Significance
Obesity is a major public health issue worldwide. Easy accessibility of junk food is considered a major contributor to the current obesity epidemic. Thus, the impact of maternal overnutrition in determining disease susceptibility in offspring has received wide attention. It has also been shown that the effects of maternal overnutrition are not limited to the immediate offspring but can also be transmitted to successive generations. Among different epigenetic marks, sperm small noncoding RNAs (sncRNAs) have recently been reported as a direct mediator of acquired traits to the progeny following postnatal trauma or paternal diet. Here, we investigate whether sperm sncRNAs contributes to the transmission of metabolic and hedonic phenotypes across generations following maternal overnutrition.
Keywords: maternal, overnutrition, obesity, epigenetic, sperm RNA
Abstract
There is a growing body of evidence linking maternal overnutrition to obesity and psychopathology that can be conserved across multiple generations. Recently, we demonstrated in a maternal high-fat diet (HFD; MHFD) mouse model that MHFD induced enhanced hedonic behaviors and obesogenic phenotypes that were conserved across three generations via the paternal lineage, which was independent of sperm methylome changes. Here, we show that sperm tRNA-derived small RNAs (tsRNAs) partly contribute to the transmission of such phenotypes. We observe increased expression of sperm tsRNAs in the F1 male offspring born to HFD-exposed dams. Microinjection of sperm tsRNAs from the F1-HFD male into normal zygotes reproduces obesogenic phenotypes and addictive-like behaviors, such as increased preference of palatable foods and enhanced sensitivity to drugs of abuse in the resultant offspring. The expression of several of the differentially expressed sperm tsRNAs predicted targets such as CHRNA2 and GRIN3A, which have been implicated in addiction pathology, are altered in the mesolimbic reward brain regions of the F1-HFD father and the resultant HFD-tsRNA offspring. Together, our findings demonstrate that sperm tsRNA is a potential vector that contributes to the transmission of MHFD-induced addictive-like behaviors and obesogenic phenotypes across generations, thereby emphasizing its role in diverse pathological outcomes.
In recent years, there has been increasing interest in how parental environmental insults can induce long-lasting physiological and behavioral changes in the progeny, and specifically how these acquired traits are transmitted from parents to subsequent generations through nongenomic mechanisms (1, 2). A better understanding of such ancestral effects may provide insight into the etiology of obesity. In addition, this will enable new preventive and therapeutic measures for various noncommunicable diseases, such as diabetes, cardiovascular, and neuropsychiatric disorders (3, 4). Changes in epigenetic marks in sperm and oocytes such as DNA methylation, histone modifications, and noncoding RNAs have been implicated in the transgenerational transmission of such heritable phenotypes (5–10).
The increasing prevalence of maternal obesity or overnutrition is a major public health concern in the present millennium as a result of long-term effects on the physiology and behavior of offspring across generations (11). Recently published works from our laboratory have shown that offspring born to 9-wk high-fat diet (HFD)-exposed dams develop an obesogenic phenotype and enhanced hedonic behaviors, such as overconsumption of palatable foods and higher sensitivity to drugs of abuse. These traits are transmitted up to the third generation (F3) via the paternal lineage without any further exposure to HFD (12, 13). In addition, a sustained reduction in striatal dopamine levels is observed across all three generations. This may imply that a hypodopaminergic state of the mesolimbic reward system is partly responsible for the altered hedonic and obesogenic phenotypes (12, 13). Similar to other early nutritional animal models, we have observed only moderate changes in CpG methylation in F1 (first-generation) and F2 (second-generation) sperm between the offspring groups (13–15). This suggests that sperm methylome might not constitute the principal mode of transmission of these phenotypes.
Although changes in DNA methylation pattern and histone modifications have been reported following a range of early-life environmental exposures (14, 16–19), a causal link between the germ cell epigenetic marks and the observed phenotypes in the offspring still remains elusive. In this context, sperm RNAs have gained wide attention recently because they can provide absolute proof of the direct dissipation of information across generations (8, 10, 20, 21). The role of sperm small noncoding RNAs (sncRNAs) in the regulation of DNA methylation, histone modifications, and mRNA transcription strongly suggests that sperm sncRNAs can be a potential vector of the gene–environment interaction (22). As mature sperm harbors a diversity of sncRNAs, such as miRNAs, PIWI-interacting RNAs (piRNAs), and tRNA-derived small RNAs (tsRNAs) (23), sperm RNA research in the context of transgenerational and intergenerational epigenetic inheritance shifted its focus to identifying the roles of specific subpopulations of sncRNAs (8, 10, 21, 24, 25). More recently, a number of studies have shown that paternal low protein or HFD exposure could alter sperm tsRNAs levels across generations (20, 26–28). These alterations in sperm tsRNAs alone are capable of transmitting paternal HFD-induced impaired glucose-tolerance phenotype in the progeny (20, 29).
We have observed persistence obesogenic and addictive-like phenotypes in offspring born to HFD-fed dams up until the third generation that could not be explained by changes in sperm methylome. Therefore, the present study aims to investigate whether sperm RNAs play a role in the inheritance of obesogenic and addictive-like traits and whether there are specific subpopulations of sperm sncRNAs that are responsible for this transmission.
Results
Hedonic and Metabolic Phenotypes in Sperm Total RNA-Injected Offspring.
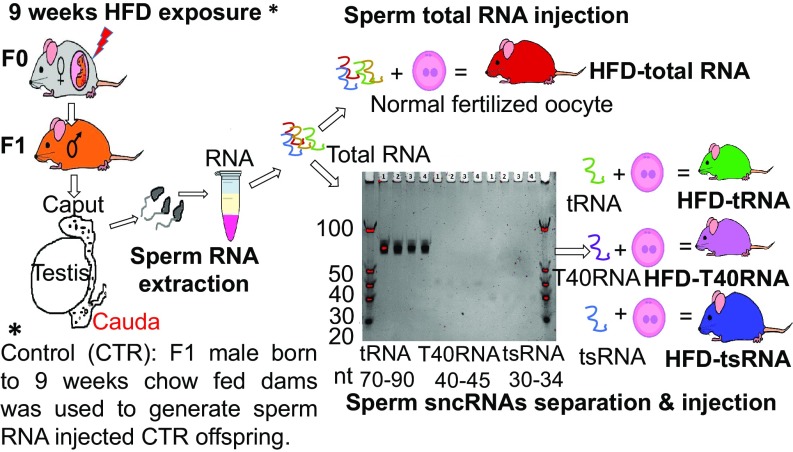
We first generated sperm total RNA-injected offspring according to the scheme outlined in Fig. 1. This allowed us to determine whether sperm RNAs could be a potential mediator of the transgenerational transmission of altered phenotypes following a maternal HFD (MHFD) challenge. F0 female mice were exposed to HFD (60% energy from fat; Kliba 2127) or regular chow diet [control (CTR); Kliba 3430] for 9 wk (3 wk before mating, 6 wk during gestation and lactation). Subsequently, F1 offspring from HFD and chow-fed (i.e., CTR) dams were exposed to chow diet and water ad libitum throughout their life. We isolated and purified total RNAs from sperm of F1-HFD and F1-CTR offspring at postnatal day (PND) 70. The purified total RNAs from both groups were injected separately into normal fertilized mouse oocytes to generate HFD-total RNA and CTR-total RNA offspring, respectively (RNA injection was normalized to the amount of six sperm). Upon weaning at PND 21, the total RNA-injected offspring were given ad libitum access to chow diet and water unless otherwise indicated. All behavioral and metabolic assessments commenced when the animal reached adulthood at PND 70.
Fig. 1.
Breeding scheme to generate sperm sncRNA-injected offspring. Female mice (F0) were fed HFD or chow diet for 3 wk preconception and 6 wk during gestation and lactation to obtain F1-HFD and F1-CTR offspring, respectively. Mature sperm was isolated from F1-HFD and F1-CTR males at PND 70. Sperm total RNAs from F1-HFD and F1-CTR males were separated on a 6% TBE gel. Sperm total RNAs as well as isolated RNA fragments at sizes ∼70–90 nt, ∼40–45 nt, and ∼30–34 nt were purified and microinjected into male pronuclei of normal fertilized eggs to generate total RNA, tRNA, T40RNA, and tsRNA offspring from both groups. Offsprings from HFD and CTR groups were on chow diet since weaning. HFD and CTR offspring groups refer to the descendants of 9 wk HFD- and chow-fed F0 dams, respectively. Colors of the mice are matched with the color codes used for the groups in subsequent graphs.
