Abstract
The FDA approved drug rapamycin can prolong lifespan in diverse species and delay the onset of age-related disease in mammals. However, a number of fundamental questions remain unanswered regarding the mechanisms by which rapamycin modulates age-related pathophysiology and lifespan. Alterations in the gut microbiota can impact host physiology, metabolism and lifespan. While recent studies have shown that rapamycin treatment alters the gut microbiota in aged animals, the causal relationships between rapamycin treatment, microbiota dynamics and aging are not known. Here, using Drosophila as a model organism, we show that rapamycin-mediated alterations in microbiota dynamics in aged flies are associated with improved markers of intestinal and muscle aging. Critically, however, we show that the beneficial effects of rapamycin treatment on tissue aging and lifespan are not dependent upon the microbiota. Indeed, germ-free flies show delayed onset of intestinal barrier dysfunction, improved proteostasis in aged muscles and a significant lifespan extension upon rapamycin treatment. In contrast, genetic inhibition of autophagy impairs the ability of rapamycin to mediate improved gut health and proteostasis during aging. Our results indicate that rapamycin-mediated modulation of the microbiota in aged animals is not causally required to slow tissue and organismal aging.
Subject terms: Protein aggregation, Ageing
Introduction
In recent years, there has been a growing appreciation of the potential impact of developing effective interventions to slow aging and prolong healthy lifespan1,2. A leading target for such interventions is the nutrient-sensing mechanistic Target of Rapamycin (mTOR) pathway3,4. The mTOR protein kinase is found in two discrete complexes, mTOR complex 1 (mTORC1) and mTORC2, each of which contains distinct protein components and phosphorylates different substrates5. Rapamycin, a small molecule that acutely inhibits mTORC1, can extend lifespan and delay the onset of age-related diseases in model organisms including flies and mice3,4,6–12. Hence, there is significant interest in elucidating the underlying mechanisms of rapamycin-mediated longevity. The accumulation of damaged and misfolded proteins due to a loss of protein homeostasis (proteostasis) is a shared hallmark of aging and numerous age-onset diseases13–15. Inhibiting mTORC1 activity can induce autophagy5,16, a cellular recycling process that can eliminate damaged proteins and organelles17,18. However, the interplay between rapamycin treatment, autophagy, proteostasis and healthy aging remains incompletely understood.
The diverse collection of commensal and symbiotic microbes (microbiota) that live in close association with many animal species can profoundly influence many aspects of host physiology, including nutrient metabolism and immune function. Indeed, there is an emerging understanding that the microbiota can influence host aging and lifespan determination19–21. Age-related alterations in microbiota composition have been reported in flies, fish, mice and humans22–27. In both flies and mice, age-onset microbial dysbiosis contributes to intestinal barrier dysfunction, a conserved pathophysiological hallmark of aging23,28. Remarkably, recolonizing the gut of middle-aged fish with bacteria from young donors prolongs lifespan and delays behavioral decline27. In addition, recent work has shown that microbial genetic composition and metabolites can positively impact host longevity29,30. Together, these findings raise the intriguing possibility that existing interventions that promote longevity do so via an interplay with the microbiota. Consistent with this idea, the lifespan-extending effects of metformin in C. elegans are eliminated when worms are cultured axenically (germ-free)31. However, there is a scarcity of data addressing the question of whether the microbiota plays an important role in mediating additional prolongevity interventions.
Rapamycin treatment has been reported to alter microbiota dynamics in both aged flies32 and mice33. More specifically, rapamycin treatment was reported to reduce bacterial load in aged fly guts, however, it was not determined whether the composition of the microbiota was altered32. Moreover, the question of whether alterations of the microbiota play a causal role in the beneficial effects associated with rapamycin treatment remains to be answered. Here, we show that rapamycin treatment leads to reduced levels of Alphaproteobacteria, a taxon that has previously been linked to health decline and mortality in aged flies23. Consistent with a previous report32, we find that rapamycin treatment maintains intestinal barrier function during aging. Importantly, we show that functional autophagy is required for the rapamycin-mediated improvement in gut function in aged flies. In addition, we show that rapamycin treatment improves proteostasis in aged muscles in an autophagy-dependent manner. Critically, however, we find that eliminating the microbiota does not prevent the anti-aging effects of rapamycin at either the tissue or organismal level. Moreover, we show that germ-free flies show improved lifespan and proteostasis upon rapamycin treatment compared to microbe-bearing control animals. Our findings demonstrate that rapamycin, provided in the food, can act directly on the host organism to maintain tissue homeostasis during aging in an autophagy-dependent fashion.
