Abstract
The fight against human disease requires a multidisciplinary scientific approach. Applying tools from seemingly unrelated areas, such as materials science and molecular biology, researchers can overcome long-standing challenges to improve knowledge of molecular pathologies. Here, custom-designed substrates composed of silicon nitride (SiN) are used to study the 3D attributes of tumor suppressor proteins that function in DNA repair events. New on-chip preparation strategies enable the isolation of native protein complexes from human cancer cells. Combined techniques of cryo-electron microscopy (EM) and molecular modeling reveal a new modified form of the p53 tumor suppressor present in aggressive glioblastoma multiforme cancer cells. Taken together, the findings provide a radical new design for cryo-EM substrates to evaluate the structures of disease-related macromolecules.
Keywords: breast cancer, cryo-electron microscopy, glioblastoma multiforme, p53, silicon nitride
Cryo-electron microscopy (cryo-EM) has revolutionized the manner in which we analyze biological processes. Early pioneers of the technique were awarded the 2017 Nobel Prize in Chemistry, bringing to light its important contribution to life sciences. Although the spatial resolution achievable with cryo-EM has skyrocketed recently, the basic principles that govern this sophisticated imaging technique remain intact. Simply, a focused stream of electrons is produced by a field emission gun source. The collimated electrons then penetrate and scatter off of frozen-hydrated molecules that are spread out upon a support film, only a few nanometers in thickness.[1] As the scattered electrons are focused by electromagnetic lenses, the amplitudes and phases of the exit waves are collected upon a detector and output as projection images. Each snapshot contains exquisite details of the sample’s molecular structures. Particle images with different angular information are used to mathematically reconstruct a 3D representation of the original object.[2–4]
Over the last decade, enormous effort in the field has been devoted to automating data collection routines and downstream computing procedures.[3] The culmination of these efforts has spurred an exciting new era for biological structural analysis. One critical opportunity for improving the current work-flow is the design of new substrates and materials to prepare fragile specimens. Typical preparation procedures involve weakly attaching macromolecules to carbon-based support films. These films are often comprised of micrometer-sized “holes” in which biological particles become frozen in time and space. More recently, alternative materials are gaining popularity in the cryo-EM field, including gold foils containing micrometer-sized holes or graphene-based substrates, both of which have proven useful for sample preparation procedures.[5] One recurring bottleneck that still confounds the field, regardless of the support material, is the grid screening process. This time-consuming step limits automation procedures due to inconsistencies in freezing conditions and ice thicknesses between specimens.
To improve upon some of the specimen-related barriers in the field, we propose the use of radically different cryo-EM substrates – precisely engineered to help control for ice thickness and particle distribution. The new custom-designed windows range from 5 to 10 μm in x- and y-dimensions and are integrated into silicon nitride (SiN) microchips (Cryo-Chips). Here, we show that Cryo-Chips hold great potential to minimize variability among frozen-hydrated specimens. Previously used microchips contained large imaging windows on the order of 200–500 μm (Figure S1, Supporting Information).[6] While sufficient for cryo-EM applications, the larger windows were more fragile and prone to break during freezing procedures.
The newly engineered microwell or micropost topographies that span the Cryo-Chip viewing window provide additional stability during freezing and enhanced particle retention on the substrate. In addition, the Cryo-Chips are more physically reinforced and resistant to bending than traditional holey substrates. This feature may also serve to sequester protein assemblies away from the harmful air–water interface. Destructive processes such as aggregation and denaturation commonly occur at this interface, rendering samples inadequate for structural studies.[5] Creating new tools to help counter these forces can streamline studies for the scientific community.
