Significance
Small-molecule metabolites from cell organelles or cytosol exert their influence on decision making processes in eukaryotic cells. However, the mechanisms underlying the regulation of cellular processes by these metabolites are not well understood. Among these small metabolites, acetyl-CoA, a most critical one, is produced in the cytosol and different cell organelles. In plants, acetyl-CoA produced from fatty acid β-oxidation in peroxisomes plays critical roles in various developmental stages. Here we found that defects in β-oxidation in peroxisomes affect both histone acetylation and DNA methylation in the nucleus. Our work provides evidence for retrograde signaling from peroxisomes to regulate nuclear epigenetic modifications in higher eukaryotes.
Keywords: β-oxidation, acetyl-CoA, histone acetylation, DNA methylation, gene silencing
Abstract
Epigenetic markers, such as histone acetylation and DNA methylation, determine chromatin organization. In eukaryotic cells, metabolites from organelles or the cytosol affect epigenetic modifications. However, the relationships between metabolites and epigenetic modifications are not well understood in plants. We found that peroxisomal acyl-CoA oxidase 4 (ACX4), an enzyme in the fatty acid β-oxidation pathway, is required for suppressing the silencing of some endogenous loci, as well as Pro35S:NPTII in the ProRD29A:LUC/C24 transgenic line. The acx4 mutation reduces nuclear histone acetylation and increases DNA methylation at the NOS terminator of Pro35S:NPTII and at some endogenous genomic loci, which are also targeted by the demethylation enzyme REPRESSOR OF SILENCING 1 (ROS1). Furthermore, mutations in multifunctional protein 2 (MFP2) and 3-ketoacyl-CoA thiolase-2 (KAT2/PED1/PKT3), two enzymes in the last two steps of the β-oxidation pathway, lead to similar patterns of DNA hypermethylation as in acx4. Thus, metabolites from fatty acid β-oxidation in peroxisomes are closely linked to nuclear epigenetic modifications, which may affect diverse cellular processes in plants.
Histone acetylation is important for neutralizing the positive charges of lysine residues and promoting chromatin relaxation; it is also required for transcription, DNA replication, histone methylation, and other histone modifications (1–4). Histones are acetylated by acetyltransferases, which transfer acetyl groups from acetyl-CoA to histone lysine residues.
Acetyl-CoA is a central metabolite that can be produced via several metabolic pathways involved in pyruvate, citrate, acetate, and fatty acid β-oxidation metabolism (5). In mammals, acetyl-CoA in mitochondria is produced from different pathways, including the fatty acid β-oxidation (6). In cytosol and nucleus, adenosine triphosphate (ATP)-citrate lyase (ACLY) cleaves citrate exported from mitochondria to regenerate acetyl-CoA that can be used for other biosynthetic processes, such as fatty acid synthesis and histone acetylation (6). In mouse, conditional loss of carnitine palmitoyltransferase 1A (CPT1A), which is required for the transfer of fatty acid into mitochondria for β-oxidation, impairs dermal lymphatic formation via histone acetylation in an ACLY-dependent manner (7). A pyruvate dehydrogenase complex can be translocated from mitochondria to nuclei to generate acetyl-CoA and mediate histone acetylation in mammalian cells in certain conditions (2, 8). In Arabidopsis, the mutations in cytosolic acetyl-CoA carboxylase (ACC1), which converts cytosolic acetyl-CoA to malonyl-CoA for elongating the plastid-produced fatty acids, lead to high accumulation of cytosolic acetyl-CoA, specifically resulting in increased H3K27 acetylation (H3K27ac) (9).
These results underscore the importance of acetyl-CoA in histone acetylation in nuclei in both mammals and plants. However, in plant cells, plastids, mitochondria, peroxisomes, and cytosol can produce acetyl-CoA (10). Whether impairment of metabolism in plant organelles will affect the nuclear epigenetic modifications is still being unraveled.
DNA methylation is a conserved epigenetic marker important in genome organization, gene expression, genomic imprinting, paramutation, and X chromosome inactivation in organisms (3, 11–13). DNA methylation patterns are coordinately determined by methylation and demethylation reactions in plants and animals (13, 14). The active removal of 5mC in Arabidopsis is carried out by a subfamily of bifunctional DNA glycosylases/lyases represented by REPRESSOR OF SILENCING 1 (ROS1) and DEMETER (DME) (15, 16). ROS1 family proteins bind DNA nonspecifically (17) and need other factors to find target genomic regions (13). Among these, ROS4/INCREASED DNA METHYLATION 1 (IDM1) is a plant homeodomain finger-containing histone acetyltransferase that catalyzes the acetylation of histone H3 lysine 18 (H3K18) and lysine 23 (H3K23) to create a favorable chromatin environment for the recruitment of ROS1 at some loci (1, 18). ROS4/IDM1, together with other factors, such as ROS5/IDM2, IDM3, methyl-CPG-binding domain 7 (MBD7), Harbinger transposon-derived protein 1 (HDP1), and HDP2, forms a complex to regulate active DNA demethylation (19–23). MET18 is a component in the cytosolic iron-sulfur cluster assembly pathway involved in the transfer of the Fe-S cluster to ROS1, which is necessary for its function (24). The expression of ROS1 is positively regulated by promoter DNA methylation, which requires a protein complex composed of Su(var)3–9 homologs (SUVHs) and SUVH-interacting DNAJ (SDJ) proteins (25–28).
To identify the components required to prevent transgene silencing and normal DNA methylation patterns in Arabidopsis, we performed a forward genetic screen for kanamycin (Kan)-sensitive mutants using the ProRD29A:LUC/Pro35S:NPTII transgenic C24 line (18). Several ROS1 alleles, multiple components of the RdDM pathway, ROS4/IDM1, ROS5/IDM2, and MBD7, were identified in this screening (1, 18, 22, 23). Here we identified an antisilencing factor, acyl-CoA oxidase 4 (ACX4), in the fatty acid β-oxidation pathway. In acx4 mutants, overall levels of H3Ac and H4Ac are reduced, and DNA methylation is increased at some genomic loci, resulting in enhanced transcriptional silencing of reporter and some endogenous genes. The mfp2 and kat2 mutants have similar DNA hypermethylation phenotypes to acx4. Our results uncover a connection between fatty acid β-oxidation and epigenetic regulation in plants.
Results
ACX4 Is a Suppressor of Transcriptional Silencing.
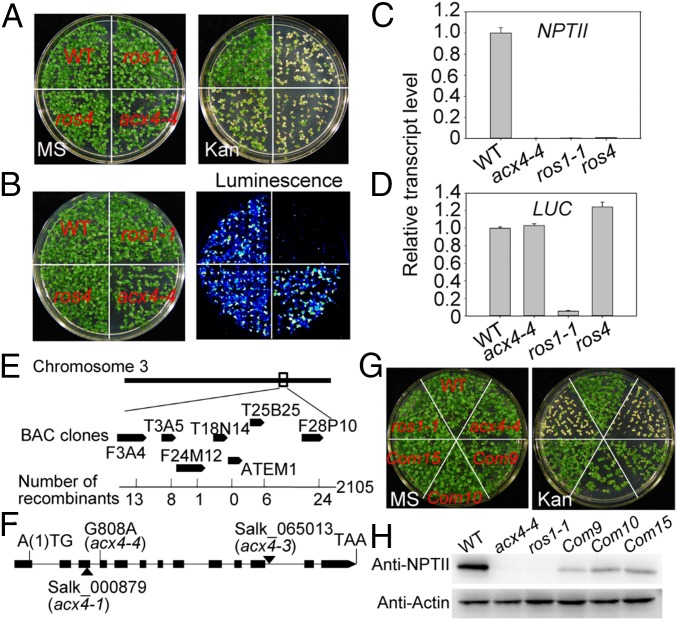
To decipher the mechanism that blocks transcriptional gene silencing, we performed a forward genetic screen using an ethyl methanesulfonate (EMS)-mutagenized population of a C24 transgenic line carrying ProRD29A:LUC and Pro35S:NPTII transgenes, both of which are actively expressed (used as the WT) in Arabidopsis (18, 22, 23). From this population, a recessive kanamycin (Kan)-sensitive mutant, acx4 (hereinafter acx4-4), was isolated. Like ros4, ros5-1, and mbd7 mutants (18, 22, 23), acx4-4 plants exhibited silenced NTPII expression (Fig. 1 A and C), but the expression of LUC was unaltered (Fig. 1 B and D), while in ros1-1, both Pro35S:NPTII and ProRD29A:LUC were silenced (Fig. 1 A–D).