Results
Rapamycin treatment improves gut health and modulates microbiota composition during aging
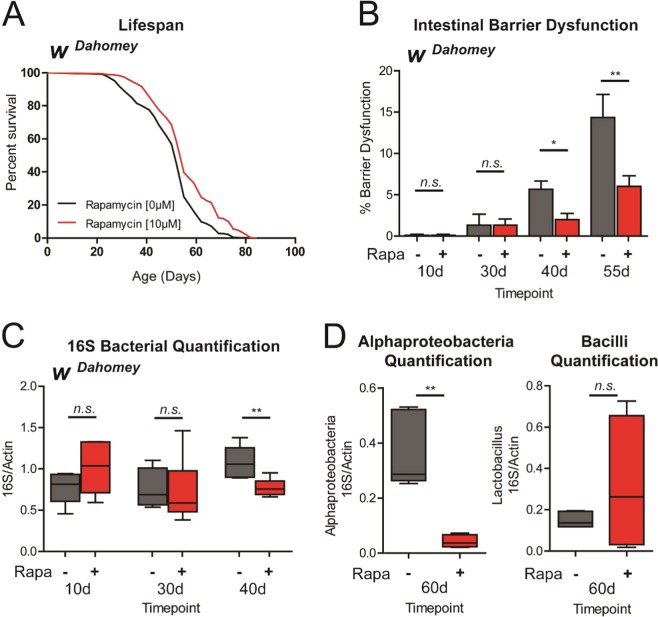
The fruit fly Drosophila melanogaster is an excellent model organism to explore the interplay between microbiota composition, intestinal health and aging21,34. In the first place, we confirmed that treating adult female flies with rapamycin supplemented in their food can prolong fly lifespan. Consistent with previous findings8, we observed that rapamycin-treated flies display a significant increase in both mean and maximum lifespan in multiple trials (Figs 1A and S1A). Intestinal barrier dysfunction is a common feature of aging organisms and has been linked to a number of human diseases35–37. Indeed, loss of intestinal barrier function has been reported in aged worms38,39, flies38,40,41, fish38, rodents28 and monkeys42. Importantly, in Drosophila at least, intestinal barrier dysfunction is a harbinger of age-onset mortality23,41. Hence, we assayed whether rapamycin could delay the onset of intestinal barrier dysfunction in aging flies. To do so, we utilized the Smurf assay to examine intestinal integrity during aging40,41. Consistent with a previous report32, we found that rapamycin treatment during adulthood delays the onset of intestinal barrier dysfunction (Figs 1B and S1B). A number of studies have demonstrated that microbial load in the Drosophila intestine increases with age22,23,43–45. It has been reported that rapamycin treatment reduces the number of colony-forming units (CFUs) in dissected guts from aged flies32. Since flies that display intestinal barrier dysfunction (Smurf flies) show increased microbial loads23, it is possible that the reported reduction in CFUs is linked to reduced numbers of Smurf flies. To better understand the impact on commensal homeostasis during aging, we utilized qPCR with universal primers to the bacterial 16S rRNA gene to characterize alterations in microbiota dynamics in response to rapamycin treatment in non-Smurf flies. Rapamycin treatment led to a significant reduction in bacterial loads (Figs 1C and S1C), with these changes occurring before detectable intestinal barrier dysfunction (in non-Smurf flies). Recent work has shown that increased microbial loads are also associated with a shift in bacterial composition toward a greater proportion of Proteobacteria species in aged flies and that this shift is more closely associated with the health status of the animal than with chronological age23. Hence, we next utilized primers to the 16S rRNA gene that are specific to the classes Bacilli and Alphaproteobacteria23. Upon rapamycin treatment, there was a decrease in Alphaproteobacteria but no significant change in Bacilli (Figs 1D and S1D,E), with this change also occurring in non-Smurf flies. Together, these findings show that rapamycin treatment limits age-related microbial dysbiosis and improves gut health in aged flies.
Figure 1.
Rapamycin treatment improves gut health and modulates microbiota composition during aging. (A) Survival curves of wDahomey females with or without rapamycin treatment from day 4 onwards (post-eclosion). p < 0.0001, log rank test; n > 225 flies. Representative result of 4 separate lifespan trials. (B) Intestinal integrity during aging of wDahomey females with or without rapamycin treatment from day 4 onwards (post-eclosion). *p < 0.05, **p < 0.01, one-way ANOVA/Bonferroni’s multiple comparisons test; n = 300 flies on day 10 per treatment. (C) Bacterial levels assayed by qPCR of the 16S rRNA gene in surface sterilized, non-smurf wDahomey females with or without rapamycin treatment from day 4 onwards (post-eclosion). **p < 0.01, Mann-Whitney U-test; n = 6 replicates of five flies per timepoint. (D) Bacterial levels assayed by taxon specific primers in surface sterilized, non-smurf wDahomey females fed with or without rapamycin from day 4 onwards. **p < 0.01, Mann-Whitney U-test; n = 6 replicates of five flies per timepoint. Rapamycin was provided in the media at a concentration of 10 μg/ml for (A–D). All error bars represent SEM.
