Abstract
The nuclear localization signal (NLS) marks proteins for transport to the nucleus and is used in various applications in many fields. NLSs are used to achieve efficient and stable transport of biomolecules. Previously, commercial vectors used in NLS studies contained three iterations of the NLS sequence, but these sequences can affect experimental results and alter protein function. Here, we investigated a new vector using a single classical NLS sequence with a mutation in pDsRed2-C1-wt to reduce experimental artifacts. In the newly constructed pDsRed2-C1-1NLS vector, the NLS sequence is placed near the multiple cloning sites of pDsRed2-C1-wt, and the multiple cloning site region was designed to facilitate insertion of the desired gene by site-directed mutagenesis. Fluorescent protein expression in the nucleus can be visually confirmed. The results show that the fluorescent protein was bound to the transport protein. The constructed vector had a cell survival rate of 89–95% and a transfection efficiency of 39–56% when introduced into animal cells, which are similar to those of other NLS vectors. Additionally, the constructed NLS vector can be used to demonstrate complementary binding between target proteins, and that the target protein is transported by the NLS transport system. Especially, we show that the vector can be useful for experiments involving the S100A10 gene. In addition, the constructed vector is useful for studies of genes and proteins that show potential for gene therapy or drug delivery applications.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1770-0) contains supplementary material, which is available to authorized users.
Keywords: Nuclear localization signal, Vector, Single construction of sequence, Viability, Transfection efficiency, S100A10
Introduction
The structure of the nuclear pore (~ 30 nm) imposes two size limits on nuclear transport (Pante and Aebi 1996). Macromolecules with more than 40 kDa in size cannot diffuse from the cytoplasm to the nucleus. Conversely, small metabolites, metal ions, and molecules smaller than ~ 40 kDa can be transported relatively freely (Wente and Rout 2010). Larger proteins require a nuclear localization signal (NLS) to be actively transported through the central channel of the pore. Colloidal gold particles of less than ~ 25 nm in diameter are transported into the nuclei of HeLa cells when coated with SV40 NLS peptide-conjugated bovine serum albumin or with nucleoplasmin, an endogenous karyophilic protein (Dworetzky et al. 1988; Feldherr and Akin 1990). Previous studies showed that small, single-stranded oligonucleotides can diffuse into the nucleus (Hagstrom et al. 1997; Chin et al. 1990; Leonetti et al. 1991). The short peptide form of the SV40 T antigen NLS (NLS/NL) was used in previous studies because it is more efficient than the long peptide form of NLS for the nuclear transport of large macromolecules such as IgM (Rihs et al. 1991; Yoneda et al. 1992). As a result, proteins larger than 40 kDa are transported from the cytoplasm to the nucleus through nuclear pores if they contain NLS.
The transport of DNA into the nucleus to regulate cellular activities is important for studies of DNA viruses, cell metabolism, and diseases (Sebestyen et al. 1998; Whittaker and Helenius 1998; Kasamatsu and Nakanishi 1998). This approach is particularly important for developing non-viral vectors for gene therapy. Studies of DNA transport have progressed as the understanding of how proteins and RNA are transported into and out of nuclei has improved (Mattaj and Englmeier 1998; Pennisi 1998). NLS tags proteins in amino acid sequences to be introduced into the nucleus of the cell by nuclear transport (Kalderon et al. 1984). NLS has been used to evaluate signal transduction pathways (Plessner et al. 2015), e.g., p62/SQSTM, to determine the relationship between the formation and reconstruction of actin and cell attachment and differentiation (Katsuragi et al. 2015). NLS is also used in metastasis-resistant prostate cancer treatment with AR-V7 protein biomarkers expressed in circulating tumor cells (Scher et al. 2017). The newly constructed NLS vector can be used in various fields, including drug delivery. Macromolecules, such as proteins over 40 kDa size, that contain NLSs or export signals bind to nuclear transport proteins (e.g., karyopherin α and β) to mediate directional movement through the nuclear pore.
Therapeutic genes, typically inserted into a plasmid, must be transported from the exterior of the cell to the nucleus and then expressed to exert their beneficial effects. Whereas naked DNA (linear or plasmid) can traverse the nuclear pores of intact nuclei (Dowty et al. 1995; Hagstrom et al. 1997), the process is inefficient compared to those of viral genomes and karyophilic proteins. An approach for increasing the efficiency of nuclear entry of foreign DNA is to link DNA to an NLS. Several groups have reported that the non-covalent association of an NLS with DNA can enhance DNA nuclear transport (Clollas and Aleström 1997; Dean 1997; Kaneda et al. 1989; Ludtke et al. 1999).
