Abstract
Epigenetic regulators are attractive targets for the development of new cancer therapies. Among them, the ATP-dependent chromatin remodeling complexes control the chromatin architecture and play important roles in gene regulation. They are often found to be mutated and de-regulated in cancers, but how they influence the cancer gene expression program during cancer initiation and progression is not fully understood. Here we show that the INO80 chromatin remodeling complex is required for oncogenic transcription and tumor growth in non-small cell lung cancer (NSCLC). Ino80, the SWI/SNF ATPase in the complex, is highly expressed in NSCLC cells compared to normal lung epithelia cells. Further, its expression, as well as that of another subunit Ino80b, negatively correlates with disease prognosis in lung cancer patients. Functionally, Ino80 silencing inhibits NSCLC cell proliferation and anchorage-independent growth in vitro and tumor formation in mouse xenografts. It occupies enhancer regions near lung cancer-associated genes, and its occupancy correlates with increased genome accessibility and enhanced expression of downstream genes. Together, our study defines a critical role of INO80 in promoting oncogenic transcription and NSCLC tumorigenesis, and reveals a potential treatment strategy for inhibiting the cancer transcription network by targeting the INO80 chromatin remodeling complex.
Keywords: Non-small cell lung cancer, INO80, Chromatin remodeling, Oncogenic transcription
Introduction
Despite advances in diagnosis and therapy, lung cancer is still the leading cause of cancer mortality throughout the world6, 22. It is often diagnosed at advanced or metastatic stage and the existing therapies can only provide limited benefit. Therefore, it remains a critical and immediate goal for lung cancer research to understand the disease and develop more-effective treatments.
Non-small-cell lung cancer (NSCLC) represents more than 85% of all lung cancer cases7. Technical improvements in high-throughput sequencing and advances from fields such as epigenetics and developmental biology have provided insights into the molecular mechanisms of lung cancer and facilitated the development of targeted therapies30, 32. For example, the identification of driver mutations in signaling pathways, such as EGFR, ALK, KRAS, as the primary oncogenic events in some NSCLCs led to a revolutionary change in the diagnosis and treatments of the disease. These targeted treatments offered a significant improvement of outcomes and disease prognosis in target populations. However, drug resistance inevitably develops after initial remission. In addition, the treatments show limited efficacy in non-targeted populations. Therefore, further investigations in lung cancer mechanisms and new therapeutic strategies are of significant scientific and social values.
Besides genetic mutations, epigenetic alterations such as aberrant DNA methylation, histone modification, or chromatin regulation, are also widely involved in tumorigenesis2, 4. Not surprisingly, epigenetic regulators play critical roles in cancer development15, 24. Among epigenetic factors, the ATP-dependent chromatin remodeling complexes have the ability to alter nucleosome composition and position to influence chromatin architecture and gene expression12, 28. By controlling the packing and unpacking of the chromatin, chromatin remodelers provide regulated DNA accessibility and have instructive roles in cell fate decision during cancer development and progression. Indeed, some chromatin remodelers show an unexpectedly high mutation rate, and the mutations may result in impaired higher-order genome structure and gene regulation13, 17, 25. However, the true significance of chromatin remodelers in tumorigenesis and tumor progression remains to be fully elucidated.
ATP-dependent chromatin remodelers can be divided into four families based on domain structures and subunit compositions, including SWI/SNF, ISWI, CHD, and INO808, 27, 34. Recent studies showed that inactivating mutations in the subunits of the SWI/SNF family chromatin remodelers are associated in various human cancers, suggesting that the SWI/SNF remodelers mostly act as tumor suppressors13, 17, 25. In comparison, the involvement of the other remodeling complex families in cancer is not well characterized. Here, we show that the INO80 chromatin remodeling complex promotes oncogenic transcription and tumor growth in lung cancer. Our study reveals a critical role of INO80 in lung cancer tumorigenesis and suggests a novel strategy for cancer treatment by targeting oncogenic transcription.
