Abstract
Introduction:
The almost exclusive use of only praziquantel for the treatment of schistosomiasis has raised concerns about the possible emergence of drug-resistant schistosomes. Consequently, there is an urgent need for new anti-schistosomal drugs. The identification of leads and the generation of high quality data are crucial steps in the early stages of schistosome drug discovery projects.
Areas covered:
Herein, the authors focus on the current developments in anti-schistosomal lead discovery, specifically referring to the use of automated in vitro target-based and whole-organism screens and virtual screening of chemical databases. They highlight the strengths and pitfalls of each of the above-mentioned approaches, and suggest possible roadmaps towards the integration of several strategies, which may contribute for optimizing research outputs and led to more successful and cost-effective drug discovery endeavors.
Expert opinion:
Increasing partnerships and access to funding for drug discovery have strengthened the battle against schistosomiasis in recent years. However, the authors believe this battle also includes innovative strategies to overcome scientific challenges. In this context, significant advances in in vitro screening as well as computer-aided drug discovery have contributed to increase the success rate and reduce the costs of drug discovery campaigns. Although some of these approaches were already used in current anti-schistosomal lead discovery pipelines, the integration of these strategies in a solid workflow should allow the production of new treatments for schistosomiasis in the near future.
Keywords: schistosomiasis, drug discovery, computer-aided molecular design, automated assays, data quality, virtual screening, perspectives
1. Introduction
Schistosomiasis is a neglected tropical disease (NTD) caused by S. mansoni, S. haematobium, and S. japonicum, three species of Schistosoma genus parasitic flatworms, accounting for the majority of human infections. Schistosomal infections cause chronic and often debilitating disease that ends up impairing development and productivity of affected individuals, and is strongly linked to extreme poverty [1]. Recent estimates of the World Health Organization suggest that around 258 million people are infected resulting up to 200,000 deaths annually. Furthermore, the disease is endemic in 78 countries worldwide, mainly in sub-Saharan Africa, Middle East, Caribbean, and South America, where the number of cases is positively correlated to poor knowledge about the disease, poor sanitation, and a lack of effective health policies [2].
Currently, the control of schistosomiasis relies on presumptive treatment or case management with a single drug, praziquantel (PZQ), which has been used in mass drug administration programs for almost four decades [3]. However, the disseminated and repeated use of this drug in endemic areas as well as high rates of reinfection raise concerns about the emergence and evolution of drug-resistant parasites [4,5]. This problem may be further aggravated by the lack of efficacy of PZQ against schistosomula and juvenile worms [6], often a potential cause of treatment failure in endemic areas. Hence, there is an urgent need for discovering new anti-schistosomal drugs. This paper focuses on current developments in anti-schistosomal lead discovery, with particular emphasis on virtual and automated in vitro target-based and whole-organism screenings. In addition, we highlight recent progress in each area and suggesting possible solutions to existing pitfalls.
2. Challenges to discovering new anti-schistosomal drugs
The long-voiced concerns associated with PZQ argue for increased efforts to identify new anti-schistosomal candidates in drug research and development (R&D) programs. However, the decades-long availability of PZQ as a well-tolerated, affordable (or donated), oral and single dose drug, associated with low financial viability of new anti-schistosomal drugs in poor resource countries does not offer an incentive to enable the high and risk-associated investments in R&D required for the discovery of new treatments for schistosomiasis. Nonetheless, assuming that ‘it is impossible develop resistance to PZQ’ is both reckless and risky, as it may minimize the interest in R&D of alternative anti-schistosomal drugs [7]. Furthermore, while public-private partnerships have been formed for some of the NTDs, e.g., the Drugs for Neglected Diseases Initiative (DNDi) focusing mainly on human African trypanosomiasis, Chagas disease, leishmaniasis, filariasis, pediatric HIV, and mycetoma, corresponding drug discovery and development programs do not yet exist for Schistosomiasis. Consequently, nowadays the number of anti-schistosomal drug candidates undergoing clinical trials is very small.
3. Schistosome drug screening strategies
The majority of anthelminthic drugs approved for human use have been derived from veterinary medicine and discovered through in vivo screening of selected compounds in animal models [8]. Since these screens are labor-intensive and expensive, attention has shifted to developing primary in vitro screens. Typically, worms are cultured for a period of days and morphological changes (e.g., shrinkage, curling, tegumental disruption, worm disintegration) and motility (e.g., increased activity, sluggishness, or paralysis) can be determined using a predetermined scale [9]. However, manual visualization of drug efficacy in vitro is subjective, laborious, and unsuitable for high-throughput screening (HTS) [10]. This limitation can be overcome by the application of automated readouts incorporated into HTS platforms, allowing very large compound collections to be screened against relevant schistosome targets or in the whole-organism, invigorating the lead discovery pipeline. Below, we will discuss the studies highlighting the progression of screening technologies developed to accelerate discovery of new lead candidates for schistosomiasis.
3.1. Target-based screens
Target-based HTS campaigns have been emphasized in last decade as a way of harvesting the investment made in parasite genomics consortiums [11]. Such efforts eventually led to translational research-based groups and platforms taking up genome sequencing of S. mansoni [12,13], S. japonicum [14] and S. haematobium [15], resulting in new information on the parasites’ biological pathways, facilitating identification of relevant targets for therapeutic intervention, and opening new possibilities to HTS on recombinant schistosome proteins. Several changes have encouraged this evolution, including price decreases of the automation and instrumentation sector, availability of commercial compound library datasets, simplification of systems and software, and recruitment of industry-experienced personnel into research groups [16–18].
Literature examples of HTS campaigns on anti-schistosomal lead discovery field are scarce, but some HTS-adapted assays for some S. mansoni targets have been published [19,20]. For instance, Simeonov and colleagues [19] developed a HTS assay for identification of thioredoxin glutathione reductase (TGR) inhibitors, i.e., a key enzyme in redox cascade pathway and a validated target for schistosomiasis [21]. As a result, oxadiazole 2-oxides were identified as TGR inhibitors and nitric oxide donors, with inhibition activities in the low micromolar to low nanomolar range. Incubation of parasites with these compounds led to rapid parasite death. Further, the treatment of schistosome-infected mice with 4-phenyl-1,2,5-oxadiazole-3-carbonitrile-2-oxide, also known as furoxan, killing all relevant stages of S. mansoni (schistosomula, juveniles, and adults), and egg-associated pathologies. The results of these studies verify the utility of oxadiazole-2-oxides as new leads in control of schistosomiasis [22].
3.2. Automated whole-organism screens
The whole-organism schistosome screen approach (phenotypic screening) is an indispensable method to discover new anti-schistosomal agents. Compared to target-based screens, whole-organism screens have, at best, a medium throughput, but validated lead compounds identified from these methods must have been able to reach the designated target within the assayed organism by crossing several biological membranes and resisting to degradation by detoxification enzymes. Conversely, compounds obtained through target-based screening are frequently found to lack activity against the whole-organism [8,23]. In view of these advantages, improvements in robotics and automated acquisition technologies over the last decade have meant that the state-of-the-art for whole-organism screening is now fast, quantifiable, and rigorous [23]. In the following subsections, we discuss advantages and limitations of state-of-the-art medium- and high-throughput methods.