Rapamycin requires autophagy to improve gut health and maintain muscle proteostasis in aged flies
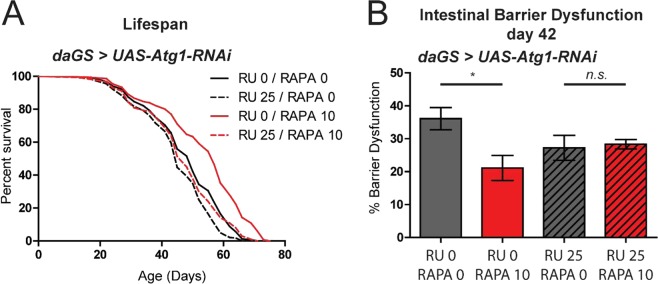
Macroautophagy, hereafter autophagy, is a degradation pathway in which cellular materials are sequestered by double-membrane vesicles known as autophagosomes, and delivered to the lysosome for degradation46. Autophagy has been shown to play important roles in aging and lifespan determination in diverse species47. More specifically, autophagy induction has been implicated as a causal factor in numerous prolongevity interventions47. Indeed, it has been reported that ubiquitous knockdown of Atg5, a gene required for autophagy, during development and adulthood prevents rapamycin-mediated lifespan extension in flies8. Here, we set out to build upon this finding by determining whether rapamycin-mediated longevity requires autophagy specifically in adult flies. To this end, we utilized the RU486-inducible, Gene-Switch expression system48 to induce adult-onset knock-down of Atg1, which is involved in the initiation of autophagosome formation18,49. We used the ubiquitous daughterless (da)GS driver line to inhibit Atg1 by RNAi in adult flies (Fig. S2A) and compared survivorship with and without rapamycin treatment. Adult-onset RNAi of Atg1 completely suppressed rapamycin-mediated longevity (Figs 2A and S2B,C). Next, we set out to determine whether the rapamycin-mediated beneficial effects on intestinal barrier function in aging flies requires functional autophagy. Critically, the ability of rapamycin to improve intestinal integrity in aged flies is dependent upon Atg1 gene activity. Feeding rapamycin to control flies improved intestinal barrier function at day 42 (Figs 2B and S2D). However, adult-onset RNAi of Atg1 prevented this rapamycin-mediated improvement in gut health (Figs 2B and S2D). Together, these findings support the idea that rapamycin requires a functional autophagy pathway, in adults, to promote gut health and longevity.
Figure 2.
Rapamycin treatment requires autophagy to improve gut health during aging. (A) Survival curves of daGS > UAS-Atg1-RNAi female flies treated with (red) or without rapamycin (black), and with (dashed lines) or without RU486 (solid lines) from day 4 onwards. p < 0.0001, log rank test; n > 205 flies. Representative result of 2 separate lifespan trials shown. (B) Intestinal integrity of day 42 daGS > UAS-Atg1-RNAi female flies treated with (red) or without (black) rapamycin, and with (dashed lines) or without (solid lines) RU486 from day 4 onwards. *p < 0.05, one-way ANOVA/Bonferroni’s multiple comparisons test; n = 180 flies per treatment. Rapamycin was provided in the media at a concentration of 10 μg/ml and RU486 at a concentration of 25 μg/ml for (A,B). All error bars represent SEM.
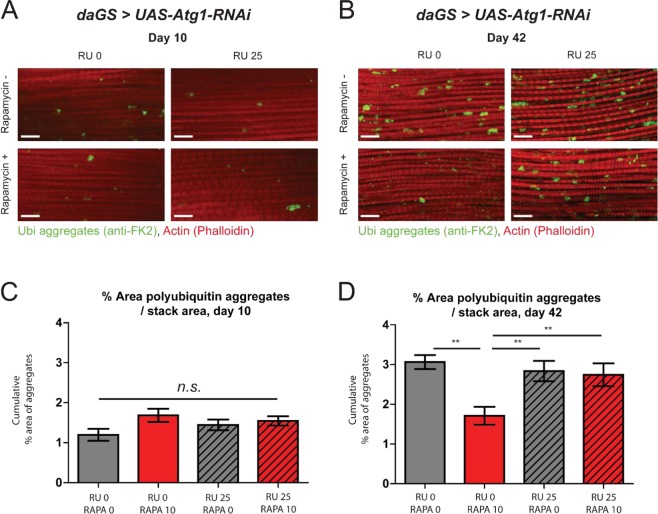
The age-related loss of muscle function and mass, sarcopenia, is a key feature of health decline in the elderly50. Studies in model organisms have revealed that loss of proteostasis is a major contributing factor to muscle aging51. Indeed, muscle aging in Drosophila is correlated with an accumulation of ubiquitinylated, insoluble protein aggregates52. We sought to determine if rapamycin was capable of improving proteostasis during muscle aging. To this end, we characterized the accumulation of ubiquitinated protein aggregates in aged flight muscles using immunofluorescence microscopy. As previously reported52–55, we observed that aged Drosophila flight muscles accumulate ubiquitinated protein aggregates consistent with a loss of proteostasis (Fig. 3A,B). Feeding rapamycin had no impact on the levels of ubiquitinated protein aggregates in young flies (Fig. 3A,C). Importantly, however, feeding rapamycin from early adulthood lead to a significant reduction in ubiquitinated protein aggregates in aged muscle (Fig. 3B,D). Autophagy has been shown to be involved in maintaining muscle proteostasis in aging organisms47,52. Hence, we examined whether the rapamycin-mediated improvement in proteostasis in aged flies requires autophagy. We used the ubiquitous daGS driver line to inhibit Atg1 by RNAi in adult flies and compared the accumulation of protein aggregates with and without rapamycin treatment. Adult-onset RNAi of Atg1 completely suppressed rapamycin-mediated improvements in proteostasis in aged muscle (Fig. 3B,D). Hence, the ability of rapamycin to delay this marker of muscle aging is dependent upon Atg1 gene activity.
Figure 3.
Rapamycin treatment maintains proteostasis during aging in an autophagy-dependent manner. (A,B) Immunostaining of indirect flight muscles from 10 and 42 day old daGS > UAS-Atg1-RNAi female flies with or without both rapamycin and RU486 treatment from day 4 onwards (post-eclosion), showing polyubiquitinated aggregates (green channel, anti-FK2) and muscles (red channel stained with phalloidin/F-Actin). Scale bar is 10 µm. (C,D) Quantifications of polyubiquitin aggregates in muscle as shown in (A,B) respectively. **p < 0.01, one-way ANOVA w/ Newman-Keuls post-test; n > 12, one fly per replicate. Rapamycin was provided in the media at a concentration of 10 μg/ml and RU486 at a concentration of 25 μg/ml for (A–D). All error bars represent SEM.