Results
INO80 expression is up-regulated in lung cancer
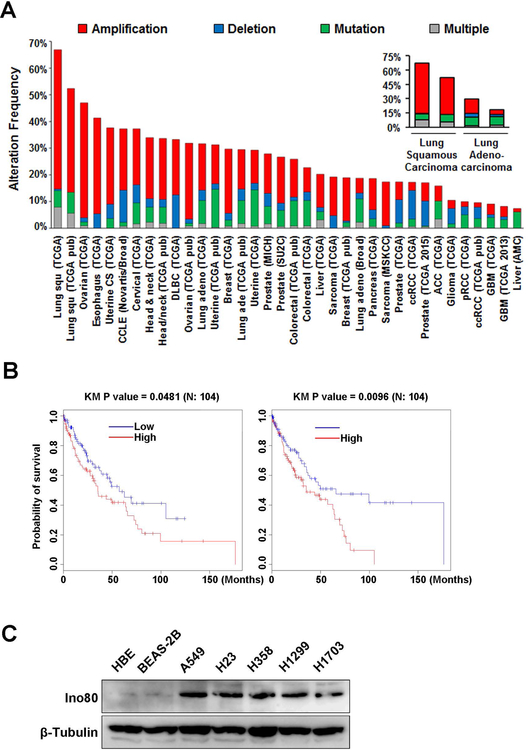
Our previous study indicates that INO80 is required for embryonic stem cell self-renewal by selectively activating pluripotency genes33. Because genes and pathways important for ESC maintenance are often re-activated in cancer5, 18, 36, and because other chromatin remodelers have been implicated in tumorigenesis13, 17, 25, we hypothesize that INO80 may also play an important role in cancer. To test the hypothesis, we first examined the alteration frequency of the INO80 subunits in various cancers based on the The Cancer Genome Atlas (TCGA) data (http://cancergenome.nih.gov/). We found that INO80 subunits show high alteration frequency in many cancer types. While different subunits are differentially altered (Figure S1), collectively INO80 complex is frequently found in lung cancers, with up to 50% of cases harboring amplifications in at least one subunit (Figure 1A). Furthermore, Kaplan-Meier survival analysis showed that patients with lower expression of the INO80 subunits Ino80 or Ino80B have significantly higher probability of survival (Figure 1B). These results suggest that INO80 expression positively correlates with lung cancer development. To provide further evidence, we examined the expression the SWI/SNF ATPase in the complex, Ino80, in human lung cancer cell lines. By western blot, we found that Ino80 expression is up-regulated in several NSCLC cell lines (A549, H23, H358, H1299, H1703) compared with the normal human bronchial epithelial cells NHBE and normal human bronchial epithelial cell line BEAS-2B (Figure 1C). Taken together, these results strongly support our hypothesis that INO80 plays an important role in NSCLC.
Figure 1. INO80 expression is elevated in lung cancer.
(A) Alteration frequency of INO80 subunits in various cancer types. Data was obtained from The Cancer Genome Atlas portal: http://www.cbioportal.org. (B) Provability of survival in patients with different levels of Ino80 or Ino80b expression. (C) Western blot showing the protein level of Ino80 in lung cancer cell lines (A549, H23, H358, H1299, H1703) and normal primary lung bronchial epithelia cells (NHBE) and cell line (BEAS-2B). β-Tubulin was used as the loading control.
INO80 is required for lung cancer cell growth
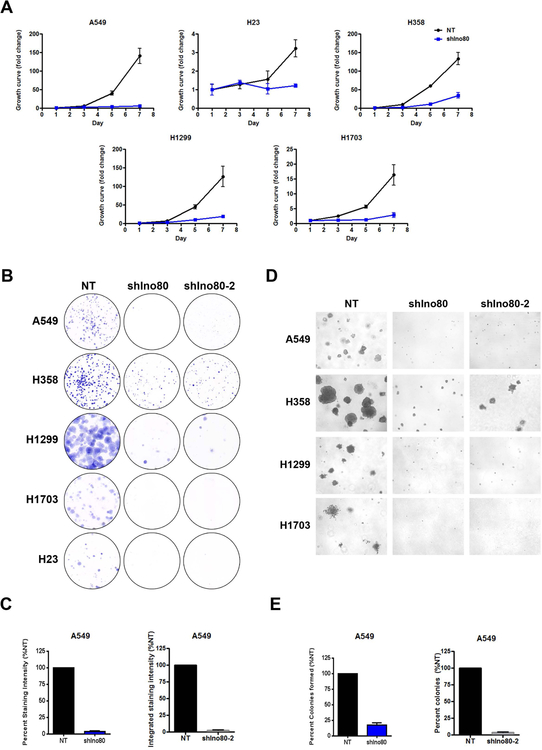
To test whether INO80 is directly required for lung cancer cell growth, we focused on the SWI/SNF ATPase, Ino80, and carried out gene silencing by RNA interference in several NSCLC lines, including A549, H1703, H358, H23, and H1299. Ino80 silencing led to a clear reduction in Ino80 expression at the protein level (Figures S2A). More importantly, Ino80 silencing significantly reduced cancer cell proliferation and/or viability as shown by the growth curves (Figure 2A). In order to minimize the off-target effect, we designed and tested additional shRNAs against Ino80. All the Ino80 shRNAs produced similar results in gene silencing as well as growth suppression in lung cancer cells (Figure S2B, S2C). In comparison, Ino80 silencing did not impair the growth and/or survival of the control NHBE or BEAS-2B cells (Figure S2D).
Figure 2. INO80 is required for lung cancer cells growth in vitro.
(A) Growth curves showing the impact of Ino80 silencing on lung cancer cell lines. Lung cancer cells were infected with lentiviral-based non-targeting (shNT) or Ino80 shRNA (shIno80) viruses and cell growth was determined at the indicated time points. Data was plotted as Mean ± SEM from 3 independent experiments. (B-C) Clonogenesis assay for NSCLC cells infected with NT (shNT), Ino80-shRNA (shIno80), or Ino80 shRNA-2 (shIno80–2) viruses. (B) Representative images. (C) Statistical analysis for A549 cells. (D-E) Anchorage-independent growth of NSCLC cells. H23 cells were not included as they did not form colonies in soft-agar. (D) Representative images. (E) Statistical analysis for A549 cells.