3.2.1. Isothermal micro-calorimetric assay
The isothermal micro-calorimetric assay effectively measures heat flow of biological processes (endo- or exothermic reactions) generated by worms using a 48-channel isothermal microcalorimeter to determine schistosome viability over the time. Sharp peaks in the heat flow occur as the organism moves, allowing the measurement of worm motility. This method reliably distinguishes real-time changes in metabolism and motility of S. mansoni schistosomula and adult worms after treatment with anti-schistosomal compounds. Although isothermal micro-calorimetry method seems to have many advantages, some drawbacks have to be noted. For instance, the heat-flow curves obtained reflect the overall activity of the worms. It is not possible to distinguish between worm contractions and increased movement, because both physiological processes result in increased metabolic activity with heat production. Other disadvantages includes the high cost of the microcalorimeter, the number of replicates (at least 400) required to achieve analyzable signals, and reduced throughput caused by extensive time for each analysis done in a real time fashion [24].
3.2.2. Impedance-based assay
Measurement of electrical impedance has been applied to assess motility of S. mansoni cercariae, adult worms, and hatching of eggs. The system named xCELLigence™ worm real-time motility assay [25], also known as xWORM assay [26], comprises a 96-well E-plate covered in gold microelectrodes which measure changes in conductivity due to contact of the worms with the electrodes. This technique has been favorably received in the field of lead discovery because of its sensitivity, broad applicability and adaptability, ability to measure motility under high-throughput, and minimal effort and training required [25,26].
3.2.3. Image-based assays
With advances of high-resolution cameras and computer-based image analysis programs, imaging also has become a powerful tool for demonstrating anti-schistosomal activity. Image-based assays have the advantage over the use of fluorescent labels in requiring fewer manipulation steps and avoiding use of potentially toxic fluorophores that may affect the outcome of the assay. In this context, high-content screening (HCS) microscopes/cameras are now able to capture high resolution images of schistosomula and adult worms in order to demonstrate phenotypic and/or motility changes [23].
HCS machines are so termed due to the large amounts of data produced requiring use of advanced automated image analysis software. Regarding HCS analysis for schistosomula, there has been recent progress in development of automated imaging algorithms utilizing video microscopy, so that time-dependent phenotypic changes following drug treatment could be monitored [27,28]. Of special interest, is the well validated high-throughput system developed by Paveley and colleagues [29]. This system includes compound stamping, robotic integration, incubation, parasite resuspension, and automated image analysis. Test compounds are cultured with schistosomula for 3 days in 384-well plates; then well content is first re-suspended by repeat aspiration to redistribute individual larvae across the field of view and to disrupt clumping of schistosomula. The HCS then collects 4-tiled objective images for phenotype analysis and five time-lapse images for motility analysis (with 4x objective). The image analysis algorithms integral to the HCS are then applied in three distinct stages. The first step in this algorithm is to identify sufficient schistosomula or to segment them from the background image using basic segmentation algorithms to detect individual larvae. Following successful segmentation, a phenotype score for individual larvae is assigned by a Bayesian classification model built using a dataset of 20,000 manually phenotyped schistosomula treated with a range of known anti-schistosomal compounds. Finally, to assess larvae motility, a score is produced for each larva based on the change in area between successive time-lapse images which is averaged across the well [23,29].
In contrast, WormAssay was developed to quantify motility of adult worms. This system is based on dark-field microscopy imaging, where plates are illuminated from the side. This enhances the contrast between the background and the worms, allowing a more efficient detection of the parasites, and consequently generates ideal images for recording and analysis. Images comprising the entirety of the plate are collected, and a further analysis assesses movement of all worms within each well by two separate algorithms based on contour velocity and area occupation. As result, WormAssay quantifies each worm’s movement simultaneously on the entire plate, with each plate taking approximately 30 seconds to 1 minute to read. The system is cost-effective and capable of screening a large number of compounds. The application supports 6-, 12-, 24-, 48-, and 96-well plates, but the assay throughput is currently limited by number of worms produced and delivered [30].
3.3. Luminescence- and fluorescence-based assays
During the last years, notable advances were obtained for detection of schistosomula viability through quantitation of luminescence and fluorescence emitted by cellular markers and metabolites. These assays can be read by an automatic plate reader, making a cheaper, simpler, more practical and more trainable read-out alternative, requiring little extra software or equipment [31]. Currently, four fluorescent assays (i.e., (i) Alamar blue [32] and (ii) resazurin [33] reagents; (iii) the fluorescein diacetate/propidium iodide multiplex assay [34]; and (iv) L-lactate commercial kit [35]) and luminescent assays based on quantification of adenosine triphosphate [31,36] have been studied. Although these assays are viable tools in anti-schistosomal lead discovery, they have some limitations. For instance, the Alamar blue assay [32] could achieve signal to concentration tests in schistosomula only after 24 hours of incubation with the marker, while the assay developed by Marxer and colleagues [33] using resazurin failed to generate dose-dependent viability curves and to distinguish between live and dead schistosomula after 72 hours of drug exposure. Fluorescein diacetate/propidium iodide assay [34] requires high number of schistosomula to assess viability in a drug sensitivity assay, which reduces its throughput. The assay developed by Howe and colleagues [35] also has a reduced throughput, since screening involves measurement of L-lactate concentrations by removal the supernatant of the well plate (without aspirating the schistosomula) and then diluting it to an acceptable fluorescence range.
4. The open data era
The current strategy to discover lead compounds consists on a data-driven process that is reliant on producing bioactivity data for large chemical libraries in high-throughput campaigns [37]. However, the creation of automatized schistosome screening platforms is beyond the reach of most researchers due to its high cost and infrastructure requirements. To make in vitro tests less expensive, researchers have used data retrieved from assays to explore computer-aided drug design (CADD) approaches. Currently, several datasets exclusively dedicated to lead discovery are freely available in publicly databases, such as PubChem Bioassay [38], BindingDB [39], and ChEMBL [40]. The data related to schistosomiasis could also be found in these databases. Deposited data include information regarding compounds, their bioactivities tested against a specific target or schistosome life stage, and the description of the biological assay extracted from the reference paper. Full exploitation of this rich source of data is of high interest to researchers in the field of CADD to explore the anti-schistosomal chemical space [41,42].
5. Virtual screening
Virtual Screening (VS) has emerged as a powerful and straightforward computational method to guide the identification of new hits from large chemical libraries. In principle, this method is often compared to a funnel, where a large number of compounds in chemical libraries (i.e., 105 to 107 compounds) are reduced by a computational tool to a smaller number that will then be tested experimentally (i.e., 101 to 103 compounds) [43,44]. In general, VS approaches do not hold the drawbacks that are characteristic of experimental approaches, such as accessible chemical space, time, automatization level, or number of worms produced. Nevertheless, the expectation that VS could completely replace in vitro or in vivo approaches is over-optimistic. In addition, typical hit rates from in vitro approaches can range between 0.01% and 0.14%, while hit rates for prospective VS typically range between 1% and 40% [45]. Thus, compounds from the subset that pass the initial VS are found to be biologically actives at a higher rate and at a lower cost. Because of their obvious advantages, VS approaches are widely employed in pharmaceutical industry and academic organizations. Nevertheless, in anti-schistosomal research, the application of VS approaches is limited by a small number of recent studies [46–50]. The main VS approaches that can be implemented in a hit identification pipeline and examples of successful applications of VS workflows leading to the identification of new anti-schistosomal hits are discussed in next sections.
