physical, computational and experimental sciences to understand complex biological systems. | https://www.nigms.nih.gov/Research/SpecificAreas/SysBio/Pages/default.aspx |
| National Cancer Institute | A systems biology approach to cancer research […] includes explicit integration of experimental biology and computational modeling to test and validate novel hypotheses or ideas. | https://grants.nih.gov/grants/guide/rfa-files/RFA-CA-15-014.html |
| National Institute of Allergy & Infectious Disease | Develop and validate predictive models of infectious disease initiation, progression, and outcomes […] derived from the study of the architecture and dynamics of systems-wide host/pathogen molecular interaction networks during infection, using integrated datasets generated from a combination of “omics” technologies. | https://www.niaid.nih.gov/research/systems-biology-infectious-diseases-research |
| National Science Foundation | A comprehensive understanding of emergent properties of biological systems through the development of an integrated theoretical framework that is guided by mathematical and physical principles and facilitated through the use of novel tools | https://www.nsf.gov/funding/pgm_summ.jsp?pims_id=504863 |
Here, we summarize the elements of engineering science that have shaped our view of systems biology and its application to fundamental knowledge, bioremediation, and human health. Already, systems bioengineering has begun to establish a core set of principles for problem solving that fuses experiments and computation.6 These principles have moved beyond the proof-of-concept stage and are ready to be tested in a real-world setting. In parallel, there is a rich opportunity to use systems bioengineering as an organizing principle for educational programs seeking an equal emphasis on science, technology, engineering, and math. To date, modern systems bioengineers are difficult to pigeonhole within the biotechnology and pharmaceutical industries, but they are poised to succeed at interfaces long recognized as barriers, such as drug-to-market failure. A long-term, engineering-themed vision of systems biology should have a sizeable impact on science, education, industry, and broader society if implemented in a cohesive way.
Combining systems biology and engineering
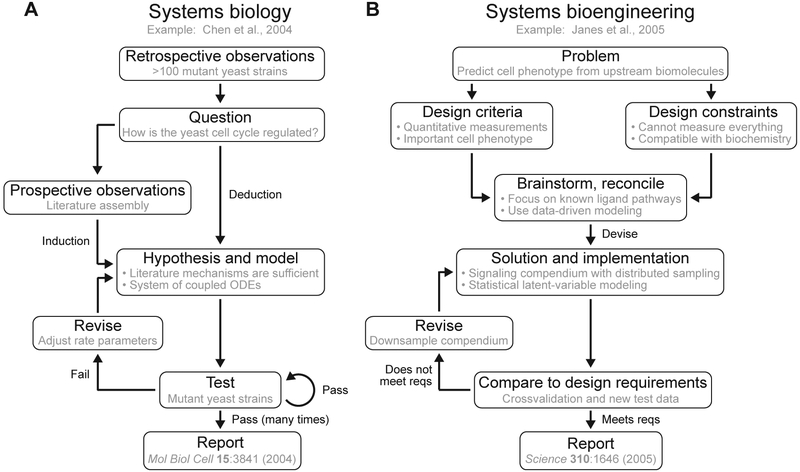
Non-engineering approaches to systems biology strongly embody the scientific method and its emphasis on generating, testing, and revising hypotheses (Fig. 1A).7 To a physicist or mathematician, a laudable goal is to derive a quantitative theory or evolutionary basis for a complex biological phenomenon, and recent examples nicely illustrate that simple principles can be found.8-10 Alternatively, those driven by technology (-omicists) or algorithms (computer scientists) favor induction to generate hypotheses after collecting and analyzing large sets of observations. Inductive systems biology is quite effective for discovery science and for synthesizing descriptions of datasets that would otherwise be intractable.11-13 Basic inductive and deductive approaches excel at framing network- or module-level questions and hypotheses about function, but the overall scope of systems biology is larger than that.
Fig. 1.
Workflows for systems biology and systems bioengineering. (A) The scientific method applied to systems biology. (B) Engineering design approach applied to systems bioengineering. The gray text provides example case studies7,14 selected from Arkin and Schaffer, 2011.15
In contrast to basic scientists, -omicists, and computer scientists, engineers design solutions according to specific problem statements, success criteria, and practical constraints. Effective solutions are synthesized from a core set of skills, which serve as the foundation for the engineering discipline. Real-world engineering problems occur with a massive number of components—suspension bridges, microprocessors, refineries—requiring integration of core principles in different combinations and at multiple scales. This integration must occur at the systems level, whereby heterogeneous inputs and outputs come together to yield a solution that is greater than the sum of its parts.6
The general paradigm of engineering-based problem solving has many parallels in systems biology, even though the details of its implementation differ in some key areas (Fig. 1B).14, 15 For example, to improve upon the design of the nose cone of an airplane, aerospace engineers can, in principle, consider any material available and be confident that the thermal properties of the material will be captured and predicted by Fourier’s Law, which states that heat flux is proportional to the temperature gradient within the material. Systems bioengineers, by contrast, are more constrained in their choice of materials, but they are freer to decide on the theoretical abstraction relative to its application, much like physicists do in the context of a defined physical system. For instance, an extracellular ligand may only bind to and signal via its cognate receptor in biology, but such autocrine-paracrine signaling can be modeled in space and time by multiple formalisms depending on the problem statement.16 By shifting design choices and constraints, systems bioengineers can still draw upon foundations of engineering while tackling important biomedical challenges.