HFD-total RNA offspring showed obesogenic phenotypes.
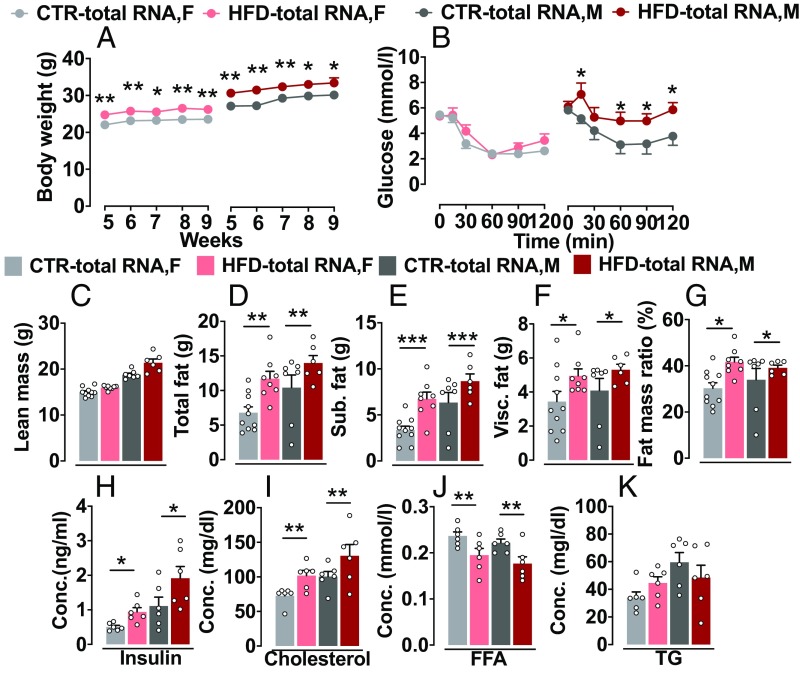
As shown in Fig. 2A, male and female HFD-total RNA offspring kept on chow diet gained significantly more weight compared with the CTR-total RNA offspring from postnatal weeks 5 to 9 (P < 0.0001). The increased weight gain was caused by an increased fat mass as measured by CT scan (Fig. 2 C–G). Male and female HFD-total RNA offspring displayed increased body fat content (P < 0.002), greater fat depot in s.c. (P < 0.001), and increased visceral fat (P < 0.03), as well as higher fat mass ratio (P < 0.02), with no significant difference in lean mass (P = 0.07), compared with their CTR littermates. Furthermore, HFD-total RNA offspring showed higher blood glucose levels (P < 0.03) than CTR following a systemic insulin injection in the insulin-tolerance test (Fig. 2B and SI Appendix, Fig. S2A). Notably, the impaired insulin sensitivity was stronger in the male HFD-total RNA offspring (P < 0.005) compared with the other groups. Consistent with these findings, HFD-total RNA offspring also displayed elevated fasted plasma insulin (P < 0.02), higher cholesterol (P < 0.009), and lower free fatty acid (FFA) levels (P < 0.002) compared with their CTR littermates (Fig. 2 H–J). No difference in plasma triglyceride level was detected between the groups (P = 0.88; Fig. 2K). These results indicate that HFD-total RNA offspring developed obesity and the metabolic syndrome-like phenotypes.
Fig. 2.
Altered metabolic phenotypes in HFD-total RNA–injected offspring. (A) Body weight: HFD-total RNA offspring showed gradually increased body weight compared with the CTR-total RNA offspring. CTR-total RNA, n = 19 (8 M, 11 F); HFD-total RNA, n = 18 (8 M, 10 F). (B) Insulin tolerance test: HFD-total RNA offspring had higher blood glucose level than CTR-total RNA following an insulin injection. HFD-total RNA male offspring showed a stronger impairment of insulin sensitivity. CTR-total RNA, n = 19 (8 M, 11 F); HFD-total RNA, n = 16 (8 M, 8 F). (C–G) Distribution of fat: HFD-total RNA offspring displayed a marked increase in total fat, s.c. fat, visceral fat, and fat mass ratio, with no difference in lean mass. CTR-total RNA, n = 17 (7 M, 10 F); HFD-total RNA, n = 14 (6 M, 8 F). (H–K) Plasma parameters: HFD-total RNA group showed higher fasted plasma insulin and cholesterol and lower FFA levels but no difference in plasma triglyceride (TG) levels compared with CTR (n = 6 M/6 F per group). Data are presented as mean ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001). F, female; M, male.
Altered hedonic-like behaviors in the HFD-total RNA offspring.
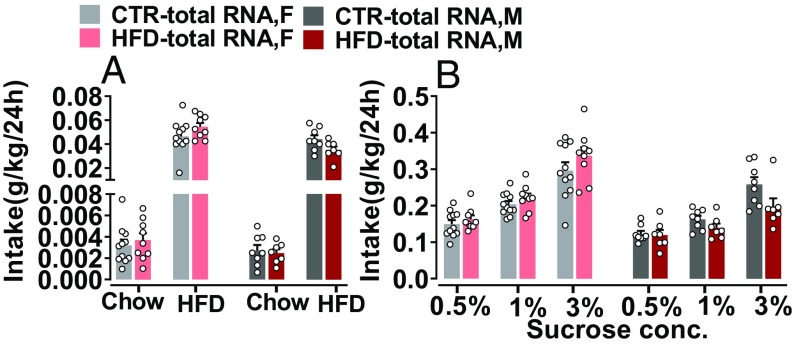
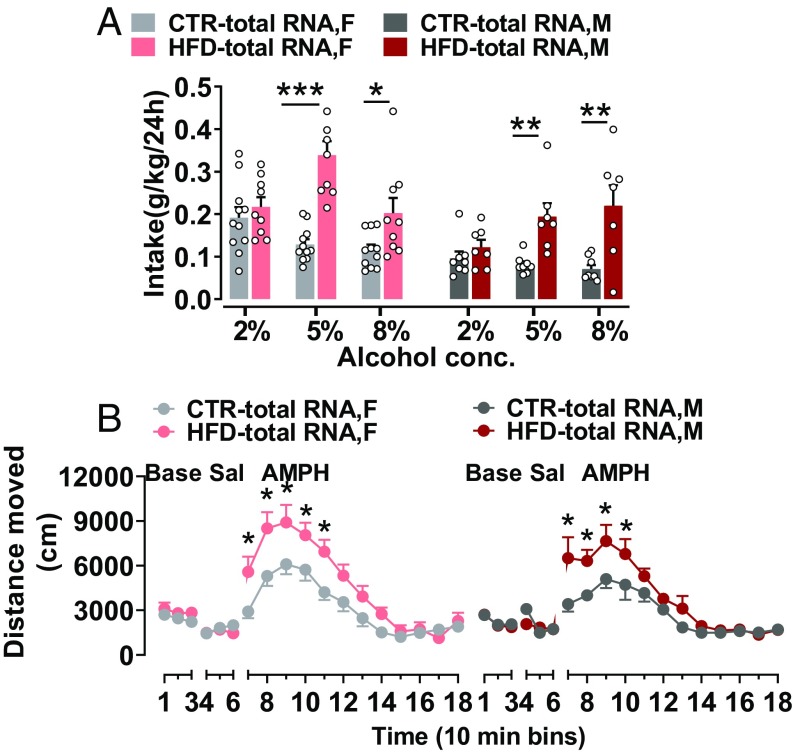
As we have shown that HFD-total RNA offspring develop an obesogenic phenotype, we were interested to investigate whether this disposition for obesity may be associated with an altered central reward system. Therefore, we next characterized offspring behavior in response to natural rewards as well as drugs of abuse (30). Consumption of appetitive stimuli has been used to index the hedonic value attributed to those stimuli (31). Therefore, we tested hedonic-like behaviors in response to natural rewards, such as HFD and sucrose, in a free-choice paradigm (32). All offspring groups consumed more HFD than chow diet (P < 0.0001), and females consumed significantly more HFD compared with the males (P < 0.01), in the HFD preference test. However, no difference was detected in HFD preference between the HFD-total RNA offspring compared with the CTR-total RNA offspring (Fig. 3A). Similarly, when offspring were given a free choice of water and sucrose solution, all offspring groups preferred the sucrose solution at all concentrations vs. water (P < 0.0001). However, no difference was observed in sucrose preference between the groups (Fig. 3B). Next, we assessed offspring’s hedonic response to drugs of abuse in an alcohol preference test and in the amphetamine sensitivity test. In the alcohol preference test, when offspring were given free choice between alcohol solution and water, all offspring groups preferred the alcohol solution at all concentrations to water (P < 0.0001). Females consumed more alcohol than males (P < 0.0001). Notably, male and female HFD-total RNA offspring showed significantly greater hedonic drinking of alcohol at higher concentrations compared with their CTR littermates, indicating an increased preference for alcohol in the HFD-total RNA offspring (P < 0.0001; Fig. 4A). Similarly, HFD-total RNA offspring show enhanced sensitivity to the acute drug stimulant effect of amphetamine. As depicted in Fig. 4B, there was no difference in baseline spontaneous locomotor activity or in response to a saline solution injection between the offspring groups. Amphetamine-induced locomotor activity was significantly increased in male and female HFD-total RNA offspring compared with their controls (P < 0.01). Together, these findings may imply that total RNA from the F1-HFD father sperm could lead to enhanced susceptibility to drug abuse in the resultant HFD-total RNA offspring.
