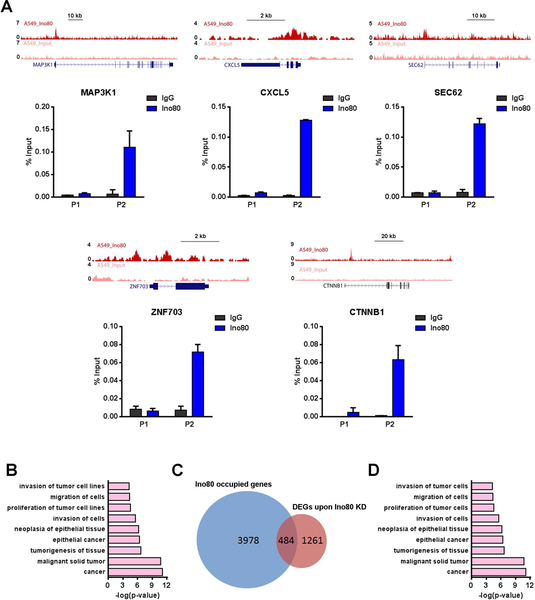
Figure 5. INO80 occupies genomic regions near lung cancer-associated genes.
(A) Genome browser tracks and ChIP-qPCR validations showing Ino80 occupancy near selected lung cancer genes MAP3K1, CXCL5, SEC62, ZNF703, CTNNB1 in A549 cells. Two genomic regions near each gene were selected for the ChIP-qPCR validations: P1 corresponds to a region that is not bound by Ino80 based on ChIP-Seq, while P2 corresponds to a region that is bound by Ino80. (B) IPA showing the enrichment of cancer-associated genes in Ino80-bound genes. Selected top categories are shown, and please see Table S4 for the complete list. (C) Venn diagram showing the overlap between DEGs after Ino80 KD and Ino80-bound genes. (D) IPA of the 580 Ino80 target genes. Selected top categories are shown, and please see Table S5 for the complete list.