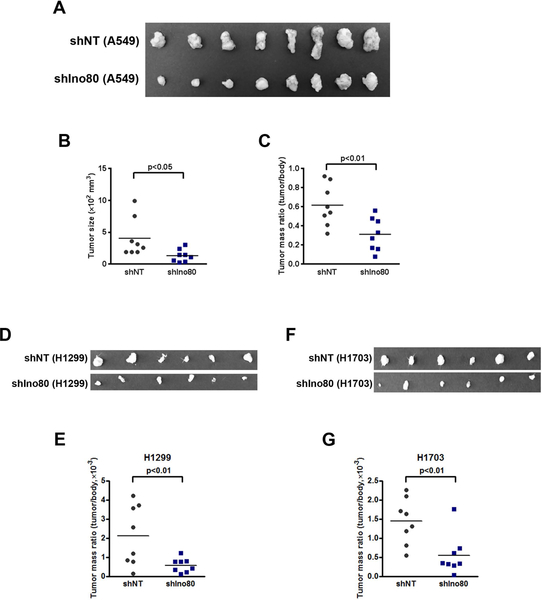
Next, we tested whether INO80 is required for the clonogenicity and anchorage-independent growth of the cancer cells, as these assays are commonly used as indicators of the tumorigenicity of transformed or cancerous cells. We found that Ino80 silencing in the A549, H1703, H358, H23, and H1299 NSCLC lines dramatically inhibited colony formation in monolayer culture and anchorage-independent growth in soft-agar (Figure 2B–E, S3). Finally, to investigate the role of INO80 in NSCLC growth in vivo, we tested the requirement for Ino80 in a mouse xenograft model. We transduced A549, H1299, and H1703 cells with lentivirus expressing either the control- or Ino80-shRNA, and injected the cells subcutaneously into immunocompromised animals. While control-shRNA transduced cells continued to grow and form tumors in the injected animals, Ino80-shRNA transduced cells show significant reduction in tumor growth, as measured by both tumor size and/or mass 4 weeks after injection (Figure 3). Collectively, the above data indicate that INO80 plays an essential role in NSCLC tumorigenesis.
Figure 3. Ino80 is required for tumor formation in mouse xenografts.
A549, H1299, and H1703 cells were infected with NT or Ino80-shRNA viruses and injected subcutaneously into immunocompromised mice. Tumors were dissected 5 weeks after the injection. (A, D, F) Representative image showing the size of the tumors. (B-C) Analysis of tumor size (B) and mass (C, E, G). Data was plotted as Mean ± SEM, and n = 8 in each group. p-value was calculated by two-tailed Student’s t-test.
INO80 promotes the expression of lung cancer-associated genes
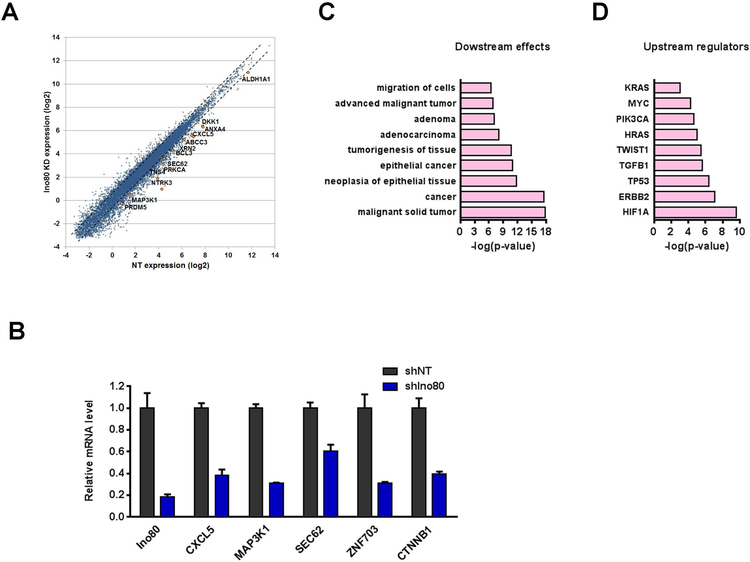
To understand how INO80 regulates lung cancer development, we first examined gene expression changes caused by Ino80 silencing in A549 by RNA-seq. Ino80 silencing led to differential expression of 1759 genes, with 1112 genes showing reduced expression and 647 genes showing increased expression. As there were significantly more genes being down-regulated (p < 0.0001), we hypothesized that INO80 may normally promote the expression of these genes (Figure 4A). Interestingly, we noticed that many known lung cancer-associated genes such as CTNNB131, Cxcl51, Map3k119, Sec6223 and Znf7033, were down-regulated after Ino80 depletion (Figure 4B). Therefore, we carried out Ingenuity pathway analysis (IPA) on the 1112 down-regulated genes, and found that indeed they were highly enriched for genes involved in cancer (Figure 4C). In addition, Ingenuity upstream regulator analysis20 showed that the gene expression changes caused by Ino80 silencing resemble those caused by the perturbations of Kras, Myc, Pik3ca, and Erbb2 (Figure 4D). As many of these predicted upstream regulators have been previously implicated in lung cancers9, 16, this result suggested that INO80 and lung cancer genes may impinge on similar downstream targets during cancer development. Together, our data support the notion that INO80 promotes the expression of lung cancer-associated genes.