5.1. Structure-based virtual screening (SBVS)
SBVS is becoming an essential tool in assisting fast and cost-efficient lead discovery. The application of SBVS strategies in schistosome lead discovery facilitates understanding the molecular basis of parasite biology and development and utilizes the knowledge of the 3D structure of the biological target in the process to select ligands with acceptable affinity and complementarity with the binding site. In this context, genomic and proteomic studies have provided basis into the identification of validated schistosome targets [51,52]. Currently, 3D structures of 35 different schistosome targets have been solved by X-ray crystallography and nuclear magnetic resonance (NMR) and stored in public databases such as the RCSB Protein Data Bank (PDB) [53], leading to attractive opportunities for the application of SBVS strategies. The main SBVS approaches are discussed below.
5.1.1. Protein-ligand docking
The most extensively used SBVS method is docking, a tool that involves fitting the ligands into the binding site of a 3D structure in order to predict their binding affinities [54]. Generally, docking calculations can be accomplished using two types of algorithms: the search algorithm and the scoring function. The search algorithm generates the various possible poses to fit the ligand into the binding pocket of the receptor. The most known search methods are: (i) Monte Carlo and genetic algorithms; (ii) molecular dynamics (MD) and energy minimization methods; and (iii) systematic methods. Scoring function ranks the different poses and locations of the ligand that are generated by the search algorithm, and orders them by a score. Commonly used scoring functions can be categorized as follows: (i) empirical scoring functions; (ii) force field-based functions; and (iii) knowledge-based functions. The discussion about search algorithms and scoring functions is beyond the scope of this review, so detailed information can be obtained elsewhere [52,54,55].
5.1.2. Structure-based pharmacophores
Structure-based pharmacophores complement docking procedures, including the same level of information, but are less computing-demanding. In structure-based pharmacophore approach, pharmacophore models are derived from the 3D structure of the protein. This method can work both with a free (apo) structure or a ligand-target complex (holo) structure. The generation of models using a holo structure allow complete exploration of interactions with binding site, and inclusion of shape and volume information derived directly from the structural data. In absence of co-crystalized ligand in binding site, the complex can also be obtained by a docking approach, where a small ligands’ binding orientation in the protein is predicted [56,57]. For apo structures, pharmacophores can also be obtained using functional groups or small fragments, also referred as molecular probes, to map possible interaction sites or hot spots within a binding site. Selected hot spots can then be converted into pharmacophoric features in order to generate a structure-based pharmacophore model [57].
5.1.3. Piggy-back
The ‘piggy-back’ is a useful strategy for exploring targets studied in other diseases (human or parasite either) once it facilitates the development of new drugs against homologous targets in other organisms. The efforts in finding validated drug targets and identification of their homologs in schistosome provide evidence of target druggability and may offer new chemical scaffolds, which can be optimized for the schistosomiasis [58]. In this context, gene ontology informations available for schistosomes can be used in order to compare targets, and consequently, identify lead candidates, based on their sequence motifs and binding sites similarities or using 3D structural information. In many cases, these approaches focus on those residues that are known from experimental molecular recognition studies or based on prediction of functional residues. For instance, we use sequence alignment to identify several S. mansoni macromolecules with homologous human and microbial targets [59]. As a result, several drugs with good bioavailability were predicted as new drugs for treatment of schistosomiasis, opening new opportunities for experimental validation [59].
5.2. Ligand-based virtual screening (LBVS)
LBVS is the approach of choice when the biological target is not known or its 3D structure is not available. In this approach, prior knowledge of a reference set of compounds with biological activity against a target is used to identify structural rules to identify new lead candidates for experimental evaluation [60]. There are four main LBVS methods: (i) similarity search; (ii) ligand-based pharmacophores; (iii) 3D shape matching; and (iv) QSAR.
5.2.1. Similarity search
Similarity search is used to identify new compounds by measuring the level of their structural similarity to the known active compounds. This approach is based on the assumption that structurally similar compounds have similar biological properties. Here the compounds are typically represented by molecular fingerprints [61], a high-dimensional vector of bits that accounts for the presence (1) or absence (0) of a fragment in a compound, turning it into a sequence of bits that can then be easily compared with other bit sequences. This comparison must then be expressed in a way that can be quantified [61]. There are many ways to assess the similarity between two vectors; the most common is Euclidean distance [62]. Whereas for molecular fingerprints, the standard estimation of similarity method is the Tanimoto coefficient, which is equal to the number of common bits set to 1 in both fingerprints divided by the total number of bits set to 1 between both fingerprints [62].
5.2.2. Ligand-based pharmacophores
Ligand-based pharmacophores are commonly used when the 3D structure of the biological target is absent. They may be explored using a set of distance constraints of known ligands to locate the relative positions of common chemical features, such as hydrogen-bond donors or acceptors; aromatic rings; partial charges; and hydrophobicity, in order to identify key common features and the relative orientations of known active ligands not shared by inactive. This method delivers good results if enough ligand information is available and if the dataset compounds are known to bind to a target in the same way. One the other hand, conformational flexibility represents one of the main difficulties in pharmacophore generation, because the same ligand could have several productive conformations, which are usually unknown [63].
5.2.3. Shape-based method
Shape-based approach is an established and effective method for identifying hits that are similar in shape and bioactivity to a reference ligand [64]. The shape of a compound refers to the part of space occupied by the chemical structure as determined by its external boundary, abstracting from other aspects the object can have such as its color, atom type, and size [65]. In biological context, shape is a fundamentally important molecular feature that often determines the fate of a compound in terms of molecular interactions with biological targets. Most ligands bind to their target through van der Waals interactions, and hence desolvation effects dominate the binding free energy. Unlike hydrogen bonds and electrostatic interactions, the strength of these hydrophobic interactions is mainly governed by the proximity of the respective electron densities. It is assumed that compounds sharing similar 3D surface shapes are likely to share similar biological activities [66]. Thus, 3D LBVS shape matching tools could offer a ‘scaffold hopping’ way to find new anti-schistosomal leads. Nonetheless, conformational flexibility remains the same obstacle as for ligand-based pharmacophores.
5.2.4. QSAR
Quantitative Structure-Activity Relationship (QSAR) analyses have been increasingly used in VS because of its high speed of screening and good hit rate. Once developed on a set of compounds with known activity, QSAR models are applied to untested chemical compounds for numerical prediction of biological activity (continuous models) or the discrimination between active and inactive compounds (binary models) [67]. In QSAR molecular fingerprints or descriptors are calculated for chemical compounds and correlated with a determined endpoint (experimental data) using a machine learning method. Currently, several machine learning methods, such as Random Forest [68] and Support Vector Machine [69], among others, are available to build QSAR models. The influence of various factors on QSAR performance is decreasing in the following row: data quality > molecular descriptors > machine learning approach [70].
5.3. Successful VS stories in anti-schistosomal hit discovery field
In the current scenario, VS remains a poorly explored strategy in anti-schistosomal R&D field. Despite the lower rate of examples available in literature, most of VS works led the discovery of hits with potency or affinity values in the low micromolar range. Below, we described the successful identification of potent anti-schistosomal hits by use of two different VS workflows.