Fig. 1.
Characterization of the acx4 mutant. (A) The acx4 mutation silenced Pro35S:NPTII, as indicated by Kan sensitivity. The acx4-4, ros1-1, and ros4 mutants were grown on MS medium or on MS medium supplemented with 50 mg/L Kan. (B) The acx4 mutation does not affect the expression of ProRD29A:LUC. The acx4-4, ros1-1, and ros4 mutants grown on MS medium were treated with 30 µM abscisic acid for 3 h, after which a luciferase assay was performed using a cold charge-coupled device camera. (C) Relative expression levels of Pro35S:NPTII in the acx4 mutant and the WT (C24 accession), ros1-1, and ros4 seedlings by qPCR. UBC28 served as an internal control. (D) Relative expression levels of ProRD29A:LUC in the acx4 mutant and WT, ros1-1, and ros4 seedlings by qPCR. UBC28 served as an internal control. (E) Diagram of the region identified as the acx4-4 mutation by map-based cloning. (F) Model structure of the AT3G51840 (ACX4) gene. A G808-to-A808 mutation occurs in the splice donor site of the third intron of the AT3G51840 gene in the acx4-4 mutant. Two T-DNA insertion mutants, acx4-1 and acx4-3, are shown. The exon and intron are marked by a black box and black line, respectively. (G) The acx4 mutant was complemented by ACX4, as tested on MS medium containing 50 mg/L Kan in three complement lines (Com9, 10, 15). (H) NPTII protein levels in WT, ros1-1, acx4-4, and three complemented lines, as indicated by immunoblotting with NPTII antibodies. ACTIN served as a loading control.
ACX4 was identified through map-based cloning. We crossed the acx4-4 mutant in the C24 accession with the WT Columbia-0 (Col-0) background and used the 2105 F2 plants for mapping. The acx4-4 mutation was localized on the bottom of chromosome 3 and then narrowed down between bacterial artificial chromosome clones T18N14 and T25B25 (Fig. 1E). By sequencing candidate genes, a G-to-A mutation was found at position 808 of AT3G51840 (from the first putative ATG), which would change the splicing acceptor site from GT to AT at the end of the third intron (Fig. 1F). By comparing the cDNAs of AT3G51840 amplified by RT-PCR from WT and acx4-4 lines, we found that the cDNA of AT3G51840 from acx4-4 was 16 bp shorter than that from WT. Furthermore, the acceptor splice site had moved to the fourth exon, which would lead to premature termination of translation and a truncated protein (SI Appendix, Fig. S1 A and B). To further examine whether the mutation in AT3G51840 causes the silencing of the Pro35S:NPTII transgene, WT AT3G51840, including the fragment 2543 bp upstream of the first putative ATG codon and 312 bp downstream of the putative stop codon TAA, was cloned and transformed into the acx4-4 mutant. Like WT, three randomly selected independent transgenic lines exhibited Kan resistance, and accumulated the NPTII protein (Fig. 1 G and H). Two additional alleles, acx4-1 and acx4-3 (Col-0 accession) (29), were obtained, representing the T-DNA insertion lines SALK_000879 and SALK_065013, with the T-DNA inserted in the third exon and 11th intron of ACX4, respectively (Fig. 1F). We introduced the acx4-1 and acx4-3 mutations into the WT transgenic plants by crossing. As expected, both mutations resulted in hypersensitivity to Kan relative to WT with greatly reduced NPTII transcripts, as determined by quantitative RT-PCR (qPCR) (SI Appendix, Fig. S2 A and B), and the expression of ProRD29A:LUC was not obviously affected in the acx4-1 and acx4-3 mutants, as indicated by luciferase and LUC transcript assays (SI Appendix, Fig. S2 A and B).
We examined the subcellular localization of ACX4 using transient assays. ACX4 was fused to the C terminus of the red fluorescent protein mCherry, CD3-990 (a mitochondrial marker) was fused to the C terminus of yellow fluorescent protein (YFP) (30), CAT2 (a peroxisomal marker) was fused to the C terminus of green fluorescent protein (GFP) (31), and nuclear localization signal (NLS) was fused to the N terminus of GFP. Mcherry-ACX4 was transiently coexpressed with each of the fused marker proteins in tobacco (Nicotiana benthamiana) epidermal cells driven by the cauliflower mosaic virus 35S promoter. ACX4 did not colocalize with CD3-990 and NLS (SI Appendix, Fig. S3 A and B) but did colocalize with GFP-CAT2 (SI Appendix, Fig. S3C), consistent with immunoelectron microscopy data from a previous study (32). Beta-glucuronidase (GUS) staining of transgenic plants expressing ProACX4:GUS revealed ACX4 expression throughout the seedling (SI Appendix, Fig. S4).
The acx4 Mutation Causes DNA Hypermethylation at the NOS Terminator of Pro35S:NPTII and Some Endogenous Genomic Loci.
To test whether silencing of the Pro35S:NPTII transgene is associated with DNA methylation, we treated the WT, acx4-4, ros1-1, and ros4 lines with the DNA methylation inhibitor 5-Aza-2′-deoxycytidine (5′-Aza). The DNA methylation inhibitor suppressed the Kan sensitivity of the acx4-4, ros1-1, and ros4 mutants (SI Appendix, Fig. S5A) and restored the expression of NPTII (SI Appendix, Fig. S5B). Furthermore, the acx4-4 ddm1 double mutant, but not the acx4-4 rdr2 double mutant, released the silencing of NPTII in acx4-4 (SI Appendix, Fig. S5 C and D). Both ddm1 and rdr2 have the same genetic background as acx4-4 (18). Silencing of NPTII in acx4-4 was suppressed by the ddm1, but not the rdr2 mutation (SI Appendix, Fig. S5 C and D), suggesting that NPTII silencing is dependent on the DDM1-mediated methylation pathway, but not the RdDM pathway. Published studies indicate that the mutations in the components of RdDM pathway cause silencing of NPTII due to the repression of ROS1 that is positively regulated by DNA methylation in its promoter region (18, 33).
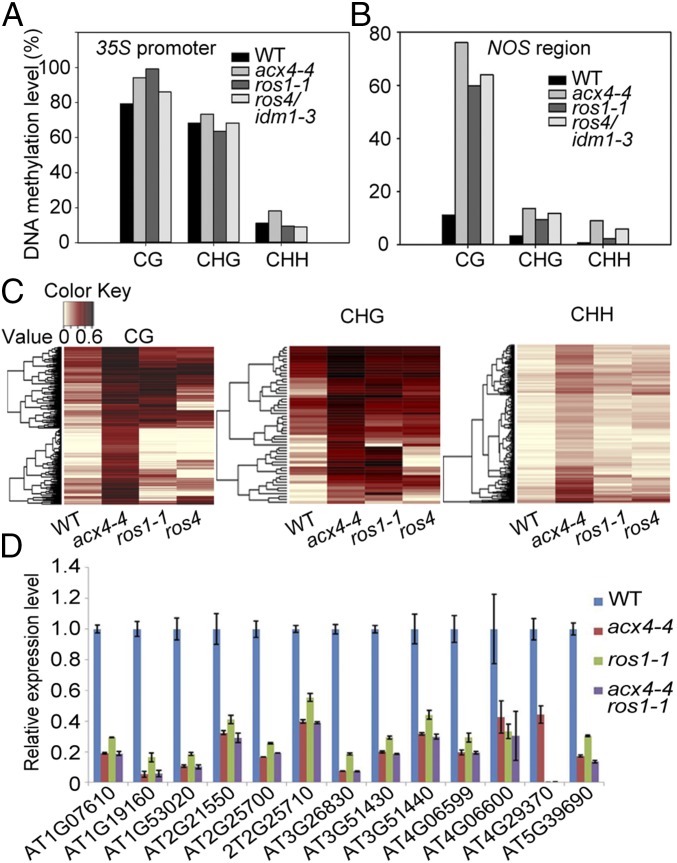
Previous studies have indicated that the ros1-1 mutation increases DNA methylation in both the RD29A promoter and the 3′-NOPALINE SYNTHASE (NOS) terminator region, but that the ros4-4, ros5-1, and mbd7 mutations increase DNA methylation mainly in the NOS terminator (22, 23). Bisulfite sequencing analyses indicated a slight increase in DNA methylation in the 35S promoter in these mutants (Fig. 2A). Like the ros1-1 and ros4 mutants, acx4-4 showed a clear increase in CG, CHG, and CHH DNA methylation (Fig. 2B) in the NOS terminator. These results suggest that the acx4 mutation leads to DNA hypermethylation in the NOS terminator and silencing of Pro35S:NPTII.