pDsRed2-C1, a vector with 4.7 kb, expresses a red fluorescent protein (RFP) with approximately 25.9 kDa, maximum excitation of 558 nm, and maximum emission of 583 nm (Remington 2002). pDsRed2-C1 enables numerous experiments to be conducted within a short time period and has a maturation time of approximately 24 h (Bevis and Glick 2002). Because of these properties, we developed pDsRed2-C1-1NLS based on pDsRed2-C1-wt. The commercially available pDsRed2-C1-3NLS vector contains three NLS repetitions and a longer NLS sequence than that of the newly constructed pDsRed2-C1-1NLS vector, which has a single NLS sequence with no repetition. The vector pDsRed2-C1-1NLS containing a monopartite sequence without repetition was obtained by slightly modifying the classical NLS sequence and multiple cloning sites (MCS) for easy insertion by restriction enzymes. To confirm the presence of functional NLS and MCS sequences in the constructed pDsRed2-C1-1NLS vector, the S100A10 gene was inserted and confirmed by electrophoresis and sequencing. Although S100A10 has been the subject of considerable research in recent years, its functions have not been fully clarified, even though several have been identified. Proteins of the S100A family are localized to the cytoplasm and/or nucleus of a wide range of cells, regulating cellular processes such as cell cycle and differentiation (Madureira and Waisman 2009). These proteins are also related to exocytosis and endocytosis by reorganization of F-actin (Ursula and Volker 2008). In addition, the S100A10 protein is linked to the transport of neurotransmitters, is found in the brain of humans and other mammals, and is involved in mood control. Because of their interaction with serotonin signaling proteins and their association with symptoms of mood swings, S100A10 proteins are a new potential target for pharmacotherapy (Per et al. 2006). Therefore, we confirmed that the newly constructed pDsRed2-C1-1NLS containing the S100A10 gene is functional.
Materials and methods
Plasmid construction
pDsRed2-C1-wt was obtained from Clontech Laboratories (Mountain View, CA, USA). The following primers were used to insert the S100A10 gene into the MCS region via restriction enzyme sites: S100A10 UP primer XhoI (CT CGA G), 5′-GGA GCT CCT CGA GCA AGC TTC ATG CCA TCT CAA ATG-3′ and S100A10 DOWN primer SalI (CAG CTG), 3′-GTC TTC CCT TTC TTC GTC GAC GGG ATC CAC CGG-5′. The PCR product was cloned into pDsRed2-C1-wt and pDsRed2-C1-1NLS using the Xho I and Sal I restriction enzymes. The cloned vectors were transformed into DH5α chemically competent E. Coli (Enzynomics, Daejeon, South Korea) and cultured in Luria–Bertani (LB) broth containing kanamycin at 37 °C for 24 h. Plasmid DNA was extracted using the AccuPrep Plasmid Mini Extraction Kit (Bioneer, Daejeon, South Korea).
Mutagenesis
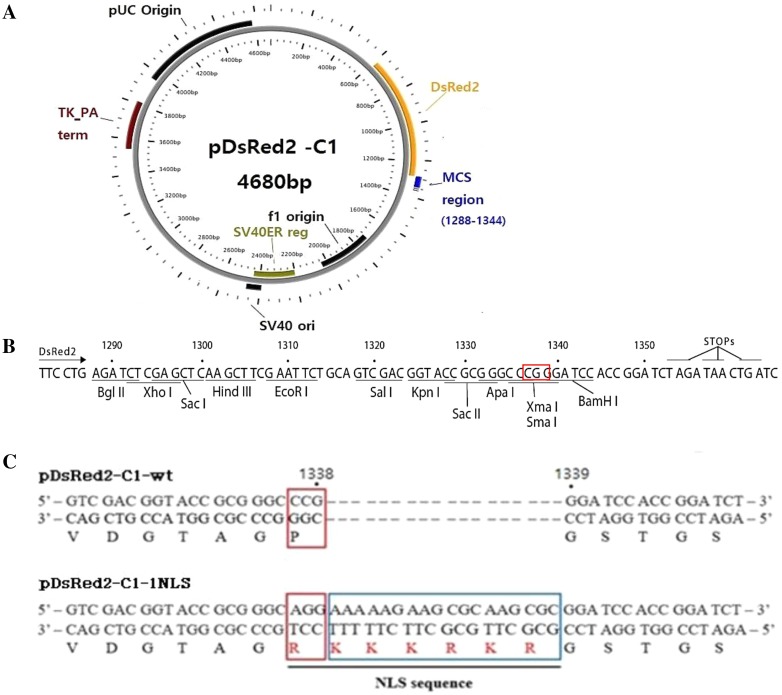
The pDsRed2-C1-wt vector was constructed from a vector containing the NLS sequence by mutating the existing sequence through site-directed mutagenesis. Mutagenesis was performed using the pDsRed2-C1-wt (Fig. 1) and EZchange Site-directed Mutagenesis Kit (Enzynomics, Daejeon, South Korea) with the following primers (Table 1): pDsRed2-C1-1NLS upstream primer 5′-GTC GAC GGT ACC GCG GGC AGG AAA AAG AAG CGC AAG CGC GGA TCC ACC GGA TCT-3′ and pDsRed2-C1-1NLS downstream primer 5′-TCT AGA TCC GGT GGA TCC GCG CTT GCG CTT CTT TTT CCT GCC CGC GGT ACC GTC-3′. The underlined primer sequences were modified by mutating the 1336CCG1338 sequence to 1336AGG1338 and then constructing the new pDsRed2-C1-1NLS by inserting 1338AAA AAG AAG CGC AAG CGC1339.
Fig. 1.
Map and MCS region of pDsRed2-C1-wt. a Vector of pDsRed2-C1-wt (Clontech), b pDsRed2-C1-wt MCS region (Clontech). c MCS regions of pDsRed2-C1-wt and pDsRed2-C1-1NLS. The NLS sequence was inserted between positions 1335 and 1339 in the sequence of pDsRed2-C1-wt (Clontech), and the CCG sequence was mutated to the CGC sequence. (a) was made by plasMapper 2.0 (https://wishart.biology.ualberta.ca/PlasMapper/index.html). For more information of pDsRed2-C1-wt, visit Clontech site (https://www.takarabio.com/assets/documents/Vector%20Documents/PT3603-5.pdf)
Table 1.