Rapamycin can extend lifespan and healthspan in the absence of a microbiota
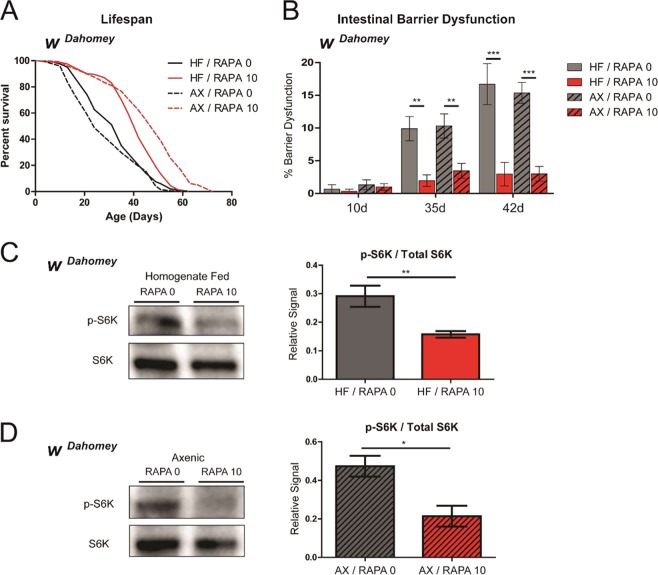
Having ascertained that alterations in microbiota dynamics are correlated with improved lifespan and tissue homeostasis upon rapamycin treatment, we next asked whether these effects were in any way microbiota-dependent. To this end, we generated cohorts of flies rendered germ-free as described previously23, and either maintained axenically or reintroduced to fly homogenate. We then examined the impact of rapamycin treatment on lifespan under both germ-free and control (homogenate-fed) conditions. Critically, we found that rapamycin was capable of extending lifespan in both the homogenate-fed and axenic conditions in multiple trials (Figs 4A and S3A), and, thus, this phenotype was not dependent upon the microbiota. Age-related microbial dysbiosis contributes to intestinal barrier dysfunction in aged animals21,23,28. As rapamycin treatment limits age-onset dysbiosis (Fig. 1C,D), we tested whether rapamycin-mediated improvements in intestinal integrity are dependent upon the microbiota. However, we found that rapamycin was able to improve intestinal barrier function during aging in both the homogenate fed and axenic conditions (Fig. 4B). Next, we examined the ability of rapamycin treatment to inhibit TOR activity in vivo in axenic and control flies. To do so, we examined phosphorylation of S6K, a well-described downstream target of TORC15, as an indicator of TOR activity. We confirmed that TOR signaling was significantly reduced by rapamycin treatment in microbe-bearing control flies, as measured by the ratios of phosphorylated S6K relative to total S6K (Figs 4C and S3C). Furthermore, we found that rapamycin treatment significantly reduced TOR signaling in axenic flies, suggesting that the efficacy of rapamycin is not dependent upon the microbiota (Figs 4D and S3C).
Figure 4.
Rapamycin treatment extends lifespan, improves barrier function and reduces TOR signaling in germ-free flies. (A) Survival curves of wDahomey female flies rendered germ free as embryos, and either maintained germ free (AX, dashed lines) or refed with a bacterial homogenate (HF, solid lines) and treated with (red) or without (black) rapamycin from day 4 onwards (post-eclosion). ***p < 0.0001, log rank test; n ≥ 200 flies. Representative result of 3 separate lifespan trials shown. (B) Intestinal integrity of day 10, 35 and 42 wDahomey flies with (HF, solid bars) or without homogenate feeding (AX, dashed bars) and with (red) or without rapamycin (black) treatment from day 4 onwards (post-eclosion). **p < 0.01 and ***p < 0.001, one-way ANOVA/Bonferroni’s multiple comparisons test; n = 300 flies at day 10 per treatment. (C) Western blot detection and densitometry of p-S6K (T398) and total S6K levels from day 10 homogenate fed (HF) wDahomey flies treated with or without rapamycin from day 4 onwards. n = 5 replicates, 10 flies per replicate; **p < 0.01; Mann-Whitney U-test. (D) Western blot detection and densitometry of p-S6K (T398) and total S6K levels from day 10 axenic (AX) wDahomey flies treated with or without rapamycin from day 4 onwards. n = 5 replicates, 10 flies per replicate; *p < 0.05; Mann-Whitney U-test. Rapamycin was provided in the media at a concentration of 10 μg/ml. All error bars represent SEM.
To further examine the role of rapamycin-mediated changes on the microbiota on aging, we extracted a bacterial homogenate from aged flies fed rapamycin, and from age-matched controls. These ‘rapamycin-fed’ and control homogenates were then refed to young axenic flies. Interestingly, however, no change in lifespan was found between these two cohorts (Fig. S3B), despite causing persistent changes in bacterial load and composition (Fig. S3D,E). Together, our findings indicate that the effects of rapamycin treatment on the microbiota are not necessary or sufficient to promote longevity.