Fig. 3.
Altered hedonic response to natural rewards in total RNA-injected offspring. (A) HFD preference: no difference in HFD consumption between HFD-total RNA and CTR-total RNA offspring was observed. (B) Sucrose preference: all offspring groups showed increased sucrose consumption at all concentrations. No difference was observed in sucrose preference between groups. HFD-total RNA, n = (7 M, 9 F); CTR-total RNA, n = (8 M, 11 F). Data are presented as mean ± SEM. F, female; M, male.
Fig. 4.
Altered hedonic response to drugs of abuse in total RNA-injected offspring. (A) Alcohol preference: HFD-total RNA offspring showed increased preference for alcohol compared with CTR. Male and female HFD-total RNA offspring consumed more alcohol at higher concentrations compared with their CTR littermates. HFD-total RNA, n = (7 M, 9 F); CTR-total RNA, n = (8 M, 11 F). (B) Amphetamine sensitivity: male and female HFD-total RNA offspring showed enhanced locomotor response to amphetamine (AMPH) compared with their CTR littermates (n = 5 M/5 F per group). Data are presented as mean ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001). Base, baseline; Sal, saline. F, female; M, male.
Characterization of Hedonic and Metabolic Phenotypes in Offspring Generated from Sperm RNA Fragment Injection.
To further investigate which subpopulations of sperm sncRNAs are responsible for the transmission of these phenotype changes, we first collected mature sperm from the cauda epididymis of F1 male mice born to HFD- and chow-fed dams. The total RNA from the sperm sample was extracted by using TRIzol-chloroform method according to the manufacturer’s instructions. The total RNA was then run on 6% Tris-borate-EDTA (TBE) gel, and the location of RNA fractions was determined by the position of standard small RNA markers by using long-wave UV light illumination of the gel. We found three prominent RNA fragments bands at sizes of ∼70–90 nt, ∼40–45 nt, and ∼30–34 nt in length. The majority of ∼70–90-nt RNAs are known to contain tRNA, so we labeled this fraction as tRNA (33). The small RNAs at the length of ∼30–34 nt in mature sperm have been reported to be enriched with fragments derived from tRNA (20, 26, 29, 33–35). We termed these RNA fractions tsRNAs as described by Chen et al. (35). The content of this fraction was later confirmed by small RNA sequencing analysis (see Fig. 8A). The RNA content at sizes of ∼40–45 nt is not known, and we therefore termed it as T40RNA. Each band at 70–90 nt, 30–34 nt, and 40–45 nt was carefully excised together with gel slice, purified, eluted, and later microinjected separately into normal fertilized oocytes at concentrations similar to the injected total ncRNAs to generate tRNA, tsRNA, and T40RNA offspring from HFD and CTR groups, respectively (Fig. 1). The resultant offspring were kept on chow diet and water unless otherwise specified.
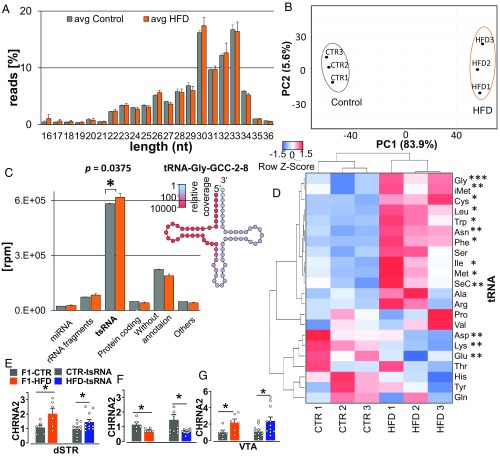
Fig. 8.
Sperm sncRNA profile in F1 fathers and differences in transcriptomes in brain of F1 father and tsRNA offspring. (A) Length distribution of sncRNA sequence reads from F1-HFD and F1-CTR males. Error bars indicate SD. (B) Principal component analysis based on 4,822 differentially expressed sncRNA species. x and y axes show principal components 1 (PC1) and 2 (PC2) that explain 83.9% and 5.6% of the total variance, respectively. (C) Composition of sncRNA transcriptomes. tsRNAs constitute the majority of sncRNAs in HFD and CTR groups. (Inset) Example of relative sncRNA coverage of tRNA-Gly-GCC-2–8. Error bars indicate SD. (D) Heat map displays relative expression of tsRNAs derived from different tRNAs across HFD and control probes. Thirteen tRNA fragments showed a significant difference in expression. Clustering bases on average linkage. The Pearson distance measurement method was used. (E) The increased level of CHRNA2 in the dSTR of F1 HFD and HFD-tsRNA offspring compared with CTR littermates. (F) Decreased levels of CHRNA2 in the Nac of F1-HFD and HFD-tsRNA offspring compared with their CTR littermates. (G) Higher expression of CHRNA2 in the VTA of F1-HFD male and HFD-tsRNA offspring compared with CTR. F1-CTR, n = 6; F1-HFD, n = 6; CTR-tsRNA, n = 12; HFD-tsRNA, n = 12. Data are presented as mean ± SEM (*P < 0.05).
Altered hedonic-like behaviors in the HFD-tsRNA offspring.
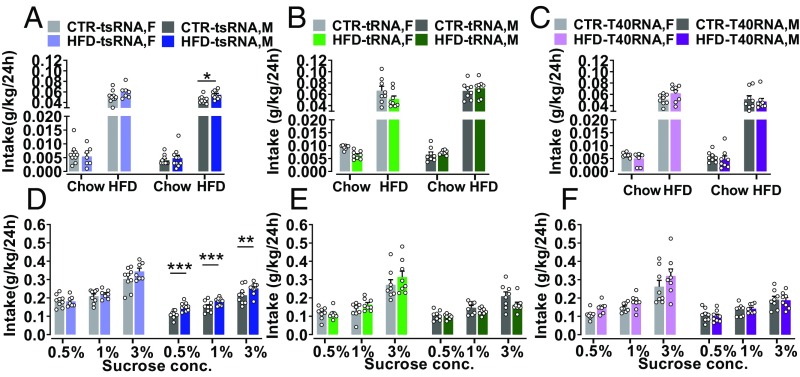
We assessed offspring’s hedonic response to natural rewards as well as drugs of abuse in the same test paradigms to identify which subgroup conserved the altered phenotypes similar to their ancestors. In the HFD preference test, a greater intake of HFD than chow diet was detected in all offspring groups (P < 0.0001). Notably, HFD-tsRNA offspring showed an increased HFD consumption compared with the CTR-tsRNA offspring (P < 0.02; Fig. 5A). Furthermore, the male HFD-tsRNA offspring showed a stronger preference to HFD compared with their CTR littermates (P < 0.03). In contrast, the offspring from HFD-tRNA (Fig. 5B) and HFD-T40 RNA (Fig. 5C) groups did not differ in their preference for HFD compared with the CTR littermates. In the sucrose preference test, all offspring groups consumed more sucrose than water (P < 0.0001). Females consumed more sucrose solution than males, as supported by a significant main effect of sex (P < 0.0001; Fig. 5 D–F). HFD-tsRNA offspring displayed greater consumption of sucrose solution compared with their controls (P < 0.02; Fig. 5D). HFD males showed increased sucrose consumption across all different concentrations compared with their CTR littermates (0.5% sucrose, P < 0.0002; 1% sucrose, P < 0.0005; 3% sucrose, P < 0.009). Neither HFD-tRNA nor HFD-T40RNA offspring showed any difference in sucrose consumption compared with their CTR littermates (Fig. 5 E and F).