Primers used for subcloning and site-directed mutagenesis
| Name of construct | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) | Enzyme | Nuclear localization | |
|---|---|---|---|---|---|
| pDsRed2-C1-1NLS | GTC GAC GGT ACC GCG GGC AGG AAA AAG AAG CGC AAG CGC GGA TCC ACC GGA TCT | TCT AGA TCC GGT GGA TCC GCG CTT GCG CTT CTT TTT CCT GCC CGC GGT ACC GTC | + | ||
| S100A10 | GGA GCT CCT CGA GCA AGCTTC ATG CCA TCT CAA ATG | GGC CAC CTA GGG CAG CTG CTT CTT TCC CTT CTG | Xho I, Sa I | _ | |
Primer of pDsRed2-C1-1NLS contains an NLS sequence. Primer of S100A10 contains the enzyme (Xho I, Sal I) sequence and is used for subcloning. pDsRed2-C1-S100A10-1NLS is constructed using two primers in sequence
Cell culture and transfection
SV40, with the classical NLS sequence, is being studied in humans and monkeys. In particular, COS cell lines are often used by biologists when studying the monkey virus, SV40. These cell lines are transfected to produce recombinant proteins for molecular biology, biochemistry, and cell biology experiments. Thus, COS1 and COS7 cells, fibroblast-like cell lines derived from kidney tissue, were used for NLS studies. COS1 and COS7 cells were purchased from the Korean Cell Line Bank (KCLB, Seoul, South Korea; No. 21650 and 21651). COS1 and COS7 cells were cultured in Dulbecco's modified Eagle’s medium (DMEM; Welgene, Gyeongsan, South Korea) supplemented with 10% (v/v) heat-inactivated fetal bovine serum and 1000 U of penicillin–streptomycin (Gibco BRL, Grand Island, NY, USA). The cells were maintained in DMEM with a humidified atmosphere (5% CO2) at 37 °C and then subcultured on a new plate once every 3 days.
COS1 and COS7 cells were transfected with pDsRed2-C1-wt, pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS via lipofectamine 3000 (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions and incubated for 36 h prior to analyses.
Fluorescence microscopy and image assay
COS1 and COS7 cells were grown overnight to approximately 75% confluence on glass coverslips in four-well plates. Cells were transfected with pDsRed2-C1-wt, pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS for the indicated times. Samples that were not processed with any reagent or transfected with vectors were used as a control. The cells were then washed with phosphate buffered saline (PBS), fixed in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 15 min at 25 °C, and permeabilized in 0.1% PBT (PBS + 0.1% Triton X-100) at 25 °C for 20 min. For fluorescent staining, the cells were blocked in PBS with 5% bovine serum albumin for 1 h at 36 °C and then incubated with Hoechst 333,342 (Thermo Fisher Scientific, USA) staining solution for 90 min at 25 °C. Finally, the cells were mounted on slides in DAKO mounting media (Sigma-Aldrich). The slides were incubated overnight or longer at 4 °C before visualization using a confocal laser scanning microscope (DE/LSM710, Zeiss, Oberkochen, Germany) at the Center for Research Instruments and Experimental Facilities of Chungbuk National University. Image analysis was performed using imaging software (Zen, black edition, Zeiss, Oberkochen, Germany) provided by the manufacturer.
Immunoblotting
Cells were harvested with Trypsin–EDTA and lysed in RIPA buffer. Protein concentrations were determined using a BCA Kit (Pierce, Rockford, IL, USA), and equal amounts of protein were separated on 12% sodium dodecyl sulfate (SDS) gels in SDS-running buffer and transferred onto polyvinylidene fluoride membranes (Invitrogen). The membranes were blocked for 90 min at 4 °C with 5% skim milk in PBS. They were then immunoblotted with anti-DsRed antibodies ((L-18) (F9), Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated anti-IgG antibodies (sc-2354, sc516102 Santa Cruz Biotechnology). Reactive bands were visualized with a ChemiDoc imaging system (Bio-Rad Laboratories, Hercules, CA) using Immobilon Western Chemiluminescent HRP Substrate (Merck, Darmstadt, Germany). Image analysis was performed using imaging software (Image Lab, Bio-Rad Laboratories, Hercules, CA).
Immunoprecipitation
COS1 and COS7 cells were transfected using Lipofectamine 3000 with pDsRed2-C1-wt, pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS. Thirty-six hours later, cells were collected with trypsin–EDTA, lysed in RIPA buffer, and incubated overnight. The lysates were centrifuged at 16,000×g for 30 min. The supernatant was incubated with anti-DsRed (Santa Cruz Biotechnology, Dallas, TX, USA) or anti-karyopherin alpha2 (Santa Cruz Biotechnology) or beta2 (Santa Cruz Biotechnology) antibodies on a rotating wheel for 1 h at 4 °C. After incubation, 30 μL of protein-A/G plus-agarose (Santa Cruz Biotechnology) was added to the antibody-coupled lysates and incubated overnight at 4 °C. The beads were washed three times with 1 mL of RIPA buffer for 5 min with low-speed pelleting between washes. The resulting bead-bound immunocomplexes were analyzed using SDS–polyacrylamide gel electrophoresis and western blotting according to standard techniques (Harlow and Lane 1988). Reactive bands were visualized on ChemiDoc imaging system (Bio-Rad Laboratories, Hercules, CA) using Immobilon Western Chemiluminescent HRP Substrate (Merck, Darmstadt, Germany). Anti-β-actin (C4) antibodies were used to detect a reference protein. Image analysis was performed using imaging software (Image Lab, Bio-Rad Laboratories, Hercules, CA).