Fig. 2.
The acx4 mutation causes hypermethylation of DNA in the NOS terminator. (A) Effects of the acx4-4, ros1-1, and ros4 mutations on DNA methylation in the Pro35S:NPTII promoter by bisulfite sequencing. (B) Effects of the acx4-4, ros1-1, and ros4 mutations on DNA methylation at the NOS region by bisulfite sequencing. (C) Heat map showing the methylation levels of the WT, acx4-4, ros1-1, and ros4 plants in hyper-DMR regions of acx4-4 in different sequence contexts. The lighter color indicates a low methylation level, and the darker color indicates a high methylation level. (D) Expression profiles of the hypermethylated genes or genes near DMRs in 7-d-old acx4-4, ros1-1, and acx4-4 ros1-1 seedlings by qPCR. Error bars represent ± SE. n = 3.
To investigate the effect of the acx4-4 mutation on genomic DNA methylation patterns, whole-genome DNA methylation profiles of acx4-4 (two biological replicates) and the WT seedlings (22, 23) were compared using next-generation sequencing after bisulfite conversion. Analysis of differentially methylated regions (DMRs) revealed 864 DMRs with increased DNA methylation (hyper-DMRs) in acx4-4, including 405 DMRs with hyper-CG methylation, 55 with hyper-CHG methylation, and 404 with hyper-CHH methylation. Heat map analysis of hyper-DMRs indicated that the levels of DNA methylation at acx4-4–specific loci were usually preferentially increased in ros1-1 and ros4 (Fig. 2C), We picked several genes to confirm their DNA methylation patterns. Individual locus bisulfite sequencing revealed increased DNA methylation levels of At4G18250, At4G29380, AT5G52400, AT1G19160, and AT3G51440 in acx4-4 and ros1-1 compared with WT (SI Appendix, Fig. S6 A and B). For AT4G29380, only the levels of CHG and CHH DNA methylation were increased in acx4-4 and ros1-1, while the level of CG methylation did not change—probably because it was already very high. Analysis of the acx4-4 ros1-1 double mutant revealed no clear additive effects on DNA methylation for AT1G19160 and AT3G51440 (SI Appendix, Fig. S6B). Whole-genome bisulfite sequencing data confirmed that, like the ros4 mutant, acx4-4 exhibited increased DNA methylation only in the NOS terminator region and not in the RD29A promoter (SI Appendix, Fig. S6C). The DNA methylation of some endogenous transposons (here we took two transposons ATREP10D and HELITRON2 as examples) was increased (SI Appendix, Fig. S6D). These results suggest that ACX4 and ROS1 work in the same pathway to regulate DNA methylation of some loci in Arabidopsis.
We next used qRT-PCR to test the expression levels of 13 genes (plus 2 kb of sequence on either side) overlapping with hypermethylated DMRs in acx4-4, ros1-1, and acx4-4 ros1-1. Transcript levels of these genes were substantially reduced in these mutants, and the acx4-4 ros1-1 double mutant had no additive effects over the single mutants (Fig. 2D). These data suggest that, like ROS1, ACX4 is critical for preventing the transcriptional silencing of some endogenous genes through DNA methylation.
Mutations in the Last Two Enzymes of the β-Oxidation Pathway Cause Pro35S:NPTII Silencing.
The first step in the fatty acid β-oxidation pathway in peroxisomes is catalyzed by ACX enzymes and produces trans-2-enoyl-CoAs (34–36). Six ACX genes have been identified in the Arabidopsis genome with different expression patterns and distinct substrate specificities (37). The second and third enzymatic reactions are catalyzed by multifunctional proteins (MFPs), which exhibit enoyl-CoA hydratase and β-hydroxyacyl-CoA dehydrogenase activities (35, 38). The last step in the pathway is catalyzed by l-3-ketoacyl-CoA thiolase (KAT), which converts l-3-ketoacyl-CoAs to acetyl-CoAs (35, 39).
We next examined whether mutations in the downstream enzymes of the β-oxidation pathway also lead to silencing of the Pro35S:NPTII transgene. Of the four catalytic activities of MFP2, two—2-trans enoyl-CoA hydratase for long-chain (C18:0) substrates and l-3-hydroxyacyl-CoA dehydrogenase for C6:0, C12:0, and C18:0 substrates—are required for β-oxidation (40). MPF2 has one homolog, abnormal inflorescence meristem 1 (AIM1), whose mutation results in an abnormal inflorescence meristem phenotype in mature plants (41). The enzyme 3-ketoacyl-CoA thiolase-2 (KAT2/PED1/PKT3) catalyzes the last step of the β-oxidation pathway to produce one molecule of acetyl-CoA in each repeat cycle; this gene has higher expression than the two redundant homologs KAT1 and KAT5 (42, 43). kat2 kat5 double mutants recapitulate the aim1 phenotype in inflorescence development and fertility (44).
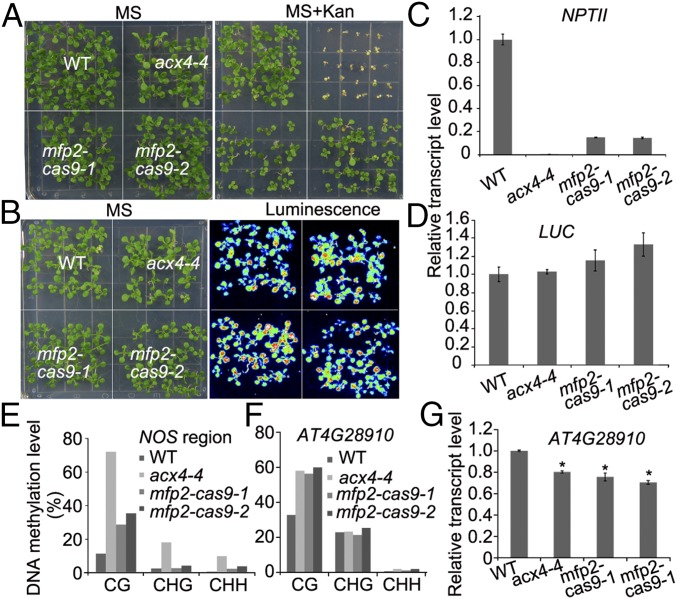
We used the egg-specific CRISPR/Cas9 system (45) to generate the mfp2-cas9-1 and mfp2-cas9-2 mutants in the ProRD29A:LUC/C24 transgenic line (WT) (SI Appendix, Fig. S7 A and D). Meanwhile, we obtained two T-DNA insertion alleles, Salk_098016 (mfp2-2) (40) and GK_787F01 (mfp2-10), and one T-DNA insertion allele, Salk_024922 (kat2-3), in which MFP2 and KAT2 expression were eliminated (SI Appendix, Fig. S7 A–C). We introduced these mutations (in Col-0 accession) into ProRD29A:LUC/C24 transgenic line by crossing. Consistent with NPTII expression levels (Fig. 3 A and C), the mfp2-cas9-1 and mfp2-cas9-2 mutants were more Kan-sensitive than the WT and less sensitive than acx4-4, but did not exhibit altered LUC expression (Fig. 3 B and D). Similarly, the mfp2-2, mfp2-10, and kat2-3 mutations caused more Kan sensitivity than the WT, but the ProRD29A:LUC transgene was not affected (SI Appendix, Fig. S8A). Bisulfite sequencing analysis showed increased DNA methylation at the NOS region of the Pro35S:NPTII gene in mfp2-cas9-1 and mfp2-cas9-2 mutants compared with the WT, although the DNA methylation at these regions was a little lower compared with that in acx4-4 (Fig. 3E). The lesser effect of mfp2 mutation on DNA methylation compared with acx4 suggests a redundancy of the MFP2 homologs in the β-oxidation pathway. Transcriptionally silent information (TSI) is regulated by the DDM1 pathway, but not by the RdDM pathway (46, 47). qPCR demonstrated that the transcript levels of TSI were reduced in mfp2-2, mfp2-10, and kat2-3 mutants compared with Col-0 plants; a similar reduction was observed in acx4-4 compared with the WT (SI Appendix, Fig. S8B). We found increased CG DNA methylation at the AT4G28910 promoter in two independent mfp2-cas9 mutant alleles, and in the acx4-4 mutant (Fig. 3F). qPCR analysis showed that AT4G28910 was expressed at lower levels in acx4-4, mfp2-cas9-1, and mfp2-cas9-2 mutants compared with the WT (Fig. 3G). Taken together, our results suggest that the β-oxidation pathway in peroxisomes regulates gene silencing and DNA methylation in Arabidopsis.
