Abstract
Myostatin (MSTN), a growth differentiation factor-8 regulates muscular development through its receptors, ACVR2A (Activin receptor type IIA) and ACVR2B (Activin receptor type IIB) by inhibiting cellular differentiation of developing somites during embryonic stage and diminishing myofibriller growth during post-embryonic period. The objective of this study was to compare the effect of knockdown of expression of myostatin, ACVR2A and ACVR2B genes on growth traits in chicken. The shRNAs for Myostatin, ACVR2A and ACVR2B genes were designed, synthesized and cloned in DEST vector. The recombinant molecules were transfected into the spermatozoa and transfected spermatozoa were inseminated artificially to the hens to obtain fertile eggs. The fertile eggs were collected, incubated in the incubator and hatched to chicks. Silencing of ACVR2B gene showed significantly higher body weight than other single, double and triple knock down of genes in transgenic birds. The carcass traits such as dressing%, leg muscle%, and breast muscle% were found with the highest magnitudes in birds with silencing of the ACVR2B gene as compared to the birds with that of other genes and control group. The lowest serum cholesterol and HDL content was found in ACVR2B silencing birds. The total RBC count was the highest in this group though the differential counts did not differ significantly among various silencing and control groups of birds. It is concluded that silencing of only one receptor of MSTN particularly, ACVR2B may augment the highest growth in chicken during juvenile stage. Our findings may be used as model for improving growth in other food animals and repairing muscular degenerative disorders in human and other animals.
Subject terms: Genetic engineering, RNAi
Introduction
Myostatin, a growth differentiating factor-8 is a member of transforming growth factor-β family expressed predominantly in the muscle tissues. Myostatin inhibits muscular development through cellular differentiation of developing somites during embryonic stage and growth of myofibrillar cells during adult stages in animals1,2. Thus, myostatin (MSTN) is regarded as the typical negative regulator of myogenesis in animals3. Earlier investigations have demonstrated biological activity of MSTN through its receptors such as activin receptor type IIB (ACVR2B, also known as ActrIIB) and activin receptor type IIA (also known as ACVR2A or ActrIIA). Both the receptors are also the members of TGF-ß super-family4. It is known that improper biological function of MSTN on account of mutation in the gene has been associated with the double muscling phenotype in cattle, suggesting that MSTN gene normally functions as a negative regulator of skeletal muscle growth in animals5. The function of MSTN also determined to be conserved across the species, as animals with genetic mutations in the MSTN gene, found in Belgian blue cattle, mouse, the whippet dog and human exhibiting hyper muscled phenotype6–10. In mice, absence of myostatin gene showed an enormous increase in skeletal muscle mass, which made animals approximately twice as those of wild type mice on account of muscle fiber hyperplasia and hypertrophy11. The enhanced expression of myostatin gene was found in humans having significant association with chronic illness, HIV infection and early aging due to muscle atrophy12–14. Thus, the MSTN has been a prime target for the development of therapies for chronic muscle degeneration (such as sarcopenia or muscle degenerative diseases), acute muscle loss (such as cachexia), and even metabolic diseases (such as obesity and Type II diabetes) in human. The discovery of biological inhibitors of MSTN, such as follistatin7, follistatin-related gene15, GDF associated serum protein-116, MSTN propeptide15, MSTN receptors17,18, ALK4 and ALK519 have offered a multifaceted approach for the treatment of muscular degenerative diseases through the neutralization of MSTN in the circulation. Exploiting these naturally occurring inhibitors or their derivatives by means of overexpression20 or gene delivery16 has produced substantial improvements in muscle mass, which implies that the regulation of skeletal muscle is not the sole responsibility of MSTN but is shared by other members of the transforming growth factor-β superfamily.
Activin IIA receptor mRNAs was first detected by in situ hybridization in embryonic spinal cord and ciliary ganglion neurons in chicken21,22. The mRNAs have also been reported to be present in the dorsal root ganglia during embryonic period, day 12–20 in rat and 12.5 day in mouse23,24. The extracellular domains of ACVR2A contained a three-fingered toxin fold25,26, which was formed by three pairs of anti-parallel β-sheets, beta1– beta2, beta3– beta4 and beta5–beta625,26. The cytoplasmic domain of ACVR2A is highly conserved and has the kinase activity. The protein has a small N-terminal lobe containing a five stranded anti-parallel β-sheet and a single α-helix, while a large C-terminal lobe containing α-helix and a loop involved in polypeptide binding. The N- and C-terminal lobes are connected by a hinge sequence, encompassing the binding site for ATP. The ACVR2A can be activated not only by activins, but also by other ligands, including myostatin and bone morphogenic proteins (BMPs). Thus, the ACVR2A is involved in a variety of biological functions including muscle growth, bone formation and viability as well as adhesion of prostatic epithelial cells27.
The activin type IIB receptor is a transmembrane serine-threonine kinase receptor for many members of the transforming growth factor-β (TGF) superfamily involved in the negative regulation of growth of muscle tissues through GDF8 pathway28. Thus, pharmacological capability of ACVR2B by blocking MSTN signaling pathway may have applications for promoting muscle growth in livestock/animals.
Rapid improvement of muscling in meat producing animals through lowering expression of MSTN and its receptors may be one of the most important approaches in livestock and poultry industry. The lowering expression of genes can be possible through many approaches of which gene silencing by shRNA has been one of them. In gene silencing, genomic modification of gene is not done but its transcribed products are degraded upon cleaving by shRNA molecules. In case of stable transfection, shRNA molecules are synthesized continuously, which has been a popular technique to achieve sequence-specific knockdown of target mRNA. Ability to obtain potent and stable RNAi silencing is critical for number of applications especially for gene knockdown of target gene validation29, functional genomic analysis30 and therapeutic use31. This approach in poultry may also be a role model for treatment of muscle decaying diseases in human. Hence, the objectives of the study were to silence the expression of MSTN and its receptors genes and to analyse its relative impact on growth and associated traits in transgenic chicken.