Cell viability assay
COS1 and COS7 cells (1.75 × 107 cells) were added to a 60-mm cell culture dish and transfected with pDsRed2-C1-wt, pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS for 36 h. The cells were washed once with PBS, treated with 0.05% trypsin–EDTA, harvested, and washed once with PBS. The harvested cells were treated with Muse Count and Viability assay kit (Merck Millipore), and reacted at room temperature for 5 min. Viable cells were measured with a Muse Cell Analyzer (Merck Millipore). Samples that were not processed with any reagent or transfected with vectors were used as a negative control, whereas those treated with only Lipofectamine 3000 reagent were used as a positive control.
Flow cytometry analysis of transfection efficiency
COS1 and COS7 cells (1.75 × 107 cells) were added to a 60-mm cell culture dish and transfected with pDsRed2-C1-wt, pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS for 36 h. Next, the cells were resuspended in 0.5 mL of PBS. Flow cytometry was performed to analyze cellular DsRed using the Guava easyCyte Flow Cytometer (Merck Millipore).
Results
Construction of NLS vectors
We constructed a new NLS vector using pDsRed2-C1-wt. Site-directed mutagenesis was performed using the MCS of pDsRed2-C1-wt with primers containing a monopartite NLS sequence (Table 1). The S100A10 gene was inserted into the wild-type and NLS vectors between Xho I and Sal I restriction enzyme sites in the MCS (Fig. 1). The NLS obtained was also used in S.Cerevisiae Ire1p NLS as a classical sequence (Goffin et al. 2006) (Fig. 1c). The production of pDsRed2-C1-1NLS and pDsRed2-C1-S100A10-1NLS was confirmed by the presence of the appropriate size band in electrophoresis (data not shown). Additional sequence analysis was performed to identify pDsRed2-C1-wt, pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt and pDsRed2-C1-S100A10-1NLS (Supplementary Fig. 1). pDsRed2-C1-1NLS and pDsRed2-C1-S100A10-1NLS contained the NLS sequence. S100A10 was inserted in pDsRed2-C1-wt and pDsRed2-C1-1NLS, which were named pDsRed2-C1-S100A10-wt and pDsRed2-C1-S100A10-1NLS, respectively.
Nuclear translocation of DsRed proteins
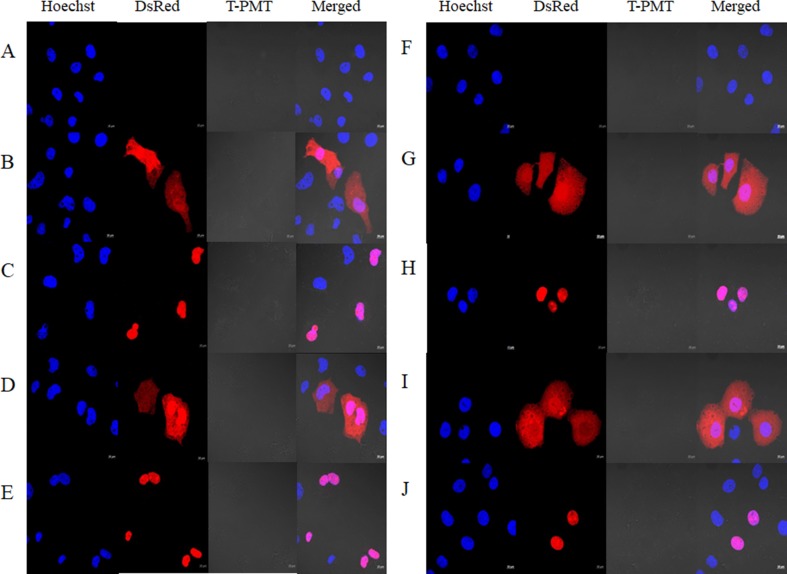
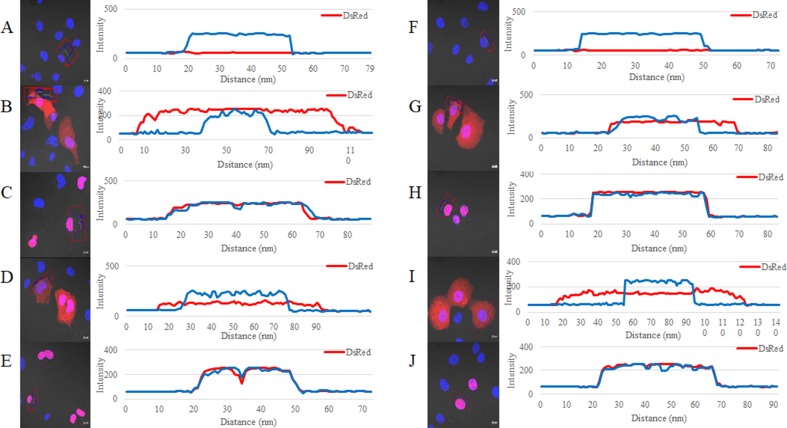
After transfection in COS1 and COS7 cells, confocal laser scanning microscopy was conducted to evaluate the subcellular localization of DsRed-wt, DsRed-S100A10-wt, DsRed-1NLS, and DsRed-S100A10-1NLS proteins (Fig. 2). DsRed-wt and DsRed-S100A10-wt were detected in the nuclei and cytoplasm (Fig. 2b, d, g, i), whereas DsRed-1NLS and DsRed-S100A10-1NLS were detected only in the nuclei (Fig. 2c, e, h, j). The cells were clearly distinguished depending on the presence or absence of the NLS sequence and based on the intensity profile results. Fluorescence analysis showed that nuclei and proteins were either clearly distinct or located in the same position (Fig. 3).