The efficacy of rapamycin treatment on proteostasis is enhanced in germ-free flies
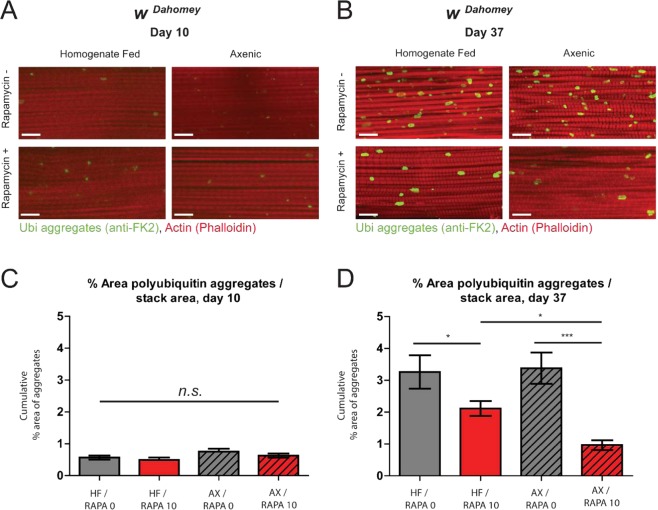
When comparing the prolongevity effects of rapamycin treatment on germ-free and control flies (Figs 4A and S3A), we noted that the magnitude of lifespan extension was more pronounced in germ-free flies. To better understand this observation, we examined the interplay between rapamycin-mediated improvements in proteostasis and the microbiota. More specifically, we examined rapamycin-mediated alterations in the accumulation of ubiquitinated protein aggregates in aged flight muscles in control and axenic flies (Fig. 5A–D). Feeding rapamycin had no impact on the levels of ubiquitinated protein aggregates in young flies (Fig. 5A,C). Importantly, we observed a significant reduction in the levels of ubiquitinated protein aggregates in axenic flies treated with rapamycin compared to microbe-bearing controls (Fig. 5B,D). Taken together, our results suggest that a microbiota is neither necessary nor sufficient to confer the beneficial effects of rapamycin at the cellular, tissue or organismal level, and in some cases is indeed more efficacious in the absence of a microbiota.
Figure 5.
The efficacy of rapamycin treatment on proteostasis is enhanced in the absence of a microbiota. (A,B) Immunostaining of indirect flight muscles from 10 and 37 day old wDahomey female flies rendered germ free as embryos, and either maintained germ free (AX) or refed a bacterial homogenate (HF), and treated with or without rapamycin from day 4 onwards, showing polyubiquitinated aggregates (green channel, anti-FK2) and muscles (red channel stained with phalloidin/F-Actin). Scale bar is 10 µm. (C,D) Quantifications of polyubiquitin aggregates in muscle as shown in (A,B) respectively. *p < 0.05, ***p < 0.001, one-way ANOVA w/ Newman-Keuls post-test; n > 13, one fly per replicate. Rapamycin was provided in the media at a concentration of 10 μg/ml for (A–D). All error bars represent SEM.
Discussion
Interventions that target the underlying aging process have the potential to transform human health. A better understanding of whether prolongevity interventions are influenced by or dependent upon microbiota composition may be critical in assessing whether these approaches are likely to prove effective in promoting healthy aging in a broad spectrum of aged individuals. Rapamycin has emerged as a leading candidate to target aging and forestall disease, extending lifespan and healthspan in a variety of organisms. It has also been reported previously that in addition to improving lifespan, rapamycin treatment induces significant changes to the host microbiota, lowering bacterial load in flies and modulating the microbiome in mammals33. We report similar changes in the wDahomey strain of Drosophila under our laboratory settings. To assess the relevance of these changes to the prolongevity effects of rapamycin, we report the first examination of the impact of rapamycin treatment on aging in germ-free animals. We observed that removal of the microbiota did not prevent the ability of rapamycin to prolong lifespan, revealing that rapamycin’s effects on the microbiota are not necessary for lifespan extension in this context. Conversely, we found that reassociation of germ-free flies with the microbiota of rapamycin-treated conventional flies had no effect on host lifespan, suggesting rapamycin-induced microbiota changes are insufficient in themselves to convey prolongevity effects. One of the physiological hallmarks of aging that we assayed was loss of intestinal barrier function. In previous work, we have shown that different strains of flies display different rates of age-onset intestinal barrier dysfunction41. Although the underlying mechanisms are not fully understood, it is plausible that genetic differences between strains and an interplay with microbiota composition result in alterations in gut homeostasis during aging. In any case, here, we report that rapamycin can delay the onset of intestinal barrier dysfunction in axenic flies. Moreover, we found the magnitude of lifespan extension observed upon rapamycin treatment was enhanced in germ-free flies compared to controls, and that rapamycin treatment leads to a greater improvement in proteostasis in germ-free flies, suggesting the presence of a microbiota may even slightly hamper the beneficial effects of rapamycin treatment. Recent studies have shown an association between microbe abundance and lifespan extension or other metabolic readouts, which is modulated by diet56–59. Taken together, these findings suggest that the ability of rapamycin to mediate lifespan extension in axenic flies might also be affected by nutrition. It is important to note that our experiments were carried out on a single diet.