Results
Knock-down chicken
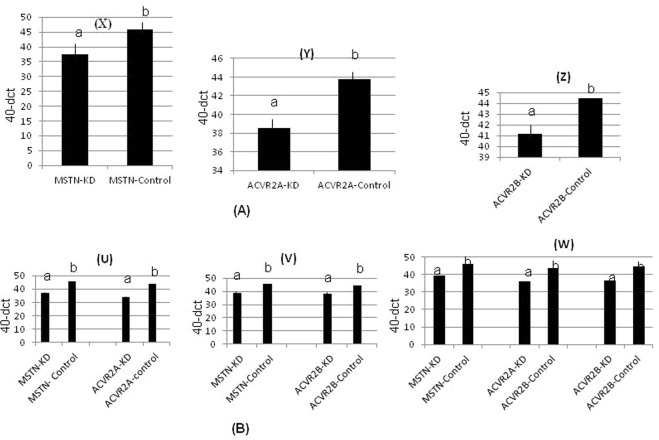
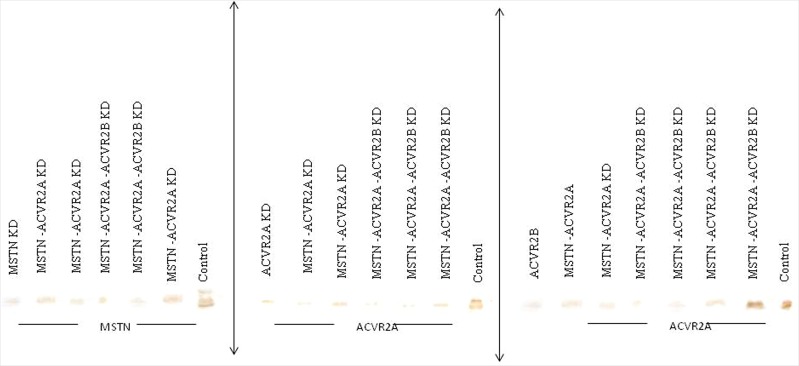
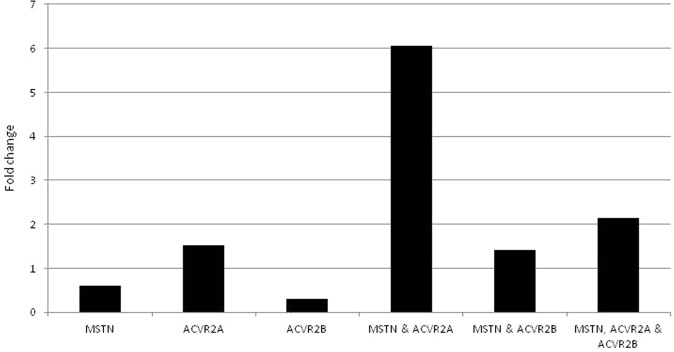
Sperm mediated gene transfer method (SMGT) was employed to develop knock down chicken. The hatching percentage varied among the treatment groups ranging from 60% in MSTN-ACVR2A-ACVR2B group to 75% in ACVR2B group (Table 1). All the birds were screened with PCR (Figs 1 and 2) and Southern blot (Fig. 3). The success rate of obtaining positive transgenic birds ranged from 6.8% in case of MSTN to 66.6% in MSTN-ACVR2A-ACVR2B group. After chicks were hatched, some of the chicks were died without any anatomical or other defects. The death rate was found to be the highest in MSTN-ACVR2A-ACVR2B group whereas the lowest rate (0%) was found in MSTN group. The expression of MSTN, ACVR2A and ACVR2B genes in transgenic group was detected by real time PCR, Western blotting and ELISA. The control group of birds showed higher mRNA expression of target genes over the single, double and triple knock down birds (Fig. 4A,B). In terms of fold change of expression, the control birds showed 337 folds higher expression of myostatin mRNA in myostatin knock down birds while the control birds had 831 and 222 folds higher expression of ACVR2A and ACVR2B mRNA than ACVR2A knock down and ACVR2B knock down birds, respectively. In double knock down birds, the control birds showed 512 and 776 folds higher expression of myostatin and ACVR2A mRNAs, respectively over the MSTN and ACVR2A double knock down birds. Likewise, the control birds showed 168 and 81 folds higher expression of MSTN and ACVR2B mRNAs, respectively over the MSTN and ACVR2B double knock down birds. In case of triple knock down, the control birds showed 93, 194 and 238 folds higher expression of MSTN, ACVR2A and ACVR2B mRNAs, respectively over the MSTN, ACVR2A and ACVR2B triple knock down birds. The Western blotting revealed the expression of the proteins in both knock down and control birds (Fig. 5). The ELISA results mentioned in the Table 1S also corroborated the lower levels of MSTN, ACVR2A and ACVR2B proteins in serum of single, double and triple knock down birds over the control group of birds. Knocking down of MSTN, ACVR2A and ACVR2B genes in the knock down birds reduced the level of mRNA expressions in the tissues, which lead to lower level of protein expression. The higher titres for MSTN, ACVR2A and ACVR2B were observed in the control group as compared to the knock down groups indicating higher serum concentrations of MSTN/ACVR2A/ACVR2B in the control group of birds. Thus, between knock down and control groups, the serum MSTN/ACVR2A/ACVR2B concentrations were diminished at the knock down birds as compared to the control groups indicating efficiency of knock down experiment to lower the level of MSTN/ACVR2A/ACVR2B protein expression. Further, we have also analysed the concurrent expression of interferon gamma gene in the knock down birds to explore the impact of shRNA on immune function. In our study, the introduction of recombinant shRNA clones did not show significant effect (P < 0.05) on expression of interferon gamma gene in knock down birds, which revealed the efficiency of shRNA molecules in development of knock down chicken (Fig. 6).
Table 1.
Hatching profile of knock down and control chicks.
| Knock down gene (s) | Number eggs incubated | Chicks hatched | Hatching% | shRNA positive chicks | Percentage of positive birds | Death percentage of positive birds |
|---|---|---|---|---|---|---|
| MSTN | 40 | 29 | 72.5 | 2 | 6.8 | − |
| ACVR2A | 40 | 27 | 67.5 | 6 | 22.2 | 16.6 |
| ACVR2B | 40 | 30 | 75.0 | 14 | 46.6 | 7.1 |
| MSTN-ACVR2A | 40 | 28 | 70.0 | 16 | 57.1 | 12.5 |
| MSTN-ACVR2B | 40 | 25 | 62.5 | 11 | 44.0 | 9.0 |
| MSTN-ACVR2A- ACVR2B | 40 | 24 | 60.0 | 16 | 66.6 | 37.5 |
| Control | 40 | 29 | 72.5 | − | − | − |
Figure 1.
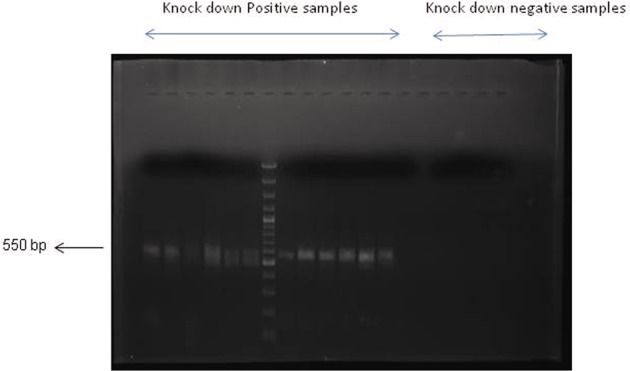
PCR based screening of knock down birds where presence of 550 bp fragment reveals the presence of recombinant DEST vector present in the genome.
Figure 2.
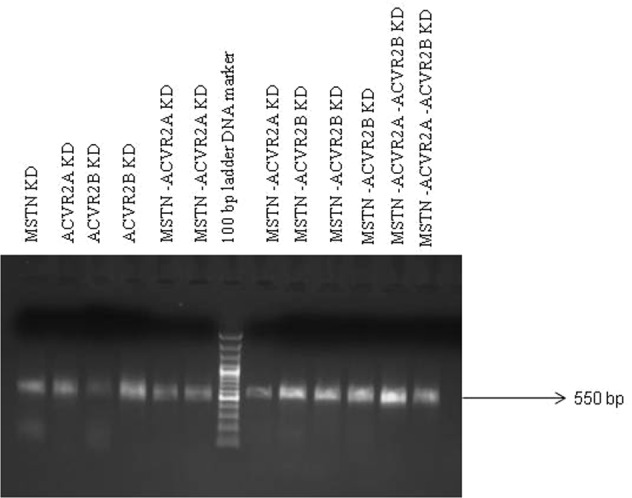
PCR based screening of knock down birds where presence of 550 bp fragment reveals the presence of recombinant DEST vector present in the genome. KD denotes knock down.
Figure 3.
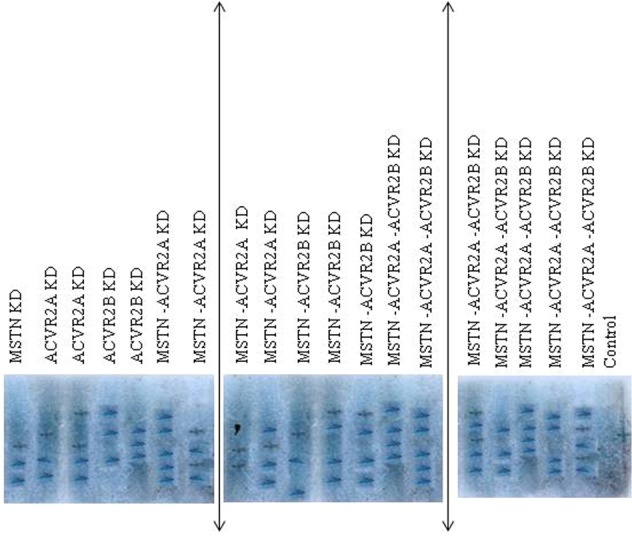
Southern blot of PstI digested genomic DNA collected from knock down and control chicken. KD denotes knock down. Blots are separated by black lines.