Among these studies, the identification of histone deacetylase 8 (HDAC8) inhibitors, a target that catalyze posttranslational modifications in schistosomes could be cited [46]. Recent studies [71,72] showed that exposure to generic histone deacetylase inhibitors led to protein acetylation and dose-dependent mortality of schistosomula and adult worms. Initially, Kannan and colleagues [46] built a homology model for S. mansoni HDAC8 based on the available X-ray structures of the human orthologue. Next, MD simulations were employed to optimize structure and evaluate the structural flexibility of the residues at the binding site of the build model. The optimized homology model was then used in a docking-based VS the drug-like subset of the ZINC database. At this stage, authors also refined chemical space of the chemical library using sub-structure filters (i.e., hydroxamate, anilinobenzamides, or thiazole-sulfonamide groups), which are known as zinc binding groups, based on information derived from human HDAC8 inhibitor complexes. Next, 75 predicted compounds were selected based on their intermolecular interactions with the zinc ion located at the binding site and docking scores. Identified hits was then tested for estimate its inhibition activity on human and S. mansoni HDAC8, resulting in the identification of eight hydroxamate derivatives as potent and lead-like inhibitors of the parasitic enzyme. Solving of the X-ray structure of S. mansoni HDAC8 with two of the VS hits, authors also confirmed the predicted binding mode.
In another study, Jacques and colleagues [50] describe a VS approach to identify inhibitors of S. mansoni NAD+ catabolizing enzyme (NACE), a target suspected to be involved in immune evasion by the parasite at the adult stage. An initial filtering biased of the chemical library toward compounds was performed to select small molecular weight compounds, in agreement with the small size of the binding site of NACE. Docking of filtered compounds into a homology model of the enzyme has led to the discovery of 1,701 virtual hits. Then, best poses were filtered using a machine learning model developed using a training set of positive and negative reference binding modes described with protein-ligand interaction fingerprints. The automated selection protocol returned 188 compounds. A visual inspection rejected 90 compounds because of high similarity between structures. Predicted compounds were then tested for estimate its inhibition activity on S. mansoni NACE, resulting in the identification of six hits active in nanomolar concentrations, accompanied by a largely enhanced selectivity for the parasitic enzyme over the human homologue CD38.
5.4. Best practices in virtual screening
Prior to performing any VS workflow, it is important to consider many pitfalls that can influence the erroneous selection of inactive compounds, also known as false positives [73,74]. It is well known that the success of any VS campaign largely depends on development of integrated approaches, combining different search methods as well as structure- and ligand-based methods, along with use of additional filters and statistical approaches to improve the selection of hits. In the following subsections, we will give some important recommendations to increase the reliability of a VS campaign.
5.4.1. Quality of dataset
Publicly available datasets contain a fraction of erroneous records, which presence is caused by lack of or incomplete data curation, measurement variations, and insufficient quality check. In short, these ‘bad’ data encompass entries containing unclear stereoisomer annotations, wrong chemical structures, experimental values, and activity units, and duplicated records [75]. The data compiled from different laboratories under possibly different experimental conditions also add certain discordance [76]. Therefore, the concordance analysis between original and compiled bioactivities as well as structure standardization procedures should be applied for the data to ensure consistency and quality of the data [77].
Pan-assay interference compounds (PAINS) could also lead the VS campaign astray. The apparent activity of PAINS is typically caused by their reactivity rather than non-covalent binding. Such compounds typically interact nonspecifically with proteins of buffer in a high percentage of bioassays [78]. Time and money are consequently wasted in attempts to optimize the biological activity of these compounds. Therefore, the use of computational filters for identifying and removing PAINS in a VS workflow is highly recommended [78].
5.4.2. Balancing the dataset
Unbalanced datasets have been widely discussed in QSAR-based VS field [79,80]. Most classifiers assume equal weighting of the classes in terms of both number of instances and the level of importance.. However, prediction a minority class of an unbalanced dataset results in a large number of erroneous predictions. Two following strategies can be used for unbalanced datasets: dataset balancing and cost-sensitive classifiers. Dataset balancing could be employed by either removing some compounds from the majority class (i.e., under-sampling) or adding some artificially-generated compounds to the minority class (i.e., over-sampling) [79,80]. The cost-sensitive classifiers were used to reduce the level of importance of majority class, a strategy in which samples are predicted to have the class that has the lowest expected cost [81].
Other VS approaches, such as structure- and ligand-based pharmacophore models, docking-based and shape-based models are highly sensitive to balanced datasets or datasets with lower rates of inactive compounds. In these specific cases, it is highly recommended the use of unbalanced datasets at a standard ratio of 36 inactive compounds for each active compound [82]. This data distribution corresponds to what would be found in a completely unbiased approach, i.e., the HTS of a large library of compounds, which could be termed as the ‘natural’ distribution.
5.4.3. Protonation and tautomerism
Ionization and tautomerism may significantly change properties in the ligand, and therefore will have an impact in descriptor calculation, definition of pharmacophore interaction points, and interaction possibilities between ligand and a target protein in docking simulations. Therefore, those phenomena could have an important effect on CADD. Consequently, accurate and relevant assignment of protonation and tautomeric states is crucial in order to match the biological system targeted in the screening process [74,83].
5.4.4. Protein flexibility
Flexibility of the target binding site is an essential but frequently overlooked aspect in structure-based studies. Proteins are flexible, which may be stabilized by ligand binding in one conformer out of an ensemble of conformations of similar energy in the unbound state. Taking into account the flexibility of the protein by docking programs is still an area of active development [84]. The ideal approach to increase protein flexibility would be screening of all possible conformations of a ligand against the full degrees of freedom of the protein structure, using MD simulations. Unfortunately, such an approach is infeasible because of the high cost of computing resources [85,86]. Currently, two streamlined strategies have been developed to solve this problem: ensemble methods and induced-fit docking. To incorporate protein flexibility, ensemble methods make use of multiple input conformations of a target using a set of different X-ray and NMR data available on PDB or extracted from multiple time steps produced by MD or Monte Carlo simulations [87,88]. The induced-fit docking allows for conformational search of the protein and the ligand is performed in parallel (i.e., both conformations are altered at the same time). To limit the combinatorial explosion of conformations that need to be generated and energetically evaluated, a small number of degrees of freedom are considered. Variables that characterize the magnitude of the conformational change along the selected degrees of protein flexibility are optimized in parallel to the ligand degrees of freedom, such as translational, rotational, and torsion changes [89].
5.4.5. Statistical validation of VS models
The quality of a VS model is mostly measured in terms of their ability to discriminate between known active and inactive compounds experimentally evaluated against a target or a whole-organism. Irrespective to the approach chosen, the VS model should be rigorously externally validated before applying it to prospective VS. The most useful statistical metrics to measure the quality of developed models for VS are: sensitivity (SE), specificity (SP), positive and negative prediction values (PPV and NPV), correct classification rate or balanced accuracy (CCR or BA), Cohen’s kappa (κ), receiver operating characteristic (ROC) curve, area under the ROC curve (AUC), determination coefficient (Q2), enrichment factor (EF), and Boltzmann-enhanced discrimination of ROC (BEDROC). The choice of these metrics depends on the approach used; for instance, CCR, SE, PPV, etc., are commonly used for QSAR, while BEDROC, AUC, EF, etc., are usually used for docking and pharmacophore modeling. We also want to remind that these metrics are just an indicator that the model could be useful for VS, but its real quality will be determined only by experimental testing of selected VS hits. In a previous paper, we have extensively discussed those and several other metrics for VS validation [90].
5.4.6. Consensus modeling
The main reason for errors for selection of inactive hits in VS lies in the usage of a single classifier or a single scoring function that very often fail to predict the activity class or binding affinity of screened compounds. This could lead to the selection of many false positives that will be erroneously placed in the top scorers of a ranked list. In an effort to optimize VS models, the consensus strategy could be applied by averaging the predicted values from the individual models, providing better statistical fit and predictive ability as compared to the individual models. This can be very useful, as it combines the advantages of different approaches and simultaneously attenuates the prediction errors of each classifier [91].