The importance of interactions between the microbiota and therapeutic treatments has been highlighted by recent work demonstrating a role for the intestinal microbiota in determining patient responses to a particular cancer therapy60–62. These interactions may take place in either direction; the impact of a given therapeutic on the microbiota may be required for its action in the host, or the microbiota may act upon the therapeutic in ways that modify its function. A number of drugs are known to be metabolized by the microbiota63, with either beneficial or detrimental effects on drug function. In addition, it has been reported that microbes can bolster or suppress the effects of fluoropyrimidines, used to treat colorectal cancers, in C. elegans64. In our experimental setting we find that the action of rapamycin on the microbiota is not required for its beneficial effects. However, our observation that the presence of a microbiota may reduce the efficacy of rapamycin treatment suggests that further investigation of the interaction between rapamycin and the microbiota is warranted. Indeed, it is possible that our findings may help explain published reports that fail to observe rapamycin-mediated lifespan extension in flies65. In conclusion, our study confirms that feeding rapamycin can impact both lifespan, and microbiota dynamics during aging. However, in our experimental setting, we have found that rapamycin’s effect on the microbiota is neither necessary nor sufficient to bring about significant lifespan gains in the host. It is of great interest whether or not our observations will be replicated in other model systems, or if they are unique to Drosophila.
Materials and Methods
Fly culture and media
The majority of this work was carried out in the standard laboratory strain wDahomey. Additional genotypes used were the da-GS GeneSwitch line received from H. Tricoire (Université Paris Diderot–Paris 7), and the UAS-Atg1-RNAi line, received from the Vienna Drosophila RNAi (VDRC) Center (stock no. 16133). Flies were cultured in a humidified, temperature-controlled incubator with 12 h on/off light cycle at 25 °C, in vials containing standard cornmeal medium (1% agar, 3% brewer’s yeast, 1.9% sucrose, 3.8% dextrose and 9.1% cornmeal; all concentrations given in wt/vol). Rapamycin (LC Labs) and RU486 (Cayman Chemical Company) were dissolved in ethanol and mixed into the media when preparing food vials. The rapamycin dose used was 10 ug/ml, and the RU486 dose was 25 ug/ml. Control food had the same total volume of ethanol vehicle (1/400, 100% ethanol) as experimental food.
Lifespan analysis
Flies that eclosed over a 36-hour period were collected and allowed to mate for approximately 60 hours. Female or male flies were collected under light nitrogen-induced anesthesia and maintained at a density of 30+/−3 flies per vial in a humidified, temperature-controlled (25 °C) incubator with a 12 hour light/dark cycle. Flies were transferred to new vials every 2–3 days and scored for the number of deaths per vial.
Intestinal barrier dysfunction assay
The ‘Smurf’/intestinal barrier dysfunction assay was performed similarly to41. Flies were aged on standard medium until the day of the Smurf assay. Dyed medium was prepared using standard medium with blue dye no. 1 (SPS Alfachem) added at a concentration of (2.5% wt/vol). Flies were then allowed to consume dyed food for 24 hours prior to quantification. A fly was counted as a Smurf when dye coloration was observed outside the digestive tract.
Genomic DNA isolation
Genomic DNA was extracted using the PowerSoil DNA isolation kit (MoBio). All flies were surface sterilized as previously described22 prior to sample preparation. To ensure consistent homogenization, whole fly samples were pre-homogenized in 150 µL of solution from the PowerSoil bead tube using a motor pestle. This homogenate was then returned to the bead tube and the manufacturers protocol was followed.
Quantitative PCR
DNA samples for qPCR of the 16S ribosomal RNA gene were prepared as described above. RNA extractions, for analysis of the knockdown of the Atg1 gene, were carried out in TRIzol (Invitrogen) as per the manufacturer’s directions. cDNA synthesis was carried out using the First Strand cDNA Synthesis Kit (Fermentas). PCR was performed with PowerUp SYBR Green Master Mix (Applied Biosystems) on a CFX96 Real Time PCR system (BioRad). Cycling conditions were as follows: 95 °C for10 minutes; 95 °C for 15 s then 60 °C for 60 s, cycled 40 times. All calculated gene expression values were normalized to the value of the loading control gene Actin5C. The following primer sequences were used:
Act5C_L –TTGTCTGGGCAAGAGGATCAG, Act5C_R - ACCACTCGCACTTGCACTTTC;
Atg1_F- GCTTCTTTGTTCACCGCTTC, Atg1_R- GCTTGACCAGCTTCAGTTCC.
Universal primers for the 16 S ribosomal RNA gene were against variable regions 1 (V1F) and 2 (V2R), as previously published66. Taxon specific 16S primers were as follows:
Bacilli_F – CGACCTGAGAGGGTAATCGGC, Bacilli_R – GTAGTTAGCCGTGGCTTTCTGG;
Alpha_F – CCAGGGCTTGAATGTAGAGGC, Alpha_R – CCTTGCGGTTCGCTCACCGGC
Immunostaining procedure for dissected indirect flight muscle
Hemithoraces were dissected and fixed for 20 minutes in PBS with 4% paraformaldehyde and 0.2% Triton X-100. After washing, samples were incubated overnight at 4 °C with an antibody detecting polyubiquitinated proteins at 1:250 mouse mAb FK2 (Enzo). Washed thoroughly and incubated with secondary anti-mouse AlexaFlour-568 (1:250), with phalloidin AlexaFluor-633. Samples were rinsed three times in PBS + 0.2% triton X-100 for 10 minutes at room temperature, then mounted in Vectashield mounting medium (Vector Labs) and imaged. For quantification of protein aggregates in hemithoraces, single-channel images were converted into grayscale and the area and total number and % area of protein aggregates was measured using ImageJ.