Fig. 5.
Altered hedonic responses to natural rewards in sperm RNA fragment-injected offspring. HFD preference: (A) all offspring groups consumed more HFD than chow diet, and females consumed more than males. HFD-tsRNA offspring showed a greater intake of HFD than CTR-tsRNA offspring. HFD-tsRNA, n = 10 M/7 F; CTR-tsRNA, n = 10 M/10 F. (B) No difference was detected in HFD preference between the groups (n = 8 M/8 F per group). (C) No difference was observed in HFD preference between the groups (n = 8 M/8 F per group). Sucrose preference: (D) HFD-tsRNA offspring showed greater sucrose consumption than CTR-tsRNA offspring. Male HFD-tsRNA offspring showed a stronger sucrose preference compared with the others at all concentrations. HFD-tsRNA, n = 10 M/7 F; CTR-tsRNA, n = 10 M/10 F. (E) No difference in sucrose preference was detected between the tRNA offspring groups (n = 8 M/8 F per group). (F) No difference in sucrose consumption between the T40RNA offspring groups was detected (n = 8 M/8 F per group). Data are presented as mean ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001). F, female; M, male.
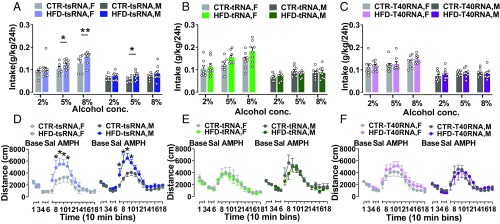
When offspring were given free access to water and alcohol, all offspring groups preferred alcohol to water (P < 0.0001). Offspring from the HFD-tsRNA group showed a marked increase in alcohol consumption compared with their controls (P < 0.02; Fig. 6A). Furthermore, we found that the female HFD-tsRNA offspring had stronger preference for alcohol at higher concentrations compared with the other groups (5% alcohol, P < 0.04; 8% alcohol, P < 0.007). On the contrary, alcohol consumption in HFD-tRNA and HFD-T40RNA offspring was comparable to that in CTR littermates (Fig. 6 B and C). In the amphetamine sensitivity test, no difference was observed in spontaneous locomotor activity and locomotor response after a saline solution injection between the groups (Fig. 6 D–F). A significant increase in locomotor activity following a systemic amphetamine injection was found in male and female HFD-tsRNA offspring compared with their CTR littermates (P < 0.009; Fig. 6D). Neither HFD-tRNA nor HFD-T40RNA offspring showed any difference in locomotor activity in response to the drug compared with their respective control groups (Fig. 6 E and F). Together, these results indicate that injection of sperm 30–34-nt RNAs from F1 HFD males predispose offspring to develop an increased preference for palatable foods as well as enhanced sensitivity to drugs of abuse similar to their fathers.
Fig. 6.
Altered hedonic responses to drugs of abuse in sperm RNA fragment-injected offspring. Alcohol preference: (A) HFD-tsRNA offspring consumed more alcohol than CTR-tsRNA offspring. Female HFD-tsRNA offspring had stronger alcohol preference compared with the others at higher concentrations. HFD-tsRNA, n = 9 M/10 F; CTR-tsRNA, n = 7 M/6 F. (B) No difference in alcohol consumption between the groups was detected (n = 8 M/8 F per group). (C) HFD-T40RNA offspring did not differ in alcohol consumption at all concentrations compared with the CTR-T40RNA offspring (n = 8 M/8 F per group). Amphetamine sensitivity: (D) male and female HFD-tsRNA offspring showed enhanced locomotor response to amphetamine (AMPH) compared with the CTR-tsRNA offspring, with no difference in locomotion at baseline (Base) and following a saline solution (Sal) injection. (E) HFD-tRNA offspring did not differ in baseline locomotor activity following a saline solution injection and after amphetamine challenge compared with the CTR-tRNA offspring. (F) No difference was observed in baseline spontaneous locomotor activity and after a saline solution injection as well as after an amphetamine challenge between the T40RNA offspring groups (n = 5 M/5 F per group). Data are presented as mean ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001). F, female; M, male.
HFD-tsRNA offspring showed altered metabolic phenotypes in normal-fed condition.
We next evaluated the metabolic phenotypes of the offspring born after different sperm RNA fragment injections. Offspring were weaned and maintained on ad libitum chow diet and water. Body weight was measured weekly from postnatal weeks 3 to 12. HFD-tsRNA offspring gained significantly more weight over the weeks compared with the CTR-tsRNA offspring (P < 0.004; SI Appendix, Fig. S1A). Furthermore, weight gain was more pronounced in male HFD-tsRNA offspring (P < 0.0001). Consistent with these findings, HFD-tsRNA offspring displayed higher blood glucose levels compared with their CTR littermates following a systemic insulin challenge, indicating an impaired insulin sensitivity in the HFD-tsRNA group (P < 0.05; SI Appendix, Figs. S1D and S2B). The effect was stronger in male HFD-tsRNA offspring compared with the other groups (P < 0.002). Offspring from HFD-tRNA as well as HFD-T40RNA showed neither differences in body weight nor alterations in blood glucose levels following an insulin challenge compared with their respective controls (SI Appendix, Fig. S1 B, C, E, and F). Together, these findings indicate that the HFD-tsRNA group developed marked weight gain associated with impaired insulin sensitivity. Interestingly, these metabolic phenotypes were stronger compared with F1-HFD fathers but similar to the phenotype observed in the HFD-total RNA offspring group.
HFD-tsRNA offspring showed altered metabolic phenotypes after chronic junk food exposure.