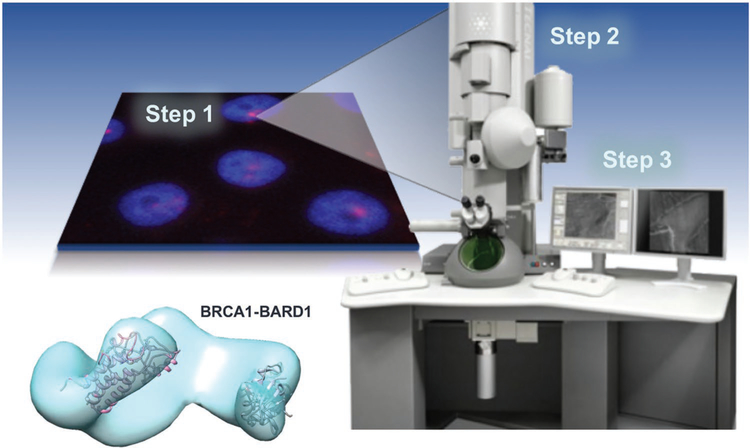
An exciting new direction for the cryo-EM field involves the structure determination of medically relevant macromolecules for therapeutic design purposes. One example of this approach includes our recent studies on breast cancer susceptibility protein (BRCA1)-protein assemblies isolated from breast cancer cells (Figure 1).[6,7] Other successful examples include the high-resolution work of Scheres and colleagues, who determined 3D structures of tau filaments isolated the brains of dementia patients.[8] Hence, engineering new tools to evaluate native protein assemblies is important to advancing the biomedical mission. SiN microchips are highly appealing for this application due to their durability, consistent flatness, and electron-transparent imaging windows (Figure S1, Supporting Information).
Figure 1.
Workflow from cancer cells to protein structure. Our workflow to capture and image native proteins isolated from human cancer cells for cryo-EM structural analysis. Data collected from cryo-EM images are used to reconstruct EM density maps, such as the BRCA1-BARD1 model shown here.
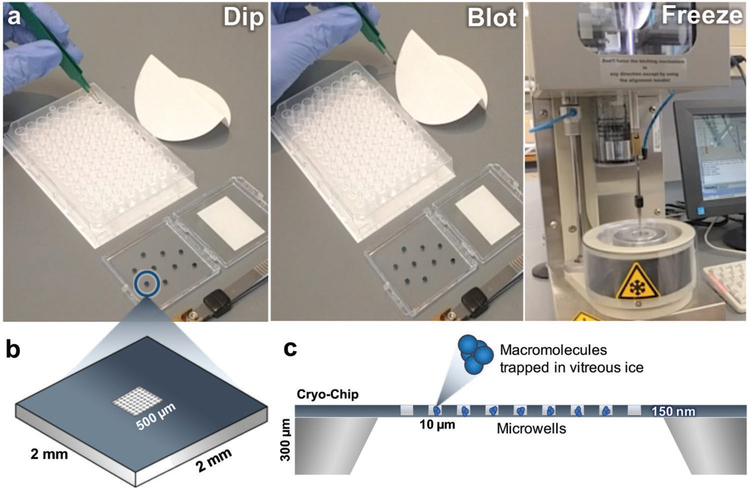
One strategy to prepare cryo-EM samples on custom microchips utilizes a 96-well microtiter plate (Figure 2 and Movie S1, Supporting Information). In this procedure, the wells of the plate may contain macromolecules of interest in a variety of activation states or buffer conditions. Glow-discharged (i.e., plasma cleaned) or surface-coated microchips are dipped into the sample and incubated in the aqueous solution (≈200–300 μL) for up to 1 min (Figure 2a). Upon removing the microchip from the sample, the excessive buffer solution is blotted onto Whatman filter paper. The microchip is then loaded onto a freezing device and plunged into a slurry of liquid ethane using standard protocols.[6,7] Optimal specimen preservation conditions on the freezing device include a blotting step of ≈2 s μL−1 of sample volume and a dwell time of ≈1 s.
Figure 2.
The preparation of frozen-hydrated macromolecules using the “cryo-EM-on-a-chip” technique. a) Microchips are dipped into the sample containing the macromolecule of interest before the excess aqueous buffer solution is wicked away using Whatman filter paper. Chips are plunge frozen into liquid ethane and used for downstream imaging and analysis. b) Schematic diagram of a Cryo-Chip with integrated microwells. The 2 mm × 2 mm chip has a central imaging region of 500 μm × 500 μm and can be engineered with a variety of microwells designs. c) Side view schematic representation of a Cryo-Chip. The height of the chip is ≈300 μm, while each individual well has a depth of 150 μm and a pitch of 10 μm. The microwells aid in the encapsulation of macromolecules in vitreous ice.