Figure 4. INO80 promotes the expression of lung cancer-associated genes.
(A) Scatter plot showing the differentially expressed genes (DEGs) after Ino80 silencing. A549 cells were infected with NT or Ino80-shRNA viruses, and gene expression changes were determined by RNA-seq 4 days after virus infection. (B) RT-qPCR showing the down-regulation of genes important in lung cancer after Ino80 silencing. Gene expression was normalized to β-actin, and values were plotted as Mean ± SEM. (C) IPA showing the enrichment of cancer-associated genes in the down-regulated genes upon Ino80 silencing. Selected top categories are shown, and please see table S3 for the complete list. (D) IPA showing the predicted upstream regulators based on the down-regulated genes upon Ino80 silencing.
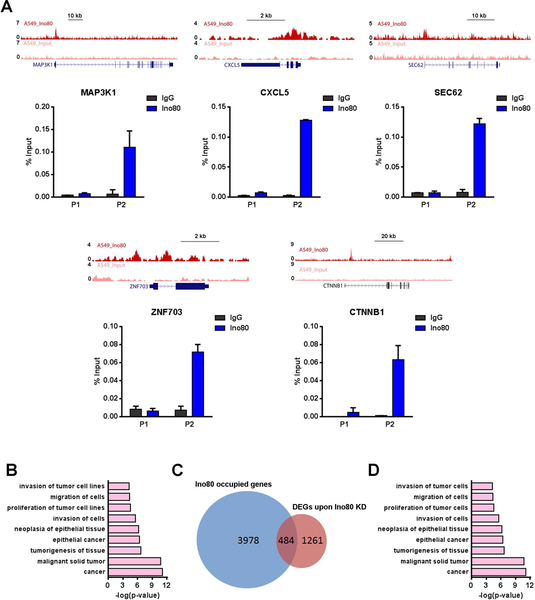
To test whether INO80 directly regulates these cancer-associated genes, we carried out chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) using Ino80 as the bait. We identified 7126 Ino80-bound peaks in A549 lung cancer cells, which were assigned to 5605 nearest genes. These Ino80-bound genes include those known lung cancer-associated genes CTNNB1, Cxcl5, Map3k1, Sec62 and Znf703, as confirmed by ChIP-qPCR (Figure 5A). Furthermore, IPA analysis showed that Ino80-bound genes are highly enriched for those involved in cancer (Figure 5B). Between the 5605 INO80-occupied genes and the 1759 differentially expressed genes after Ino80 silencing, 580 were in common and are likely direct targets of INO80 (Figure 5C). Again, these 580 potential INO80 targets are highly enriched for genes involved in cancer development (Figure 5D). Together, the above results suggest that INO80 may activate the expression of lung cancer-associated genes to promote lung cancer tumorigenesis.
Figure 5. INO80 occupies genomic regions near lung cancer-associated genes.
(A) Genome browser tracks and ChIP-qPCR validations showing Ino80 occupancy near selected lung cancer genes MAP3K1, CXCL5, SEC62, ZNF703, CTNNB1 in A549 cells. Two genomic regions near each gene were selected for the ChIP-qPCR validations: P1 corresponds to a region that is not bound by Ino80 based on ChIP-Seq, while P2 corresponds to a region that is bound by Ino80. (B) IPA showing the enrichment of cancer-associated genes in Ino80-bound genes. Selected top categories are shown, and please see Table S4 for the complete list. (C) Venn diagram showing the overlap between DEGs after Ino80 KD and Ino80-bound genes. (D) IPA of the 580 Ino80 target genes. Selected top categories are shown, and please see Table S5 for the complete list.
INO80 occupies enhancers near lung cancer-associated genes
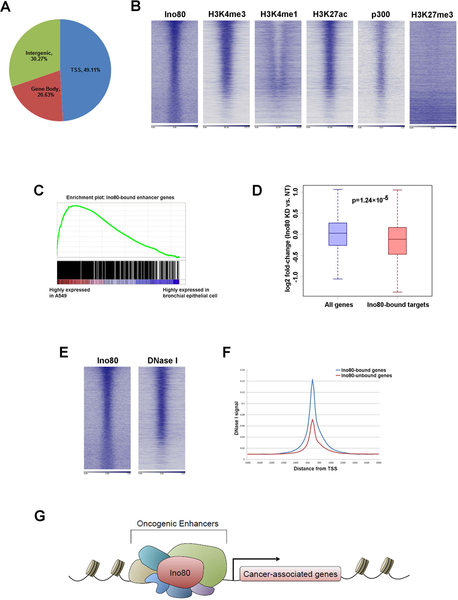
To understand how INO80 regulates gene expression, we compared the genomic occupancy of Ino80 and histone markers. We found that the Ino80 peaks were enriched in gene-rich chromosomal regions and were often found near transcription start sites (Figure 6A). Further, Ino80 binding strongly correlated with those of active enhancer markers, including H3K4me1, H3K27ac and p300 (Figure 6B). In contrast, Ino80 bound-regions show minimal occupancy by the repressive marker H3K27me3. Interestingly, gene set enrichment analysis (GSEA) indicated that genes near Ino80-bound enhancers are highly enriched for those that show elevated expression in in A549 (Figure 6C). Therefore, these results suggest that INO80 occupies enhancers near cancer-associated genes. To investigate the functional consequence of INO80 occupancy, we examined the impact of Ino80 silencing on those cancer-associated genes whose enhancers are occupied by INO80. Compared to all genes, the cancer-associated genes with Ino80 occupancy showed more significant down-regulation after Ino80 silencing (Figure 6D), suggesting that INO80 normally activates the enhancers and promotes downstream gene expression.