Educational and philosophical foundations of engineering
Established engineering disciplines are brought together by their educational core and the principles of conservation of mass, energy, and momentum. For instance, chemical engineering is unified by its three pillars of thermodynamics (energy), kinetics (mass), and transport (momentum). All mechanical engineers take courses in statics and dynamics (momentum) plus heat transfer and thermodynamics (energy). The core and its principles instill undergraduates with a set of theoretical tools, grounded in practice, which can be applied to any problem an engineer faces. This perspective contrasts biology, for example, where physical conservation laws are generally less helpful for understanding and prediction, leaving the coursework in biochemistry, cell biology, and genetics no choice but to emphasize facts and methods over core principles.
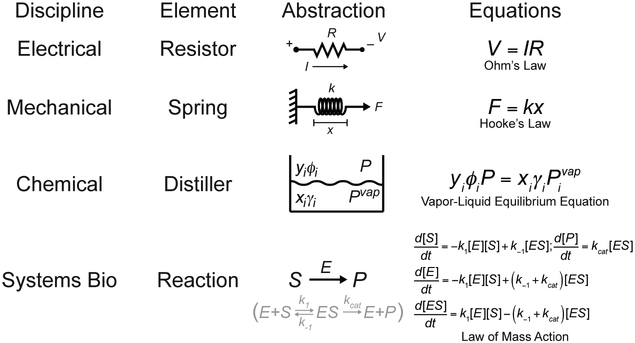
Systems biology seeks a framework to formalize such principles and resonates with engineers for precisely this reason, building upon the components and connections identified by molecular biologists over many decades. Indeed, molecular biology comprises part of the indispensible “engineering core” for systems biology. Just as electrical engineers require the language of resistors, capacitors, and op amps to capture and predict circuit performance, systems bioengineers speak of receptors, adaptors, kinases, and transcription factors within the cell. However, whereas circuit or mechanical elements are simplified operators with reliable input-output properties (Fig. 2, upper), biomolecules within cells exhibit noisy, complex, and context-specific behavior that is difficult to characterize across extended operating regimes. In addition, equilibrium or pseudo-steady state assumptions— valid for many engineered chemical systems—do not generally hold for intracellular reactions (Fig. 2, lower). Accordingly, systems-level abstractions that are modular17 rather than biomolecular may prove more useful if such modules and their transfer functions can be cleanly defined.
Fig. 2.
Representative operational elements of engineering. In electrical engineering, Ohm’s Law states that current (I) is proportional to voltage (V) according to the resistance (R) of the element. In mechanical engineering, Hooke’s Law states that displacement (x) is proportional to applied force (F) according to the spring constant (k) of the element. In chemical engineering, the vapor-liquid equilibrium equation relates pressure (P) to the mole fraction of the ith component in the liquid (xi) and vapor (yi) phases based on its fugacity coefficient (φi), activity coefficient (γi), and vapor pressure (pivap). In systems bioengineering, the law of mass action is required to describe the conversion of substrate (S) to product (P) catalyzed by an enzyme (E) and involving an enzyme-substrate intermediate (ES). The kinetics of the reaction involve a system of differential equations with rate parameters describing reversible ES formation (k1, k−1) and catalytic conversion (kcat).
A second, complementary engineering core for systems biology is predictive modeling. Models are obviously used by systems biologists and engineers alike, but the breadth of approaches invoked by systems bioengineers stands apart. Although systems biology models are diverse overall, there is a notable bias toward differential-equation models by physicists, regression or Bayesian models by statisticians, and machine learning approaches by computer scientists. Engineers, by contrast, are trained to evaluate the problem statement and constraints before deciding on a specific approach. The view is that deterministic, probabilistic, causal, and statistical models all have the potential to play a role.18 Even relative to other engineering fields, the scope of approaches that can be brought to bear on systems biology is striking. Mechanical engineers may use lumped-component modeling in one setting and finite element analysis in others, but systems bioengineers often apply different theoretical formalisms within a single research project.19-26 A spectrum of approaches is necessary and important, because the time-length scales and granularity relevant for one facet of work may change in the next. Systems bioengineers make a habit of stepping back at each point to reassess their assumptions, approaches, and envisioned execution before moving ahead to the next phase of work, which usually involves transitioning from model to experiment or vice versa. There are direct parallels to other engineering disciplines: for example, in the breakdown of Fourier’s law at the nanoscale.27
The impact of an engineering core culminates in the capstone of design. Reconciling design criteria subject to constraints is emblematic of engineering. It is critical to consider design goals together with cost-benefit analyses, feasibility assessments, and milestone-driven timelines. Engineers are asked to navigate the various dimensions of design concurrently, which are required to translate science to a product on an industrial scale. The backdrop of engineering design has a profound impact on one’s approach to systems biology. Pragmatism dominates, with the goal of identifying a workable solution or starting point now in lieu of a perfect one that may never arrive. For example, incomplete information about the earliest molecular events in receptor-activated signal transduction should not disqualify physicochemical models that seek to discover when known mechanisms fail.28-30 The complexity of models and experiments must be tailored to the problem statement, and there is a natural back-and-forth between the two. In contrast to signal transduction, receptor-activated cell phenotypes are poorly encoded by physicochemical models, and predictions are often more successful with statistical, data-driven approaches.31-33 The engineering goal is not to build models, but to use models as a means for gaining sufficient understanding of a system that it can be controlled and manipulated by perturbations (e.g., drugs) in a predictable way. The applied science of modeling requires moving forward under reasonable assumptions while lacking absolute certainty about every last pathway and mechanism. Models become another type of experiment en route to achieving what one originally set out to accomplish. On its own, systems biology is driven more by questions than product development (Fig. 1A), but the philosophy of engineering design minimizes undirected exploration and hones forward thinking toward tangible outcomes (Fig. 1B).