Figure 4.
(A) The mRNA expression level in knock down and control birds. (X) MSTN knock down group where MSTN-KD denotes level of MSTN mRNA in MSTN knock down birds and MSTN-Control denotes level of MSTN mRNA in control birds. (Y) ACVR2A knock down group where ACVR2A-KD denotes level of ACVR2A mRNA in ACVR2A knock down birds and ACVR2A-Control denotes level of ACVR2A mRNA in control birds. (Z) ACVR2B knock down group where ACVR2B-KD denotes level of ACVR2B mRNA in ACVR2B knock down birds and ACVR2B-Control denotes level of ACVR2B mRNA in control birds. Different superscripts indicate significance at P < 0.05. (B) The mRNA expression level in knock down and control birds. (U) Double knock down (MSTN & ACVR2A) group where MSTN-KD denotes level of MSTN mRNA in double knock down birds; MSTN-Control denotes level of MSTN mRNA in control birds; ACVR2A-KD denotes level of ACVR2A mRNA in double knock down birds and ACVR2A-Control denotes level of ACVR2A mRNA in control birds. (V) Double knock down (MSTN & ACVR2B) group where MSTN-KD denotes level of MSTN mRNA in double knock down birds; MSTN-Control denotes level of MSTN mRNA in control birds; ACVR2B-KD denotes level of ACVR2B mRNA in double knock down birds and ACVR2B-Control denotes level of ACVR2B mRNA in control birds. (W) Tripple knock down (MSTN, ACVR2A & ACVR2B) group where MSTN-KD denotes level of MSTN mRNA in tripple knock down birds; MSTN-Control denotes level of MSTN mRNA in control birds, ACVR2A-KD denotes level of ACVR2A mRNA in tripple knock down birds and ACVR2A-Control denotes level of ACVR2A mRNA in control birds; ACVR2B-KD denotes level of ACVR2B mRNA in tripple knock down birds and ACVR2B-Control denotes level of ACVR2B mRNA in control birds. Different superscripts indicate significance at P < 0.05.
Figure 5.
Western blot of myostatin/ACVR2A/ACVR2B expressed in breast muscle tissue collected from knock down and control groups of chicken. Blots are separated by black lines. KD denotes knock down.
Figure 6.
The fold change of mRNA expression of IFN-gamma in control group as compared to knock down birds. The expression was not differed significantly among the groups.
Effect of knock down on growth traits
The silencing of myostatin and its two receptors namely, ACVR2A and ACVR2B significantly (P < 0.05) affected body weights at different ages in chicken (Table 2). At day old age, silencing of MSTN gene showed the highest body weight in chicken. At day 28, silencing of only ACVR2B and MSTN, and combination of MSTN, ACVR2A and ACVR2B showed higher body weights than the control birds. However, silencing of ACVR2B gene in transgenic birds had the highest body weight at 28th (454.2 ± 18.7 g), 35th (703.4 ± 20.6 g) and 42nd (779.1 ± 21.4 g) day of age. In case of MSTN knock down and triple knock down of MSTN, ACVR2A and ACVR2B genes, respectively, the body weight at 35th day (668.3 ± 12.2 and 657.2 ± 32.4 g) and 42nd day (735.9 ± 14.0 and 732.1 ± 32.8 g) was significantly higher than the control group (581.5 ± 8.8 and 639.1 ± 10.3 g).
Table 2.
Growth traits in terms of body weights in knock down and control chicken. Column-wise different superscripts indicate significance at P < 0.05.
| Knock down gene (s) | Average body weight (g) at | ||||
|---|---|---|---|---|---|
| Day old | 2nd week | 4th week | 5th week | 6th week | |
| MSTN | 45.9 ± 2.5b | 158.9 ± 0.5 | 450.1 ± 28.7b | 668.3 ± 12.2b | 735.9 ± 14.0b |
| ACVR2A | 42.9 ± 0.4ab | 143.9 ± 5.7 | 414.7 ± 22.0ab | 615.2 ± 20.3a | 705.7 ± 27.4ab |
| ACVR2B | 41.2 ± 0.8ab | 158.8 ± 5.6 | 454.2 ± 18.7b | 703.4 ± 20.6b | 779.1 ± 21.4b |
| MSTN-ACVR2A | 37.7 ± 1.3a | 147.8 ± 5.0 | 377.6 ± 13.1a | 585.0 ± 19.0a | 662.3 ± 25.0a |
| MSTN-ACVR2B | 41.4 ± 0.9ab | 152.0 ± 6.2 | 401.7 ± 19.5ab | 640.7 ± 34.5ab | 697.9 ± 31.2a |
| MSTN-ACVR2A-ACVR2B | 40.4 ± 0.6a | 159.3 ± 3.4 | 450.3 ± 16.3b | 657.2 ± 32.6b | 732.1 ± 32.8b |
| Control | 39.0 ± 0.4a | 143.2 ± 2.8 | 356.3 ± 6.3a | 581.5 ± 8.8a | 639.1 ± 10.3a |
Evaluation of muscle fibre
The electron microscopic views of muscle fibre of knock down and control birds have been presented in Fig. 7. The diameter of muscle fibres were significantly (P < 0.05) affected by silencing of ACVR2B and MSTN genes in transgenic chicken. The highest fibre diameter was observed in birds with silencing of ACVR2B gene (27.9 ± 0.8 µm) whereas the smallest fibre width was observed in control birds (16.1 ± 1.1 µm). In case of myofibril numbers in breast muscle, we observed significantly (P < 0.05) higher number of myofibrils in ACVR2B, MSTN and MSTN- ACVR2A- ACVR2B group as compared to control (Table 3). The highest number of myofibrils was found in ACVR2B group (4.1 ± 0.25). The silencing of MSTN and MSTN-ACVR2A-ACVR2B also showed significantly (P < 0.05) higher number of myofibrils (3.7 ± 0.22 and 3.4 ± 0.29, respectively) as compared to the control group (2.2 ± 0.23). We estimated correlation coefficient between body weights at different ages and muscle fibre number. The correlation between muscle fibre number and body weights at day old, 2nd, 4th, 5th and 6th week of age were 0.5, 0.8, 0.9, 0.9 and 0.9, respectively.
Figure 7.
Electron microscopic view (SEM) of breast muscle fibres in the control and knock down birds for MSTN, ACVR2A and ACVR2B genes individually as well as in combination.
Table 3.
Muscle fibre parameters in knock down and control chicken. Row-wise different superscripts indicate significance at P < 0.05.
| Parameter | Knock down gene (s) | ||||||
|---|---|---|---|---|---|---|---|
| MSTN | ACVR2A | ACVR2B | MSTN-ACVR2A | MSTN-ACVR2B | MSTN-ACVR2A-ACVR2B | control | |
| Muscle fibre size (µm) | 19.9 ± 1.6a | 26.1 ± 2.4b | 27.9 ± 0.8b | 26.1 ± 2.9b | 27.7 ± 1.1b | 26.2 ± 3.7b | 16.1 ± 1.1a |
| Muscle fibre No. | 3.7 ± 0.22b | 2.8 ± 0.31a | 4.1 ± 0.29c | 2.5 ± 0.34a | 2.6 ± 0.27a | 3.4 ± 0.29b | 2.2 ± 0.23a |
Evaluation of carcass traits
The carcass quality traits such as dressing%, leg muscle% and breast muscle% were significantly (P < 0.05) affected by silencing of MSTN, ACVR2A and ACVR2B genes (Table 4). The dressing% was significantly higher in birds with silencing of ACVR2B, MSTN-ACVR2A, MSTN-ACVR2B and MSTN-ACVR2A-ACVR2B genes as compared to the control birds. In case of breast muscle%, the highest magnitude was observed in birds with silencing of MSTN-ACVR2B gene (38.4 ± 2.9%). In case of leg muscle%, silencing of ACVR2B gene had the best (27 ± 0.5%) performance over other groups.