Figure 6. INO80 occupies enhancers near lung cancer-associated genes and activates oncogenic transcription.
(A) Ino80 peak distribution in the genome. (B) Heat maps showing Ino80, H3K4me3, H3K4me1, H3K27ac, p300 and H3K27me3 occupancy in A549 cells. Genes were sorted based on Ino80 peak FDR. (C) GSEA showing that the Ino80-bound enhancer genes are enriched for those that are highly expressed in A549 vs. normal branchial epithelial cells. (D) Box plot showing that Ino80 silencing results in down-regulation of Ino80-bound lung cancer-associated genes. (E) Heat maps showing Ino80 occupancy and DNase I hypersensitive sites in A549 cells. Genes were sorted based on Ino80 peak FDR. (F) DNase I hypersensitivity at genes bound or not bound by Ino80. (G) Proposed model for INO80 in lung cancer: INO80 occupies enhancers near lung cancer-associated genes and promotes oncogenic transcription.
It has been reported that INO80 catalyzes the removal of unacetylated H2A.Z from chromatin to facilitate gene transcription in yeast29. Furthermore, acetylation of histone H3 on lysine 56 (H3K56ac) enhances the H2A.Z replacement activity of INO8035. Thus, we carried out H2A.Z and H3K56ac ChIP-seq in A549 cells transduced with non-targeting or Ino80 shRNA viruses. We found that Ino80 silencing did not drastically change H2A.Z or H3K56ac occupancy at Ino80-bound promoter regions (Figure S4), suggesting that H3K56ac modification or H2A.Z eviction may not account for the gene expression changes caused by Ino80 silencing in A549.
Finally, because INO80 is a chromatin remodeler, we hypothesized that it may regulate gene expression by regulating chromatin structure and genome accessibility. To test the hypothesis, we compared Ino80 occupancy to a published dataset of DNase I hypersensitivity sites in A549 cells. DNase I hypersensitive sites are genomic regions that are sensitive to nuclease cleavage, and are often used as indicators of open chromatin regions. We found that there is a strong overlap between INO80-bound regions and DNase I hypersensitive sites (Figure 6E). Furthermore, INO80-bound regions showed significantly higher DNase I sensitivity compared to INO80-unbound regions (Figure 6F). Collectively, our data support the following model: in lung cancer, INO80 occupies enhancers near cancer-associated genes. Its binding leads to a more open chromatin state and enhanced genome accessibility, thereby activating oncogenic transcription to promote tumorigenesis (Figure 6G).
Discussion
Originally discovered in yeast, The INO80 chromatin remodeling complex has been implicated in transcription regulation, DNA replication and repair, and embryonic stem cell maintenance8, 27, 34. However, unlike the SWI/SNF family remodeler, its involvement in cancer was not well understood. In this study, we show that INO80 is required for lung cancer cell growth in vitro and tumor formation in mouse xenografts. We propose that INO80 binds enhancer regions near cancer-associated genes and promotes their expression by increasing genome accessibility. Our study uncovered a novel function of INO80 in cancer development, and revealed a critical role of chromatin remodeling in the regulation of enhancers and oncogenic transcription.
It has been suggested that the activity of certain molecular pathways in ESCs and cancers may be closely related, as the acquired ability of cancer cells to divide perpetually and support tumor growth resembles that of ESCs. Indeed, genes that are essential for ESC self-renewal are often found to play important roles in cancer5, 18, 36, and the gene expression signature and higher order chromatin structure show some resemblance between ESCs and cancers10. Since INO80 also regulates the ESC state, our results on INO80 in NSCLC add additional support to these earlier findings. Further, it is possible that INO80 may contribute to the potential parallels in the regulation of the chromatin state in ESCs and cancer cells.
Chromatin remodeling complexes use the energy of ATP hydrolysis to modulate nucleosome composition and positioning. They can regulate the chromatin structure and accessibility of genomic regions where they reside to influence the binding of transcription factors or transcription machineries, and thereby control gene expression12, 28. We found that INO80 occupies enhancer regions near cancer-associated genes. Furthermore, its occupancy strongly correlates with increased DNase I hypersensitivity. These results suggested that INO80 may facilitate the establishment and/or maintenance of an open chromatin state at enhancers to promote the expression of downstream genes. Our findings are consistent with previous reports that chromatin remodeling regulates enhancer activities14, 21. Interestingly, Ino80 depletion was reported to inhibit proliferation and anchorage-independent growth of oncogene-transformed mouse embryonic fibroblasts. However, Ino80 haploinsufficiency did not change tumor incidence and latency in p53 deletion mice26. It is possible that Ino80 expression in the heterozygote mice is still high enough to allow tumorigenesis, and different cancers or different cell types may show different dependency on the expression level of Ino80.