Fig. 2.
Confocal microscopy experiments. Vectors transfected in COS1 (a–e) and COS7 (f–j) cells. a Control b pDsRed2-C1-wt (Clontech) c pDsRed2-C1-1NLS d pDsRed2-C1-S100A10-wt e pDsRed2-C1-S100A10-1NLS f control g pDsRed2-C1-wt (Clontech) h pDsRed2-C1-1NLS i pDsRed2-C1-S100A10-wt j pDsRed2-C1-S100A10-1NLS. Controls (a) and (f) correspond to non-treated COS1 and COS7 cells
Fig. 3.
Fluorescence density analysis. Vectors transfected in COS1 (a–e) and COS7 (f–j) cells. a Control b pDsRed2-C1-wt (Clontech) c pDsRed2-C1-1NLS d pDsRed2-C1-S100A10-wt e pDsRed2-C1-S100A10-1NLS f control g pDsRed2-C1-wt (Clontech) h pDsRed2-C1-1NLS i pDsRed2-C1-S100A10-wt j pDsRed2-C1-S100A10-1NLS. Blue line represents the fluorescence intensity in the nucleus and red represents the fluorescence intensity of DsRed. Controls (a) and (f) correspond to non-treated COS1 and COS7 cells
Interaction of NLS signal sequence with importin
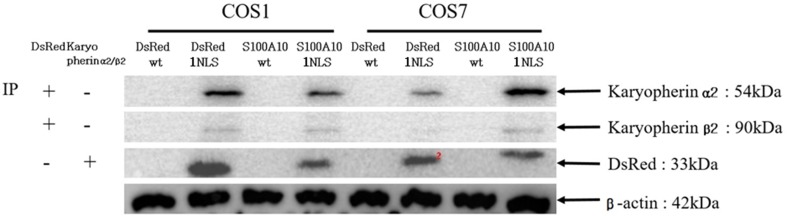
The constructed pDsRed2-C1-1NLS and pDsRed2-C1-S100A10-1NLS vectors contained a monopartite NLS sequence, which transports proteins through the process of binding to and separating from importin α (karyopherin α) and importin β (karyopherin β) via their NLS motif. COS1 and COS7 cells were transfected with the pDsRed2-C1-wt, pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS vectors. Immunoprecipitation was conducted to confirm the binding of importin α and importin β using antibodies (Fig. 4). DsRed-wt and DsRed-S100A10-wt did not interact with importin in COS1 and COS7 cells. In contrast, DsRed-1NLS and DsRed-S100A10-1NLS interacted with importin.
Fig. 4.
Immunoprecipitation of modified pDsRed2-C1-1NLS and pDsRed2-C1-S100A10-1NLS. Immunoprecipitation was performed using COS1 and COS7 cell lines. pDsRed2-C1-wt (Clontech), pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS vectors were transfected into each cell line for 24 h. Cells were lysed after transfection and immunoprecipitation was performed in the lysate using DsRed, karyopherin α2/β2 antibodies, and protein A/G plus-agarose (Santa Cruz). Samples that transfected with pDsRed2-C1-wt (Clontech), pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, pDsRed2-C1-S100A10-1NLS were identified using antibodies that detected karyopherin α2/β2 protein. Samples evaluated for karyopherin α2/β2 were confirmed by western blotting using antibodies for DsRed protein. Anti-β-actin (C4) antibody (Santa Cruz) was used to detect a reference protein in whole cell lysates prior to immunoprecipitation
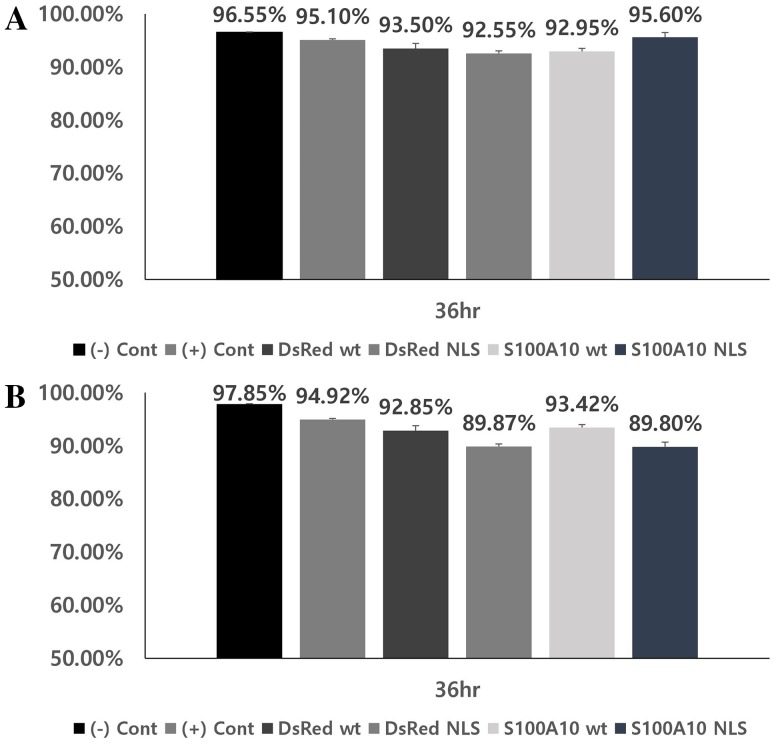
Cell viability and transfection efficiency after transfection with the new NLS vector
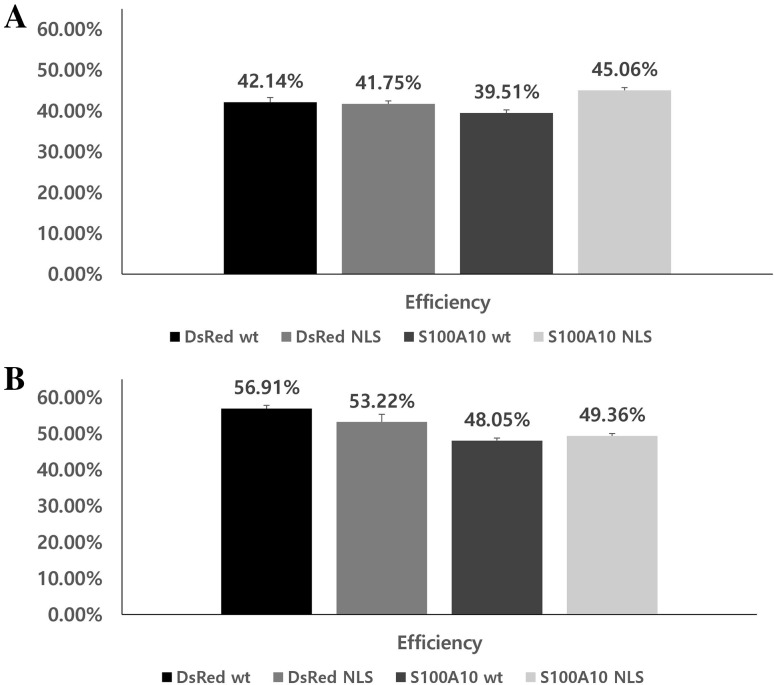
We compared the commercially available vector with the constructed pDsRed2-C1-1NLS, pDsRed2-C1-S100A10-wt, and pDsRed2-C1-S100A10-1NLS vectors. Cells were transfection for 36 h. Cell viability was evaluated to determine the effect of the constructed vectors. pDsRed2-C1-wt resulted in 93.5% and 92.85% survival rates for COS1 and COS7 cells, respectively (Fig. 5). Viability of COS1 and COS7 cells were 92.55% and 89.87% for pDsRed2-C1-1NLS, 92.95% and 93.42% for pDsRed2-C1-S100A10-wt, and 95.60% and 89.80% for pDsRed2-C1-S100A10-1NLS, respectively (Fig. 5). The transfection efficiencies for COS1 and COS7 cells were 42.14% and 56.91%, respectively, using pDsRed2-C1-wt, and 41.75% and 53.22%, using pDsRed2-C1-1NLS (Fig. 6). The efficiencies of pDsRed2-C1-S100A10-wt were 39.51% and 48.05%, whereas those of pDsRed2-C1-S100A10-1NLS were 45.06% and 49.36%, respectively (Fig. 6).