Table 4.
Carcass characteristics in knock down and control chicken. Column-wise different superscripts indicate significance at P < 0.05.
| Knock down gene (s) | Dressing% | Neck% | Breast% | Leg% | Spleen% | Bursa% | Back% | Heart% |
|---|---|---|---|---|---|---|---|---|
| MSTN | 68.7 ± 6.8a | 5.0 ± 0.3 | 25.0 ± 3.2a | 27.1 ± 3.1b | 0.2 ± 0.001 | 0.2 ± 0.001 | 30.5 ± 4.2 | 0.6 ± 0.006 |
| ACVR2A | 70.1 ± 3.2a | 5.2 ± 0.3 | 31.0 ± 1.5ab | 23.9 ± 0.9ab | 0.2 ± 0.1 | 0.1 ± 0.001 | 25.7 ± 1.0 | 0.8 ± 0.1 |
| ACVR2B | 75.3 ± 1.8b | 4.0 ± 0.3 | 31.4 ± 0.7ab | 27.0 ± 0.5b | 0.2 ± 0.001 | 0.2 ± 0.001 | 24.0 ± 0.3 | 0.6 ± 0.1 |
| MSTN-ACVR2A | 81.3 ± 3.0b | 4.4 ± 0.3 | 33.9 ± 1.9b | 22.3 ± 1.1a | 0.2 ± 0.001 | 0.1 ± 0.001 | 23.5 ± 1.1 | 0.7 ± 0.0 |
| MSTN-ACVR2B | 77.6 ± 4.2b | 3.9 ± 0.2 | 38.4 ± 2.9b | 20.3 ± 1.7a | 0.2 ± 0.001 | 0.1 ± 0.001 | 21.2 ± 1.5 | 0.6 ± 0.1 |
| MSTN-ACVR2A-ACVR2B | 80.4 ± 1.7b | 3.5 ± 0.3 | 27.6 ± 2.2a | 20.7 ± 1.3a | 0.2 ± 0.001 | 0.1 ± 0.001 | 22.8 ± 0.9 | 0.5 ± 0.0 |
| Control | 69.5 ± 1.2a | 3.3 ± 0.3 | 27.6 ± 2.1a | 19.3 ± 1.3a | 0.3 ± 0.001 | 0.2 ± 0.001 | 25.9 ± 1.7 | 0.7 ± 0.1 |
The serum cholesterol, triglycerides and HDL content were significantly (P < 0.05) affected by silencing of ACVR2A, ACVR2B, MSTN-ACVR2A and MSTN-ACVR2B genes (Table 5). In case of serum cholesterol content, the birds with silencing of ACVR2B gene had the lowest magnitude (72.1 ± 8.6 mgh/dL) over other groups of birds including control group (91.5 ± 3.6 mg/dL). In case of serum triglycerides, birds with silencing of ACVR2A, ACVR2B and MSTN-ACVR2B genes had significantly (P < 0.05) higher quantity (62.7 ± 16.8, 63.3 ± 28.6 and 55.8 ± 12.6mg/dL, respectively) in serum over the control group of birds (30.6 ± 4.4 mg/dL). In case of HDL, birds with silencing of ACVR2B and MSTN-ACVR2A genes showed significantly (P < 0.05) higher quantity (46.8 ± 3.1 and 50.5 ± 4.0 mg/dL, respectively) than the control group of birds (20.0 ± 3.4 mg/dL).
Table 5.
Serum biochemical parameters in knock down and control chicken. Column-wise different superscripts indicate significance at P < 0.05.
| Knock down gene (s) | Cholesterol (mg/dL) | Triglycerides (mg/dL) | HDL (mg/dL) |
|---|---|---|---|
| MSTN | 97.5 ± 20.1b | 22.8 ± 7.8a | 11.2 ± 3.4a |
| ACVR2A | 98.5 ± 19.1b | 62.7 ± 16.8b | 22.9 ± 5.2a |
| ACVR2B | 72.1 ± 8.6a | 63.3 ± 28.6b | 46.8 ± 3.1b |
| MSTN-ACVR2A | 131.8 ± 9.2c | 26.7 ± 3.9a | 50.5 ± 4.0b |
| MSTN-ACVR2B | 97.1 ± 13.0b | 55.8 ± 12.6b | 24.7 ± 7.0a |
| MSTN-ACVR2A-ACVR2B | 86.5 ± 6.3b | 37.6 ± 11.9a | 25.5 ± 6.2a |
| Control | 91.5 ± 3.6b | 30.6 ± 4.4a | 20.0 ± 3.4a |
Evaluation of blood cell profile
The blood cell profile was explored in knock down and control birds where significantly different total RBC and WBC count were observed among the knock down and control groups (Table 2S). The significantly higher number of RBC was found in birds silenced with ACVR2B (5.35 ± 0.15 million/ml) and MSTN-ACVR2A-ACVR2B (5.09 ± 0.23million/ml) genes. In all the groups of knock down birds except for MSTN, the total RBC was found to be higher than control group. In case of WBC count, the number was significantly reduced in all knock down birds as compared to the control one. However, the highest reduction over control group was detected in the birds of ACVR2A silencing (5884 ± 444 vs 9459 ± 898). The birds of MSTN-ACVR2A and MSTN-ACVR2A-ACVR2B groups did not show significant changes of total WBC count as compared to that of the control group of birds. Other blood cells did not show any significant change between knock down and control groups.
Discussion
The transgenic knock down chicken was produced by employing sperm mediated gene transfer method where the average success rate of 40.5% was quite encouraging. In earlier studies, we standardized the techniques of production of transgenic knock down chicken, where we observed that the sperm mediated method was quite efficient with variable success rate in the range of 30.5 to 40.9% to produce knock down birds31. In another study, authors reported sperm transfection assisted gene editing (STAGE) an alternative technique to produce knock out or transgenic chicken32. However, the present sperm mediated method has been successful to develop knock down/transgenic chicken for MSTN and its receptors. For each gene, 2 best shRNA molecules tested previously in cell culture were used in vivo to produce knock down transgenic chicken33–35. The silencing of three genes namely, MSTN and its receptors, ACVR2A and ACVR2B were analysed separately as well as in different combinations such as MSTN-ACVR2A, MSTN-ACVR2B and MSTN-ACVR2A-ACVR2B. The knock down birds did not show any anatomical changes, but we found 7.1 to 37.5% dead chicks in all knock down groups except MSTN knock down group. The highest death percentage was observed in knocking down of combination of all three genes. This may be due to improper muscular development and physiological inefficiency of embryo on account of knocking down of MSTN, ACVR2A and ACVR2B genes. However, healthy knock down chicks were sacrificed at 6 weeks of age on account of collection of tissues and assessing carcass quality traits.
Important physiological parameters such as blood cell profiles reveals that the birds of ACVR2B group had 25.5% higher number of RBC than that of the control group, which may be due to mitigating growing requirement of cellular metabolism in relatively large sized body of transgenic chicken compared to the control birds. We suggest that silencing of MSTN alone cannot increase the number of RBC but in presence of silencing of ACVR2A and ACVR2B, the number of total RBC have been increased to meet the body requirement such as supply of oxygen to the cells. The other blood cell such as WBC count was also influenced by gene silencing. The total WBC count was significantly decreased in single knock down of MSTN, ACVR2A and ACVR2B genes as compared to the control group. It is known that WBCs are important in conferring immunity to the animals36. Lowering the number of WBCs on account of gene knock down may have deleterious effect in sustaining immunity in the birds.