There are other foundational tools that shape the engineering psyche and the approach to systems biology. In biochemical networks, rules of thumb place biophysical limits on reaction rates and enable order-of-magnitude estimates of time and length scales that govern the system.34 For models involving differential equations or optimization, numerical methods are not viewed as part of a default pipeline but rather as decision points that must be deliberated with respect to speed and suitability. Follow-on analyses of model uncertainty,35 error,36 and failure33, 37 can turn out to be more informative than the computational predictions themselves. Engineers sweat the details of both their models and their measurements, recognizing that nothing should be taken for granted.38-42 The educational and conceptual roots of engineering create a systems biology that is distinctively multifaceted, methodologic, inventive, and extensible.
Engineered biological systems vs. systems bioengineering
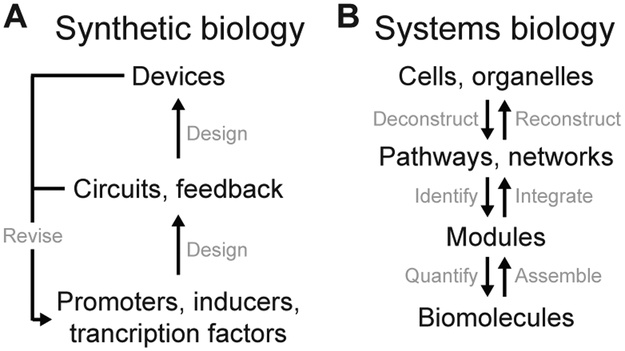
To some, an engineering approach to bioscience implies synthetic biology more often than systems biology.43 Synthetic biology undoubtedly brings a forward-engineering perspective with its focus on design specification, biological parts, and mechanistic abstraction.44 By contrast, the essence of systems biology combines reverse engineering with an emphasis on feedback, adaptation, and dynamics (Fig. 3)—can we learn how an unfamiliar device works to such an extent that we can fix it?45 Here, mathematical models serve as powerful deductive tools that test current understanding and alternative hypotheses through falsifiable predictions.46 Model-experiment failures are not dead-ends but suggestions that new regulatory mechanisms remain to be defined.47-49 By contrast, synthetic-biology computations ought not to fail if they are to be useful as design tools or principles.50, 51 For biological understanding and application, synthetic biologists build and systems biologists model.
Fig. 3.
Different intersections of biology and engineering. (A) Bottom-up approach of synthetic biology. (B) Top-down and middle-out approaches of systems biology.
What roles do systems bioengineers play in deciphering an evolved—rather than engineered—biological system? One role is as a knowledge integrator, synthesizing individual molecular and cellular mechanisms into a collective whole that is both explanatory and predictive. Mathematical models provide a mechanism for integration, recognizing that the exact modeling formalism depends critically on the biological question and state of the science.18 Various modeling approaches may be required to synthesize different facets of a single, overarching research project that incorporates different data types from a team of investigators.19-24 A systems-level synthesis never succeeds on the first pass but is worthwhile for the discrepancies and voids in understanding that it reveals. Pragmatic “gap-filling” methods have even been developed to identify voids systematically and propose usable workarounds.52
Another more-underappreciated role for systems bioengineering lies in experimental design. Systems-level research is not entirely inductive. Systems models have been used to formulate multiple competing hypotheses and design the experiments that best distinguish among these.48, 53 It is possible to pose and test complex hypotheses rigorously through –omics experiments that yield multivariate data. However, no form of retrospective modeling can salvage a big dataset with poorly defined objectives and a litany of confounding variables. A systems model cannot swoop in and save the end of the day, but when conceived at the onset, a model can sharpen thinking as to how data should be collected. For systems bioengineers who are bilingual in quantitative modeling and hypothesis-driven experimentation, it is natural to plan one while envisioning applications of the other.
Systems bioengineering and biomedical Big Data
Systems bioengineering naturally intertwines data generation with predictive modeling and quantitative analysis. This perspective ostensibly runs against ongoing computational and informatic efforts in biomedical Big Data, which seek to foster the creative reuse of large, accessible repositories by novel algorithms.54 We see no conflict between the two approaches, however, instead viewing systems models and Big Data algorithms as part of a complementary sequence. By engaging at an early phase of data generation, systems bioengineers can locally shape a study design at inception and, importantly, assess progress toward its goals while the data are being collected. Big Data science subsequently focuses on the more-global permanence and interoperability of the observations for applications beyond those originally anticipated.55 Both steps are critical for ensuring the long-term value of experimental outlays.
As biomedical Big Data begins to satiate some fields, there is a second opportunity after bioinformatic reductions to revisit predictive modeling with a focus on causal mechanisms. For example, the ~1000 breast tumors sequenced by The Cancer Genome Atlas provide 90% statistical power to detect 90% of driver genes that are mutated at a frequency of at least 2%.56 An integrated multiplatform analysis of these data beautifully shows that breast cancer is two strikingly different molecular diseases.57 Nevertheless, it remains mysterious why certain mutations co-occur and why the overall spectrum differs among molecular subtypes.58 Mechanistic models focusing on mutated driver networks may shed light on the relationship between molecular states and cancer susceptibility as they relate to therapeutic vulnerabilities. Beyond human disease, the National Science Foundation has recognized among its 10 Big Ideas‡ that molecular-level Big Data should be leveraged to predict biological phenotypes. Going forward, systems bioengineering stands to benefit from data science as much as it contributes to it.