Fig. 3.
MFP2 prevents transcriptional gene silencing of Pro35S:NPTII. (A) The mfp2 CRISPR/Cas9 mutants are more sensitive to Kan than the WT, but more resistant to Kan than acx4-4. Seeds were planted on MS medium supplemented with 150 mg/L Kan and cultured for 2 wk before being photographed. (B) The mfp2 CRISPR/Cas9 mutants do not affect ProRD29A:LUC. (C) The mfp2 CRISPR/Cas9 mutants and acx4-4 reduced Pro35S:NPTII expression levels compared with the WT, as determined by qPCR analyses. UBC28 was used as an internal control. (D) qPCR analysis of the expression levels of ProRD29A:LUC in the indicated genotypes. UBC28 served as an internal control. (E) Compared with the WT, increased methylation levels were observed in the NOS region of mfp2 CRISPR/Cas9 and acx4-4 mutants, according to bisulfite sequencing analysis. (F) Bisulfite sequencing data showing methylation levels of the AT4G28910 promoter in the WT and indicated mutant plants. (G) qPCR analysis of expression levels of the AT4G28910 gene in the WT and different mutant plants. UBC28 served as an internal control. Data are means ± SE (n = 3) with three replicate in one experiment. Three independent experiments were done with similar results. *P < 0.05.
MFP2 and KAT2 Target Some Common Loci as ACX4 for DNA Methylation.
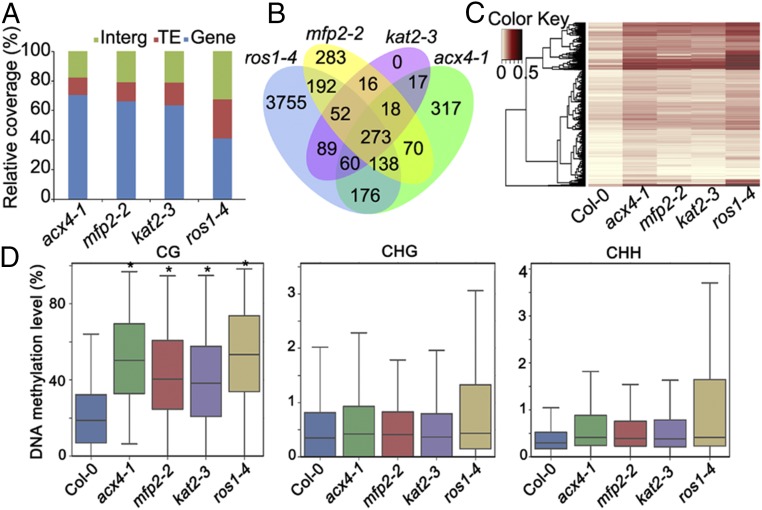
To determine whether MFP2 and KAT2 also function in modulating DNA methylation at the whole genome level, we performed bisulfite sequencing analyses and identified 1,153 DMRs in acx4-1, 1,135 DMRs in mfp2-2, and 686 DMRs in kat2-3 compared with the WT (all in the Col-0 background), respectively. Among these, 1,069 were hypermethylated and 84 were hypomethylated in acx4-1, 1,045 were hypermethylated and 90 were hypomethylated in mfp2-2, and 623 were hypermethylated and 63 were hypomethylated in kat2-3 (SI Appendix, Fig. S9). More than 60% of these hyper-DMRs were located in genic regions in acx4-1, mfp2-2, and kat2-3, whereas only 40% were located in genic regions in ros1-4 (Col-0) (Fig. 4A and SI Appendix, Fig. S9). Furthermore, 60.5% of 1,069 hyper-DMRs in acx4-1, 60.7% of 1,045 hyper-DMRs in mfp2-2, and 76.1% of 623 hyper-DMRs in kat2-3 overlapped with those in ros1-4 (Fig. 4B). Approximately 50% of the 1,069 hyper-DMRs in mfp2-2 and 60% of the 623 hyper-DMRs in kat2-3 were hypermethylated in acx4-1 (Fig. 4B).
Fig. 4.
MFP2 and KAT2 target some common loci as ACX4 for DNA methylation. (A) Comparison of the genomic regions of hyper-DMRs in acx4-1, mfp2-2, kat2-3, and ros1-4 mutants. All mutants are from the Col-0 accession. TE, transposable element. (B) Numbers of overlapping hyper-DMRs in acx4-1, mfp2-2, kat2-3, and ros1-4 mutants. (C) Heat map comparing the methylation levels in acx4-1 hyper-DMRs with the same regions in mfp2-2, kat2-3, and ros1-4 mutants. Columns represent the indicated genotypes; rows represent differentially methylated loci. Hyper-DMRs in acx4-1 relative to the WT were used to compare DNA methylation levels with the same regions in mfp2-2, kat2-3, and ros1-4. Light yellow indicates a low methylation level, and black indicates a high methylation level. (D) Boxplots representing methylation levels of Col-0, acx4-1, mfp2-2, kat2-3, and ros1-4 mutants in those regions that are hyper-DMRs in acx4-1 in different sequence contexts. Two-tailed Student’s t test, *P < 0.05.
Profiling the methylation levels of mfp2-2, kat2-3, and ros1-4 in the loci that were hypermethylated in acx4-1 revealed increased DNA methylation levels at these loci compared with Col-0 (Fig. 4C). Boxplots indicate that at those hyper-DMRs that appear specific for acx4-1, the average DNA methylation levels in the CG context but not the CHG or CHH contexts were higher in the mfp2-2, kat2-3, and ros1-4 mutants than in Col-0 plants (Fig. 4D), suggesting that fatty acid β-oxidation preferentially affects genomic regions in the CG context. These data indicate that mutations in ACX4, MFP2, and KAT2 affect the DNA methylation levels of some common genomic regions (SI Appendix, Fig. S10 A–E).
The acx4 Mutant Has Reduced Histone Acetylation.
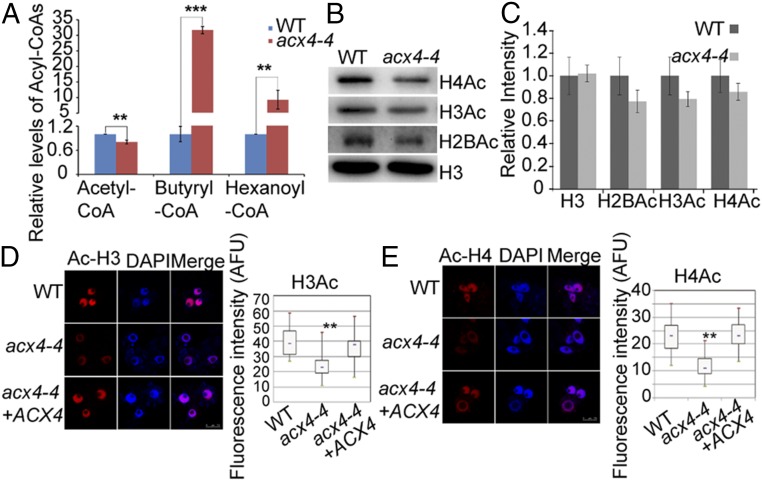
ACX4 catalyzes the conversion of fatty acyl-CoAs to trans-2-enoyl CoA. This is thought to be the predominant way by which the rate of acetyl-CoA flux is controlled through β-oxidation in peroxisomes. We asked whether the level of acetyl-CoA is reduced in acx4 mutants. High-performance liquid chromatography tandem mass spectrometry (HPLC-MS/MS) was used to measure the accumulation of acetyl-CoA and its related metabolites butyryl-CoA and hexanoyl-CoA, the main substrates for ACX4. Both butyryl-CoA and hexanoyl-CoA accumulated to much higher levels, but the level of acetyl-CoA was significantly decreased in acx4-4 compared with the WT (Fig. 5A).