We compared the growth performance of the chicks derived from knock down and control groups. The body weights at different ages during juvenile stages were significantly affected by silencing of MSTN, ACVR2A and ACVR2B genes in chicken. At day old age, the birds with silencing of MSTN gene had the highest body weight in knock down group with 17.6% superiority over the control group of birds. At 35 and 42 days of age, silencing of ACVR2B gene took upper hand having the highest body weights with 20.8 and 21.9% superiority, respectively over the control group. In addition, carcass traits which are very important in chicken were also analyzed in transgenic and non-transgenic control birds. The dressing% which has been the major indicator of carcass quality was found as significantly lower in control group of birds as compared to ACVR2B, MSTN-ACVR2A, MSTN-ACVR2B and MSTN-ACVR2A-ACVR2B knock down birds where the trend of dressing % was MSTN-ACVR2A > MSTN-ACVR2A-ACVR2B > MSTN-ACVR2B > ACVR2B > MSTN. The MSTN-ACVR2A, MSTN-ACVR2A-ACVR2B, MSTN-ACVR2B and ACVR2B silenced birds had 16.9%, 15.6%, 11.6% and 8.3% superiority of dressing% over the control group birds.
Other important carcass trait such as breast muscle and leg muscle% were also in higher side in knock down chicken over the control birds. The significantly higher magnitudes were observed in MSTN-ACVR2B (38.4 ± 2.9%) and MSTN-ACVR2A (33.9 ± 1.9%) knock down birds for breast muscle and MSTN (27.1 ± 3.1%) and ACVR2B (27.0 ± 0.5%) knock down birds for leg muscle over control group of birds (27.6 ± 2.1 and 19.3 ± 1.3%, respectively). The similar trend of muscular hypertrophy was observed in MSTN knock down in Zebra fish37, catfish38 and human39 revealing importance gene silencing for MSTN for enhancing growth.
In meat animals, another major factor for meat consumption is serum cholesterol which compels us to eat chicken products in limited quantity. Higher serum cholesterol will make chicken produce rich in cholesterol content, which is the most undesired component of chicken egg and meat. Cholesterol is the most important predisposing factor for cardiac ailment. In this study, the serum cholesterol content was significantly lowest in ACVR2B knock down chicken over the control group emphasizing acceptability of poultry produce developed through this technique. The HDL content in serum is very important as HDL has been the good lipid required in the body for maintaining different physiological functions such as causing reverse cholesterol transport from the periphery to the liver and controls pleiotropic effects on inflammation, haemostasis and apoptosis leading to minimizing coronary heart diseases in human40, muscular dystrophy and type II diabetes41. In our study, we have found significantly (P < 0.05) higher serum HDL content in the birds of ACVR2B and MSTN-ACVR2A group as compared to the control one. The ACVR2B and MSTN- ACVR2A group had 134% and 152.5% higher HDL content in serum, respectively over the control birds. As the HDL content was high and serum cholesterol content was low in birds with ACVR2B silencing, it may be considered that meat of ACVR2B silenced birds may be of good quality with nutritionally best suited to the consumers with heart and high blood pressure ailments. Thus, the enhancement of quality of chicken meat can be possible through silencing of these genes both individually and in combination. We suggest that out of these three negative factors for controlling growth, single knock down for ACVR2B gene might be the best approach for improving growth in chicken. By knocking down only one factor, ACVR2B is sufficient to obtain optimum results as MSTN would not find its receptor to act negatively on growth. Thus, we can suggest that RNAi technology may be applied for augmenting production performance of animals and treatment of diseases in human and animals42.
It is concluded that single knock down of MSTN and its receptors (ACVR2A and ACVR2B) had potential to enhance body weight as compared to the combinations of MSTN and its receptors. These novel findings may help to utilize this technique for enhancing body mass in other food animals and to explore this technique in curing muscular decaying diseases in human being.
Methods
Animals
The broiler based PD-1 chicken line maintained at the Institute farm, ICAR-Directorate of Poultry Research, Hyderabad, India was included in the present study. A total of 280 fertile eggs were collected from 134 hens inseminated with the pooled semen prepared by collecting from 35 cocks. The experimental design has been detailed in Table 1. The whole study has been approved by the Institute Animal Ethics committee (IAEC) and Institute Biosafety Committee (IBSC) of ICAR-Directorate of Poultry Research, Hyderabad, India. The work was also noted by the RCGM, Dept. of Biotechnology, Govt. of India and the bio-safety guidelines of RCGM were also followed during the experiment. The birds were sacrificed following the standard protocol of cervical dislocation approved by the IAEC committee of the Institute. The animal welfare measures like ad lib feeding, watering and management of the hatched birds were taken care off by providing required space during the experiment.
Designing and cloning of shRNA oligos
The shRNA molecules were designed from the coding sequence of chicken MSTN (Accession No. AF346599), ACVR2A (Accession No. NM_205367.1) and ACVR2B genes (Accession No. NC_204317) with Invitrogen Block-iT RNAi designer programme (Table 6). Two best shRNA molecules were identified based on our previous study conducted under in vitro cell culture system for MSTN31, ACVR2A32 and ACVR2B genes33. In this study, BLOCK-iT U6 RNAi Entry Vector (Invitrogen) and pBLOCK-iT DEST6 vector (Invitrogen) were used to generate permanent expression clones. Earlier, the pBLOCK-iT Dest vector was also reported to be used for stable RNA interference against CXCR7 gene43. The pBLOCK-iT™6-DEST vector is promoterless Gateway® destination vector into which the RNAi cassette is transferred. The DEST vector allows stable expression of short hairpin RNA (shRNA) in dividing eukaryotic cells for RNAi analysis. Double-stranded oligo encoding an shRNA to the target gene was cloned in U6 promoter guided pENTR™/U6 vector to prepare RNAi cassette. The entire RNAi cassette is recombined to the pBLOCK-iT™-DEST vector to generate the expression clone so that shRNA molecules are synthesized in the body (Fig. 8).
Table 6.
shRNA molecules against MSTN, ACVR2A and ACVR2B genes transfected for production of knock down chicken.
| Gene | Molecule name | shRNA molecules (5′-3′) | NCBI Accession No. | Location in the gene | Reference |
|---|---|---|---|---|---|
| MSTN | shRNA3 | CACCGACTGTGATGAGCACTCAACGCGAACGTTGAGTGCTCATCACAGTC | AF346599 | 298 | Bhattacharya et al.33 |
| AAAAGACTGTGATGAGCACTCAACGTTCGCGTTGAGTGCTCATCACAGTC | |||||
| shRNA4 | CACCGCTCCGGAGAATGTGAATTTGCGAACAAATTCACATTCTCCGGAGC | 811 | |||
| AAAAGCTCCGGAGAATGTGAATTTGTTCGCAAATTCACATTCTCCGGAGC | |||||
| ACVR2A | shRNA1 | CACCGCAGTTGCTAGAGATCAAAGCCGAAGCTTTGATCTCTAGCAACTGC | NM_205367.1 | 89 | Satheesh et al.34 |
| AAAAGCAGTTGCTAGAGATCAAAGCTTCGGCTTTGATCTCTAGCAACTGC | |||||
| shRNA2 | CACCGGATGTACAGACATCACAAGCCGAAGCTTGTGATGTCTGTACATCC | 476 | |||
| AAAAGGATGTACAGACATCACAAGCTTCGGCTTGTGATGTCTGTACATCC | |||||
| ACVR2B | shRNA1 | CACCGCAACTACTGCAATGAGAAATCGAAATTTCTCATTGCAGTAGTTGC | NC_204317 | 317 | Guru Vishnu (2016) |
| AAAACAACTACTGCAATGAGAAATTTCGATTTCTCATTGCAGTAGTTGC | |||||
| shRNA3 | CACCGGTGGGAACAAGGAGGTATATCGAAATATACCTCCTTGTTCCCACC | 836 | |||
| AAAAGTGGGAACAAGGAGGTATATTTCGATATACCTCCTTGTTCCCACC |
Figure 8.