Engineering principles for systems biology
The synergy between systems biology and engineering will likely define a new set of engineering-inspired principles for the life sciences.59 We envision that these principles will diverge from those of conventional engineering disciplines, although commonalities may be found in the area of complex systems. Biology lacks governing equations like those of Maxwell or Navier-Stokes, which form the bedrock of many engineering problem statements. In biological systems, the most apt governing assumption, and even the most suitable mathematical formalism, can and should be debated.60, 61 Biology is not purely chemical, electrical, or mechanical but all of these together, and so grafting restricted theories onto biomolecules, cells, and tissues will succeed in some focused regimes but fail in others. This trust-but-verify principle applies equally to systems-biology models and experiments, reinforcing model-experiment iterations as an important crosscheck for engineers.
Another principle that circles back to engineering pragmatism is the dual use of correlation and causation. Correlative experiments are not intrinsically bad when recognized for what they are.62 Lack of correlation can exclude hypotheses more rapidly than an alternative set of mechanistic experiments,63 and shrewd correlative designs can hedge toward results that ultimately prove causative.59 Analogously, engineers recognize a valuable place for systems-biology models that are more explanatory than predictive. Explanatory models iterate less directly with experiment but can be useful for describing observations in the context of a working theory.64 By embracing correlation, causation, explanation, and prediction, systems bioengineers can balance the pros and cons of each approach and adapt tools to the problem at hand.
Systems bioengineering requires analytic versatility, because biological regulatory networks are moving targets. Mutation and adaptation to changing environments mean that systems biology must work with unknown unknowns. A two-state model of ligand binding and receptor activation might hold under most normal circumstances, for instance, but what about when the receptor is mutated or the lipid microenvironment is perturbed? The fragility of many systems models stands in contrast to the reliability of cell-cell communication itself, as during development.65 Instead of using robustness as catch-all organizing principle,66 it may be more productive to rally around incomplete knowledge as a persistent engineering hurdle to circumvent.
Systems bioengineering as a conduit for engineering education
In an academic setting, it is a joy to teach and work with fledgling systems bioengineers. The field attracts individuals interested in STEM careers that encompass science, technology, engineering, and math. For this student body, no topic is intellectually off limits, which enables numerical algorithms and data handling to be treated as deeply as experimental design and molecular biology. Systems biology thereby acts as an effective gateway for quantitatively inclined students in the life sciences to incorporate an engineering mindset.67 It also shows experimentally inclined engineers the value of theory and computation. Perhaps because of the relative newness of the field, our experience is that systems bioengineering students are refreshingly gender balanced, although more work needs to be done to promote racial, ethnic, and socioeconomic diversity. Critical to this effort is reaching precollege and underserved students early and often (Table 2), dispelling the false choices that many perceive in their plan of study: math or science, engineering or medicine, and so forth.
Table 2.
Engaging students and trainees in systems bioengineering.
| Educational level | Exposure to systems bioengineering |
|---|---|
| Precollege | • Generate and handle quantitative life-science data • Introduce the concept of a “model” from biological and computational standpoints • Demonstrate how simple rule-based models lead to complex patterns and behaviors |
| Undergraduate | • Highlight connections between differential equations and biochemistry, linear algebra and statistics, and structured programming and numerical methods, among others • Provide introductory elective courses in systems bioengineering • Introduce systems bioengineering research in a laboratory setting • Construct, assess, and refine a predictive systems model for a senior Capstone design co-developed with feedback from industry |
| Graduate | • Provide enhanced educational electives in systems bioengineering • Master systems bioengineering research in a laboratory setting • Mentor undergraduates in systems bioengineering research • Introduce systems bioengineering research in an industrial setting |
| Postdoctoral | • Lead systems bioengineering research in a laboratory setting • Learn grantsmanship for systems bioengineering research proposals • Create opportunities to master systems bioengineering research in an industrial setting • Co-mentor graduate students in systems bioengineering research • Co-teach or guest lecture in systems bioengineering elective courses |
The scope of material relevant to systems bioengineering is so large and interwoven that there is great flexibility in how content can be delivered (Box 1). As an educational program, biological engineering carries a notable advantage of being able to incorporate systems biology into its core. At MIT, the bioengineering Ph.D. track and applied biosciences track within the Department of Biological Engineering converge on two required courses—20.420 and 20.440—that encompass predictive modeling, -omics data generation, and systems analysis. Alternatively, broader bio(medical) engineering programs may provide a suite of systems-relevant electives at the undergraduate¶ and graduate levels. For the past decade, the Department of Biomedical Engineering at the University of Virginia has offered elective courses that integrate various modeling formalisms or modeling with experimentation (Box 1). The academic cross-training of students through these electives has directly spawned new research directions in the department.21, 22, 68-70 In classical engineering departments, appending considerable systems coursework is challenging because of a perceived departure from that department’s educational core (see above). Graduate-level training grants incentivize co-listed or joint-taught classes and could broaden systems-level offerings while minimizing distraction from core teaching needs.
Box 1. Current and future courses relevant to systems bioengineering.