Fig. 5.
The acx4 mutation reduces histone acetylation. (A) Relative levels of acetyl-CoA, butyryl-CoA, and hexanoyl-CoA determined by HPLC-MS/MS in 10-d-old seedlings. SD ± SE (n = 3) of three independent biological repeats is shown. Two-tailed Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. (B) Overall levels of H2BAc, H3Ac, and H4Ac as determined by immunoblotting. Total proteins extracted from the WT and acx4-4 seedlings were used for immunoblotting with anti-H2BAc, -H3Ac, and -H4Ac antibodies. H3 was detected as a loading control. (C) Quantified mean data normalized to H3 showing decreased levels of acetylated H2B, H3, and H4 in acx4-4 compared with the WT. Error bars represent ± SE from three independent experiments (n = 3). (D and E) Levels of acetylated H3 (D) and H4 (E) in WT, acx4-4, and the complemented acx4 line 9, as indicated by immunofluorescence under confocal microscopy. Acetylated-H3/H4 colocalized with the nuclear stain DAPI (blue). Representative images (Left) and quantified mean data (Right) are shown. Values are means ± SE, n = 40. **P < 0.01.
We next conducted immunoblotting assays to assess whether defective β-oxidation in the acx4-4 mutant would change the level of histone acetylation in vivo. Compared with WT, levels of all three acetylated (Ac) core histones (H2B, H3, and H4) were decreased in the acx4-4 mutant (Fig. 5 B and C). Immunostaining nuclei using H3Ac or H4Ac antibodies showed that levels of these acetylated histones were significantly reduced in acx4-4 compared with the WT or complemented acx4-4 (Fig. 5 D and E). Furthermore, chromatin immunoprecipitation (ChIP)-PCR assays with antibodies for the acetylated histones H2B, H3, and H4 revealed that levels of H3Ac and H4Ac, but not of H2BAc, were substantially reduced at both the 35S promoter and NOS regions (SI Appendix, Fig. S11). These results indicate that histone acetylation is reduced in the acx4-4 mutant and suggest that peroxisome-derived acetyl-CoA is important for histone acetylation.
Decreased Histone H3K18Ac Is Associated with DNA Hypermethylation in acx4 Mutant.
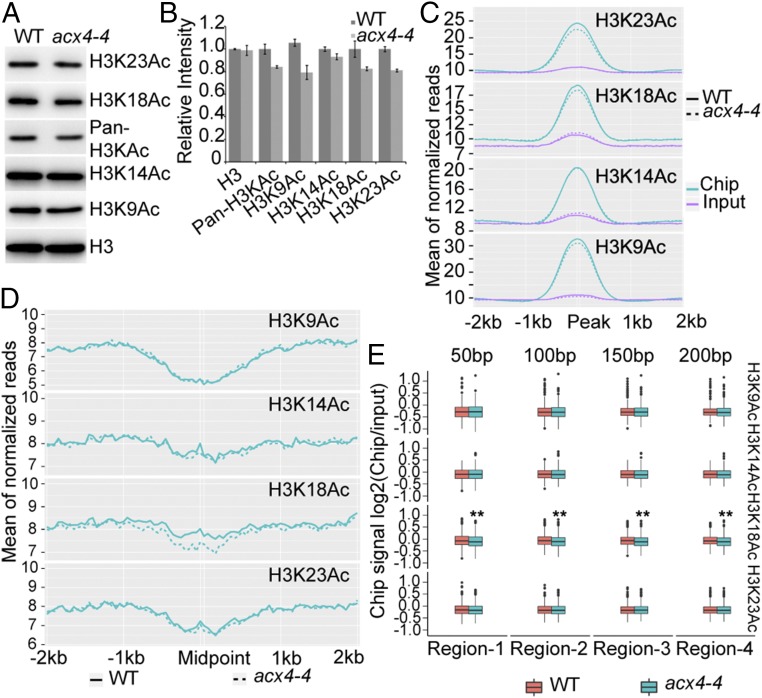
Recent studies have demonstrated that the DNA demethylation complex recognizes acetylated H3K18 and H3K23 at some target loci for DNA demethylation, suggesting that histone acetylation is required for active DNA demethylation (1, 20–23). To better evaluate whether ACX4-dependent generation of acetyl-CoA selectively affects acetylation of histone H3 at lysine residues, we analyzed the levels of histone H3 acetylation at K9, K14, K18, K23, and K27 in acx4-4 and WT by immunoblotting. The levels of H3K9Ac, H3K18Ac, and H3K23Ac were moderately reduced, but those of H3K14Ac and H3K27Ac were unchanged (Fig. 6 A and B and SI Appendix, Fig. S12). To determine which histone acetylation marks are associated with DNA hypermethylation in acx4 mutant, we conducted ChIP followed by sequencing (ChIP-seq) in WT and acx4-4 using antibodies against H3K9Ac, H3K14Ac, H3K18Ac, and H3K23Ac. For H3K14Ac, H3K18Ac, and H3K23Ac ChIP-seq experiments, two biological replicates were performed. Pearson’s correlation between two replicates are shown in SI Appendix, Fig. S13. In our analysis, 15,421 H3K9Ac, 8,460 H3K14Ac, 9,040 H3K18Ac, and 14,884 H3K23Ac peaks were identified in the WT. Consistent with the immunoblotting results, we found that ChIP-seq reads on H3K9Ac, H3K18Ac, and H3K23Ac peaks, but not those on H3K14Ac peaks, were decreased in acx4-4 relative to WT (Fig. 6C).
Fig. 6.
The hyper-DMRs in acx4-4 are associated with reduced H3K18Ac. (A) The acx4-4 mutant reduces histone acetylation. Histone proteins were extracted and used for immunoblotting with different antibodies. Histone H3 served as a loading control. (B) Quantified mean data normalized to H3 showing the relative levels of different H3 acetylation modifications in acx4-4 compared with the WT. Error bars represent ± SE from three independent experiments. n = 3. (C) Comparison of ChIP-seq reads with different histone acetylation antibodies between acx4-4 and the WT. −2 kb and +2 kb represent 2 kb upstream and 2 kb downstream of the peak middle point site, respectively. (D) Correlation analyses of DMRs with different histone acetylation modifications between acx4-4 and the WT. Only the enrichment levels of H3K18Ac are found to be decreased in acx4-4 relative to the WT. −2 kb and +2 kb represent 2 kb upstream and 2 kb downstream of middle point site of the hyper-DMR region, respectively. (E) Boxplots showing that the H3K18Ac levels of nearby hyper-DMRs are significantly lower in acx4-4 than in WT. 50 bp, 100 bp, 150 bp, and 200 bp represent the distances upstream and downstream of the middle point site of the hyper-DMR region. Two-tailed Student’s t test, **P < 0.01.