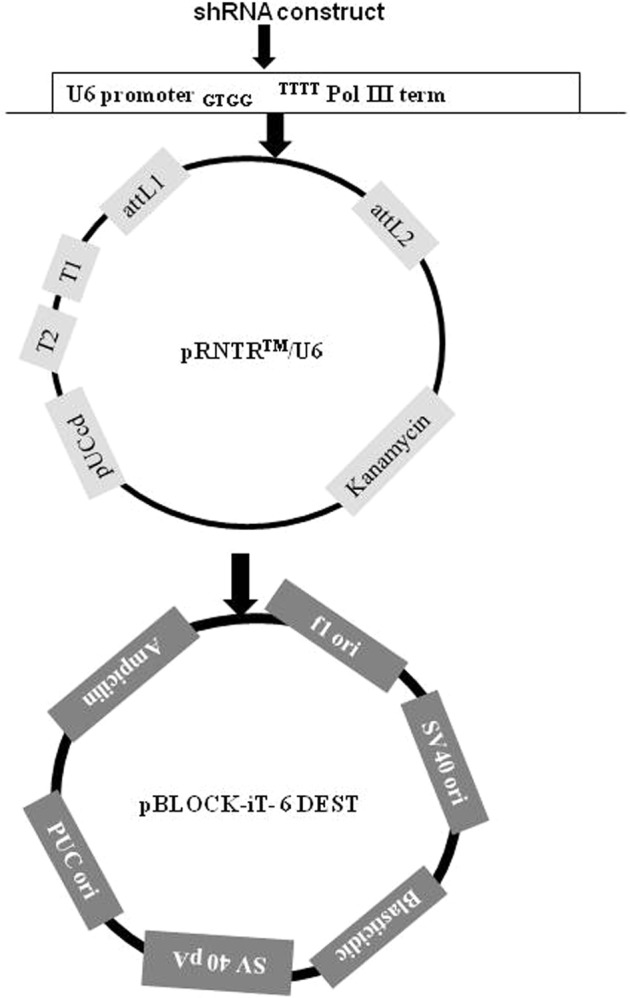
Schematic diagram of maps of two vectors (pRNTRTM/U6 and pBLOCK-iT- 6 DEST).
Sperm transfection with shRNA molecules
A total of 35 cocks of PD-1 broiler chicken line were randomly selected for collection of semen. The semen collected from all the animals were pooled and centrifuged at 500 g for 10 min to remove seminal plasma. The sperms were washed with HT ‘High Temperature’ buffer (NaCl-0.8 g, TES buffer-1.37 g and glucose-0.69 g) for three times. The total number of sperm was counted in the Neubeur chamber. The sperms were transfected with 10 µg shRNA of myostatin, ACVR2A and ACVR2B clones by electroporation with Gene Pulser (Biorad) at 160 mV for 25 milisecond for 1 pulse. A volume of 0.25 ml HT buffer containing about 100 Million sperm was inseminated to each hen and accordingly, the total quantity of sperm was divided into 6 knock down groups and 1 control group (Only electrical impulse without DNA was given to the sperm before insemination). Accordingly, the sperm allotted to each group was transfectd separately with by electroporation. The transfected sperms were inseminated to each hen under each treatment group. The insemination with same protocol was repeated on the consecutive days and from 3rd day onwards, eggs were collected and kept at 4 °C temperature till incubation. The eggs were incubated at the incubator for 18 days at 98–100 °F with 78–80% relative humidity and turning 6 times a day. Then, eggs were candled on 19th day and the fertile eggs were further kept in the hatcher for 3 days at 98–100 °F with 78–80% relative humidity. The chicks were hatched and and all chicks were marked with wing band. The chicks were maintained in the brooder house upto 6 weeks of age by providing ad lib feeding, watering and lighting to warm the house.
Screening of positive knock-down birds by PCR and Southern blotting
Blood samples were collected from all the birds under study alongwith the control group. Genomic DNA was isolated following standard protocol44. The genomic DNA was used for PCR and Southern blotting. For PCR, a pair of primers (DESTFQ: ACGTCGACGGATCGGGACGAGATC DESTRQ: CGCTAGGGCGCTGGCAAGTG) were designed from the DEST vector for amplification of 550 bp fragment of the vector encompassing 50 bp shRNA molecules of the genes. Through PCR, the positive birds were screened first and then, all the positive birds were also subjected to Southern blotting by digesting with PstI restriction enzyme for confirmation. Probes were prepared from DEST vector, which were lebeled with biotin-streptavidin conjugated to alkalin phosphatase and detected with NBT/BCIP for spot hybridization.
Real time PCR
For real time PCR, total RNA was isolated from the breast muscle tissues of birds of the treatment as well as and control groups using Trizol according to the Manufacturer’s instruction (Sigma). The total RNA was reverse transcribed with oligo dT primer to synthesize single stranded cDNA. The real time PCR of the respective treatment and control groups was performed in duplicate for myostatin, ACVR2A, ACVR2B and GAPDH genes with SYBR Green chemistry. The pairs of primers designed for MSTN (Accession No.AF346599), ACVR2A (Accession No. NM_205367.1) and ACVR2B (Accession No. NC_204317) and GAPDH (Accession No. NM_204305.1) genes for real time PCR were MYTQF: GCA AAA GCT AGC AGT CTA TG and MYTQR: TCC GTC TTT TTC AGC GTT CT for myostatin, 5′-GATCCTGGACCACCACCGC-3′ and 5′-CTGGATAGGGAATATTTTGACTG-3′ for ACVR2A, F: 5′-AGGGCAACTACTGCAATGAG-3′ and R:5′-CACTGACAGGACAGCGATG-3′ for ACVR2B and QGAPDHF: 5′-CTGCCGTCCTCTCTGGC-3′ and QGAPDHR: 5′-GACAGTGCCCTTGAAGTGT-3′ for GAPDH genes. The GAPDH was used as reference gene. The amplification and comparative analysis of mRNA expression was performed by calculating 40-Δct in both knock down and control groups according to the method mentioned in the literature45, where, ∆Ct = Average Ct of target gene – Average Ct of reference gene.
Fold change of expression = 2−∆∆Ct 46, where
Western blotting
The birds of all the groups at the age of 6 weeks were slaughtered by cervical dislocation method approved by the Institute Animal Ethics Committee of ICAR-Directorate of Poultry Research, Hyderabad, India. The breast muscle tissues of all the birds were collected and the crude protein was prepared from the tissues. The protein samples were electrophoresed in Tris-Glysin-SDS buffer (pH 8.3) at 60 V in 12% polyacrylamide gels of which one set of gels were stained with coomassie brilliant blue for visualization of resolved protein bands and another set of gels were used to transfer proteins onto PVDF membrane in the semidry transfer apparatus (Biorad) at 100 V for 1 hour. Then, the membrane was treated with anti rat IgG-HRP conjugate (Sigma) diluted to 1:1000 in TBS Tween buffer and visualized by treatment with DAB substrate using standard protocol45.
Elisa
The ELISA of serum samples of the experimental birds were carried out to detect the level of MSTN, ACVR2A and ACVR2B proteins in the knock down and control birds. Detection of myostatin, ACVR2A and ACVR2B are based on standard sandwich enzyme-linked immuno-sorbent assay technology. An antibody specific for MSTN or ACVR2A or ACVR2B had been pre-coated onto a 96-well plate (12 × 8 Well Strips) and blocked. Standards or test samples are added to the wells, incubated and removed. HRP detector antibody specific for MSTN or ACVR2A or ACVR2B was added, incubated and followed by washing. HRP-Peroxidase Conjugate was then added, incubated and unbound conjugate was washed away. An enzymatic reaction was produced through the addition of TMB substrate which was catalyzed by HRP generating a blue color product that changed yellow after adding acidic stop solution. The density of yellow coloration read by absorbance at 450 nm in ELISA reader was quantitatively proportional to the amount of sample MSTN or ACVR2A or ACVR2B captured in the well. The absorbance values for MSTN, ACVR2A and ACVR2B protein content in the serum of knock down and control group of birds estimated through ELISA were compared between knock down and control groups, and between titres within each group following LSD test (SPSS 20.0 software).