5.4.7. ADME/Tox filters
It is well established that poor pharmacokinetics and toxicity (ADME/Tox) properties represent one of the main reasons drug candidates failing in clinical trials. However, during the last decade, combinatorial chemistry and HTS have significantly increased the number of compounds for which early data on ADME/Tox are needed, and, therefore, has increased efforts and costs in the lead discovery process [92]. Obviously, the use of computational tools cannot replace the experimental screens, but they reveal an early ADME/Tox profile in VS campaigns. In this context, our group has been working to overcome or reduce these failures by developing computational tools and web services for the early prediction of some ADME/Tox properties [74,93,94]. Our knowledge points to the importance of these filters in the early stages of hit-to-lead discovery campaigns, which helps in the selection of virtual hits before acquisition and biological evaluation. However, usually the application of all possible ADMETox and other possible structural filters could result in elimination of all VS hits [95]. At first glance, this statement seems to be paradoxical, however, all computational approaches are just an instrument developed to help the researcher but not to substitute his expertise and knowledge.
5.4.8. Integrated VS workflows
The development of integrated workflows, i.e., combining several VS methods and computational tools, is also a useful approach to increase hit rates during experimental validation. Compilation of these workflows could be either in sequential or parallel forms. In the sequential approach, different methods are used in a pipeline to sequentially filter the number of hits retrieved, until the number is small enough for extensive biological testing. Often, methods like pharmacophore screening are used in the beginning of the multi-step screening process. As the number of hits decreases, computationally more expensive methods can be applied to further filter the retrieved compounds, such as docking. In the parallel approach, different methods are run independently and the predicted hits in both screens are combined in a consensus approach for biological testing. The methods used should be complementary and ideally should include ADME/Tox filters, pharmacophore models, QSAR models, similarity-based models, as well as docking-based models [96,97].
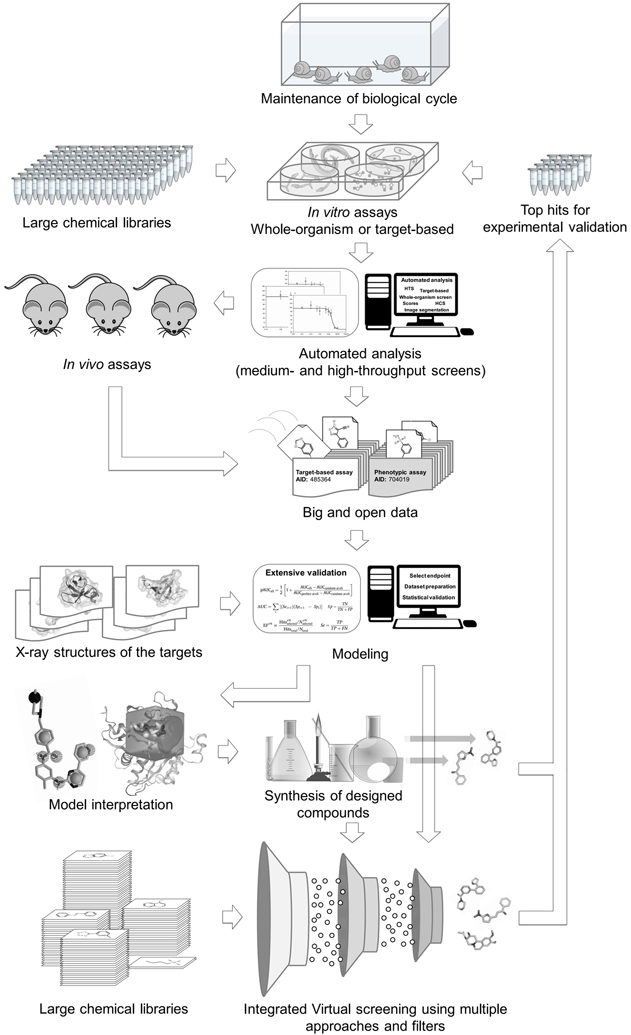
6. Conclusions
In conclusion, we emphasize the importance of integrating modern experimental and computational approaches to accelerate schistosomiasis drug discovery. We propose a workflow, presented in Figure 1, showing the possible road map to integrate such approaches. Combinatorial chemistry and organic synthesis can provide a large number of compounds to be screened using automated assays, for the whole organism in different life stages or for selected schistosome targets. The large amount of data generated from these HTS/HCS assays and public availability of this information started the era of big and open data. These data boost the use of computer-assisted technologies for VS, allowing generation of predictive models for prioritization of VS hits or structural design of novel compounds with desired properties. Further, these computational hit are assayed in the target-based, whole-organism, luminescence or fluorescence-based platforms. Successful integration of all these technologies can accelerate the discovery of new anti-schistosomal leads that could ultimately enter clinical trials.
Figure 1.
Suggested antischistosomal lead discovery workflow in open data era.
7. Expert opinion
Despite the urgent need in finding a new treatment for schistosomiasis and the relative success that has been recently achieved, defeating this NTD may still be far reaching. There are multiple potential ways of battling schistosomiasis. The first one is the development a vaccine. At present, there are ongoing clinical trials regarding several vaccines against various Schistosoma strains [98]. To this purpose, a vaccine against Schistosoma haematobium is under Phase III of clinical trials in Senegal and Niger and two vaccines against Schistosoma mansoni are under Phase I of clinical trials in US and Brazil [98]. However, despite this, there are numerous challenges in vaccine development related to protective antigen discovery, product development, and the preclinical and clinical testing stages. For instance, reverse vaccinology approaches that work well for small bacterial and viral genomes have not yet been successful for eukaryotic parasites because of the greater complexity of the parasite genome and the requirement to employ eukaryotic expression systems to reliably produce soluble and properly folded antigens. In addition to scientific challenges, socioeconomic issues also present a bottleneck for the development and implementation of new vaccines that should be available to those who need them most, especially in developing countries in South America, Africa, and Asia.
Another way of tackling schistosomiasis is the development of new drug(s) which, ideally, should be active against all the different species of Schistosoma. Thus, such drug(s) need(s) to be promiscuous enough to cover a variety of species and selective enough not to hit other targets in human cells, because the interaction with some of them could lead to strong undesirable side effects. Although this is likely to be achievable, as in the case of vaccines, similar socioeconomic challenges could limit the development and distribution of such drug in poor resource countries. Another interesting possibility is the use of combination therapy, which could help overcome scientific bottlenecks such as drug resistance, toxicity, etc. Despite the fact that combinatorial therapy may eventually lead to undesired drug-drug interactions, careful planning of drug combinations will easily circumvent this issue
One of the solutions to decrease the potential economic burden related to drug R&D for schistosomiasis is the use of computational techniques for targeted design of new chemical entities that will decrease both the time and costs associated. Expected future development of combined therapy will in turn boost the development and/or optimization of reliable computational tools for handling and analyzing mixtures [99]. In addition, these approaches should take into account both synergistic and antagonistic effects. We strongly believe that socioeconomic battle against schistosomiasis emerged with London Declaration of 2012 as evidence of willingness to advance through partnerships and provision of funding to R&D [100], but the future battle may include novel approaches that will help to overcome scientific challenges. In conclusion, the era of big and open data has just started and data is increasingly accumulating. Integration and curation of these data will greatly improve the success of drug discovery efforts. Furthermore, development of novel computational approaches capable of handling millions of compounds is essential, in order to avoid that the amount of data generated exceeds the amount of data that can be processed. In turn, these improved tools may also contribute to reducing the number of animals that are required for in vivo testing of new compounds. Finally, we note that even the best computational and VS approaches are just the tools and the overall success of the project still depends only on the ability of a scientist to think critically, to understand the advantages and disadvantages of computational and other tools, to perceive the final goals of his research, and to interpret the consequences of his discoveries.