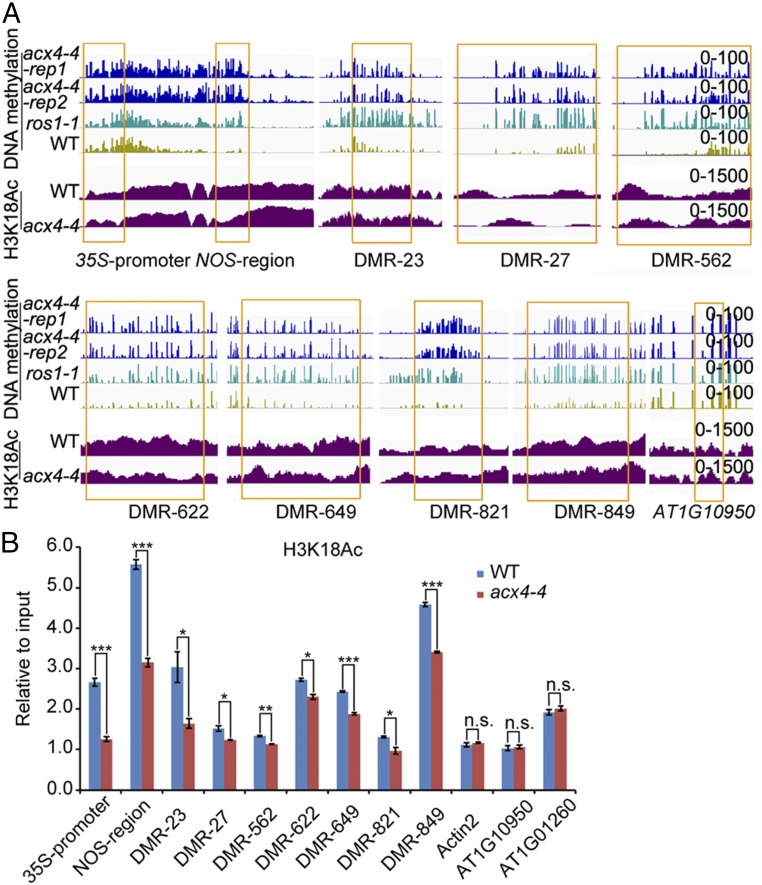
We next plotted ChIP-seq reads on acx4-4 hyper-DMRs in WT and acx4-4 and found that the enrichment levels of H3K18Ac around hyper-DMRs, but not other histone acetylation makers, were clearly decreased in acx4-4 relative to WT (Fig. 6D). In agreement with this finding, boxplots show much lower H3K18Ac levels of nearby hyper-DMRs in acx4-4 compared with WT (Fig. 6E). Several hyper-DMRs shared by ros1-1 and acx4-4 were selected for further investigation by ChIP-qPCR (qPCR) assays, which revealed lower H3K18Ac levels in the acx4-4 mutant compared with WT (Fig. 7 A and B). As negative controls, no changes were found at ACTIN2, AT1G10950, and AT1G01260. These results suggest that reduced histone H3K18 acetylation in acx4-4 is correlated with DNA hypermethylation.
Fig. 7.
Comparison of hyper-DMRs and reduced H3K18Ac at some loci in acx4, ros1-1, and WT. (A) DMRs and H3K18Ac levels in different loci among acx4-4 and WT. The DNA methylation levels in ros1-1 were included. (B) ChIP-qPCR confirmation of H3K18Ac levels in different DMRs between acx4-4 and WT. Values are means ± SE of three replicates in one experiment. Three independent experiments were done with similar results. Two-tailed Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001, n.s., no significant difference.
Overexpression of ATP-Citrate Lyase A and B Together Can Rescue the Kanamycin Sensitivity of acx4-4.
Acetyl-CoA is impermeable to membranes. The foregoing data suggest that mutations of genes in the β-oxidation pathway reduce the acetyl-CoA level in peroxisomes, which may in turn decrease the acetyl-CoA level in cytosol and affect histone acetylation and DNA methylation or affect other metabolisms, such as the reduced production of some phytohormones, as some products of β-oxidation may go to different metabolite pathways to produce MeJA, SA, or auxin (48, 49). Under different hormone treatments, such as NAA, SA, or MeJA, the Kan sensitivity of acx4-4 mutant was not changed (SI Appendix, Fig. S14A), indicating that Pro35S:NPTII silencing is not regulated by these phytohormones.
We further added some possible precursors for acetyl-CoA biosynthesis, such as succinate, acetate, oxoglutarate, and citric acid, to the growth medium to see whether these chemicals could rescue the Kan-sensitive phenotype of acx4-4, but found that they could not. We then searched for a strategy to increase cytosolic acetyl-CoA in vivo. In the cytosol, ATP-citrate lyase (ACL) is the sole key enzyme for generating acetyl-CoA (50). Unlike in animals, in which ACL has a monomeric structure, plant ACL consists of two distinct subunits, ACLA (45 kDa) and ACLB (65 kDa), and the heterooctomer holoenzyme is expected to have an A(4)B(4) configuration (50). We speculated that if ACL is overexpressed in acx4-4 mutant, more acetyl-CoA will be produced, which may compromise the Kan-sensitive phenotype of acx4-4. We expressed ACLA1-GFP (AT1G10670) and ACLB2-MYC (AT5G49460) under the control of 35S promoter in acx4-4. We obtained transgenic lines and tested their Kan sensitivity, but did not find any difference between acx4-4 and acx4-4 expressing ACLA1-GFP or ACLB-MYC (SI Appendix, Fig. S14 B and C), suggesting that overexpressing one subunit of ACL does not affect the silenced Pro35S:NPTII.
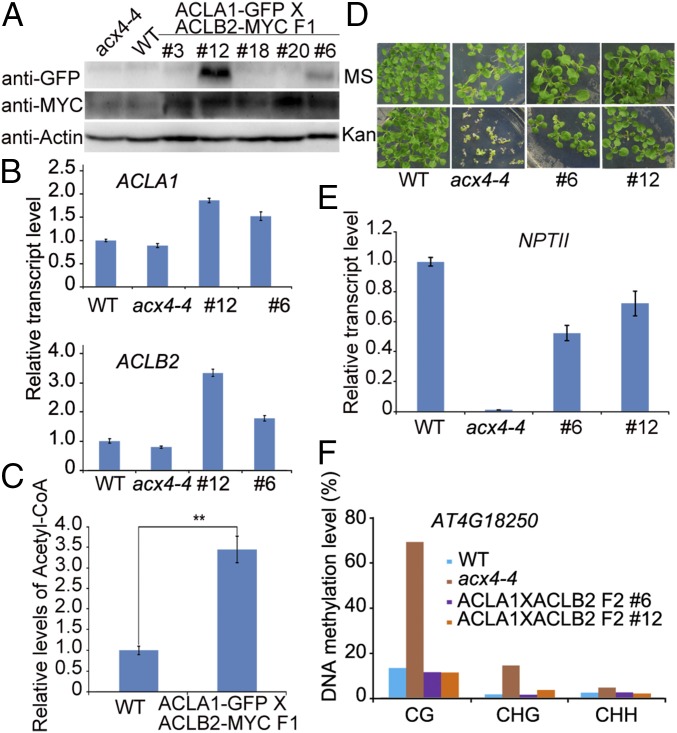
We then crossed different transgenic lines of ACLA1-GFP with those of ACLB2-MYC and obtained F1 seeds. Protein immunostaining using GFP and MYC antibodies and qPCR indicated that the ACLA1-GFP/ACLB2-MYC F1 #12 and #6 lines showed high levels of both ACLA1-GFP and ACLB2-MYC protein and high transcript levels of the two genes (Fig. 8 A and B). The acetyl-CoA content in ACLA1-GFP/ACLB2-MYC F1 #12 was three times that in the WT (Fig. 8C). Both lines #12 and #6 had the Kan-sensitive phenotype of acx4-4 rescued and expressed higher levels of NPTII than acx4-4 (Fig. 8 D and E). Bisulfite sequencing revealed that the DNA methylation of endogenous target gene AT4G18250 was recovered to WT level in both lines #6 and #12 in F2 Kan-resistant seedlings (Fig. 8F). These results indicate that increasing acetyl-CoA content rescued the acx4-4 Kan sensitivity.
Fig. 8.