Growth and carcass traits
All the experimental birds including control were included in the study for measuring body weights at day1 and 2nd, 4th, and 6th week of age. The growth rates between day1 to day14, day15 to day28 and day29 to day42 were calculated for all the birds. Feed efficiency of all the birds were calculated by amount of feed consumed for attaining 1 kg body weight at 6 weeks of age. All the experimental birds were sacrificed by cervical dislocation. Different slaughter parameters such as carcass weight, dressing%, breast%, leg%, back%, wing%, neck% and heart% were measured.
Blood cell profiles and biochemical parameters
Blood samples were collected from the knock-down, scrambled and control birds. The blood cells were counted by hemocytometer for all the samples. For RBC counting, blood was taken upto 0.5 mark in the RBC pipette and then, Hyaem’s fluid was sucked upto 101 mark in that pipette. Then, blood and Hyaem’s fluid was mixed properly and initial few drops of blood mixture was thrown out. Then, the blood mixture was charged in the Neubeur chamber of the Haemocytometer and RBC was counted in the chamber with 40X magnification under microscope. Total RBC concentration was calculated following the standard formula.
For total WBC counting, blood was taken upto 0.5 mark in the WBC pipette and then, Truck’s fluid (WBC dilution fluid) was sucked upto 11 mark in that pipette. Then, blood and Truck’s fluid was mixed properly and initial 3 to 4 drops of mixture was thrown out. Then, the blood mixture was charged in the Neubeur chamber of the Haemocytometer and WBC was counted in the chamber with 10X magnification under microscope. Total WBC concentration was calculated following the standard formula.
For differential count, blood smear was prepared on a glass slide and fixed with methanol. The slide was stained first with Field stain-B (Eosin- 1 g; Disodium hydrogen phosphate-5g; Potassium dihydrogen phosphate-6.25 g; Distilled water-500 ml) and then, with Field stain-A (Azrure I- 0.5 g; Disodium hydrogen phosphate-5g; Potassium dihydrogen phosphate-6.25 g; Methylene blue- 0.8 g; Distilled water-500 ml). The slide was washed in water, dried and observed under the Microscope to visualize the WBCs (Neutrophil, Eosinophil, Basophil, Lymphocyte and Monocyte), where cytoplasm took acidic stain and nucleus took basic stain.
The blood samples without anti-coagulant were used to collect serum of all the birds. The serum samples were used to estimate triglyceride, total cholesterol and HDL in Blood analyzer with the analysis kit (Jeev Diagnostics Pvt. Ltd, India) following Manufacturer’s protocol.
Electron Microscopy
Breast muscle sample of two birds from each experimental group including control were fixed in 2.5% gluteradehyde in 0.1 M phosphate buffer for 30 minutes (ph 7.2) and were mounted over cover glass placed on the double sided carbon conductivity tape of the aluminum stubs. After fixing, cover slips were put on 1% aqueous osmium tetraoxide fumes for 30 mints, and exposed to 80% and 100% alcohol fumes each 15 minutes and dried under vacuum desiccation for 15 minutes. A thin layer of gold coating over the samples were done by using an automated sputter coater (Model-JEOL JFC-1600) for 3 minutes and scanned under Scanning Electron Microscope (SEM-Model:JOEL-JSM 5600) at required magnification as per the standard procedure at RUSKA Lab, College of Veterinary Science, PVNRTVU, Rajendranagar, Hyderabad, India47. The muscle fibre diameter (in millimeter unit) was measured at different location of the fibre with Vernier calipers and then, divided by the magnification observed under the microscope to arrive the average diameter of the muscle fibre. The diemeter estimation was also performed at RUSKA Lab, College of Veterinary Science, PVNRTVU, Rajendranagar, Hyderabad, India.
Immuno-histochemistry
The breast muscle tissues were collected from knock-down and control birds. The tissues were kept in formalin (10%) overnight. Then, the tissues were cleaned with running tap water water. The tissues were treated with Xyline and put in paraffin wax at −10 °C. On next day, slides (6 µm) were prepared using microtome machine (Medimeas: Model No. MRM-ST) and kept for drying at 40 °C for 2 to 3 hours. The slides were stained with Haematoxyline-Eosine stain. The slides were mounted with DPX and examined under microscope for counting muscle fibres.
Statistical analysis
The effect of shRNA molecules of MSTN, ACVR2A and ACVR2B genes on growth traits, carcass traits, blood profiles, serum biochemical parameters, and number as well as diameter of muscle fibres were analysed by GLM procedure with SPSS20.0 software. The shRNA class and sex of the birds were used as fixed effects. The model used for analyzing effects was Y = µ + Ri + Sj + Eijk
Where, Y = Trait; µ = Overall mean; Ri = Fixed effect of ith shRNA molecule; Sj = Fixed effect of jth sex and Eijk = kth residual effect
The Duncan’s multiple range test (DMRT) was performed to determine the effect of each group. The correlation coefficient between number of muscle fibres and body weights at different ages were estimated following standard statistical method (SPSS20.0).
Supplementary information
Acknowledgements
The work was conducted under National fellow project and first author convey deep sense of gratitude to ICAR for providing financial support to carry out the experiment. The first author also convey thanks to the Director, ICAR-Directorate of Poultry Research for helping to conduct animal experiment.
Author Contributions
T.K. Bhattacharya designed the experiment, designed oligos, analysed data and prepared the manuscript. R. Shukla conducted wet lab experiments. R.N. Chatterjee prepared the tables and graphs. S.K. Bhanja maintained and regenerated the animals for the experiment.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44217-z.
References
- 1.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci (USA) 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickford JGH, et al. Polymorphisms in the ovine myostatin gene (MSTN) and their association with growth and carcass traits in New Zealand Ronney sheep. Anim Genet. 2009;41:64–72. doi: 10.1111/j.1365-2052.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- 3.Sato F, Kurokawa M, Yamauchi N, Hattori M. Gene silencing of myostatin in differentiation of chicken embryonic myoblasts by small interfering RNA. American J Cell Physiol. 2006;291:C538–C545. doi: 10.1152/ajpcell.00543.2005. [DOI] [PubMed] [Google Scholar]
- 4.Lee SJ, et al. Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci (USA) 2005;102:18117–18122. doi: 10.1073/pnas.0505996102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kambadur R, Sharma M, Smith TPL, Bass JJ. Mutations in myostatin (GDF8) in Double-Muscled Belgian Blue and Piedmontese Cattle. Genome Research. 1997;7:910–915. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 6.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci (USA) 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amthor H, et al. Lack of myostatin results in excessive muscle growth but impaired force generation. Proc Natl Acad Sci (USA) 2007;104(6):1835–1840. doi: 10.1073/pnas.0604893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosher DS, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007;3:e79. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuelke M, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. New England. Journal Medicine. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 10.Grobet L, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nature Genetics. 1997;17:4–5. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 11.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Cadavid NF, et al. Organization of the human myostatin gene and expression in healthy men and HIV-infected men with muscle wasting. Proc Natl Acad Sci (USA) 1998;95:14938–14943. doi: 10.1073/pnas.95.25.14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivey FM, et al. Effects of age, gender, and myostatin genotype on the hypertrophic response to heavy resistance strength training. J Gerontol A Biol Sci Medical Sci. 2000;55:M641–M648. doi: 10.1093/gerona/55.11.M641. [DOI] [PubMed] [Google Scholar]