Fig. 5.
Cell viability in cells transfected with vectors. Cell viability of transfected vectors in a COS1 and b COS7 cells. Values are presented as the mean ± SEM (n = 3) and expressed as percentage decrease in cell viability. (−) Controls correspond to non-treated COS1 and COS7 cells. ( + ) Controls correspond to COS1 and COS7 cells treated only with transfection reagents
Fig. 6.
Transfection efficiency in cells transfected with vectors. Transfection efficiency of transfected vectors in a COS1 and b COS7 cells. Values are presented as the mean ± SEM (n = 3) and expressed as percentage in cell efficiency
Discussion
The DsRed protein emits red–orange fluorescence when excited and is present throughout the cell. Fluorescent molecules, including green fluorescent protein, yellow fluorescent protein, and blue fluorescent protein, can be problematic because of the long time required between protein synthesis and fluorescence emission. On the contrary, DsRed is particularly useful because monomers can be generated through genetic engineering. Moreover, it has a maturation time of approximately 24 h (Piatkevich and Verkhisha 2011). The constructed NLS vector minimized the deformation of MCS to maintain the function of the vector. To minimize functional and structural changes in the inserted protein, the classical NLS sequence inserted at the distal end of the MCS was minimized. A monopartite NLS sequence (KKKRKR) consisting of six amino acids was inserted immediately after the mutation site (Fig. 1). The newly constructed pDsRed2-C1-1NLS vector was detected in the nucleus along the importin transport system. A protein containing the NLS motif can form a complex with domains of importins α and β for transport into the nucleus (Kim et al. 2017). The classical NLS motif binds to importin alpha in an expanded form with the principal chain antiparallel to the direction of the import α. Importin α then recognizes and binds to the domain of importin β. Importin beta mediates interactions with the nuclear pore during translocation. When transferred into the nucleus through the nuclear pore, RanGTP binds to the complex and transforms the structure of the importin β. The action of Nup2 and Ces1 separates the binding of NLS motif and importin α, and which is transferred to the cytoplasm again through the nucleus (Lange et al. 2007). Based on this, we constructed a new pDsRed2-C1-NLS vector. DsRed-wt was expressed in both the cytoplasm and nucleus (Fig. 2b, g), whereas DsRed-1NLS developed a red color only in the nucleus (Fig. 2c, h). Particularly, DsRed-S100A10-wt was distributed throughout the cell (Fig. 2d, i). In contrast, DsRed-S100A10-1NLS was detected only in the nucleus (Fig. 2e, j).