Article highlights box.
The possible evolution of praziquantel-resistant schistosomes and the lack of new effective drugs call for a paradigm shift from contemporary to modern anti-schistosomal lead discovery strategies;
The main advances in high- and medium-throughput lead screening technologies have opened up new possibilities for anti-schistosomal drug discovery pipelines.
Data produced on extract bioactivity from medium- and high-throughput assays, i.e., open data available on publicly databases, can now be explored in a computational data-driven process.
Both experimental (i.e., target-based assays, automated whole-organism assays, and luminescence- and fluorescence-based assays) and computational (i.e., structure-based and ligand-based strategies) drug discovery approaches have strengths and pitfalls, but carefully planned integration of these strategies could lead to new treatments for schistosomiasis in the near future.
Declaration of interest
The authors would like to thank Brazilian funding agencies, CNPq, CAPES, and FAPEG, for financial support and fellowships. E Muratov thanks the financial support from NIH (GM 096967 and GM66940), CNPq (grant 400760/2014–2), and UNC for Junior Faculty Development Award. CH Andrade and PVL Cravo are Research Fellows in productivity of CNPq. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Abbreviations
- NTD
neglected tropical disease
- PZQ
praziquantel
- R&D
research and development
- DNDi
Drugs for Neglected Diseases Initiative
- HTS
high-throughput screening
- TGR
thioredoxin glutathione reductase
- HCS
high-content screening
- CADD
computer-aided drug design
- SBVS
structure-based virtual screening
- LBVS
ligand-based virtual screening
- HDAC8
histone deacetylase 8
- NACE
NAD+ catabolizing enzyme
- QSAR
quantitative structure-activity relationships
- PAINS
pan-interference assay compounds
- MD
molecular dynamics
- CCR
correct classification rate
- ROC
receiver operating characteristic
- AUC
area under the ROC curve
- EF
enrichment factor
- BEDROC
Boltzmann-enhanced discrimination of ROC
- ADME/Tox
pharmacokinetics and toxicity
Bibliography
Papers of special note have been highlighted as either of interest (●) or of considerable interest (●●) to readers.
- [1].Colley DG, Bustinduy AL, Secor WE, et al. Human schistosomiasis. Lancet 2014;383:2253–64 [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● Very concise and clear description of schistosomiasis.
- [2].World Health Organization. Schistosomiasis [ONLINE]. Available at: http://www.who.int/mediacentre/factsheets/fs115/en/ [Last accessed 3 January 2016]
- [3].Gönnert R, Andrews P. Praziquantel, a new board-spectrum antischistosomal agent. Z Parasitenkd 1977;52:129–50 [DOI] [PubMed] [Google Scholar]
- [4].Ismail M, Metwally A, Farghaly A, et al. Characterization of isolates of Schistosoma mansoni from Egyptian villagers that tolerate high doses of praziquantel. Am J Trop Med Hyg 1996;55:214–8 [DOI] [PubMed] [Google Scholar]
- [5].Fallon PG, Sturrock RF, Niang AC, et al. Short report: diminished susceptibility to praziquantel in a Senegal isolate of Schistosoma mansoni. Am J Trop Med Hyg 1995;53:61–2 [PubMed] [Google Scholar]
- [6].Wang W, Wang L, Liang Y. Susceptibility or resistance of praziquantel in human schistosomiasis: a review. Parasitol Res 2012;111:1871–7 [DOI] [PubMed] [Google Scholar]
- [7].Caffrey CR, Secor WE. Schistosomiasis: from drug deployment to drug development. Curr Opin Infect Dis 2011;24:410–7 [DOI] [PubMed] [Google Scholar]
- [8].Geary TG, Sakanari J a, Caffrey CR. Anthelmintic drug discovery: into the future. J Parasitol 2015;101:125–33 [DOI] [PubMed] [Google Scholar]
- [9].Ramirez B, Bickle Q, Yousif F, et al. Schistosomes: challenges in compound screening. Expert Opin Drug Discov 2007;2:S53–61 [DOI] [PubMed] [Google Scholar]
- [10].Keiser J In vitro and in vivo trematode models for chemotherapeutic studies. Parasitology 2010;137:589–603 [DOI] [PubMed] [Google Scholar]
- [11].Nwaka S, Hudson A. Innovative lead discovery strategies for tropical diseases. Nat Rev Drug Discov 2006;5:941–55 [DOI] [PubMed] [Google Scholar]; ●● Overview of drug discovery for neglected tropical diseases.
- [12].Berriman M, Haas BJ, LoVerde PT, et al. The genome of the blood fluke Schistosoma mansoni. Nature 2009;460:352–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Protasio AV, Tsai IJ, Babbage A, et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis 2012;6:e1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou Y, Zheng H, Chen Y, et al. The Schistosoma japonicum genome reveals features of host–parasite interplay. Nature 2009;460:345–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Young ND, Jex AR, Li B, et al. Whole-genome sequence of Schistosoma haematobium. Nat Genet 2012;44:221–5 [DOI] [PubMed] [Google Scholar]; ●● Schistosome genomics data available to start drug discovery projects.
- [16].Frearson JA, Collie IT. HTS and hit finding in academia – from chemical genomics to drug discovery. Drug Discov Today 2009;14:1150–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rees S The promise of open innovation in drug discovery: an industry perspective. Future Med Chem 2015;7:1835–8 [DOI] [PubMed] [Google Scholar]
- [18].Dahlin JL, Walters MA. The essential roles of chemistry in high-throughput screening triage. Future Med Chem 2014;6:1265–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Simeonov A, Jadhav A, Sayed AA, et al. Quantitative high-throughput screen identifies inhibitors of the Schistosoma mansoni redox cascade. PLoS Negl Trop Dis 2008;2:e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kuhn I, Kellenberger E, Said-Hassane F, et al. Identification by high-throughput screening of inhibitors of Schistosoma mansoni NAD(+) catabolizing enzyme. Bioorg Med Chem 2010;18:7900–10 [DOI] [PubMed] [Google Scholar]
- [21].Kuntz AN, Davioud-Charvet E, Sayed AA, et al. Thioredoxin glutathione reductase from Schistosoma mansoni: an essential parasite enzyme and a key drug target. PLoS Med 2007;4:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sayed AA, Simeonov A, Thomas CJ, et al. Identification of oxadiazoles as new drug leads for the control of schistosomiasis. Nat Med 2008;14:407–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Paveley RA, Bickle QD. Automated imaging and other developments in whole-organism anthelmintic screening. Parasite Immunol 2013;35:302–13 [DOI] [PubMed] [Google Scholar]
- [24].Manneck T, Braissant O, Haggenmuller Y, et al. Isothermal Microcalorimetry To Study Drugs against Schistosoma mansoni. J Clin Microbiol 2011;49:1217–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Smout MJ, Kotze AC, McCarthy JS, et al. A novel high throughput assay for anthelmintic drug screening and resistance diagnosis by real-time monitoring of parasite motility. PLoS Negl Trop Dis 2010;4:e885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rinaldi G, Loukas A, Brindley PJ, et al. Viability of developmental stages of Schistosoma mansoni quantified with xCELLigence worm real-time motility assay (xWORM). Int J Parasitol Drugs Drug Resist 2015;5:141–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Asarnow DE, Singh R. Segmenting the Etiological Agent of Schistosomiasis for High-Content Screening. IEEE Trans Med Imaging 2013;32:1007–18 [DOI] [PubMed] [Google Scholar]
- [28].Asarnow D, Rojo-Arreola L, Suzuki BM, et al. The QDREC web server: determining dose-response characteristics of complex macroparasites in phenotypic drug screens. Bioinformatics 2015;31:1515–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Paveley R a, Mansour NR, Hallyburton I, et al. Whole Organism High-Content Screening by Label-Free, Image-Based Bayesian Classification for Parasitic Diseases. PLoS Negl Trop Dis 2012;6:e1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marcellino C, Gut J, Lim KC, et al. WormAssay: a novel computer application for whole-plate motion-based screening of macroscopic parasites. PLoS Negl Trop Dis 2012;6:e1494. [DOI] [PMC free article] [PubMed] [Google Scholar]; ● A concise and clear description of three different HCS assays.