In recent years, it has become increasingly obvious that understanding the epigenetic regulations in lung cancer development may open new avenues for clinical intervention4, 15, 24. Compared to genetic alterations, epigenetic modifications are usually reversible. Moreover, they often simultaneously impact multiple genes and pathways essential for cancer cell survival and proliferation. Thus, targeting epigenetic mechanisms in cancer can provide unique opportunities for the development of more effective therapeutic strategies. Among epigenetic changes, chromatin remodeling is critical to the modulation of gene expression across a variety of physiological processes including cancer13, 17, 25. Growing evidence indicates that chromatin remodeling complexes have a widespread role in tumorigenesis. However, most chromatin remodelers studied so far typically show loss-of-function mutations in cancers and are not directly targetable. In comparison, we found that INO80 promotes oncogenic transcription and tumorigenesis. Indeed, amplifications of INO80 subunits were widely found in various cancers including lung cancer, and correlated with poor disease prognosis. Therefore, our findings suggest a potential new strategy for cancer treatment by inhibiting the INO80-dependent oncogenic transcription.
Materials and methods
Cell culture
A549, H1703, H358, H23, and H1299 cell lines were obtained from ATCC, and cultured in DMEM supplemented with 10% fetal bovine serum (FBS). Normal lung bronchial epithelial cells NHBE and cell line BEAS-2B was from Lonza, and was cultured using the BEGM BulletKit (Lonza).
For lentiviral shRNA infections, cells were plated at ~1 × 105 / well in 12-well plates. On the next day, shRNA lentiviruses were added to the cells, with fresh medium and polybrene at 5 μg/ml. The plate was centrifuged at 1000 x g for 45 min at room temperature, and cells were returned to the incubator and cultured.
For growth curves, cells were transduced with either shNT or shIno80 lentiviruses, and subsequently re-plated in 24-well plates. Cell number was counted at the indicated time points. Growth curves were generated from the average of three experiments, and plotted as Mean ± SEM.
For clonogenic assays, cells were plated in triplicate at appropriate cell densities for each cell line (100 – 500 cells/well) in 6-well plates, and cultured for 2–4 weeks for colony formation. Resulting colonies were washed once with PBS, fixed and stained with 0.05% (w/v) crystal violet in 50% methanol and 10% acetic acid for 30 min at room temperature. Colonies were carefully rinsed with distilled water, until background staining of the wells was minimal. Plates were air-dried and scanned. Staining intensities were quantified with ImageJ.
For anchorage-independent growth in soft agar, cells were resuspended in 0.4% agar in complete medium at 1 × 104/well in 6-well plates and layered on top of 0.6% agar as described previously. After 14 days, colonies were stained with 2 mg/ml MTT dissolved in PBS, for 1 h at 37 ℃. Plates were scanned, and colonies were quantified with ImageJ.
Mouse xenografts
All animal experiments were conducted following protocols approved by the Animal Care and Use Committee of the National Institute of Environmental Health Sciences. A549 cells were infected with shNT or shIno80 lentiviruses. Two days later, 2×106 cells in 0.1mL of phosphate-buffered saline (PBS) were injected subcutaneously into six-week old male SCID-BEIGE mice (Charles River Laboratories). Injected mice were divided into the shNT and shIno80 groups and mixed randomly within each group. 5 weeks after injection, mice were sacrificed without blinding and tumors formed from the injection sites were removed for analysis.
Western blot
Cells were lysed in cell lysis buffer (Thermo Scientific) containing the protease inhibitor cocktail (Roche). The lysates were sonicated and quantified by BCA assay (Life Technologies). Forty μg of total protein was loaded and separated by SDS-PAGE on 4%−12% Bis-Tris gel (Life Technologies) and transferred to nitrocellulose membranes. The membranes were blocked by 5% milk in TBS-T and then incubated with Ino80 antibody (Proteintech 18810–1-AP) overnight at 4℃. Afterwards, membranes were incubated with secondary antibodies and developed using ECL Detection Kit (GE Healthcare Bio-Sciences).
RNA isolation, RT-qPCR, and RNA-seq
Total RNA was isolated from cells using the GeneJet RNA purification kit (Thermo Scientific), and 0.5 μg total RNA was reverse transcribed to generate cDNA using the iScript cDNA Synthesis Kit (Bio-Rad) according to manufacturer’s instructions. qPCRs were performed using the SsoFast EvaGreen Supermix (Bio-Rad) on the Bio-Rad CFX-384 or CFX-96 real-time PCR System. Actin was used for normalization. Primers used in the study are listed in Table S1.
For RNA-seq, One microgram total RNA was used for sequencing library generation using the Truseq RNA Library Prep Kit V2 (Illumina) according to the manufacturer’s instructions. All libraries were sequenced on the NextSeq sequencer (Illumina) using the 75 nt paired-end sequencing protocol. All sequencing data will be deposited to the GEO database and made available to the public.