The custom-designed microchips illustrated here contain integrated SiN imaging windows (500 μm × 500 μm) having a pitch of 5–10 μm between windows (Figure 2b). Custom window designs can vary in spacing and pitch, and they are incised down to produce 50 nm thick microwells or areas spanning the 50 nm thick film with 150 nm posts. The presence of the microwells or posts across the membrane may serve to improve sample retention and control for uniform ice distribution across the imaging array (Figure 2c). Cryo-Chips can also be tailored to accommodate specimen-specific properties, such as particle size and sample thicknesses up to ≈5 μm by controlling the spacer height, depending upon the desired application. Eukaryotic cells cultured upon SiN have shown to be useful for whole cell tomography applications.[9]
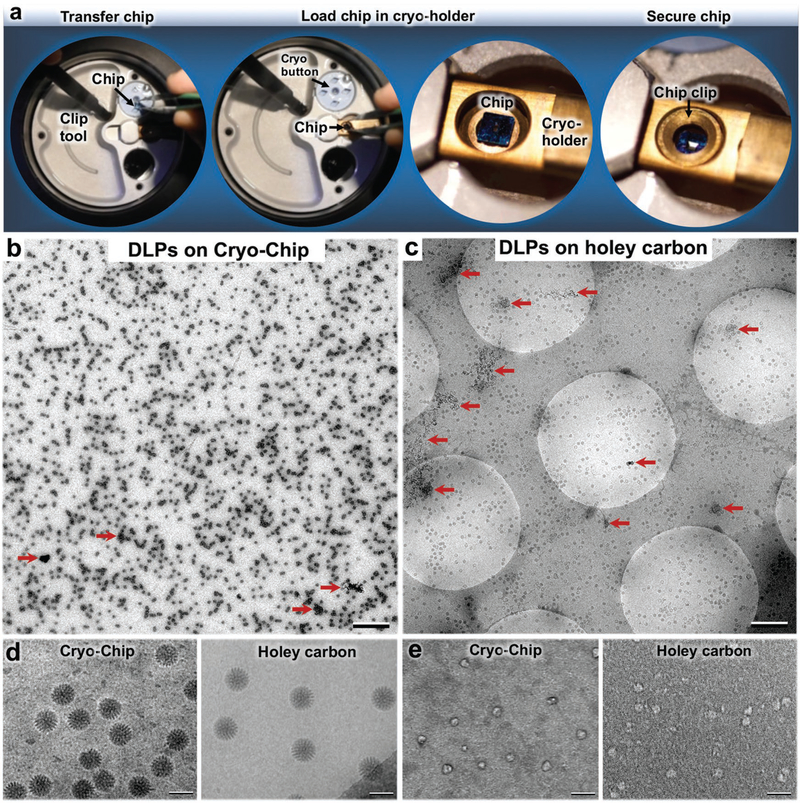
As the frozen-hydrated specimens are prepared, they are easily transferred under liquid nitrogen from storage vessels to an EM specimen holder (Figure 3a and Movie S2, Supporting Information). The chip transfer procedure entails its careful removal from a cryogenic storage buttons (Ted Pella, Inc.) and centering the specimen into the tip of the holder, similar to the procedures used for conventional EM grids. The chip is secured in the holder using a bronze grid clip with minor modifications. The specimen holder with the secured sample is transferred into the EM while maintaining a temperature of ≈−180 °C.
Figure 3.
Cryo-Chip specimen transfer and image comparisons. a) Using a clip tool, Cryo-Chips are transferred from cryogenic storage buttons to the tip of a EM specimen holder, then secured in place using a modified clip ring. A comparison of DLPs prepared on b) Cryo-Chips and c) holey carbon grids revealed a consistent ice layer and a greater number of usable particles on Cryo-Chips. AWIPs are indicated by red arrows. Scale bar is 0.5 μm in panels (b) and (c). d) Samples prepared using SiN substrates demonstrated higher visual contrast for active DLPs prepared on holey carbon films, as previously demonstrated.[19] e) Enhanced image contrast was noted for Cryo-Chips specimens of smaller molecules such as BRCA1-associated protein assemblies. Scale bar is 100 nm.