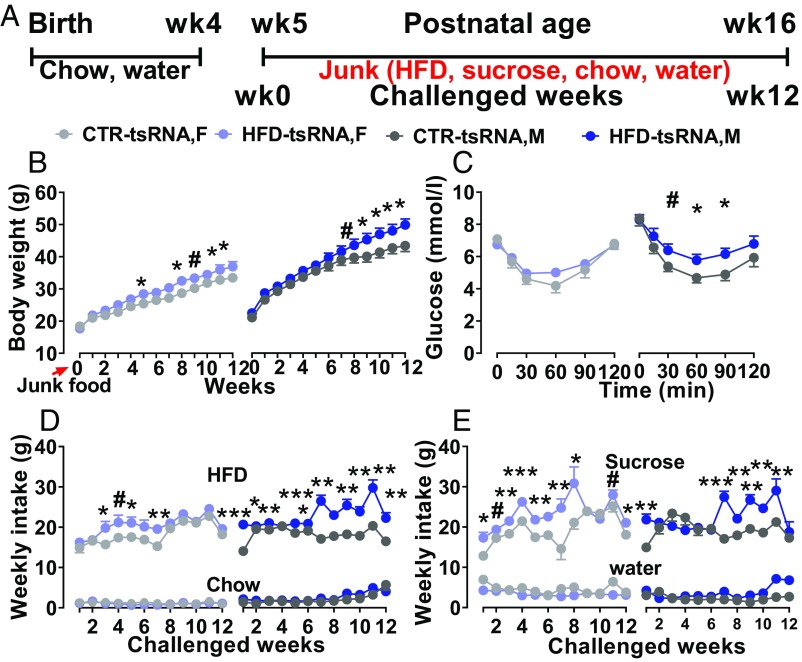
HFD-tsRNA offspring maintained on standard chow diet gained significantly more weight and showed increased preference for palatable foods, such as HFD and sucrose solution. We next evaluated their feeding behavior as well as food preference by giving the mice free access to palatable foods (HFD and 1% sucrose solution) and regular chow diet and water for 12 wk (from postnatal week 5 to postnatal week 16) in their home cages and characterized their metabolic phenotypes (Fig. 7A). Independent control groups of HFD-tsRNA and CTR-tsRNA offspring were given ad libitum chow and water during this time period. Body weight and food intake were measured weekly during the experimental period. As depicted in Fig. 7B, at the beginning of the chronic junk food exposure, no difference in body weight was observed between the groups. After 5 wk of junk food exposure, female HFD-tsRNA offspring showed a significant increase in body weight compared with the CTR littermates, and the difference progressively increased over the 12-wk junk food exposure (P < 0.009). In the males, a significant body weight difference commenced after 9 wk of junk food exposure. Weight gain was significantly stronger in the HFD-tsRNA group compared with their controls (P < 0.004). At the end of the experiment, male HFD-tsRNA offspring weighed 14.99% more than their CTR littermates, which was associated with increased total fat content (P < 0.004; SI Appendix, Fig. S3B), s.c. fat (P < 0.008; SI Appendix, Fig. S3C), visceral fat depots (P < 0.002; SI Appendix, Fig. S3D), and fat mass ratio (P < 0.01; SI Appendix, Fig. S3E), with no difference in lean body mass (P = 0.08; SI Appendix, Fig. S3A). However, no difference in fat mass distribution was detected between the female HFD-tsRNA and CTR-tsRNA groups. Furthermore, male HFD-tsRNA offspring developed impaired insulin sensitivity at the end of the experiment (P < 0.008; Fig. 7C and SI Appendix, Fig. S2C). Consistent with these findings, male HFD-tsRNA offspring showed elevated fasted plasma insulin (P < 0.04; SI Appendix, Fig. S3F), plasma leptin (P < 0.002; SI Appendix, Fig. S3H), and cholesterol levels (P < 0.01; SI Appendix, Fig. S3G). The weekly food intake measurements indicated that offspring exposed to junk food consumed more HFD and drank more from the sucrose solution compared with chow food and water, respectively (Fig. 7 D and E). No difference in chow food and water consumption was detected between the groups. Notably, male and female HFD-tsRNA offspring showed significantly higher HFD (P < 0.0001; Fig. 7D) and sucrose consumption (P < 0.0001; Fig. 7E) compared with their CTR littermates during the junk food choice period. In the control group (chow/water-fed cohort), no difference was detected in consumption of chow (mean, HFD-tsRNA = 25.33 ± 0.34; CTR-tsRNA = 24.51 ± 0.35) and water (mean, HFD-tsRNA = 30.99 ± 0.59; CTR-tsRNA = 29.73 ± 0.48) between HFD-tsRNA and CTR-tsRNA offspring (SI Appendix, Fig. S4 A and B). We next measured their energy expenditure in the metabolic cages. As shown in SI Appendix, Fig. S5 A–E, the junk food-exposed HFD-tsRNA offspring did not show any difference in O2 consumption (P = 0.89), CO2 production (P = 0.68), heat production (P = 0.86), respiratory exchange ratio (P = 0.11), or general locomotor activity (P = 0.91) compared with the CTR-tsRNA offspring. Together, these findings demonstrate that, upon free choice of chronic junk food, the HFD-tsRNA offspring exhibited greater preference for palatable foods compared with the CTR-tsRNA offspring. The long-term preference and excessive consumption of palatable foods further exacerbated the obesity state and other features of the metabolic syndrome in the HFD-tsRNA group. This obesogenic phenotype was more pronounced in male HFD-tsRNA offspring.
Fig. 7.
Altered metabolic phenotypes in HFD-tsRNA offspring following junk food choice test. (A) Experimental design: tsRNA offspring from HFD and CTR groups were given free access to HFD and 1% sucrose along with chow and water for 12 wk, starting 2 wk after weaning (at postnatal week 5). (B) Body weight: HFD-tsRNA offspring gradually gained more weight compared with the CTR littermates following junk food choice exposure. The weight gain was more marked in male HFD-tsRNA offspring compared with the others. CTR-tsRNA, n = 21 (12 M, 9 F); HFD-tsRNA, n = 23 (13 M, 10 F). (C) Insulin tolerance test: HFD-tsRNA offspring had persistently higher blood glucose level than CTR-tsRNA offspring following insulin injection. A stronger impairment of insulin sensitivity was detected in male HFD-tsRNA offspring. CTR-tsRNA, n = 17 (9 M, 8 F); HFD-tsRNA, n = 22 (11 M, 11 F). (D) Food intake: male and female HFD-tsRNA offspring showed marked increase in HFD consumption over the weeks compared with the CTR-tsRNA offspring. (E) Solution intake: male and female HFD-tsRNA offspring consumed more sucrose solution compared with the CTR-tsRNA offspring. No difference was observed in chow food and water consumption between the groups. CTR-tsRNA, n = 20 (12 M, 8 F); HFD-tsRNA, n = 23 (13 M, 10 F). Data are presented as mean ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001). F, female; M, male.
Sperm sncRNA Profiling of HFD and CTR F1 Father and Functional Correlation.
To confirm whether sperm tsRNA is the major mediator of the altered hedonic and metabolic phenotypes in the progeny, we next investigated the sperm sncRNA profiles (16–40 nt) of the HFD and CTR F1 fathers by small RNA deep-sequencing analysis. Analysis of the sequencing data revealed that the size distribution of sequencing reads is similar between the HFD and CTR sperm (Fig. 8A). Principal component analysis of differentially expressed small RNAs showed clear clustering of the sample groups (Fig. 8B). In addition, sperm of both groups contain significantly higher amounts of tsRNAs (∼28–34 nt) compared with the other sncRNA population. Comparative analysis of sncRNAs between HFD and CTR F1 sperm revealed significantly higher amounts of tsRNAs in the HFD vs. their controls (P < 0.04; Fig. 8C), predominantly 5′ tRNA halves (SI Appendix, Fig. S6A). A difference in the expression of several tRNA fragments was detected between HFD and control sperm. The difference between source tRNAs as a whole was statistically significant for 13 tRNAs (Fig. 8D). Next, to understand how sperm tsRNAs could program increased hedonic-like behaviors and metabolic abnormalities in the HFD-tsRNA offspring, we screened for putative target transcripts of the differentially expressed tsRNAs. We predicted 3,306 transcripts to be up-regulated and 2,711 transcripts to be down-regulated in the HFD group compared with their controls by assigning score values to each transcript based on the different amount of targeting tsRNA molecules between both conditions (SI Appendix, Fig. S7A). We then selected the top 50 predicted targets from the up-regulated and down-regulated transcripts and performed an intensive literature search to identify the most promising candidates based on their implications in development, epigenetic regulation, central dopamine signaling pathways, metabolic function, addiction pathology, obesity, and related metabolic syndrome. We identified seven potential targets (CHRNA2, VAV3, ZCCHC11, EEFA1, DHRS3, DAB2IP, GRIN3A) related to neuronal development and addiction pathology and assessed their expression levels in the dorsal striatum (dSTR), nucleus accumbens (Nac), and ventral tegmental area (VTA) of F1 fathers and tsRNA offspring. These brain structures were selected because of their role in initiation and maintenance of hedonic-driven behaviors (36). One of the candidate genes, CHRNA2, which is known to be involved in nicotine and alcohol addiction (37, 38), was increased in the dSTR of F1-HFD fathers (P < 0.05) and tsRNA-HFD offspring compared with the CTR groups (P < 0.03; Fig. 8E). Similarly, the level of CHRNA2 was found to be higher in the VTA of F1-HFD fathers (P < 0.05) and tsRNA-HFD offspring (P < 0.03; Fig. 8G) compared with their CTR littermates. In contrast, the expression of CHRNA2 was significantly lower in the Nac of F1-HFD (P < 0.05) and tsRNA-HFD offspring (P < 0.04; Fig. 8F) compared with the CTR groups. Further, another potential target gene, GRIN3A, that has been implicated in neuronal plasticity associated with drug addiction (39, 40), showed altered expression in the dSTR of F1-HFD and tsRNA-HFD offspring (SI Appendix, Fig. S7B). The level of GRIN3A was higher in the F1-HFD males compared with the F1-CTR males (P < 0.02), whereas the expression was lower in the HFD-tsRNA offspring compared with their CTR littermates (P < 0.05). Several other candidates (VAV3, EEFA1, ZCCHC11, DHRS3, and DAB2IP) that play important roles in neurite growth and differentiation during the early phase of development showed altered expression patterns in the dSTR and Nac of F1-HFD fathers and HFD-tsRNA offspring compared with CTR littermates (SI Appendix, Fig. S8 A–D), with no changes in VTA (SI Appendix, Fig. S8 E and F). Together, these findings suggest a possible functional association between altered sperm tsRNAs levels in F1-HFD fathers and the observed altered hedonic phenotypes in the resulting HFD-tsRNA offspring.