ChIP-qPCR and ChIP-Seq Sample Preparation
Ino80 ChIP were performed as described previously. Briefly, A375 or SK-MEL-147 cells were fixed using 1% formaldehyde for 10 min, and 0.125 M glycine was added to stop the fixation. Cells were harvested, and DNA was fragmented to 300–500 bp by sonication with a micro-tip attached to a Misonix 3000 sonicator. Immunoprecipitation was performed with Ino80 antibody (Proteintech 18810–1-AP) conjugated to Dynabeads protein G beads (Life Technology). ChIP DNA was eluted, reverse cross-linked, extracted by phenol/chloroform, and precipitated. For ChIP-seq, 1 ng ChIP DNA or input DNA was used to generate sequencing libraries using the Nextera XT DNA sample preparation Kit (Illumina). Libraries were sequenced on the NextSeq sequencer (Illumina) using the 75 nt paired-end sequencing protocol. All sequencing data will be deposited to the GEO database and made available to the public
TCGA data analysis
TCGA data were processed as previously described11. In brief, TCGA mRNA expression data were obtained from TCGA Synapse portal (syn300013). We obtained patient overall survival time from TCGA marker papers, or TCGA data portal (https://tcgadata.nci.nih.gov/tcga/). We used the univariate Cox test to examine whether the expression level was significantly correlated with patient survival (if you used cox P-value). We divided the patients into two groups based on their gene expression values (cutoff as median expression value), and then used logrank test to examine whether the expression level was significantly correlated with patient survival and displayed it in Kaplan-Meier Survival curves (if you used KM P-value).
RNA-Seq Analysis
Raw reads were filtered to only include those with a mean Phred quality score of 20 or greater. Adapter was trimmed from the reads using Trim Galore version 0.2.8. The filtered reads were aligned to the hg19 assembly using Tophat2 version 2.0.4. For the alignment and all subsequent analyses that require a gene model, a RefSeq gene model downloaded from the UCSC browser on February 5, 2013 was used. To get raw read counts for each gene, the htseq-count tool in the HTSeq package version 0.6.0 was used. The read counts were normalized and differentially expressed genes (DEGs) were identified using the DESeq R package version 1.14.0. DEGs were required to have an FDR of 0.05 or less and a fold change of 1.5 or greater. To obtain FPKM measurements for each gene, Cufflinks version 2.0.2 was used. A single measurement for each gene symbol was obtained by taking the sum of all RefSeq genes that have the same gene symbol. Any gene with an FPKM of less than 0.1 in any sample was removed.
ENCODE ChIP-Seq Data
For all analyses of H3K4me3, H3K4me1, H3K27ac, p300, H3K27me3, and DNase I, publically available ENCODE ChIP-Seq data was downloaded from the UCSC browser. Please see Table S6 for datasets used for the analysis.
ChIP-Seq Analysis
Adapter was removed using Cutadapt version 1.2.1. Reads were filtered to only include those with a mean Phred quality score of 20 or greater. The filtered reads were aligned to the hg19 assembly using Bowtie version 0.12.8 with the following paramters: -v 2 -m 1 --best --strata -I 15 -X 1000. The aligned reads were deduplicated by removing any read pairs where both mates aligned to the same genomic positions as another read. To obtain genome coverage tracks, the genomecov tool from the BEDTools suite version 2.17.0 was used. To normalize the coverage, the density was multiplied by 10 million and divided by the number of aligned, deduplicated read pairs. To call peaks, SICER version 1.1 was used, with the following paramters: REDUNDANCY_THRESHOLD=100, WINDOW_SIZE=200, GAP_SIZE=800, SPECIES=hg19, EFFECTIVE_GENOME_FRACTION=0.77, FDR=0.00001. The peaks were categorized as a TSS peak if it overlapped with a RefSeq TSS, or a gene body peak if it did not overlap with a TSS but did overlap with a RefSeq gene body. If it did not overlap with either, then it was categorized as intergenic. The RefSeq gene model used for this categorization and all subsequent analyses that require a gene model was downloaded from the UCSC browser on February 5, 2013. Ino80-bound genes were identified by finding the closest RefSeq TSS to each Ino80 peak center. Peaks were only included if it had a fold change in signal compared to Input of 2 or greater, and if the TSS was within 20 kb of the peak center. Heatmaps were generated by calculating the average normalized ChIP-seq signal in 100 nt bins in the region 5 kb upstream and downstream of the Ino80 peak centers. The rows were sorted by the FDR values associated with the peaks. The DNase I metagene was created by first generating a heatmap of the average normalized DNase I signal in 100 nt bins in the region 5 kb upstream and downstream of all RefSeq TSSs. Then, the genes in the heatmap were split into two categories, based on whether or not the TSS overlapped with an Ino80 peak. Finally, the metagenes were generated by taking the average of the genes within each category for all bins. Ino80-bound enhancer peaks were defined as Ino80 peaks with a fold change over Input of 2 or greater that also overlap with an H3K4me1 peak and an H3K27ac peak and a p300 peak. Ino80-bound enhancer genes were defined based on the closest TSS to the Ino80-bound enhancer peaks.