To better understand performance differences between Cryo-Chip supports and commercially available holey carbon grids (Protochips, Inc.), we used transcriptionally active rotavirus double-layered particles (DLPs, ≈2.5 MDa) as a model system. Simian DLPs (SA11 strain) were purified as previously described and contained in aqueous buffer solution (100 × 10−3 M Tris-HCl pH 7.5, 6 × 10−3 M MgAc, 4 × 10−3 M DTT, 2 × 10−3 M each of ATP, GTP, CTP, UTP, and 1 μL RNasin (Promega Corp., Madison, WI)). Samples (2–3 μL aliquots) from the same biochemical preparation were frozen on the two respective substrates using the same conditions and protocols. Images were recorded using an Eagle 2k HS charge-coupled device (CCD) camera integrated into a (FEI Company) Spirit Bio-Twin transmission electron microscope (TEM) equipped with a LaB6 filament and operating at 120 kV (Figure 3b–d).
In comparing the overall landscape of DLPs prepared on Cryo-Chips and holey carbon grids, we found overall consistencies in ice thickness and enhanced particle retention on the Cryo-Chips (Figure 3b,c). Typically, only particles that distribute in holes are used for structural analysis, which immediately excludes the complexes that distribute on the carbon film. Depending upon the dimensions of the carbon matrix that spans between holes, this selection step may exclude more than half of the total particles per specimen. By contrast, we found a greater number of useful particles encompassing the same surface area on Cryo-Chips than on holey carbon grids. We attributed this observation to a higher propensity for particle retention due to the fixed z-spacer size (≈150 nm) and to the lack of carbon matrix that inherently limits the number of particles for analysis (Figure 3b,c).
Denatured proteins or patches of aggregation were also visibly present in the DLP specimens prepared on both substrates (Figure 3b,c; red arrows). These nonoptimal areas limit the number of protein assemblies that can be included in downstream image analysis. Such effects are notably due to molecular disruptions in protein complexes that occur at the air–water interface (AWIPs, air–water interface particles). Although this comparison study is currently being validated using a variety of different specimens and statistical measures, these initial observations suggest a potential means to advance specimen viability through the use of custom-designed microchips.
We further assessed images of DLPs recorded at a magnification of 30 000 × at a defocus value of ≈−1 μm using an electron dose of ≈5 electrons Å−2. We observed enhanced visual contrast in the DLPs prepared on SiN in comparison to the holey carbon substrate, as previously noted (Figure 3d).[10] One possible explanation for this enhanced contrast is minimal charging effects with Cryo-Chips in comparison to holey carbon substrates. Specimens prepared on holey carbon substrates are suspended in ice within carbon films that coat copper foil grids. As each of these items has variable thermal expansion rates, beam-induced effects are prevalent. Specimens prepared in ice layers over a single semiconductive material, such as SiN, experience less variability in thermal rates. As such, less inherent beam-induced movements may be observed during image acquisition.
Building upon the finding of enhanced visual contrast, we investigated whether the same effect held true for samples isolated from human cancer cells. We prepared cryo-EM samples of the BRCA1-associated protein assemblies isolated from breast cancer cells (HCC1937 line). A full description of the biochemical purification scheme used to obtain the BRCA1 complexes was recently published.[7] This protein specimen is an appealing model as its molecular weight and particle diameter (≈300 kDa, 100 Å) is much smaller than that of rotavirus assemblies (≈2.5 MDa, 800 Å). Purified BRCA1 complexes were prepared in aqueous buffer solution containing 50 × 10−3 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.5), 150 × 10−3 M NaCl, 10 × 10−3 M MgCl2, 10 × 10−3 M CaCl2. Aliquots (≈2 μL each) of the purified complexes were added to either Cryo-Chips or conventional holey carbon grids, each coated with 20% nickel-nitrilotriacetic acid (Ni-NTA) lipid mono layers (please see Materials and Methods in the Supporting Information). Chips contained custom designed microwells having a 500 μm × 500 μm window array and a 10 μm pitch (Figure 2b,c). The same cryogenic preservation procedures were used for each specimen and samples were examined under low-dose conditions (≈5 electrons Å−2).
Specimens were examined using the same FEI Spirit Bio-Twin EM equipped with a LaB6 filament and operating at 120 kV. Images were recorded using a magnification of 50 000 × at a defocus value of ≈−1 μm. Images of Cryo-Chip specimens consistently revealed particles with higher visible contrast compared with images of holey carbon samples (Figure 3c). Contrast values were inverted in the images of BRCA1 assemblies displayed in Figure 3c for ease in particle identification. Protein assemblies identified on Cryo-Chip substrates contained stronger edge boundaries (dark halos) that facilitate particle detection in automated image processing routines, such as those implemented in the RELION software package.[3,11] This halo was not present in particles images prepared on holey carbon films. Similarly, an enhanced white halo effect surrounded the viral assemblies displayed in Figure 3d, the Cryo-Chip samples. This halo effect was not present in the samples prepared on holey carbon films. Moreover, specimens prepared on holey carbon drifted more during imaging routines than Cryo-Chip specimens examined at the same magnification in the same session.