Discussion
The present study provides evidence that obesogenic and hedonic phenotypes in the progeny following MHFD exposure are mediated through sperm tsRNAs. Among different subgroups of sperm sncRNAs, microinjection of sperm tsRNAs from F1 HFD male into the naïve zygote could induce obesogenic phenotypes and addictive-like behaviors, such as increased preference for palatable foods and enhanced sensitivity to drugs of abuse in the resulting offspring. These findings indicate that sperm tsRNAs are essential for the transmission of such acquired traits to subsequent generations. Furthermore, deep-sequencing analysis of sperm sncRNAs of F1 HFD and CTR fathers revealed that the HFD sperm had a larger proportion of sperm tsRNAs, predominantly 5′ tRNA halves, compared with CTR sperm. Together, these results provide a direct link between sperm tsRNAs and the inheritance of MHFD-induced hedonic and metabolic traits in the progeny.
Sperm tsRNA Mediated Transmission of Altered Hedonic Phenotypes.
The present study shows that microinjection of total RNAs or tsRNAs from F1-HFD male sperm into naïve-fertilized oocytes could induce an increased response to drugs of abuse in the resultant offspring. We observed an increased sensitivity to amphetamine in male and female HFD-total RNA as well as HFD-tsRNA offspring, similar to the F1-HFD (12, 41, 42) and the F2-HFD offspring (13) conceived through conventional means. In addition, male and female HFD-total RNA as well as HFD-tsRNA offspring showed an increased preference for alcohol compared with the CTR littermates, similar to their ancestors (12, 41, 42). Conversely, HFD-total RNA offspring did not show any difference in response to natural rewards, such as HFD and sucrose, compared with their CTR littermates, unlike the phenotypes observed in HFD-F1 ancestors (12, 43, 44). Injection of isolated tsRNA from the HFD-F1 sperm could recapitulate the enhanced preference for natural rewards in male HFD-tsRNA offspring. A similar pattern of phenotypic variation across generations was reported in several other models of parental diet-induced metabolic abnormalities (21, 35, 45). In a paternal Western diet (WD), the immediate offspring derived from the microinjection of sperm miR19 of WD father did not develop obesity, whereas the subsequent descendant showed the complete obesogenic phenotype (21). There may be several possible explanations for these phenotypic variations among the progeny. First is the involvement of other regulatory machineries such as amplification of RNAi reactions, RNA-dependent RNA polymerases, or other sncRNAs that can silence or induce target mRNA expression in different tissues (46). Second is the “rheostat effect” for imprinted genes, which defines the interindividual variability of gene expression with respect to epigenetic alterations (47).
Sperm tsRNA Mediated Transmission of Altered Metabolic Phenotypes.
There is a growing body of evidence that long-term paternal dietary insult such as low-protein diet (26), HFD (29, 35), or WD (21) exposure can induce sperm sncRNA-mediated transmission of metabolic phenotypes in the offspring. However, very little is known about whether maternal overnutrition affects offspring sperm RNA profile and its association with acquired phenotypes. We believe this is the first work that shows that sperm total RNA- and tsRNA-mediated transmission of MHFD induced a strong metabolic phenotype in the progeny. Similar to other parental nutrition models (21, 35), variation in metabolic phenotypic inheritance was also evident in offspring generated through injection of sperm RNAs from F1-HFD males. Notably, here, the obesogenic phenotype, including marked adiposity, insulin insensitivity, and altered circulating metabolic parameters, was more pronounced in the HFD-total RNA and HFD-tsRNA offspring reared on chow diet compared with the mild phenotype observed in the HFD-F1 offspring (12). Human daily food choice and consumption is challenged by the selection of energy-dense, highly palatable, heavily processed junk foods over a healthy, balanced diet. The easy availability and increased preference for palatable foods aggravate the risk of obesity and the metabolic syndrome in some individuals. We and others have shown that a free-preference choice of a calorie-dense diet could induce overconsumption of palatable foods that facilitated the development of obesity and diabetes in the offspring (12, 48, 49). To simulate human daily food preference and consumption in the tsRNA offspring group, we enabled free access to highly palatable foods (HFD and sucrose solution) as well as to a healthy diet (chow and water) for a prolonged period. This led to increased consumption of palatable foods in male and female HFD-tsRNA compared with the CTR-tsRNA offspring. Interestingly, male HFD-tsRNA offspring were more prone to develop an obese trait than their female littermates. Similar pattern of sex-specific phenotypic inheritance has been noticed in several recent models of MHFD (7, 13, 50, 51), but the contribution of sex in different phenotypic outcomes remains elusive (50).
Altered Sperm tsRNA Content of F1 Fathers and Functional Association with the Observed Phenotypes.
Among different subpopulations of sperm RNAs, tsRNAs have been reported to constitute the major portion of sncRNAs (34). Although the biological function is still not clear, a potential link between higher sperm tsRNA levels and obesity has been observed in human and animal studies. A recent human study reported greater abundance of several tRNA fragments in the sperm of obese men compared with healthy lean subjects (52). Similarly, increased levels of tsRNAs were also observed in adult rodents following long-term HFD or low protein exposure (26, 29, 35). In one study, Chen et al. (35) reported that a subset of tsRNAs showed an altered expression profile and RNA modifications in HFD-fed mice. Moreover, the authors showed altered gene expression of metabolic pathways in early embryo and islets of the resultant offspring (35). Sharma et al. (26) showed that protein restriction in male mice affects the tsRNAs of mature sperm, particularly 5′ fragments of glycine-tRNA. These affected tsRNAs could repress the genes associated with the endogenous retroelements (MERVL) in the early embryo that might underlie the emergence of obesogenic phenotypes during adulthood (26). Consistent with their observations, we also found a significantly higher tsRNA content in F1 HFD sperm, with the predominance of 5′-tRNA halves, compared with F1 CTR. Further, injection of tsRNAs from HFD-F1 males induced obesogenic and addictive-like phenotypes in the resultant offspring, similar to their fathers. Notably, in the paternal HFD and protein-restricted models, the adult male mice (F0) were exposed to the dietary insults for a period of 6 wk to 6 mo and the metabolic effects were observed in the immediate offspring (F1) (26, 29, 35). Taken together, the added significance of our findings is threefold. (i) F1 male sperm were exposed to HFD exclusively during the early stages of development, which encompasses the intrauterine and early postnatal weeks. Even such a short HFD exposure could induce significant alterations in the tsRNA content. (ii) The altered sperm tsRNAs were able to transfer MHFD-induced abnormal phenotypes in subsequent generations (F2), indicating a transgenerational inheritance. (iii) In addition to the inheritance of metabolic traits, we have shown here that sperm tsRNAs from F1-HFD males could also mediate addictive-like phenotypes in the resultant offspring.