Biomolecular Kinetics & Cellular Dynamics (20.420, graduate BE core [MIT]): Presents quantitative analysis of complex biochemical and biophysical interactions among biomolecules involved in a wide range of intracellular and extracellular contexts, including metabolic, regulatory, structural, and communication processes. This analysis is deeply grounded in thermodynamic and kinetic principles. There is strong emphasis on hands-on construction and application of prior knowledge-based, or hypothesis-driven, computational models for application problems arising in microbiology, cell biology, and pharmacology, rooted in real data from journal publications. Focus is on predicting how changes in molecular-level properties result in changes in higher-level phenotypic behaviors. Relevant experimental measurement and manipulation techniques are featured to ensure understanding of the sources of data being modelled.
Analysis of Biological Networks (20.440, graduate BE core [MIT]): Presents integrated description of and perspective on experimental -omic methods across spectrum of levels, including genomic, transcriptomic, epigenomic, proteomic, and functional genomics, with emphasis on identifying and employing appropriate data-driven computational frameworks for gaining insights from the ‘big data’ sets obtained from these experimental methods. There is strong emphasis on mathematical underpinnings, and hands-on use, of computational techniques for analyzing each of the kinds of -omics data as well as their integration, in the context of real data from journal publications.
Systems Bioengineering Modeling & Experimentation (BME 4550, undergraduate BME elective [UVA]): Introduces techniques for constructing mathematical and computational models of vascular biological processes and utilizing experimental methods to validate those models at many levels of organizational scale, from genome to whole-tissue. Students rotate through three modules where they attend lectures, read literature, and participate in discussions focused on various modeling and experimental validation techniques. In each module, they work in teams of three to complete group modeling projects that apply the modeling techniques specific to the particular module. Teams may also conduct experiments relevant to the biological question of each module. Topics to be covered include choice of modeling techniques appropriate to addressing particular biological problems at different scales, quantitative characterization of biological properties, assumptions and model simplification, parameter estimation and sensitivity analysis, model verification and validation, and integration of computational modeling with experimental approaches.
Computational Systems Bioengineering (BME 8315, graduate BME elective [UVA]): Introduces techniques for constructing mathematical and computational models of biological processes at many levels of organizational scale—from genome to whole-tissue. Students will learn: 1) How existing and emerging experimental techniques can provide quantitative data for validating biological models. 2) Which modeling techniques are best suited for addressing specific biological problems. 3) How complex problems in biology can be simplified while maintaining biological relevance. In each module, students will work in teams and gain hands-on experience in the different simulation environments while allowing them to address relevant biological problems of different length scales using appropriate modeling techniques.
Modeling of Biological Processes in Environmental Systems (CHE 8820, graduate elective [UVA]): Use of mathematical models to describe processes such as biological treatment of chemical waste, including contaminant degradation and bacterial growth, contaminant and bacterial transport, and adsorption. Engineering analyses of treatment processes such as biofilm reactors, sequenced batch reactors, biofilters, and in situ bioremediation. May include introduction to hydrogeology, microbiology, transport phenomena and reaction kinetics relevant to environmental systems; application of material and energy balances in the analysis of environmental systems; and dimensional analysis and scaling. Guest lectures by experts from industry, consulting firms and government agencies to discuss applications of these bioremediation technologies.
Images, Numbers, and Visualization (high school or undergraduate elective [envisioned]): This hybrid course introduces the coupling among optical microscopy, digital image analysis, and data visualization. The precollege level covers the basic principles of fluorescence imaging (optics, aberration, filtering), segmentation (dark-frame and flat-field corrections, thresholding), and high-content data display (hierarchical clustering, data tracks, circle maps). The undergraduate elective covers more advanced topics, including super-resolution imaging, algorithm-based segmentation pipelines, and t-stochastic neighbor embedding.
Programming Biology (high school elective [envisioned]): This course introduces AP-level biology topics through an open-source, visual computer programming language.§ Students apply the fundamentals of structured programming to simulate key biological principles (mutation-selection, diffusion, competition) and test hypotheses beyond their intuition. Advanced topics include cell behavior, tissue patterning, and population dynamics.
Systems biology provides a powerful launching point for didactic creativity and innovation. Faculty must challenge themselves to see connections outside of conventional engineering or biology courses and create content that emphasizes the multidisciplinarity of systems bioengineering (Box 1, envisioned). In doing so, instructors lead by example and reinforce how different facets of systems biology come together in interesting and exciting ways. This is especially important at the precollege level, where coursework is highly compartmentalized and resources are more limited. There are burgeoning efforts to create cost-efficient open learning platforms for systems biology †. Ultimately, reducing the barrier for entry will require carefully crafted systems-biology courses with enough standalone content that prerequisites are minimal.