- 14.Reardon KA, Davis J, Kapsa RM, Choong P, Byrne E. Myostatin, insulin-like growth factor-1, and leukemia inhibitory factor mRNAs are upregulated in chronic human disuse muscle atrophy. Muscle Nerve. 2001;24:893–899. doi: 10.1002/mus.1086. [DOI] [PubMed] [Google Scholar]
- 15.Hill JJ, et al. The myostatin propeptide and the follistatin-related gene are inhibitory binding proteins of myostatin in normal serum. J Biol Chem. 2002;277:40735–40741. doi: 10.1074/jbc.M206379200. [DOI] [PubMed] [Google Scholar]
- 16.Haidet AM, et al. Long-term enhancement of skeletal muscle mass and strength by single gene administration of myostatin inhibitors. Proc Natl Acad Sci (USA) 2008;105:4318–4322. doi: 10.1073/pnas.0709144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogdanovich S, Perkins KJ, Krag TO, Whittemore LA, Khurana TS. Myostatin propeptide-mediated amelioration of dystrophic pathophysiology. FASEB J. 2005;19(6):543–549. doi: 10.1096/fj.04-2796com. [DOI] [PubMed] [Google Scholar]
- 18.Fauteck SP, Kandarian SC. Sensitive detection of myosin heavy chain composition in skeletal muscle under different loading conditions. Am J Physiol-Cell Ph. 1995;268(2):419–424. doi: 10.1152/ajpcell.1995.268.2.C419. [DOI] [PubMed] [Google Scholar]
- 19.Grootenhuis MA, De Boone J, Van Der Kooi AJ. Living with muscular dystrophy: health related quality of life consequences for children and adults. Health Qual Life Outcomes. 2007;5:3. doi: 10.1186/1477-7525-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatani M, et al. Transgenic expression of a myostatin inhibitor derived from follistatin increases skeletal muscle mass and ameliorates dystrophic pathology in mdx mice. FASEB J. 2008;22(2):477–487. doi: 10.1096/fj.07-8673com. [DOI] [PubMed] [Google Scholar]
- 21.Ohuchi H, et al. Expression pattern of the activin receptor typeIIA gene during differentiation of chick neural tissues, muscle and skin. FEBS Let. 1992;303:185–189. doi: 10.1016/0014-5793(92)80515-I. [DOI] [PubMed] [Google Scholar]
- 22.Kos K, Coulombe JN. Activin receptor mRNA expressionby neurons of the avian ciliary ganglion. J. Neurobiol. 1997;32:33–44. doi: 10.1002/(SICI)1097-4695(199701)32:1<33::AID-NEU4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.Roberts VJ, Barth SL. Expression of messenger ribonucleicacids encoding the inhibin/activin system duringmid- and late-gestation rat embryogenesis. Endocrinology. 1994;134:914–923. doi: 10.1210/endo.134.2.8299586. [DOI] [PubMed] [Google Scholar]
- 24.Feijen A, Goumans MJ, van den Eijnden-van Raaij AJ. Expression of activin subunits, activin receptors and follistatin in postimplantation mouse embryos suggests specific developmental functions for different activins. Development. 1994;120:3621–3637. doi: 10.1242/dev.120.12.3621. [DOI] [PubMed] [Google Scholar]
- 25.Greenwald J, Fischer WH, Vale WW, Choe S. Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase. Nature Struc. Biol. 1999;6:18–22. doi: 10.1038/4887. [DOI] [PubMed] [Google Scholar]
- 26.Thompson TB, Woodruff TK, Jardetzky TS. Structures of an ActrIIB: activinA complex reveal a novel binding mode for TGF-beta ligand: receptor interactions. EMBO J. 2003;22:1555–1566. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lotinun S, Evans GL, Turner RT, Oursler MJ. Deletion of membrane-bound steel factor results in osteopenia in mice. J. Bone Mineral Res. 2005;20:644–652. doi: 10.1359/JBMR.041209. [DOI] [PubMed] [Google Scholar]
- 28.De Caestecker M. The transforming growth factor-beta superfamily of receptors. Cytokine Growth Factor Rev. 2004;15:1. doi: 10.1016/j.cytogfr.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Kumar D, Gustafsson C, Klessig DF. Validation of RNAi silencing specificity using synthetic genes: salicylic acid-binding protein 2 is required for innate immunity in plants. Plant J. 2006;45:863–868. doi: 10.1111/j.1365-313X.2005.02645.x. [DOI] [PubMed] [Google Scholar]
- 30.Ganesan AK, et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 2008;4(12):e1000298. doi: 10.1371/journal.pgen.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaser T, et al. RNAi-mediated gene silencing in tumour tissue using replication-competent retroviral vectors. Gene Ther. 2011;18(10):953–60. doi: 10.1038/gt.2011.48. [DOI] [PubMed] [Google Scholar]
- 32.Cooper CA, et al. Generation of gene edited birds in one generation using sperm transfection assisted gene editing (STAGE) Transgenic Research. 2017;26:331–347. doi: 10.1007/s11248-016-0003-0. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharya TK, Shukla R, Chatterjee RN, Dushyanth K. Knock down of the myostatin gene by RNA interference increased body weight in chicken. Journal of Biotechnology. 2016;241:61–68. doi: 10.1016/j.jbiotec.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Satheesh P., Bhattacharya T. K., Kumar P., Chatterjee R. N., Dhara S. K., Paswan C., Shukla R., Dushyanth K. Gene expression and silencing of activin receptor type 2A (ACVR2A) in myoblast cells of chicken. British Poultry Science. 2016;57(6):763–770. doi: 10.1080/00071668.2016.1219693. [DOI] [PubMed] [Google Scholar]
- 35.Guru Vishnu, P. Molecular characterization and in vitro silencing of ACTR-IIB gene in chicken. Ph.D Thesis, submitted to ICAR-Indian veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, India (2016).
- 36.Yadav SP, et al. In vivo cell-mediated immune, hemaglutination inhibition response, hematological and biochemical values in native vs. exotic chicken breeds. Poultry Science. 2018;97:3063–3071. doi: 10.3382/ps/pey182. [DOI] [PubMed] [Google Scholar]
- 37.Gao, Y. et al. Depletion of myostatin b promotes somatic growth and lipid metabolism in Zebrafish. Frontiers in Endocrinology, 10.3389/fendo.2016.00088 (2016). [DOI] [PMC free article] [PubMed]
- 38.Khalil, K. et al. Generation of Myostatin Gene-Edited Channel Catfish (Ictalurus punctatus) via Zygote Injection of CRISPR/Cas9 System. Scientific Reports 7 (2017). [DOI] [PMC free article] [PubMed]
- 39.Mosler S, Relizani K, Mouisel E, Amthor H, Diel P. Combinatory effects of siRNA‐induced myostatin inhibition and exercise on skeletal muscle homeostasis and body composition. Physiological Reports. 2014;2(3):e00262. doi: 10.1002/phy2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ali KM, Wonnerth A, Huber K, Wojta J. Cardiovascular disease risk reduction by raising HDL cholesterol – current therapies and future opportunities. British Journal of Pharmacology. 2012;167:1177–1194. doi: 10.1111/j.1476-5381.2012.02081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan T, et al. Silencing Myostatin Using Cholesterol-conjugated siRNAs Induces Muscle Growth. Molecular Therapy- Nucleic Acids. 2016;5(8):e342. doi: 10.1038/mtna.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novobrantseva T, et al. Systemic RNAi-mediated gene silencing in nonhuman primate and rodent myeloid cells. Molecular Therapy–Nucleic Acids. 2012;1:e4. doi: 10.1038/mtna.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miao Z, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proceedings of National Academy of Science. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharya TK, Chatterjee RN, Priyanka M. Polymorphism of Pit-1 gene and its association with growth traits in chicken. Poultry Science. 2012;91:1057–1064. doi: 10.3382/ps.2011-01990. [DOI] [PubMed] [Google Scholar]
- 45.Bhattacharya TK, Chatterjee RN, Dushyanth K, Shukla R. Cloning, characterization and expression of myostatin (growth differentiating factor-8) gene in broiler and layer chicken (Gallus gallus) Molecular Biology Reports. 2014;42(2):319–327. doi: 10.1007/s11033-014-3753-x. [DOI] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Bozzola J. J. & Russel, L. D. In: Electron Microscopy Principles and Techniques for Biologist 2nd edn. Jones and Bartlett Publishers, Sudbury, Massachusetts. pp., 19–24, 54–55 and 63–67 (1998).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.