Our study further identified predicted target transcripts of the differentially expressed tsRNAs between F1-HFD and F1-CTR sperm. Seven of these candidate genes, CHRNA2, GRIN3A, VAV3, ZCCHC11, EEFA1, DHRS3, and DAB2IP, were functionally related to neuronal growth, differentiation, axonal guidance, dendritic spine formation, and maturation in the developing brain (53–59). These genes showed altered expressions in the Nac, dSTR, and VTA of the F1-HFD and tsRNA-HFD offspring. Notably, expression of CHRNA2 and GRIN3A is known to be highly regulated during the early postnatal period (PND 3–21), indicating a critical window for early-life insults (53, 54, 60). In addition, a mutation in CHRNA2 (also known as nicotine acetylcholine receptor-α subunit 2; nAChrs-α) in humans has been reported to increase the risk of nicotine and alcohol addiction (61, 62). Global KO of CHRNA2 in mice was shown to potentiate nicotine self-administration, food-reinforcement behaviors, and withdrawal symptoms via the alteration of GABA, glutamate, and dopamine signaling in the interpedencular nucleus, habenula, and mesoaccumbal pathways (37, 63, 64). The persistent alteration of CHRNA2 expression observed in the dSTR, Nac, and VTA of F1-HFD fathers and HFD-tsRNA offspring could cause similar circuit-level adaptations in the dopaminergic and glutamatergic pathways. This, in turn, can lead to the manifestation of addictive-like behaviors. In parallel, we also observed altered expression of GRIN3A in the dSTR of F1-HFD and HFD-tsRNA offspring. GRIN3A is highly enriched in the glutamatergic projections of mesocorticolimbic dopaminergic circuits in the adult brain (65). The protein encoded by GRIN3A, i.e., NMDA receptor subtype 2, is known to modulate dopamine signaling in drug addiction (65, 66). Up- or down-regulation of the GRIN3A subunit of the NMDA receptors in the dopamine circuits have been reported in rodents to alter the glutamatergic tone and stimulate drug-induced synaptic plasticity. This was shown to trigger behavioral sensitization, self-administration, and drug-seeking behavior in advanced stages of cocaine and methamphetamine addiction (39, 40, 67). This suggests that altered GRIN3A expression might also contribute to the addictive phenotypes developed in our model. Given that tsRNA alters gene expression via posttranscriptional modifications (68), it may imply that MHFD exposure induced increased sperm tsRNA levels. Thus, we could hypothesize that alterations of these genes’ expression during early development may have had an impact on neuronal development as well as central circuit-level function. These changes led to the predisposition of hedonic-like behaviors in the offspring. Although recent evidence suggests significant contributions of brain miRNAs and long noncoding RNAs (lncRNAs) in drug addiction (69), the role of tsRNA in the brain is currently unknown. Our findings can only provide a possible functional association of altered sperm tsRNAs and addictive-like behaviors in the offspring. However, more studies are still needed to address the complex mechanisms through which sperm tsRNAs induce such phenotypes.
An important observation of the present study is that the obesogenic traits and hedonic behaviors observed in the tsRNA-HFD offspring are quite subtle compared with the strong effects detected in HFD-total RNA-injected offspring. This clearly suggests that a single epimutation (i.e., altered sperm tsRNAs) is highly unlikely to be responsible for the inheritance of the whole phenotypes; rather, a complex interaction between sperm ncRNAs, DNA methylation, histone modifications, and chromatin remodeling might play a role in the transmission of the whole phenotype (70). Some potential targets in the total RNA pool can be miRNAs, piRNAs, and lncRNAs. For instance, alterations in sperm miRNA profiles have been reported in response to various environmental insults such as early-life stress (8, 24) and dietary perturbations (21, 27, 28). Considering the involvement of miRNA families (miR-let 7, miR-10, miR19) in the regulation of metabolic homeostasis and sperm miRNA-mediated transmission of paternal WD-induced obesogenic traits in the offspring, miRNAs could also contribute to the acquired traits observed in our MHFD model (21, 28, 71). piRNAs are known to regulate transposon silencing and rearrangement of transposon elements in the gametic genome (72) and therefore can also play a role in intergenerational and transgenerational epigenetic inheritance. Studies in invertebrate species have described piRNA-mediated transmission of RNAi memory across multiple generations (73, 74), suggesting that they might be involved in the inheritance of metabolic state. Similar to piRNAs, lncRNAs are also involved in genomic imprinting and early embryonic development, and may therefore play a critical role in transmission (75). It has recently been reported in a mouse model of postnatal stress that microinjection of sperm lncRNAs or sncRNAs from male mice exposed to postnatal stress could recapitulate some of the risk-taking behaviors and metabolic alterations similar to the phenotypes observed in natural offspring of posttraumatic fathers (10). However, it is necessary to combine sperm lncRNAs and sncRNAs by injecting total RNAs to reproduce all phenotypes in adulthood, suggesting a synergistic action of different sperm RNA fractions. Similar cross-talk between sperm microRNAs and chromatin remodeling during early embryogenesis has been demonstrated in a transgenerational mouse model of gigantism (21). The microinjection of miR-124 into fertilized oocytes led to accelerated growth of the progeny across three generations. However, the maintenance of the giant phenotype in the progeny depended on the increased repressive histone marks (H3K9me2 and H3K9me3) at sox-9 locus in F0 and F1 embryos following initial miR-124 exposure, suggesting sperm RNA-mediated chromatin remodeling during embryogenesis. Therefore, we could speculate that a complex interaction between different sperm RNA fractions and other epigenetic factors such as DNA methylation, histone modifications, and chromatin remodeling might exist for the inheritance of such complex phenotypes in our model.
In conclusion, our findings demonstrate that sperm tsRNA is a potential vector that partly contributes to the transmission of MHFD-induced addictive-like and obesogenic phenotypes across generations, thereby emphasizing its role in diverse pathological outcomes. However, how and at which stages of sperm development this epigenetic mark is affected by MHFD exposure, and how it could carry the information of metabolic and hedonic traits to the next generation, remain to be elucidated. In a recent report, Zhang et al. (29) showed that DNMT2 (DNA methyltransferase 2)-mediated elevated RNA modifications (m5C and m2G) could increase the level of sperm tsRNA following long-term paternal HFD exposure and thereby contribute to the transmission of paternal metabolic traits in the offspring. It will therefore be of great interest to examine whether such RNA modification machinery or other regulatory pathways, such as interplay between sperm tsRNA and other epigenetic marks (76), are involved in MHFD-induced alterations of sperm tsRNA profile and maintenance of such epigenetic memory in the subsequent generations.
Materials and Methods
Animals, feeding and breeding design, sperm collection, RNA extraction, small RNA isolation and sperm RNA microinjection, experimental design, small RNA library preparation, sequencing analysis and tsRNA target prediction, gene-expression analysis, Western blot, behavioral and metabolic tests, and statistical analyses are described in the SI Appendix. All animal experiments and procedures were approved by the Zurich Cantonal Veterinarian’s Office, Switzerland.
Supplementary Material
Acknowledgments
This work has been supported by a Swiss Federal Institute of Technology (ETH Zurich) Grant ETH-15 13-1 (to D.P.-R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820810116/-/DCSupplemental.