Overexpression of ACLA1 and ACLB2 together can rescue the Kan-sensitive phenotype of acx4-4. (A) Protein levels detected with different antibodies in different F1 seedlings of acx4-4 expressing both ACLA1-GFP and ACLB2-MYC. The F1 seedlings were obtained by crossing the different lines of acx4-4 expressing ACLA1-GFP with those expressing ACLB2-MYC. ACTIN served as a loading control. (B) Relative expression levels of ACLA1 and ACLB2 in two F1 lines as determined by qPCR. (C) Relative levels of Acetyl-CoA in F1 seedlings. Values are means ± SE (n = 3) of three independent biological repeats is shown. Two-tailed Student’s t test, **P < 0.01. (D) Comparison of Kan-resistant phenotypes among WT, acx4-4, and two F1 lines on MS containing 25 mg/L Kan. (E) Relative transcriptional levels of NPTII in WT, acx4-4, and two F1 lines. (F) Bisulfite sequencing analyses of DNA methylation for endogenous gene AT4G18250 in WT, acx4-4, and two acx4-4 lines expressing both ACLA1 and ACLB2 that were isolated from F2 seedlings with Kan resistance.
Discussion
Acetyl-CoA is a central metabolite produced from fatty acids, glucose, and amino acids. It is a donor of single acetyl groups for histone acetylation and is crucial for cell growth and proliferation (39, 51, 52). Because it is impermeable to membranes, acetyl-CoA can be produced in different organelles, such as peroxisomes, mitochondria, and plastids (5). In these organelles, acetyl-CoA must be condensed to other molecules before being exported to the cytosol. However, whether organellar acetyl-CoA can affect nuclear DNA methylation in pants is unknown. In this study, we provide genetic and molecular evidence showing that a deficiency of short-acid β-oxidation can reduce histone acetylation and increase DNA methylation at a subset of genomic loci in Arabidopsis, suggesting a close connection between organelle metabolites and nuclear epigenetic modifications.
DNA methylation and histone modifications are primary epigenetic markers for the regulation of chromatin stability and transcription (12, 53). In our genetic screen, we identified the ROS4/IDM1, ROS5/IDM2, and MBD7 genes for their roles in DNA demethylation in the ProRD29A:LUC/C24 transgenic line (18, 22, 23). The current favored model suggests that MBD7 binds to genomic regions with high CG methylation density and physically associates with ROS4/IDM1 to create acetylated H3K18 and H3K23 marks, which in turn facilitate recruitment of the DNA demethylation enzyme ROS1 (1, 18). These studies suggest that histone acetylation is required for DNA demethylation enzymes to target genomic loci to prevent DNA hypermethylation and gene silencing.
Using the same ProRD29A:LUC screening system, we identified ACX4 and found that compared with WT, acx4 mutants have greatly reduced histone acetylation levels. In acx4 hyper-DMRs, the levels of H3K18Ac were reduced relative to the WT (Fig. 6 D and E), suggesting a close connection between DNA hypermethylation and histone acetylation at these loci. A comparison of DNA hypermethylated loci showed that in acx4, hyper-DMRs partially overlap with those identified in the ros1 mutant (Fig. 4B), supporting the notion that ACX4 functions in ROS1-mediated DNA demethylation. The presence of other hyper-DMRs in the acx4 mutant that did not increase in the ros1 mutant (Figs. 2C and 4C) suggests that in these loci, DNA demethylation is controlled by ROS1 paralogs, or histone acetylation directly affects DNA methylation, as seen in had6 mutants (54). The expression levels of hypermethylated genes, or genes located near hyper-DMRs, were reduced in the acx4-4 and ros1-1 mutants, and the acx4-4 ros1-1 double mutant had no additive effect on gene expression relative to the single mutants (Fig. 2D), suggesting the importance of ACX4 in preventing transcriptional gene silencing by DNA demethylation.
We found that acx4-1, mfp2-2, and kat2-3 mutants have similar hyper-DMRs, suggesting that β-oxidation in peroxisomes is crucial for modulating DNA methylation in nuclei. In plants and microbes, β-oxidation occurs in peroxisomes, while in mammalian cells, it occurs mainly in mitochondria. In mammalian cells, the addition of butyrate, a short-chain fatty acid that can be oxidized in mitochondria, induces ACL-mediated histone acetylation (55). Both cytosolic and mitochondrial acetyl-CoA synthetases (ACSS2 and ACSS1) are required for acetate-induced histone acetylation under hypoxia in cancer cells (56). Acetyl-CoA produced by the β-oxidation pathway in peroxisomes may have a similar role in modulating nuclear histone acetylation in plants, which in turn would affect DNA methylation.
We found that the levels of all three acetylated core histones (H2B, H3, and H4) were down-regulated in acx4 compared with the WT (Fig. 5 B and C), demonstrating that ACX4 is important for histone acetylation. However, along with affecting DNA methylation, histone acetylation usually has a generally positive effect on gene expression. The globally reduced histone acetylation in acx4 mutants would also affect the expression of genes not related to DNA methylation, which merits future exploration. Since acetyl-CoA can be produced in various metabolic pathways in plant cells (5) and other organisms (39), further studies are needed to demonstrate whether these other pathways are also involved in histone acetylation and DNA demethylation.
In plants, the β-oxidation of storage fatty acids is required to establish seedlings during germination. This is coupled with the glyoxylate cycle to convert acetyl-CoA into succinate. Succinate can be used for amino acid biosynthesis or can be converted into sucrose by gluconeogenesis. Sucrose can then be catabolized into pyruvate by glucose oxidation and converted to acetyl-CoA by the pyruvate dehydrogenase complex in mitochondria. Acetyl-CoA can enter the tricarboxylic acid cycle in mitochondria to produce citrate, which can be exported to the cytosol and converted to acetyl-CoA by ACL (10). However, in later developmental stages, neither β-oxidation nor the glyoxylate cycle is considered essential, because photosynthesis can provide the carbohydrates and precursors required for protein and nucleic acid biosynthesis. In our study, we used Murashige and Skoog (MS) medium supplemented with 2% sucrose (sufficient for heterotrophic seedling growth) to isolate the acx4 mutant. Contrary to previous studies indicating that exogenous application of sucrose can largely rescue the growth defects of various mutants in the β-oxidation pathways (29, 32, 57), our study indicates that Kan-sensitive acx4 phenotypes could not be rescued by sucrose or by any other possible precursors for acetyl-CoA biosynthesis, such as succinate, acetate, oxoglutarate, and citric acid. Peroxisomal β-oxidation not only functions in fatty acid catabolism, but also is required for the metabolism of hormones and amino acids (58). Similarly, exogenous application of hormones, including jasmonate, salicylate, and NAA, could not rescue the phenotype of Kan sensitivity in acx4 (SI Appendix, Fig. S14A). These results suggest that exogenous addition of these components cannot bypass the β-oxidation defect in the acx4 mutant to rescue the silencing phenotype. We speculate that the metabolites provided by β-oxidation in cells might be used more efficiently than exogenous sources for biosynthesis of the acetyl-CoA donor for histone acetylation, or that active DNA demethylation is a slow process that cannot be reflected so quickly after addition of sucrose or other compounds in the medium. Another possibility is that active DNA demethylation is preferentially active during early development, such that once high levels of DNA methylation are established during seed development, they cannot be decreased by later addition of the precursors for acetyl-CoA to the seedlings. We found that when acx4-4 expressed both ACLA1-GFP and ACLB2-MYC, more acetyl-CoA was produced, and the Kan-sensitive phenotype and DNA hypermethylation were rescued, suggesting that acetyl-CoA plays a critical role in mediating DNA demethylation and antisilencing.
The status of cellular metabolism is closely connected to epigenetic modifications (59, 60). Nutrition is well known to affect development and diseases through metabolism that modulates epigenetic modifications in mammals (61). Further elucidation of the connections between peroxisomal metabolism and chromatin regulation will enhance our understanding of epigenetic regulation and development.
Methods
Plant Growth Conditions and Mutant Screening.
Seeds sterilized with 0.5% NaClO were sown onto MS medium plates containing 2% (wt/vol) sucrose and 0.8% (wt/vol) agar. After 3–4 d at 4 °C, the plates were transferred to a growth chamber at 22 °C under long-day conditions (23-h light/1-h dark). Seven-day-old seedlings were transferred to soil and cultivated in a greenhouse at 20 °C under long-day conditions (16-h light/8-h dark).
The WT C24, ros1-1, ros4, rdr2, ddm1, acx4-4, mfp2-cas9-1, and mfp2-cas9-2 mutants (C24 background) mentioned in this study all carried the RD29A-LUC and Pro35S:NPTII transgenes (16, 18). The T-DNA lines ros1-4, SALK_000879 (acx4-1), SALK_065013 (acx4-3), SALK_098016 (mfp2-2), GK_787F01 (mfp2-10), and SALK_024922 (kat2-3) were obtained from the Arabidopsis Stock Center.