Systems bioengineers and industry
A recognized systems-level problem for the pharmaceutical and biotechnology industries is drug-to-market failure. The goal is to treat complex, slowly evolving diseases—involving many cell types and genetic predispositions—with acute molecular perturbations (drugs). In pharmaceutical research and development, drug failure is wastefully slow.71, 72 Many drug failures occur biologically at the systems level. In cancer and infectious disease, for example, stochastic or feedback-triggered adaptations cause drug tolerance and resistance.73, 74 Reciprocally, disease networks show nonobvious vulnerabilities that can be targeted therapeutically. Systems-level sensitivity analysis of a receptor tyrosine kinase signaling network highlighted an enzyme-deficient receptor as a lead therapeutic target,75 which is now in clinical trials for lung cancer (NCT02387216). Systems biology therefore has the promise to streamline target identification and failure diagnoses,76 most meaningfully in the setting of newer organotypic cultures that reflect multicellular organization more accurately.77, 78
As individuals, systems bioengineers are underutilized by many large companies. Systems bioengineers obtain deep mastery in two or three concrete areas, making it a challenge to figure out where such individuals “fit” in a traditional organization. They reside at the intersection of molecular biology, modeling, and data science, which are often siloed teams in big corporations. Pharmaceutical companies that first piloted standalone systems-biology research divisions a decade ago eventually closed them for strategic reasons, recognizing that systems biology conforms less well to the modular, input-output performance of medicinal chemistry, ADME-tox, and others. Exploiting systems biology in industry does not require another department but instead a cultural shift that encourages individuals to operate as liaisons between departments. Smaller biotechnology companies are beginning to recognize the nimbleness of systems bioengineers and have sought to maximize value by seeding such individuals throughout an organization. Success depends critically on buy-in from both the leadership team and the employees that work alongside systems bioengineers.
For achieving industry goals in systems biology, an alternative to cross-fertilization is contracting and consulting. Multiple firms now offer mechanistic bio-modeling, applied bio-math, and related services to larger companies on a contractual basis. Our conversations with these firms indicate that work requests outstrip working capacity, and the demand for systems-biology modelers at those firms exceeds supply. Contracting or consulting systems biologists will be most successful when provided unfettered access to the internal data needed to develop high quality models. Tailoring undergraduate and Ph.D.-level educational pipelines for industry-focused systems bioengineers could provide fertile career paths akin to the consulting options available in mechanical or civil engineering.
Conclusions and future outlook
Early in the modern era of systems biology, engineers were not lacking theories or approaches but rather quantitative data to test them.15 Now, large-scale biological datasets are commonplace, but they are more likely to be impenetrable than insightful. Even highly accessed public data repositories are used far more often as lookup tables than as foundations for integrative knowledge.79 While the situation is improving,55 it remains true for most studies that no one will know the data better or analyze it more deeply than the data generator. Tools for genome editing create an unprecedented means to test hypotheses and solve problems related to regulatory networks, but their implementation is still rate limiting. Consequently, there must be a renewed focus on data efficiency—how can data generation be scaled such that understanding per unit data is maximized? As engineers, we believe that the answer lies in coupling experiments early with predictive models (Fig. 1B).
Systems bioengineering represents a remarkable training opportunity for academics. Armed with a suite of multidisciplinary tools, systems bioengineers should provide a competitive advantage to employers in industry. Key toward realizing this vision is close communication between educators and open-minded industrial partners, with both thinking creatively outside of their respective disciplines. Our past experiences partnering with biologists and clinicians have shown that partners frequently come with misconceptions about what a systems bioengineer can do. Successful partnerships result when both parties move closer to one another, into new territory that neither had originally envisioned. There is a vast educational and scientific landscape yet to explore at the interface of systems bioengineering, biotechnology, and drug development.
The 21st century systems biologists of the first generation are now directing research groups as tenured faculty across the world. The successful outcomes of new academic departments, educational initiatives, and scientific journals indicate that systems biology is here to stay. But, this is no time to become inward-looking or complacent—we should be no more satisfied with the state of systems modeling than electrical engineers were with the vacuum tube. And just as the iron lung was a workable-but-suboptimal engineering solution for polio, we recognize that engineering must partner with biology rather than fight against it.80 Systems bioengineers create a multi-talented ecosystem that will provide long-term benefits to science, health, and society if properly nurtured for who they are.
Acknowledgements
We thank Steve Wiley, Feilim Mac Gabhann, John Stankovic, and Pamela Norris for comments on the manuscript. This work was supported by a Research Innovation Award (#R38) from University of Virginia School of Engineering & Applied Science.