- [31].Panic G, Flores D, Ingram-Sieber K, et al. Fluorescence/luminescence-based markers for the assessment of Schistosoma mansoni schistosomula drug assays. Parasit Vectors 2015;8:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mansour NR, Bickle QD. Comparison of microscopy and Alamar blue reduction in a larval based assay for schistosome drug screening. PLoS Negl Trop Dis 2010;4:e795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Marxer M, Ingram K, Keiser J. Development of an in vitro drug screening assay using Schistosoma haematobium schistosomula. Parasit Vectors 2012;5:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Peak E, Chalmers IW, Hoffmann KF. Development and validation of a quantitative, high-throughput, fluorescent-based bioassay to detect schistosoma viability. PLoS Negl Trop Dis 2010;4:e759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Howe S, Zöphel D, Subbaraman H, et al. Lactate as a Novel Quantitative Measure of Viability in Schistosoma mansoni Drug Sensitivity Assays. Antimicrob Agents Chemother 2015;59:1193–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lalli C, Guidi A, Gennari N, et al. Development and Validation of a Luminescence-based, Medium-Throughput Assay for Drug Screening in Schistosoma mansoni. PLoS Negl Trop Dis 2015;9:e0003484. [DOI] [PMC free article] [PubMed] [Google Scholar]; ● A concise and clear description of several fluorescence/luminescence-based assays.
- [37].Nantasenamat C, Prachayasittikul V. Maximizing computational tools for successful drug discovery. Expert Opin Drug Discov 2015;10:321–9 [DOI] [PubMed] [Google Scholar]
- [38].Wang Y, Xiao J, Suzek TO, et al. PubChem’s BioAssay Database. Nucleic Acids Res 2012;40:D400–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu T, Lin Y, Wen X, et al. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res n.d;35:D198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 2012;40:D1100–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Goldmann D, Montanari F, Richter L, et al. Exploiting open data: a new era in pharmacoinformatics. Future Med Chem 2014;6:503–14 [DOI] [PubMed] [Google Scholar]; ● Importance of open data in drug discovery.
- [42].Karthikeyan M, Vyas R. Role of Open Source Tools and Resources in Virtual Screening for Drug Discovery. Comb Chem High Throughput Screen 2015;18:528–43 [DOI] [PubMed] [Google Scholar]
- [43].Tanrikulu Y, Krüger B, Proschak E. The holistic integration of virtual screening in drug discovery. Drug Discov Today 2013;18:358–64 [DOI] [PubMed] [Google Scholar]
- [44].Kar S, Roy K. How far can virtual screening take us in drug discovery? Expert Opin Drug Discov 2013;8:245–61 [DOI] [PubMed] [Google Scholar]
- [45].Zhu T, Cao S, Su P-C, et al. Hit identification and optimization in virtual screening: practical recommendations based on a critical literature analysis. J Med Chem 2013;56:6560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kannan S, Melesina J, Hauser A-T, et al. Discovery of inhibitors of Schistosoma mansoni HDAC8 by combining homology modeling, virtual screening, and in vitro validation. J Chem Inf Model 2014;54:3005–19 [DOI] [PubMed] [Google Scholar]
- [47].Liu J, Dyer D, Wang J, et al. 3-oxoacyl-ACP reductase from Schistosoma japonicum: integrated in silico-in vitro strategy for discovering antischistosomal lead compounds. PLoS One 2013;8:e64984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liu J, Dyer DH, Cheng J, et al. Aldose reductase from Schistosoma japonicum: crystallization and structure-based inhibitor screening for discovering antischistosomal lead compounds. Parasit Vectors 2013;6:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Postigo MP, Guido RVC, Oliva G, et al. Discovery of new inhibitors of Schistosoma mansoni PNP by pharmacophore-based virtual screening. J Chem Inf Model 2010;50:1693–705 [DOI] [PubMed] [Google Scholar]
- [50].Jacques SA, Kuhn I, Koniev O, et al. Discovery of Potent Inhibitors of Schistosoma mansoni NAD + Catabolizing Enzyme. J Med Chem 2015;58:3582–92 [DOI] [PubMed] [Google Scholar]
- [51].Ferreira LG, Oliva G, Andricopulo AD. Target-based molecular modeling strategies for schistosomiasis drug discovery. Future Med Chem 2015;7:753–64 [DOI] [PubMed] [Google Scholar]
- [52].Lionta E, Spyrou G, Vassilatis DK, et al. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem 2014;14:1923–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rose PW, Prlić A, Bi C, et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res 2015;43:D345–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ferreira L, dos Santos R, Oliva G, et al. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015;20:13384–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cerqueira NMFSA, Gesto D, Oliveira EF, et al. Receptor-based virtual screening protocol for drug discovery. Arch Biochem Biophys 2015;582:56–67 [DOI] [PubMed] [Google Scholar]
- [56].Pirhadi S, Shiri F, Ghasemi JB. Methods and applications of structure based pharmacophores in drug discovery. Curr Top Med Chem 2013;13:1036–47 [DOI] [PubMed] [Google Scholar]
- [57].Caporuscio F, Tafi A. Pharmacophore modelling: a forty year old approach and its modern synergies. Curr Med Chem 2011;18:2543–53 [DOI] [PubMed] [Google Scholar]
- [58].Njoroge M, Njuguna NM, Mutai P, et al. Recent Approaches to Chemical Discovery and Development Against Malaria and the Neglected Tropical Diseases Human African Trypanosomiasis and Schistosomiasis. Chem Rev 2014;114:11138–63 [DOI] [PubMed] [Google Scholar]
- [59].Neves BJ, Braga RC, Bezerra JCB, et al. In Silico Repositioning-Chemogenomics Strategy Identifies New Drugs with Potential Activity against Multiple Life Stages of Schistosoma mansoni. PLoS Negl Trop Dis 2015;9:e3435. [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● Example of state-of-the-art strategy for drug repurposing.