The newly constructed pDsRed2-C1-NLS vector works well by interacting with domains of importins α and β in the nuclear transport of proteins (Fig. 4). The survival rates of cell transfected with the pDsRed2-C1-1NLS and pDsRed2-C1-S100A10-1NLS were 89.80–95.60% in COS1 and COS7 cell lines, indicating that the survival rate and toxicity are similar to those of the wild-type vector, with survival rates of 92.85–93.50% (Fig. 5). The transfection efficiency rates of pDsRed2-C1-1NLS and pDsRed2-C1-S100A10-1NLS were 41.75–53.22% in COS1 and COS7 cell lines. These results show little difference from the wild-type vector transfection rate of 39.51–56.91%.
S100A10 is approximately 300 bp in length and is found as a heterogeneous tetramer composed of a free complex, homodimers, or complexes with two molecules of annexin 2. S100A10 is a key scaffolding molecule in the cell and interacts with plasma membrane proteins via annexin 2 (Rety et al. 1999). It also interacts with cytoplasmic and periplasmic membrane-associated proteins such as AHNAK, during intracellular development. S100A10 plays a role in the transport of proteins related to mood regulation, nociception, and cell polarization. It is predominately expressed in the lungs and kidneys, but is found in all cells of the body. The transported S100A10 proteins are mostly cell surface receptors in signaling pathways and ion channels (Rescher and Gerke 2008). Although the exact mechanism is unclear, S100A10 is essential for controlling serotonin signaling in the brain (Warner et al. 2009). S100A10 deficiency causes depression-like behaviors (Hagstrom et al. 1997) and S100A10 is a candidate therapeutic target for depression and anxiety, as it has similar effects to antidepressants administered to mice (Svenningsson et al. 2006). The newly constructed NLS vector will enable studies of how various protein function and reach their subcellular locations. Additionally, The S100A10 interacts with various proteins, such as cell surface receptors and the circular monoclonal calcium potassium channel (Svenningsson and Greengard 2007). The S100A10 protein, which does not have an NLS sequence, was expressed uniformly throughout the cell in the wild-type vector, but was clearly observed only in the nucleus when inserted into the NLS vector.
The transfection efficiency of S100A10-containing vectors in this study was slightly lower than that of the vector without the inserted gene because of the additional 300-bp sequence. However, the transfection efficiencies of known cell lines and transfection reagents were approximately 40–45% for COS1 cells and 48–57% for COS7 cells, indicating no difference in efficiency using the vector. Our results demonstrate that the newly constructed NLS vector had minimal impact on DNA and protein levels in cell lines when the NLS sequences were not repeated. In this study, the pDsRed2-C1-S100A10-1NLS vector was constructed as a model, but the newly constructed pDsRed2-C1-1NLS vector can be used to clone several types of genes. This vector will enable genetics-based studies of diseases and may be useful for drug delivery.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by the Basic Science Research Program, through the National Research Foundation of Korea (NRF), funded by the Ministry of Science ICT & Future Planning (NRF-2014R1A1A1002349). This article was revised by a professional English proofreader (Editage).
Author contributions
HSY, YJO, EJL and SSK performed the conceptualization; HSY, YJO and EJL performed the data curation and formal analysis; SSK and SHH performed funding and acquisition and resources; HSY, YJO, EJL and SSK performed the experiments including of investigation, methodology, software; HSY wrote the manuscript including of data visualization, original draft, review and editing; SHH performed project administration, supervision and validation. All authors read and approved the final manuscript.
Funding
This research was supported by the Basic Science Research Program, through the National Research Foundation of Korea (NRF), funded by the Ministry of Science ICT & Future Planning (NRF-2014R1A1A1002349).
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest (commercial or otherwise) to declare regarding this study.
Contributor Information
Hee Sang You, Email: yhs1532@nate.com.
Yeon Jeong Ok, Email: oyj5798@naver.com.
Eun Jeong Lee, Email: babbo89@hanmail.net.
Sang Sun Kang, Email: jin95324@chungbuk.ac.kr.
Sung Hee Hyun, Email: hyunsh@eulji.ac.kr.