Footnotes
references
- 1.Ideker T, Galitski T and Hood L, Annu Rev Genomics Hum Genet, 2001, 2, 343–372. [DOI] [PubMed] [Google Scholar]
- 2.Kirschner MW, Cell, 2005, 121, 503–504. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald JB, Schoeberl B, Nielsen UB and Sorger PK, Nat. Chem. Biol, 2006, 2, 458–466. [DOI] [PubMed] [Google Scholar]
- 4.Civelek M and Lusis AJ, Nat. Rev. Genet, 2014, 15, 3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ideker T, Winslow LR and Lauffenburger DA, Ann. Biomed. Eng, 2006, 34, 1226–1233. [DOI] [PubMed] [Google Scholar]
- 6.Lauffenburger DA, Integr. Biol. (Camb.), 2012, 4, 9. [DOI] [PubMed] [Google Scholar]
- 7.Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B and Tyson JJ, Mol. Biol. Cell, 2004, 15, 3841–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai L, Dalal CK and Elowitz MB, Nature, 2008, 455, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilfinger A, Norman TM, Vinnicombe G and Paulsson J, Phys. Rev. Lett, 2016, 116, 058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mugler A, Levchenko A and Nemenman I, Proc. Natl. Acad. Sci. U. S. A, 2016, 113, E689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JC, Alvarez MJ, Talos F, Dhruv H, Rieckhof GE, Iyer A, Diefes KL, Aldape K, Berens M, Shen MM and Califano A, Cell, 2014, 159, 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, Pelechano V, Styles EB, Billmann M, van Leeuwen J, van Dyk N, Lin ZY, Kuzmin E, Nelson J, Piotrowski JS, Srikumar T, Bahr S, Chen Y, Deshpande R, Kurat CF, Li SC, Li Z, Usaj MM, Okada H, Pascoe N, San Luis BJ, Sharifpoor S, Shuteriqi E, Simpkins SW, Snider J, Suresh HG, Tan Y, Zhu H, Malod-Dognin N, Janjic V, Przulj N, Troyanskaya OG, Stagljar I, Xia T, Ohya Y, Gingras AC, Raught B, Boutros M, Steinmetz LM, Moore CL, Rosebrock AP, Caudy AA, Myers CL, Andrews B and Boone C, Science, 2016, 353. [Google Scholar]
- 13.Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E, Pe'er D and Nolan GP, Science, 2014, 346, 1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA and Yaffe MB, Science, 2005, 310, 1646–1653. [DOI] [PubMed] [Google Scholar]
- 15.Arkin AP and Schaffer DV, Cell, 2011, 144, 844–849. [DOI] [PubMed] [Google Scholar]
- 16.Lander AD, Cell, 2007, 128, 245–256. [DOI] [PubMed] [Google Scholar]
- 17.Hartwell LH, Hopfield JJ, Leibler S and Murray AW, Nature, 1999, 402, C47–52. [DOI] [PubMed] [Google Scholar]
- 18.Janes KA and Lauffenburger DA, Curr. Opin. Chem. Biol, 2006, 10, 73–80. [DOI] [PubMed] [Google Scholar]
- 19.Garmaroudi FS, Marchant D, Si X, Khalili A, Bashashati A, Wong BW, Tabet A, Ng RT, Murphy K, Luo H, Janes KA and McManus BM, Proc. Natl. Acad. Sci. U. S. A, 2010, 107, 17053–17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinway SN, Biggs MB, Loughran TP Jr., Papin JA and Albert R, PLoS Comput. Biol, 2015, 11, e1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virgilio KM, Martin KS, Peirce SM and Blemker SS, Interface Focus, 2015, 5, 20140080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang CC, Bajikar SS, Jamal L, Atkins KA and Janes KA, Nat. Cell Biol, 2014, 16, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B, Sedlak JC, Srinivas R, Creixell P, Pritchard JR, Tidor B, Lauffenburger DA and Hemann MT, Cell, 2016, 165, 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saucerman JJ, Healy SN, Belik ME, Puglisi JL and McCulloch AD, Circ. Res, 2004, 95, 1216–1224. [DOI] [PubMed] [Google Scholar]
- 25.Valdes-Parada FJ, Porter ML, Narayanaswamy K, Ford RM and Wood BD, Adv Water Resour, 2009, 32, 1413–1428. [Google Scholar]
- 26.Ford RM and Harvey RW, Adv Water Resour, 2007, 30, 1608–1617. [Google Scholar]
- 27.Chang CW, Okawa D, Garcia H, Majumdar A and Zettl A, Phys. Rev. Lett, 2008, 101, 075903. [DOI] [PubMed] [Google Scholar]
- 28.Aldridge BB, Burke JM, Lauffenburger DA and Sorger PK, Nat. Cell Biol, 2006, 8, 1195–1203. [DOI] [PubMed] [Google Scholar]
- 29.Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA and Sorger PK, Mol. Syst. Biol, 2009, 5, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy RJ, Gajadhar AS, Swenson EJ, Rothenberg DA, Curran TG and White FM, Proc. Natl. Acad. Sci. U. S. A, 2016, 113, 3114–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janes KA and Yaffe MB, Nat. Rev. Mol. Cell Biol, 2006, 7, 820–828. [DOI] [PubMed] [Google Scholar]
- 32.Miller-Jensen K, Janes KA, Brugge JS and Lauffenburger DA, Nature, 2007, 448, 604–608. [DOI] [PubMed] [Google Scholar]
- 33.Janes KA, Reinhardt HC and Yaffe MB, Cell, 2008, 135, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papin JA, Hunter T, Palsson BO and Subramaniam S, Nat. Rev. Mol. Cell Biol, 2005, 6, 99–111. [DOI] [PubMed] [Google Scholar]
- 35.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR and Sethna JP, PLoS Comput. Biol, 2007, 3, 1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelhardt B, Frohlich H and Kschischo M, Sci. Rep, 2016, 6, 20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdi A, Tahoori MB and Emamian ES, Sci Signal, 2008, 1, ra10. [DOI] [PubMed] [Google Scholar]