To further test the robustness of the Cryo-Chips, we sought to acquire structural information of a novel protein assembly isolated from a different cancer source. The tumor suppressor protein, p53, participates in DNA damage response and is often called the “guardian of the genome.”[12] During its life cycle, modifications to p53 can stimulate its repair response in the nucleus through the process of ubiquitination. As p53 is mutated in approximately half of all human tumors, it is important to investigate its structure–function relationship in the context of human disease.[12,13] To facilitate this opportunity, we used Cryo-Chip supports having integrated microwells to prepare p53 assemblies isolated from glioblastoma multiforme (GBM) cells (U87MG line). A full description of protein isolation and imaging procedures is provided in the Materials and Methods in the Supporting Information.
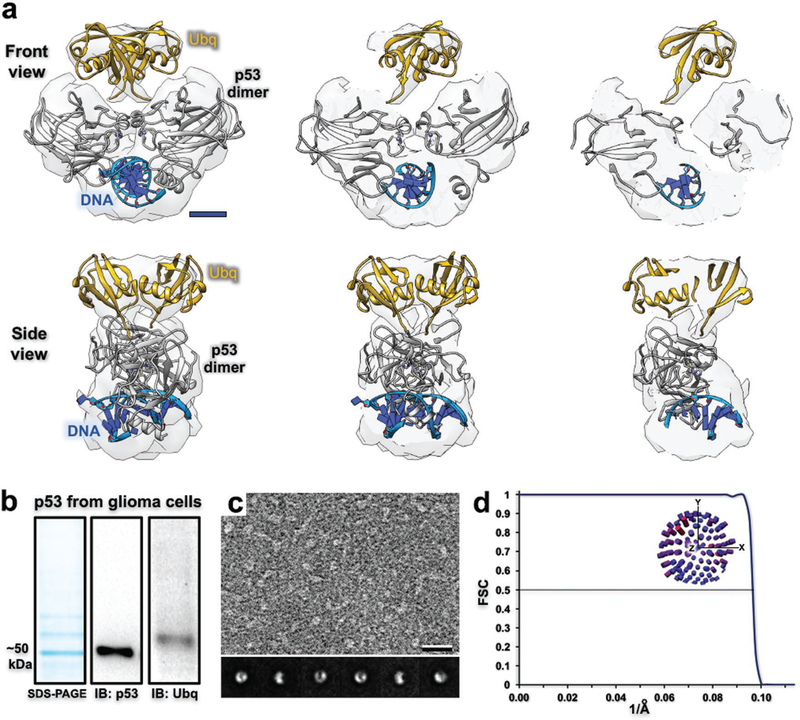
Based on the characteristic morphology and physical dimensions of the particles noted in our EM images, we identified a dimeric form of ubiquitinated-p53 (≈116 kDa, 60 Å diameter) in our biochemical preparation (Figure 4a and Movies S3, S4, Supporting Information). Particles of p53 assemblies were difficult to identify in specimens prepared on holey carbon grids compared to Cryo-Chips (Figure S2, Supporting Information). The p53 protein naturally forms dimers and tetramers upon DNA binding in the nucleus of the cells during stress response activities.[14] According to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blot analysis, wild-type p53 migrated at ≈50 kDa and multiple bands on the gel suggested post-translational modifications were present on p53 (Figure 4b). A positive signal for ubiquitin modifications was confirmed by western blot analysis, accounting for much of the higher molecular weight material. As p53 is subject to phosphorylation at many different residues, other modifications are likely to occur in glioblastoma cells during disease progression.
Figure 4.