References
- Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- Chin DJ, Green GA, Zon G, Szoka F, Jr, Straubinger R. Rapid nuclear accumulation of injected oligodeoxyribonucleotides. New Biol. 1990;2:1091–1100. [PubMed] [Google Scholar]
- Collas P, Aleström P. Nuclear localization signals: a driving force for nuclear transport of plasmid DNA in zebrafish. Biochem Cell Biol. 1997;75:633–640. doi: 10.1139/o97-044. [DOI] [PubMed] [Google Scholar]
- Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- Dowty ME, Williams P, Zhang G, Hagstrom JE, Wolff JA. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc Natl Acad Sci. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworetzky SI, Lanford RE, Feldherr CM. The effects of variations in the number and sequence of targeting signals on nuclear uptake. J Cell Biol. 1988;107:1279–1287. doi: 10.1083/jcb.107.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr CM, Akin D. The permeability of the nuclear envelope in dividing and nondividing cell cultures. J Cell Biol. 1990;111:1–8. doi: 10.1083/jcb.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin L, Vodala S, Fraser C, Ryan J, Timms M, Meusburger S, Catimel B, Nice EC, Silver PA, Xiao CY, Jans DA, Gething MJH. The unfolded protein resonse transducer Ire1p contains a nuclear localization sequence recognized by multiple β importins. Mol Biol Cell. 2006;17:5309–5323. doi: 10.1091/mbc.e06-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom JE, Ludtke JJ, Bassik MC, Sebestyén MG, Adam SA, Wolff JA. Nuclear import of DNA in digitonin-permeabilized cells. J Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) A laboratory manual. Cold Spring Harbor Laboratory, New York, pp 579
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39(3):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kaneda Y, Iwai K, Uchida T. Increased expression of DNA cointroduced with nuclear protein in adult rat liver. Science. 1989;243:375–378. doi: 10.1126/science.2911748. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H, Nakanishi A. How do animal DNA viruses get to the nucleus? Ann Rev Microbiol. 1998;52:627–686. doi: 10.1146/annurev.micro.52.1.627. [DOI] [PubMed] [Google Scholar]
- Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM 1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- Kim YH, Han M-E, Oh S-O. The molecular mechanism for nuclear transport and its application. Anat Cell Biol. 2017;50:77–85. doi: 10.5115/acb.2017.50.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin α. J Biol Chem. 2007;282(8):5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc Natl Acad Sci. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke JJ, Zhang G, Sebestyén MG, Wolff JA. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J Cell Sci. 1999;112:2033–2041. doi: 10.1242/jcs.112.12.2033. [DOI] [PubMed] [Google Scholar]
- Madureira PA, Waisman DM. S100A10 (S100 calcium binding protein A10) Atlas Genet Cytogenet Oncol Haematol. 2009;13(9):657–659. [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Ann Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Pante N, Aebi U. Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science. 1996;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- Pennisi E. The nucleus's revolving door. Science. 1998;279:1129–1131. doi: 10.1126/science.279.5354.1129. [DOI] [PubMed] [Google Scholar]
- Per S, Karima C, Ilan R, Marc F, Xiaoqun Z, Malika EY, et al. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311(5757):77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- Piatkevich KD, Verkhusha VV. Guide to red fluorescent proteins and biosensors for flow cytometry. Methods Cell Biol. 2011;102:431–461. doi: 10.1016/B978-0-12-374912-3.00017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R. Nuclear F-actin formation and reorganization upon cell spreading. J Biol Chem. 2015;290:11209–11216. doi: 10.1074/jbc.M114.627166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington SJ. Negotiating the speed bumps to fluorescence. Nat Biotechnol. 2002;20:28–29. doi: 10.1038/nbt0102-28. [DOI] [PubMed] [Google Scholar]
- Rescher U, Gerke V. S100A10/p11: family, friends and functions. Pflügers Archiv. 2008;455:575–582. doi: 10.1007/s00424-007-0313-4. [DOI] [PubMed] [Google Scholar]
- Réty S, Sopkova J, Renouard M, Osterloh D, Gerke V, Tabaries S, et al. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Mol Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- Rihs H-P, Jans D, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 1991;10:633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Graf RP, Schreiber NA, McLaughlin B, Lu D, Louw J, et al. Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur Urol. 2017;71:874–882. doi: 10.1016/j.eururo.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyen MG, Ludtke JJ, Bassik MC, Zhang G, Budker V, Lukhtanov EA, et al. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nat Biotechnol. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, Greengard P. p11 (S100A10)—an inducible adaptor protein that modulates neuronal functions. Curr Opin Pharmacol. 2007;7:27–32. doi: 10.1016/j.coph.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, Chergui K, Rachleff I, Flajolet M, Zhang X, El Yacoubi M, et al. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- Ursula R, Volker G. S100A10/p11: family, friends and functions. Pflügers Arch. 2008;455(4):575–582. doi: 10.1007/s00424-007-0313-4. [DOI] [PubMed] [Google Scholar]
- Warner-Schmidt JL, Flajolet M, Maller A, Chen EY, Qi H, Svenningsson P, et al. Role of p11 in cellular and behavioral effects of 5-HT4 receptor stimulation. J Neurosci. 2009;29:1937–1946. doi: 10.1523/JNEUROSCI.5343-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harbor Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker GR, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- Yoneda Y, Semba T, Kaneda Y, Noble RL, Matsuoka Y, Kurihara T, et al. A long synthetic peptide containing a nuclear localization signal and its flanking sequences of SV40 T-antigen directs the transport of IgM into the nucleus efficiently. Exp Cell Res. 1992;201:313–320. doi: 10.1016/0014-4827(92)90279-H. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.