References
- 1.Bohacek J, Mansuy IM (2015) Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nat Rev Genet 16:641–652. [DOI] [PubMed] [Google Scholar]
- 2.Sales VM, Ferguson-Smith AC, Patti ME (2017) Epigenetic mechanisms of transmission of metabolic disease across generations. Cell Metab 25:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrero-Bosagna C, Jensen P (2015) Globalization, climate change, and transgenerational epigenetic inheritance: Will our descendants be at risk? Clin Epigenetics 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanson MA, Gluckman PD (2014) Early developmental conditioning of later health and disease: Physiology or pathophysiology? Physiol Rev 94:1027–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miska EA, Ferguson-Smith AC (2016) Transgenerational inheritance: Models and mechanisms of non-DNA sequence-based inheritance. Science 354:59–63. [DOI] [PubMed] [Google Scholar]
- 6.Bale TL. (2015) Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci 16:332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn GA, Bale TL (2011) Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology 152:2228–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gapp K, et al. (2014) Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 17:667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Y, et al. (2014) Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci USA 111:1873–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gapp K, et al. (October 30, 2018) Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol Psychiatry, 10.1038/s41380-018-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godfrey KM, et al. (2017) Influence of maternal obesity on the long-term health of offspring. Lancet Diabetes Endocrinol 5:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peleg-Raibstein D, et al. (2016) Enhanced sensitivity to drugs of abuse and palatable foods following maternal overnutrition. Transl Psychiatry 6:e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarker G, et al. (2018) Transgenerational transmission of hedonic behaviors and metabolic phenotypes induced by maternal overnutrition. Transl Psychiatry 8:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radford EJ, et al. (2014) In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 345:1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shea JM, et al. (2015) Genetic and epigenetic variation, but not diet, shape the sperm methylome. Dev Cell 35:750–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richetto J, et al. (2017) Genome-wide DNA methylation changes in a mouse model of infection-mediated neurodevelopmental disorders. Biol Psychiatry 81:265–276. [DOI] [PubMed] [Google Scholar]
- 17.Franklin TB, et al. (2010) Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry 68:408–415. [DOI] [PubMed] [Google Scholar]
- 18.Campos EI, Stafford JM, Reinberg D (2014) Epigenetic inheritance: Histone bookmarks across generations. Trends Cell Biol 24:664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber-Stadlbauer U, et al. (2017) Transgenerational transmission and modification of pathological traits induced by prenatal immune activation. Mol Psychiatry 22:102–112. [DOI] [PubMed] [Google Scholar]
- 20.Chen Q, Yan W, Duan E (2016) Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 17:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandjean V, et al. (2015) RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci Rep 5:18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12:136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casas E, Vavouri T (2014) Sperm epigenomics: Challenges and opportunities. Front Genet 5:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodgers AB, Morgan CP, Leu NA, Bale TL (2015) Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci USA 112:13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster A, Skinner MK, Yan W (2016) Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet 2:dvw001, and erratum (2016) 2:dvw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma U, et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Castro Barbosa T, et al. (2015) High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab 5:184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cropley JE, et al. (2016) Male-lineage transmission of an acquired metabolic phenotype induced by grand-paternal obesity. Mol Metab 5:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. (2018) Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 20:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wise RA, Bozarth MA (1982) Action of drugs of abuse on brain reward systems: An update with specific attention to opiates. Pharmacol Biochem Behav 17:239–243. [DOI] [PubMed] [Google Scholar]
- 31.Wise RA. (2008) Dopamine and reward: The anhedonia hypothesis 30 years on. Neurotox Res 14:169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willner P, Towell A, Sampson D, Sophokleous S, Muscat R (1987) Reduction of sucrose preference by chronic unpredictable mild stress, and its restoration by a tricyclic antidepressant. Psychopharmacology (Berl) 93:358–364. [DOI] [PubMed] [Google Scholar]
- 33.Schimmel P, Ribas de Pouplana L (1995) Transfer RNA: From minihelix to genetic code. Cell 81:983–986. [DOI] [PubMed] [Google Scholar]
- 34.Peng H, et al. (2012) A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 22:1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, et al. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351:397–400. [DOI] [PubMed] [Google Scholar]
- 36.Everitt BJ, Robbins TW (2013) From the ventral to the dorsal striatum: Devolving views of their roles in drug addiction. Neurosci Biobehav Rev 37:1946–1954. [DOI] [PubMed] [Google Scholar]
- 37.Lotfipour S, et al. (2013) Targeted deletion of the mouse α2 nicotinic acetylcholine receptor subunit gene (Chrna2) potentiates nicotine-modulated behaviors. J Neurosci 33:7728–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker MO, et al. (2016) Moderate alcohol exposure during early brain development increases stimulus-response habits in adulthood. Addict Biol 21:49–60. [DOI] [PubMed] [Google Scholar]
- 39.Yuan T, et al. (2013) Expression of cocaine-evoked synaptic plasticity by GluN3A-containing NMDA receptors. Neuron 80:1025–1038, and erratum (2015) 88:432. [DOI] [PubMed] [Google Scholar]
- 40.Ortinski PI. (2014) Cocaine-induced changes in NMDA receptor signaling. Mol Neurobiol 50:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bocarsly ME, et al. (2012) Effects of perinatal exposure to palatable diets on body weight and sensitivity to drugs of abuse in rats. Physiol Behav 107:568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avena NM. (2010) The study of food addiction using animal models of binge eating. Appetite 55:734–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naef L, et al. (2011) Maternal high-fat intake alters presynaptic regulation of dopamine in the nucleus accumbens and increases motivation for fat rewards in the offspring. Neuroscience 176:225–236. [DOI] [PubMed] [Google Scholar]
- 44.Ong ZY, Muhlhausler BS (2011) Maternal “junk-food” feeding of rat dams alters food choices and development of the mesolimbic reward pathway in the offspring. FASEB J 25:2167–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carone BR, et al. (2010) Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houri-Zeevi L, Rechavi O (2017) A matter of time: Small RNAs regulate the duration of epigenetic inheritance. Trends Genet 33:46–57. [DOI] [PubMed] [Google Scholar]
- 47.Beaudet AL, Jiang YH (2002) A rheostat model for a rapid and reversible form of imprinting-dependent evolution. Am J Hum Genet 70:1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayol SA, Farrington SJ, Stickland NC (2007) A maternal ‘junk food’ diet in pregnancy and lactation promotes an exacerbated taste for ‘junk food’ and a greater propensity for obesity in rat offspring. Br J Nutr 98:843–851. [DOI] [PubMed] [Google Scholar]
- 49.la Fleur SE, Luijendijk MC, van der Zwaal EM, Brans MA, Adan RA (2014) The snacking rat as model of human obesity: Effects of a free-choice high-fat high-sugar diet on meal patterns. Int J Obes (Lond) 38:643–649. [DOI] [PubMed] [Google Scholar]
- 50.Dunn GA, Morgan CP, Bale TL (2011) Sex-specificity in transgenerational epigenetic programming. Horm Behav 59:290–295. [DOI] [PubMed] [Google Scholar]
- 51.Huypens P, et al. (2016) Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat Genet 48:497–499. [DOI] [PubMed] [Google Scholar]
- 52.Donkin I, et al. (2016) Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab 23:369–378. [DOI] [PubMed] [Google Scholar]
- 53.Son JH, Winzer-Serhan UH (2006) Postnatal expression of alpha2 nicotinic acetylcholine receptor subunit mRNA in developing cortex and hippocampus. J Chem Neuroanat 32:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kehoe LA, et al. (2014) GluN3A promotes dendritic spine pruning and destabilization during postnatal development. J Neurosci 34:9213–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauzeau V, et al. (2010) Vav3 is involved in GABAergic axon guidance events important for the proper function of brainstem neurons controlling cardiovascular, respiratory, and renal parameters. Mol Biol Cell 21:4251–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung Y, et al. (2016) An epigenetic mechanism mediates developmental nicotine effects on neuronal structure and behavior. Nat Neurosci 19:905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khalyfa A, et al. (2001) Characterization of elongation factor-1A (eEF1A-1) and eEF1A-2/S1 protein expression in normal and wasted mice. J Biol Chem 276:22915–22922. [DOI] [PubMed] [Google Scholar]
- 58.Veenvliet JV, Smidt MP (2014) Molecular mechanisms of dopaminergic subset specification: Fundamental aspects and clinical perspectives. Cell Mol Life Sci 71:4703–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salami F, Qiao S, Homayouni R (2015) Expression of mouse Dab2ip transcript variants and gene methylation during brain development. Gene 568:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong HK, et al. (2002) Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J Comp Neurol 450:303–317. [DOI] [PubMed] [Google Scholar]
- 61.Dash B, Lukas RJ, Li MD (2014) A signal peptide missense mutation associated with nicotine dependence alters α2*-nicotinic acetylcholine receptor function. Neuropharmacology 79:715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuo L, et al. (2016) Associations of rare nicotinic cholinergic receptor gene variants to nicotine and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet 171:1057–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Upton M, Lotfipour S (2015) α2-Null mutant mice have altered levels of neuronal activity in restricted midbrain and limbic brain regions during nicotine withdrawal as demonstrated by cfos expression. Biochem Pharmacol 97:558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsumoto M, Hikosaka O (2007) Lateral habenula as a source of negative reward signals in dopamine neurons. Nature 447:1111–1115. [DOI] [PubMed] [Google Scholar]
- 65.Pérez-Otaño I, Larsen RS, Wesseling JF (2016) Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci 17:623–635. [DOI] [PubMed] [Google Scholar]
- 66.Hemby SE, Horman B, Tang W (2005) Differential regulation of ionotropic glutamate receptor subunits following cocaine self-administration. Brain Res 1064:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang X, et al. (2017) Methamphetamine abuse impairs motor cortical plasticity and function. Mol Psychiatry 22:1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez G, Choudury SG, Slotkin RK (2017) tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 45:5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sartor GC, St Laurent G 3rd, Wahlestedt C (2012) The emerging role of non-coding RNAs in drug addiction. Front Genet 3:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gapp K, Bohacek J (2018) Epigenetic germline inheritance in mammals: Looking to the past to understand the future. Genes Brain Behav 17:e12407. [DOI] [PubMed] [Google Scholar]
- 71.Frost RJ, Olson EN (2011) Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA 108:21075–21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764. [DOI] [PubMed] [Google Scholar]
- 73.Ashe A, et al. (2012) piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grentzinger T, et al. (2012) piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res 22:1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X, et al. (2017) Systematic identification and characterization of long non-coding RNAs in mouse mature sperm. PLoS One 12:e0173402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma U, Rando OJ (2017) Metabolic inputs into the epigenome. Cell Metab 25:544–558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.