Microarray Analysis
Publically available microarray expression data from A549 cells and normal lung cells were downloaded from GEO (accession numbers GSM185869 and GSM185872). To obtain a single expression measurement for each gene, the average of all probes associated with a given gene was calculated. GSEA analysis was performed using the GseaPreranked tool in GSEA version 2.1.0 on the log2 fold change between the A549 cells and normal lung. Ino80-bound targets used for the boxplot were defined as any Ino80-bound enhancer genes that were also upregulated with a fold change of 2 or greater between the A549 cells and normal lung. The p-value for the difference between all genes and Ino80-bound targets was calculated using a two-sample Student’s t-test.
Supplementary Material
Acknowledgements:
We thank the NIEHS Animal, Epigenomics, and Bioinformatics core facilities for assistance with various techniques and experiments. This study was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences Z01ES102745 (to GH) and by National Institutes of Health Grants CA042978 and CA161494 (to ADC).
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Arenberg DA, Keane MP, DiGiovine B, Kunkel SL, Morris SB, Xue YY et al. (1998). Epithelial-neutrophil activating peptide (ENA-78) is an important angiogenic factor in non-small cell lung cancer. The Journal of clinical investigation 102: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M (2012). Epigenetic protein families: a new frontier for drug discovery. Nature reviews Drug discovery 11: 384–400. [DOI] [PubMed] [Google Scholar]
- 3.Baykara O, Bakir B, Buyru N, Kaynak K, Dalay N (2015). Amplification of chromosome 8 genes in lung cancer. Journal of Cancer 6: 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belinsky SA (2015). Unmasking the lung cancer epigenome. Annual review of physiology 77: 453–474. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics 40: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender E (2014). Epidemiology: The dominant malignancy. Nature 513: S2–3. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK (2014). Non-small-cell lung cancers: a heterogeneous set of diseases. Nature reviews Cancer 14: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conaway RC, Conaway JW (2009). The INO80 chromatin remodeling complex in transcription, replication and repair. Trends in biochemical sciences 34: 71–77. [DOI] [PubMed] [Google Scholar]
- 9.El-Telbany A, Ma PC (2012). Cancer genes in lung cancer: racial disparities: are there any? Genes & cancer 3: 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M (2011). Open chromatin in pluripotency and reprogramming. Nature reviews Molecular cell biology 12: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han L, Diao L, Yu S, Xu X, Li J, Zhang R et al. (2015). The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer cell 28: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hargreaves DC, Crabtree GR (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell research 21: 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helming KC, Wang X, Roberts CW (2014). Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer cell 26: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q et al. (2011). Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome research 21: 1650–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakopovic M, Thomas A, Balasubramaniam S, Schrump D, Giaccone G, Bates SE (2013). Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Frontiers in oncology 3: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P et al. (2010). Genomic aberrations in lung adenocarcinoma in never smokers. PloS one 5: e15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadoch C, Crabtree GR (2015). Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Science advances 1: e1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Orkin SH (2011). Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome medicine 3: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolluri SK, Bruey-Sedano N, Cao X, Lin B, Lin F, Han YH et al. (2003). Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Molecular and cellular biology 23: 8651–8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer A, Green J, Pollard J Jr., Tugendreich S (2014). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H et al. (2015). Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennon FE, Cianci GC, Cipriani NA, Hensing TA, Zhang HJ, Chen CT et al. (2015). Lung cancer-a fractal viewpoint. Nature reviews Clinical oncology 12: 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linxweiler M, Linxweiler J, Barth M, Benedix J, Jung V, Kim YJ et al. (2012). Sec62 bridges the gap from 3q amplification to molecular cell biology in non-small cell lung cancer. The American journal of pathology 180: 473–483. [DOI] [PubMed] [Google Scholar]
- 24.Liu SV, Fabbri M, Gitlitz BJ, Laird-Offringa IA (2013). Epigenetic therapy in lung cancer. Frontiers in oncology 3: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O (2015). SWI/SNF chromatin remodeling and human malignancies. Annual review of pathology 10: 145–171. [DOI] [PubMed] [Google Scholar]
- 26.Min JN, Tian Y, Xiao Y, Wu L, Li L, Chang S (2013). The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell research 23: 1396–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison AJ, Shen X (2009). Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nature reviews Molecular cell biology 10: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T (2013). Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144: 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF (2013). Management of non-small-cell lung cancer: recent developments. Lancet 382: 709–719. [DOI] [PubMed] [Google Scholar]
- 31.Stewart DJ (2014). Wnt signaling pathway in non-small cell lung cancer. Journal of the National Cancer Institute 106: djt356. [DOI] [PubMed] [Google Scholar]
- 32.Viktorsson K, Lewensohn R, Zhivotovsky B (2014). Systems biology approaches to develop innovative strategies for lung cancer therapy. Cell death & disease 5: e1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X et al. (2014). INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell stem cell 14: 575–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe S, Peterson CL (2010). The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harbor symposia on quantitative biology 75: 35–42. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL (2013). A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 340: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY (2008). Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell stem cell 2: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.