Generation of axenic and re-associated flies
To generate axenic (germ-free) flies, embryos were treated by bleach and ethanol as described previously67. Briefly, <12-h-old embryos were dechorionated in 3% sodium hypochlorite (50% v/v regular bleach) for 20 min, rinsed in 70% ethanol for 5 min, and then washed three times with 1 × PBS + 0.01% Triton X-100. Axenic embryos were transferred to autoclaved media vials in a laminar flow cabinet. Axenic conditions within each vial were confirmed by plating swabs from spent vials on MRS agar before flies were taken as samples or pooled into a lifespan, as per68. To generate flies associated with microbes as embryos, whole fly homogenate (10 fly equivalent: 600 μL of conventionally reared fly homogenate glycerol stock/bottle) was added to medium containing axenic embryos.
Preparation of fly homogenate for re-association and adult feeding
Conventionally reared adult flies were surface sterilized by 70% ethanol prior to homogenization to ensure only internal microbes were present in the homogenate. Surface sterile flies were homogenized with a motor pestle in 1.5 mL tube with 200 μL of sterile PBS (50 flies/tube). Homogenates were then pooled and sterile PBS added to adjust to one fly equivalent in 50 μL PBS. For storage 1/5 volume of 80% sterile glycerol was added and aliquots were stored at −80 °C until use. For adult feeding, freshly prepared homogenate (one fly equivalent in 50 μL PBS) was added to standard food vials and allowed to dry. Unless otherwise noted, homogenate from 10 day post-eclosion wDahomey flies was used for homogenate-fed controls. To test the effects of rapamycin-induced changes to the microbiota on host lifespan, homogenate was prepared from 42 day post-eclosion wDahomey females reared either on rapamycin media or ethanol control media as outlined above.
Western blot
Samples for western blot were obtained from 10 day old female flies (head and thorax only). Samples were homogenized and lysates were separated by SDS page using standard procedures. Membranes were probed with primary antibodies against anti-phospho-p70 S6K T398 (dilution 1:2000, Cell Signaling, 9209) and anti-total S6K 1:300 (dilution 1:500, Santa Cruz, C-18, SC-230). The rabbit antibodies were detected using horseradish peroxidase-conjugated anti-rabbit IgG antibodies at 1:2000 (Sigma) and the mouse antibodies were detected using horseradish peroxidase-conjugated anti-mouse IgG antibodies 1:2000 dilution (Sigma). Amersham ECL chemiluminescent/chemifluorescent reagent (GE Healthcare) was used to visualize horse radish peroxidase activity, and the chemifluorescence was detected using a PXi Touch scanner (Syngene).
Statistics
All statistical tests were implemented in GraphPad Prism. The comparison of survival curves was done using the log-rank test. Comparison of Smurf proportion per time point was carried out using one-way ANOVA with Bonferroni’s multiple comparison’s test. Comparisons of ubiquitin aggregates were tested for significant differences using one-way ANOVA with the Newman-Keuls post-test. All other data comparisons were tested for significant differences using the Wilcoxon-Mann-Whitney U-test. All statistical tests are two-sided.
Supplementary information
Acknowledgements
We thank H. Tricoire, the Vienna Drosophila RNAi Center, and the Drosophila Stock Center (Bloomington) for fly stocks; Brenda Chan for help with fly work. This work was supported by NIH grants (R01AG037514, R01AG049157, and R01AG040288) to D.W.W. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This research was conducted while D.W.W. was a Julie Martin Mid‐Career Awardee in Aging Research supported by The Ellison Medical Foundation and AFAR.
Author Contributions
J.S. and A.R. designed, performed and analyzed experiments and wrote the manuscript. R.I.C. and W.W.J. designed and analyzed experiments. D.W.W. designed and analyzed experiments, acquired funding and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44106-5.