- 38.Ebrahim A, Almaas E, Bauer E, Bordbar A, Burgard AP, Chang RL, Drager A, Famili I, Feist AM, Fleming RM, Fong SS, Hatzimanikatis V, Herrgard MJ, Holder A, Hucka M, Hyduke D, Jamshidi N, Lee SY, Le Novere N, Lerman JA, Lewis NE, Ma D, Mahadevan R, Maranas C, Nagarajan H, Navid A, Nielsen J, Nielsen LK, Nogales J, Noronha A, Pal C, Palsson BO, Papin JA, Patil KR, Price ND, Reed JL, Saunders M, Senger RS, Sonnenschein N, Sun Y and Thiele I, Mol. Syst. Biol, 2015, 11, 831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayenga HN, Thorne BC, Peirce SM and Humphrey JD, Ann. Biomed. Eng, 2011, 39, 2669–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janes KA, Sci Signal, 2015, 8, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monast CS, Furcht CM and Lazzara MJ, Biophys. J, 2012, 102, 2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soltis AR and Saucerman JJ, Bioinformatics, 2011, 27, 2888–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endy D, Nature, 2005, 438, 449–453. [DOI] [PubMed] [Google Scholar]
- 44.Way JC, Collins JJ, Keasling JD and Silver PA, Cell, 2014, 157, 151–161. [DOI] [PubMed] [Google Scholar]
- 45.Lazebnik Y, Cancer Cell, 2002, 2, 179–182. [DOI] [PubMed] [Google Scholar]
- 46.Albeck JG, Burke JM, Spencer SL, Lauffenburger DA and Sorger PK, PLoS Biol, 2008, 6, 2831–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altan-Bonnet G and Germain RN, PLoS Biol., 2005, 3, e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuepfer L, Peter M, Sauer U and Stelling J, Nat. Biotechnol, 2007, 25, 1001–1006. [DOI] [PubMed] [Google Scholar]
- 49.Tsui R, Kearns JD, Lynch C, Vu D, Ngo KA, Basak S, Ghosh G and Hoffmann A, Nat Commun, 2015, 6, 7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brophy JA and Voigt CA, Mol. Syst. Biol, 2016, 12, 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danino T, Mondragon-Palomino O, Tsimring L and Hasty J, Nature, 2010, 463, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blais EM, Chavali AK and Papin JA, Methods Mol. Biol, 2013, 985, 61–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sample V, DiPilato LM, Yang JH, Ni Q, Saucerman JJ and Zhang J, Nat. Chem. Biol, 2012, 8, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, Guyer M and Green ED, J. Am. Med. Inform. Assoc, 2014, 21, 957–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiley HS, Sci Signal, 2011, 4, pe9. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES and Getz G, Nature, 2014, 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MD, Niu B, McLellan MD, Uzunangelov V, Zhang J, Kandoth C, Akbani R, Shen H, Omberg L, Chu A, Margolin AA, Van't Veer LJ, Lopez-Bigas N, Laird PW, Raphael BJ, Ding L, Robertson AG, Byers LA, Mills GB, Weinstein JN, Van Waes C, Chen Z, Collisson EA, N. Cancer Genome Atlas Research, Benz CC, Perou CM and Stuart JM, Cell, 2014, 158, 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.T. C. G. A. Network, Nature, 2012, 490, 61–70.23000897 [Google Scholar]
- 59.Janes KA and Lauffenburger DA, J. Cell Sci, 2013, 126, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fisher J and Henzinger TA, Nat. Biotechnol, 2007, 25, 1239–1249. [DOI] [PubMed] [Google Scholar]
- 61.Walpole J, Papin JA and Peirce SM, Annu. Rev. Biomed. Eng, 2013, 15, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vilela M and Danuser G, Nat. Cell Biol, 2011, 13, 1011. [DOI] [PubMed] [Google Scholar]
- 63.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA and Sorger PK, Mol. Cell. Proteomics, 2005, 4, 1569–1590. [DOI] [PubMed] [Google Scholar]
- 64.Shmueli G, Stat Sci, 2010, 25, 289–310. [Google Scholar]
- 65.Lander AD, Science, 2013, 339, 923–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kitano H, Nat. Rev. Genet, 2004, 5, 826–837. [DOI] [PubMed] [Google Scholar]
- 67.Wingreen N and Botstein D, Nat. Rev. Mol. Cell Biol, 2005, 7, 829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rouillard AD and Holmes JW, J. Physiol, 2012, 590, 4585–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benedict KF, Mac Gabhann F, Amanfu RK, Chavali AK, Gianchandani EP, Glaw LS, Oberhardt MA, Thorne BC, Yang JH, Papin JA, Peirce SM, Saucerman JJ and Skalak TC, Ann. Biomed. Eng, 2011, 39, 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin KS, Blemker SS and Peirce SM, J. Appl. Physiol, 2015, 118, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR and Schacht AL, Nat. Rev. Drug Discov, 2010, 9, 203–214. [DOI] [PubMed] [Google Scholar]
- 72.Pammolli F, Magazzini L and Riccaboni M, Nat. Rev. Drug Discov, 2011, 10, 428–438. [DOI] [PubMed] [Google Scholar]
- 73.Chandarlapaty S, Cancer Discov, 2012, 2, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maisonneuve E and Gerdes K, Cell, 2014, 157, 539–548. [DOI] [PubMed] [Google Scholar]
- 75.Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie L, Linggi B, Kalra A, Paragas V, Bukhalid R, Grantcharova V, Kohli N, West KA, Leszczyniecka M, Feldhaus MJ, Kudla AJ and Nielsen UB, Sci Signal, 2009, 2, ra31. [DOI] [PubMed] [Google Scholar]
- 76.Butcher EC, Nat. Rev. Drug Discov, 2005, 4, 461–467. [DOI] [PubMed] [Google Scholar]
- 77.Clevers H, Cell, 2016, 165, 1586–1597. [DOI] [PubMed] [Google Scholar]
- 78.Wang CC, Jamal L and Janes KA, Wiley Interdiscip. Rev. Syst. Biol. Med, 2012, 4, 51–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones AR, Overly CC and Sunkin SM, Nat. Rev. Neurosci, 2009, 10, 821–828. [DOI] [PubMed] [Google Scholar]
- 80.Brent R, Nat. Biotechnol, 2004, 22, 1211–1214. [DOI] [PubMed] [Google Scholar]