Structural and biochemical analysis of p53 assembly isolated from human cancer cells. a) Cross sections through the 3D density map of the p53 dimer (gray structure; pdb code, 2A0C) isolated from GBM cells (U87MG line). The density located proximal to the dimer structure fits two ubiquitin monomers (yellow; pdb code, 1UBQ) and suggests a biologically relevant configuration to attach to each p53 subunit. Scale bar is 15 Å. b) p53 migrated to ≈50 kDa according to a denaturing gel and western blot analysis. Extra bands that migrated slower than ≈50 kDa appear to be ubiquitinated-p53 (Ubq) according to western blot detection. c) EM image of the p53 preparation. Corresponding class averages show consistent features and alternative views of the protein assembly. Scale bar is 100 Å; box size is 200 Å. d) According to the 0.5-FSC criteria, the ubiquitinated-p53 density map resolves to 10 Å and particles were not limited in their angular distribution (inset distribution plot).
We posit that p53 dimers and monomers were both present in our biochemical preparation, albeit difficult to visualize individual monomers in the EM images. The presence of the p53 monomers did not affect image processing procedures as their dimensions were minimal (≈20 Å diameter) in comparison to dimeric assemblies (≈60 Å diameter). Class averages of the dimeric particles were well defined with consistent features and particles adopted multiple views in the images (Figure 4c). The final EM reconstruction refined to ≈10 Å according to the 0.5 Fourier shell correlation (FSC) criteria and the data were not limited in the angular distribution of particles (Figure 4d). Molecular models from a crystal structure of a p53 multimer engaging DNA were used to interpret the structure (pdb code, 2AC0).[15] The ubiquitin model (pdb code, 1UBQ[16]) was placed in the density map in a biologically relevant manner that enables the proper residues of ubiquitin to be in proper proximity to the p53 subunits (Figure 4a). Overall, these results demonstrate the useful nature of Cryo-Chip supports to prepare native p53-DNA repair assemblies derived from human cancer cells. This technical advancement provides the foundation for future work to analyze functional differences among wildtype and mutated forms tumor suppressors as disease-related molecular targets.
As the production of carbon support films can be highly variable, a valuable asset for the EM community is the design of new flat substrates that are transparent to the electron beam. Using custom-designed SiN microchips, we observed enhanced image contrast in biological specimens compared with similarly prepared conventional holey carbon films. Improved contrast in particle images correlates with more accurate computing procedures and the potential to produce EM structures with less time-consuming technical barriers. Due to the difficulty in manufacturing uniformly flat and durable holey carbon film, charging effects may ensue during imaging, which cause beam-induced movements. Adding amorphous carbon layers to blanket the perforated film may mitigate charging effects, although these measures may become obsolete through the use of Cryo-Chips. Other new support materials were also proven effective at reducing beam-induced movements, which may cause resolution-limiting effects.[10,17,18]
We also demonstrate that custom-designed SiN microchips are a powerful tool to harvest biological samples and prepare them for cryo-EM structural studies. Enhanced contrast was seen in two case studies involving highly symmetric virus assemblies and nonsymmetric cancer-related proteins. As these model samples were effortless to prepare and assess using the custom-designed substrates, we turned to a more challenging complex that is often mutated in aggressive and incurable brain tumors.
Frozen-hydrated specimens of p53 assemblies were prepared from GBM cells. Corresponding biochemical analysis and downstream computing procedures indicated a dimeric p53 complex engaging DNA. The 10 Å structure of the native protein assembly was ubiquitinated in a manner that is consistent with DNA damage response. As post-translational modifications on p53 are important for its activation and nuclear repair processes, the captured assemblies suggest a snapshot of the tumor suppressor performing one of its most essential functions. This conformational view of p53 is important as certain ubiquitinated forms of the protein enable erroneous DNA repair events, supporting the growth and proliferation of cancer cells. Taking together, the use of custom-designed cryo-chips may transform our view of medically relevant protein targets in the context of human health and disease.
Supplementary Material
Acknowledgements
N.A.A. and A.C.V. contributed equally to this work. This work was supported by the National Institutes of Health and the National Cancer Institute [R01CA193578, R01CA227261, and R01CA219700 to D.F.K.]. Additional support was provided by the University of Virginia-Virginia Tech Carilion Seed Fund Award and the Cartledge Charitable Foundation. The EM reconstruction for the dimeric p53 assembly was deposited in the EMdatabank, code EMD-0378, and is freely available for download.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Nick A. Alden, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA Department of Biomedical Engineering, Pennsylvania State University, University Park, PA 16802, USA; Center for Structural Oncology, Pennsylvania State University, University Park, PA 16802, USA.