References
- 1.Fontana L, Kennedy BK, Longo VD, Seals D, Melov S. Medical research: treat ageing. Nature. 2014;511:405–407. doi: 10.1038/511405a. [DOI] [PubMed] [Google Scholar]
- 2.Kaeberlein M, Rabinovitch PS, Martin GM. Healthy aging: The ultimate preventative medicine. Science. 2015;350:1191–1193. doi: 10.1126/science.aad3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy BK, Lamming DW. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab. 2016;23:990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson JE, et al. Rapamycin slows aging in mice. Aging Cell. 2012;11:675–682. doi: 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjedov I, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai DF, et al. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13:529–539. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn JM, et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12:851–862. doi: 10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majumder S, et al. Lifelong rapamycin administration ameliorates age-dependent cognitive deficits by reducing IL-1beta and enhancing NMDA signaling. Aging Cell. 2012;11:326–335. doi: 10.1111/j.1474-9726.2011.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spilman P, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5:e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor, R. C. & Dillin, A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol3, 10.1101/cshperspect.a004440 (2011). [DOI] [PMC free article] [PubMed]
- 14.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilchez D, Saez I, Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun. 2014;5:5659. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 20.Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell. 2014;156:408–411. doi: 10.1016/j.cell.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark RI, Walker DW. Role of gut microbiota in aging-related health decline: insights from invertebrate models. Cell Mol Life Sci. 2018;75:93–101. doi: 10.1007/s00018-017-2671-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Clark RI, et al. Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell Rep. 2015;12:1656–1667. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langille MG, et al. Microbial shifts in the aging mouse gut. Microbiome. 2014;2:50. doi: 10.1186/s40168-014-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claesson MJ, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 27.Smith, P. et al. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife6, 10.7554/eLife.27014 (2017). [DOI] [PMC free article] [PubMed]
- 28.Thevaranjan N, et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host Microbe. 2017;21:455–466. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han B, et al. Microbial Genetic Composition Tunes Host Longevity. Cell. 2017;169:1249–1262. doi: 10.1016/j.cell.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khanna A, et al. A genome-wide screen of bacterial mutants that enhance dauer formation in C. elegans. Sci Rep. 2016;6:38764. doi: 10.1038/srep38764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabreiro F, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan X, et al. Rapamycin preserves gut homeostasis during Drosophila aging. Oncotarget. 2015;6:35274–35283. doi: 10.18632/oncotarget.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bitto, A. et al. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. Elife5, 10.7554/eLife.16351 (2016). [DOI] [PMC free article] [PubMed]
- 34.Jasper H. Exploring the physiology and pathology of aging in the intestine of Drosophila melanogaster. Invertebr Reprod Dev. 2015;59:51–58. doi: 10.1080/07924259.2014.963713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu DJ, Jasper H. Epithelia: Understanding the Cell Biology of Intestinal Barrier Dysfunction. Curr Biol. 2017;27:R185–R187. doi: 10.1016/j.cub.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 36.Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Annual rev of pathol. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- 37.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dambroise E, et al. Two phases of aging separated by the Smurf transition as a public path to death. Sci Rep. 2016;6:23523. doi: 10.1038/srep23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelino S, et al. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PLoS Genet. 2016;12:e1006135. doi: 10.1371/journal.pgen.1006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rera M, et al. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci USA. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell EL, et al. Reduced Intestinal Motility, Mucosal Barrier Function, and Inflammation in Aged Monkeys. J Nutr Health Aging. 2017;21:354–361. doi: 10.1007/s12603-016-0725-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio. 2014;5:e01117–14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 Promotes Gut Immune Homeostasis to Limit Commensal Dysbiosis and Extend Lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen, M., Rubinsztein, D. C. & Walker, D. W. Autophagy as a promoter of longevity-insights from model organsisms. Nat Rev Mol Cell Biol 19, 579–593, 10.1038/s41580-018-0033-y (2018). [DOI] [PMC free article] [PubMed]
- 48.Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 50.Doherty TJ. Invited review: Aging and sarcopenia. J Appl Physiol (1985) 2003;95(1727):1717. doi: 10.1152/japplphysiol.00347.2003. [DOI] [PubMed] [Google Scholar]
- 51.Demontis F, Piccirillo R, Goldberg AL, Perrimon N. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis Model Mech. 2013;6:1339–1352. doi: 10.1242/dmm.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rana A, et al. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat Commun. 2017;8:448. doi: 10.1038/s41467-017-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci USA. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulgherait M, Rana A, Rera M, Graniel J, Walker DW. AMPK Modulates Tissue and Organismal Aging in a Non-Cell-Autonomous Manner. Cell Rep. 2014;8:1767–1780. doi: 10.1016/j.celrep.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada, R., Deshpande, S. A., Bruce, K. D., Mak, E. M. & Ja, W. W. Microbes Promote Amino Acid Harvest to Rescue Undernutrition in Drosophila. Cell Rep, 10, 865-872, 10.1016/j.celrep.2015.01.018 (2015). [DOI] [PMC free article] [PubMed]
- 57.Keebaugh ES, Yamada R, Obadia B, Ludington WB, Ja WW. Microbial Quantity Impacts Drosophila Nutrition. Development, and Lifespan. iScience. 2018;4:247–259. doi: 10.1016/j.isci.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tefit MA, Leulier F. Lactobacillus plantarum favors the early emergence of fit and fertile adult Drosophila upon chronic undernutrition. J Exp Biol. 2017;220:900–907. doi: 10.1242/jeb.151522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong AC, Dobson AJ, Douglas AE. Gut microbiota dictates the metabolic response of Drosophila to diet. J Exp Biol. 2014;217:1894–1901. doi: 10.1242/jeb.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 61.Gopalakrishnan V, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matson V, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yip LY, Chan EC. Investigation of Host-Gut Microbiota Modulation of Therapeutic Outcome. Drug Metab Dispos. 2015;43:1619–1631. doi: 10.1124/dmd.115.063750. [DOI] [PubMed] [Google Scholar]
- 64.Scott TA, et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell. 2017;169:442–456 e418. doi: 10.1016/j.cell.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harrison B, Tran TT, Taylor D, Lee SD, Min KJ. Effect of rapamycin on lifespan in Drosophila. Geriatr Gerontol Int. 2010;10:110–112. doi: 10.1111/j.1447-0594.2009.00569.x. [DOI] [PubMed] [Google Scholar]
- 66.Claesson MJ, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bakula M. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J Invertebr Pathol. 1969;14:365–374. doi: 10.1016/0022-2011(69)90163-3. [DOI] [PubMed] [Google Scholar]
- 68.Resnik-Docampo M, et al. Tricellular junctions regulate intestinal stem cell behaviour to maintain homeostasis. Nat Cell Biol. 2017;19:52–59. doi: 10.1038/ncb3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.