- [60].Ripphausen P, Nisius B, Bajorath J. State-of-the-art in ligand-based virtual screening. Drug Discov Today 2011;16:372–6 [DOI] [PubMed] [Google Scholar]
- [61].Muegge I, Mukherjee P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin Drug Discov 2015:1–12 [DOI] [PubMed] [Google Scholar]
- [62].Cereto-Massagué A, Ojeda MJ, Valls C, et al. Molecular fingerprint similarity search in virtual screening. Methods 2015;71:58–63 [DOI] [PubMed] [Google Scholar]
- [63].Vuorinen A, Schuster D. Methods for generating and applying pharmacophore models as virtual screening filters and for bioactivity profiling. Methods 2015;71:113–34 [DOI] [PubMed] [Google Scholar]
- [64].Koes DR, Camacho CJ. Shape-based virtual screening with volumetric aligned molecular shapes. J Comput Chem 2014;35:1824–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kortagere S, Krasowski MD, Ekins S. The importance of discerning shape in molecular pharmacology. Trends Pharmacol Sci 2009;30:138–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ghemtio L, Perez-Nueno VI, Leroux V, et al. Recent trends and applications in 3D virtual screening. Comb Chem High Throughput Screen 2012;15:749–69 [DOI] [PubMed] [Google Scholar]
- [67].Cherkasov A, Muratov EN, Fourches D, et al. QSAR modeling: where have you been? Where are you going to? J Med Chem 2014;57:4977–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]; ● Perspectives of QSAR evolution.
- [68].Breiman L Random forests. Mach Learn 2001;45:5–32 [Google Scholar]
- [69].Vapnik V The Nature of Statistical Learning Theory. 2nd ed. New York: Springer; 2000 [Google Scholar]
- [70].Muratov EN, Artemenko AG, Varlamova EV, et al. Per aspera ad astra: application of Simplex QSAR approach in antiviral research. Future Med Chem 2010;2:1205–26 [DOI] [PubMed] [Google Scholar]
- [71].Dubois F, Caby S, Oger F, et al. Histone deacetylase inhibitors induce apoptosis, histone hyperacetylation and up-regulation of gene transcription in Schistosoma mansoni. Mol Biochem Parasitol 2009;168:7–15 [DOI] [PubMed] [Google Scholar]
- [72].Marek M, Kannan S, Hauser A-T, et al. Structural basis for the inhibition of histone deacetylase 8 (HDAC8), a key epigenetic player in the blood fluke Schistosoma mansoni. PLoS Pathog 2013;9:e1003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Scior T, Bender A, Tresadern G, et al. Recognizing pitfalls in virtual screening: a critical review. J Chem Inf Model 2012;52:867–81 [DOI] [PubMed] [Google Scholar]; ● A concise description of problems, shortcomings, failures, and technical traps of VS methods.
- [74].Braga RC, Alves VM, Silva AC, et al. Virtual screening strategies in medicinal chemistry: the state of the art and current challenges. Curr Top Med Chem 2014;14:1899–912 [DOI] [PubMed] [Google Scholar]; ●● State-of-the-art, challenges, and perspectives of VS strategies in medicinal chemistry.
- [75].Williams AJ, Ekins S. A quality alert and call for improved curation of public chemistry databases. Drug Discov Today 2011;16:747–50 [DOI] [PubMed] [Google Scholar]
- [76].Fourches D, Muratov E, Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model 2010;50:1189–204 [DOI] [PMC free article] [PubMed] [Google Scholar]; ● Handbook of chemical data curation.
- [77].Fourches D, Muratov E, Tropsha A. Curation of chemogenomics data. Nat Chem Biol 2015;11:535. [DOI] [PubMed] [Google Scholar]
- [78].Baell JB, Holloway G a. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 2010;53:2719–40 [DOI] [PubMed] [Google Scholar]; ●● Importance of PAINS in drug discovery.
- [79].Chang CY, Hsu MT, Esposito EX, et al. Oversampling to overcome overfitting: Exploring the relationship between data set composition, molecular descriptors, and predictive modeling methods. J Chem Inf Model 2013;53:958–71 [DOI] [PubMed] [Google Scholar]
- [80].Zakharov AV, Peach ML, Sitzmann M, et al. QSAR modeling of imbalanced high-throughput screening data in PubChem. J Chem Inf Model 2014;54:705–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Schierz AC. Virtual screening of bioassay data. J Cheminform 2009;1:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Huang N, Shoichet BK, Irwin JJ. Benchmarking Sets for Molecular Docking. J Med Chem 2006;49:6789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● Overview of docking approaches.
- [83].Kalliokoski T, Salo HS, Lahtela-Kakkonen M, et al. The Effect of Ligand-Based Tautomer and Protomer Prediction on Structure-Based Virtual Screening. J Chem Inf Model 2009;49:2742–8. [DOI] [PubMed] [Google Scholar]
- [84].Lin J-H. Accommodating protein flexibility for structure-based drug design. Curr Top Med Chem 2011;11:171–8 [DOI] [PubMed] [Google Scholar]
- [85].Salsbury FR. Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr Opin Pharmacol 2010;10:738–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. J Comput Aided Mol Des 2012;26:15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hou X, Li K, Yu X, et al. Protein Flexibility in Docking-Based Virtual Screening: Discovery of Novel Lymphoid-Specific Tyrosine Phosphatase Inhibitors Using Multiple Crystal Structures. J Chem Inf Model 2015;55:1973–83 [DOI] [PubMed] [Google Scholar]
- [88].Liao C, Sitzmann M, Pugliese A, et al. Software and resources for computational medicinal chemistry. Future Med Chem 2011;3:1057–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Xu M, Lill MA. Induced fit docking, and the use of QM/MM methods in docking. Drug Discov Today Technol 2013;10:e411–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Braga RC, Andrade CH. Assessing the performance of 3D pharmacophore models in virtual screening: how good are they? Curr Top Med Chem 2013;13:1127–38 [DOI] [PubMed] [Google Scholar]
- [91].Tropsha A Best Practices for QSAR Model Development, Validation, and Exploitation. Mol Inform 2010;29:476–88 [DOI] [PubMed] [Google Scholar]
- [92].van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov 2003;2:192–204 [DOI] [PubMed] [Google Scholar]
- [93].Braga RC, Alves VM, Silva MFB, et al. Pred-hERG: A Novel web-Accessible Computational Tool for Predicting Cardiac Toxicity. Mol Inform 2015;34:698–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Alves VM, Muratov E, Fourches D, et al. Predicting chemically-induced skin reactions. Part I: QSAR models of skin sensitization and their application to identify potentially hazardous compounds. Toxicol Appl Pharmacol 2015;284:262–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kubinyi H The Changing Landscape in Drug Discovery Royal Society of Chemistry: London, 2007 [Google Scholar]
- [96].Drwal MN, Griffith R. Combination of ligand- and structure-based methods in virtual screening. Drug Discov Today Technol 2013;10:e395–401 [DOI] [PubMed] [Google Scholar]
- [97].Lavecchia A, Di Giovanni C. Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 2013;20:2839–60 [DOI] [PubMed] [Google Scholar]
- [98].Hotez PJ, Bottazzi ME, Strych U. New Vaccines for the World’s Poorest People. Annu Rev Med 2016;67:405–17 [DOI] [PubMed] [Google Scholar]
- [99].Muratov EN, Varlamova EV., Artemenko AG, et al. Existing and Developing Approaches for QSAR Analysis of Mixtures. Mol Inform 2012;31:202–21 [DOI] [PubMed] [Google Scholar]
- [100].Uniting to Combat NTDs. The London Declaration [ONLINE]. Available from: http://unitingtocombatntds.org/resource/london-declaration [Last accessed 16 March 2016]