Dr. A. Cameron Varano, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA Department of Biomedical Engineering, Pennsylvania State University, University Park, PA 16802, USA; Center for Structural Oncology, Pennsylvania State University, University Park, PA 16802, USA; Translational Biology, Medicine, and Health Graduate Program, Virginia Tech, Blacksburg, VA 24061, USA; Huck Institutes of the Life Sciences, Pennsylvania State University, University Park PA 16802, USA.
Dr. William J. Dearnaley, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA Department of Biomedical Engineering, Pennsylvania State University, University Park, PA 16802, USA; Center for Structural Oncology, Pennsylvania State University, University Park, PA 16802, USA; Huck Institutes of the Life Sciences, Pennsylvania State University, University Park PA 16802, USA.
Maria J. Solares, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA Center for Structural Oncology, Pennsylvania State University, University Park, PA 16802, USA; Translational Biology, Medicine, and Health Graduate Program, Virginia Tech, Blacksburg, VA 24061, USA; Huck Institutes of the Life Sciences, Pennsylvania State University, University Park PA 16802, USA.
William Y. Luqiu, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA
Yanping Liang, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA.
Prof. Zhi Sheng, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA
Prof. Sarah M. McDonald, Department of Biology, Wake Forest University, Winston-Salem, NC 27109, USA
Dr. John Damiano, Application Science, Protochips Inc. Morrisville, NC 27560, USA
Dr. Jennifer McConnell, Application Science, Protochips Inc. Morrisville, NC 27560, USA
Dr. Madeline J. Dukes, Application Science, Protochips Inc. Morrisville, NC 27560, USA
Prof. Deborah F. Kelly, Virginia Tech Carilion Research Institute, Virginia Tech, Roanoke, VA 24016, USA Department of Biomedical Engineering, Pennsylvania State University, University Park, PA 16802, USA; Center for Structural Oncology, Pennsylvania State University, University Park, PA 16802, USA; Huck Institutes of the Life Sciences, Pennsylvania State University, University Park PA 16802, USA.
References
- [1].Taylor KA, Glaeser RM, Science 1974, 186, 1036. [DOI] [PubMed] [Google Scholar]
- [2].Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A, J. Struct. Biol 1996, 116, 190. [DOI] [PubMed] [Google Scholar]
- [3].Scheres SH, J. Struct. Biol 2012, 180, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].De Rosier DJ, Klug A, Nature 1968, 217, 130. [DOI] [PubMed] [Google Scholar]
- [5].Glaeser RM, Nat. Methods 2016, 13, 28. [DOI] [PubMed] [Google Scholar]
- [6].Winton CE, Gilmore BL, Tanner JR, Varano AC, Sheng Z, Kelly DF, Microsc. Today 2017, 25, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liang Y, Dearnaley WJ, Varano AC, Winton CE, Gilmore BL, Alden NA, Sheng Z, Kelly DF, Sci. Adv 2017, 3, e1701386. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [8].Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SHW, Nature 2017, 547, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ring EA, Peckys DB, Dukes MJ, Baudoin JP, de Jonge N, J. Microsc 2011, 243, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tanner JR, Dukes MJ, Melanson LA, McDonald SM, Kelly DF, J. Anal. Mol. Technol 2013, 1, 6. [Google Scholar]
- [11].Scheres SH, J. Struct. Biol 2015, 189, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Muller PA, Vousden KH, Science 1990, 250, 1233.1978757 [Google Scholar]
- [13].Friedman PN, Chen X, Bargonetti J, Prives C, Proc. Natl. Acad. Sci. USA 1993, 90, 3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z, Mol. Cell 2006, 22, 741. [DOI] [PubMed] [Google Scholar]
- [15].Vijay-Kumar S, Bugg CE, Cook WJ, J. Mol. Biol 1987, 194, 531. [DOI] [PubMed] [Google Scholar]
- [16].Rhinow D, Kuhlbrandt W, Ultramicroscopy 2008, 108, 698. [DOI] [PubMed] [Google Scholar]
- [17].Yoshioka C, Carragher B, Potter CS, Microsc. Microanal 2010, 16, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kam J DA, Tanner JR, McDonald SM, Kelly DF, Technology 2014, 02, 44. [Google Scholar]
- [19].Gilmore BL, Varano AC, Dearnaley W, Liang Y, Marcinkowski BC, Dukes MJ, Kelly DF, Meth. Mol. Biol 2018, 1764, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.