The acx4-4 mutant was identified from an EMS-mutagenized WT population as described previously (18). In brief, since WT seedlings can grow on MS medium containing 50 mg/L Kan, mutants that did not grow well on this medium were selected and transferred to MS medium for recovery. Their Kan-sensitive phenotype was confirmed in the next generation. Putative mutants were crossed with a Columbia accession (gl1), and Kan-sensitive plants from the F2 seedlings were assayed by PCR (using primers Pro35S:NPTII-F and Pro35S:NPTII-R). Mutants (approximately 2,000) containing the T-DNA insertion were selected for mapping.
For the complementation experiment, the genomic sequence of ACX4 from −2,543 to +3,546 bp was cloned into the SacI and SmaI (XmaI) sites of pCAMBIA1300; this sequence contains the 2,543-bp promoter, the coding region, and the 190-bp 3′ region. Using Agrobacterium tumefaciens GV3101, this construct was transformed into the acx4-4 mutant. Transgenic plants were selected and analyzed for sensitivity to Kan.
Individual Loci DNA Methylation Analysis.
Genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit. DNA methylation at individual loci was analyzed using bisulfite sequencing.
For bisulfite sequencing, 500 ng of genomic DNA was treated with the EZ Methylation-Gold Kit (Zemo Research), following the manufacturer’s protocol. Approximately 75 ng of bisulfite-treated DNA was used for PCR with the specific primers listed in SI Appendix, Table S1. PCR products were cloned into the pMD18-T vector (Takara Bio), and at least 15 independent clones from each sample were sequenced for each region.
Real-Time qPCR.
Total RNA was extracted from 7-d-old seedlings using TRIzol reagent (Invitrogen), and contaminating DNA was removed using RNase-free DNase I (Takara Bio). Approximately 4 μg of mRNA was used for first-strand cDNA synthesis using Moloney Murine Leukemia Virus Reverse Transcriptase (Promega) in a 20-μL reaction volume, and 5 μL of a 1:20 dilution of the cDNA reaction mixture was used as a template in a 20-µL PCR with SYBR Green Master Mix (Takara Bio) on a Step One Plus machine (Applied Biosystems) with three technical replicates. UBC28 served as an internal control. RNA transcript levels were determined by qPCR in a 20-µL reaction mixture, with TUB8 as an internal control. The primers used for PCR are listed in SI Appendix, Table S1.
Histochemical GUS Staining.
The ACX4 promoter fragment (from −2,620 to −1 bp) and the GUS-coding fragment were cloned into the pCAMBIA1391 vector. The recombinant plasmid was introduced into A. tumefaciens strain GV3101 and then transformed into C24 WT plants. Histochemical GUS staining was conducted on the T2 transgenic plants as described previously (62).
Histone Extraction and Immunoblotting.
Ten-day-old seedlings were ground to a fine powder in liquid nitrogen and suspended in nuclear isolation buffer (250 mM sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM Pipes pH 6.8, and 0.8% Triton X-100). The mixture was centrifuged, and the pellet was resuspended in 0.4 M H2SO4 and incubated for at least 1 h on ice. The preparation was centrifuged again, followed by the addition of a 20-fold volume of acetone to precipitate the histone protein. The preparation was kept at −20 °C overnight, after which the proteins were dissolved in 4 M urea and then used for protein blot analysis with the following antibodies: anti-H2BAc (ab1759; Abcam), anti-H3 (17–10046; EMD Millipore), anti-H3Ac (7–615; EMD Millipore), anti–Pan-H3Ac (ab47915; Abcam), anti-H3K9ac (07–352; EMD Millipore), anti-H3K14ac (07–353; EMD Millipore), anti-H3K18ac (ab1191; Abcam), anti-H3K23ac (07-355; EMD Millipore), and anti-H4Ac (17–630; EMD Millipore). Anti-H3 immunoblot was used as a loading control.
Immunofluorescence Assay.
An immunofluorescence assay was performed using 10-d-old seedlings. Nuclei were fixed in 4% paraformaldehyde and blocked with 3% BSA in PBS. The primary antibody H3Ac (7–615; EMD Millipore) was diluted at 1:100, and H4Ac (17–630; EMD Millipore) was diluted at 1:200, incubated overnight at 4 °C, and finally incubated with rabbit Alexa Fluor 594 (A11012m; Invitrogen)-conjugated secondary antibodies for 2 h at 37 °C. Chromatin was counterstained with DAPI in Prolong Gold (Invitrogen). Images were acquired with a Leica TCS SP8 STED 3X confocal microscope at 100× magnification.
ChIP Assays.
ChIP assays were performed as described previously (63) using 10-d-old seedlings and the following antibodies: anti-H2BAc (ab1759; Abcam), anti-H3Ac (17–615; EMD Millipore), anti-H4Ac (17–630; EMD Millipore), anti-H3K9ac (07–352; EMD Millipore), anti-H3K14ac (07–353; EMD Millipore), anti-H3K18ac (ab1191; Abcam), anti-H3K27ac (07-360; EMD Millipore), and anti-H3K23ac (07-355; EMD Millipore). ChIP products were eluted into 50 μL of Tris-EDTA buffer and diluted to a ratio of 1:5, and then a 2-μL aliquot of this dilution was used for each qPCR reaction. The primers used for qPCR are listed in SI Appendix, Table S1.
ChIP-Seq and Data Analysis.
The ChIP DNA concentration was determined using a fluorescence-based quantification method (Qubit 3.0 system; Life Technologies). High-throughput sequencing libraries were prepared using the NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (E6240; New England BioLabs) according to the manufacturer’s instructions. Sequencing was performed on an Illumina HiSEq 2500 instrument with single-end 50-bp reads. The reads were aligned to the TAIR10 Arabidopsis reference genome using Bowtie (64) with default parameters. Reads that perfectly and uniquely mapped to the genome were retained for further analysis. For each ChIP-seq, mapped reads were pooled from both replicates. Wiggle (WIG) format was generated using MACS program (65) and visualized using IGV (66). Acetylation-enriched peaks were identified using SICER (67) with a window size setting of 200 bp, gap size of 200 bp, false discovery rate (FDR) ≤0.05, and fold change ≥1.5. The genome-wide occupancies, boxplots, and Pearson correlations were performed with R packages.
Localization of the mCherry-ACX4 Fusion Protein.
The red fluorescent protein mCherry was fused to the N terminus of ACX4 under the control of the 35S promoter, and this construct was cloned into a modified pCAMBIA1300 vector. The vector carrying YFP-CD3-990 was generated in a previous study (30). NLS was fused to the N terminus of GFP under the control of the 35S promoter and cloned into the pCAMBIA1300 vector to generate NLS-GFP. The peroxisomal marker GFP-CAT2 was obtained from a previous study (31). The transient expression assay in tobacco and image acquisition were also described previously (68).
Whole-Genome Bisulfite Sequencing and Data Analysis.
For plants in the C24 background, genomic DNA was isolated from 7-d-old seedlings using the Qiagen DNeasy Plant Mini Kit and sent to BGI (Shenzhen, China) for bisulfite treatment, library preparation, and sequencing using the Illumina HiSeq 2000 sequencing system. Raw reads obtained from sequencing were trimmed using SolexaQA software (69). Clean reads were mapped to the Arabidopsis reference sequence (TAIR 10) using Bismark, allowing two mismatches. DMR identification was performed as described previously (23). Bins applied in this study were 100 bp from the reference genome. DMR identification used the standard of an absolute difference in methylation levels in the CG, CHG, and CHH contexts of at least 0.4, 0.2, and 0.1, respectively, and the Benjamini–Hochberg-corrected FDR was <0.01 (Fisher’s exact test).
For plants in the Col-0 background, genomic DNA was extracted from 14-d-old seedlings using the DNeasy Plant Mini Kit and sent to BGI for bisulfite treatment, library preparation, and sequencing using the Illumina HiSeq 4000 system. For library preparation, 5 µg of genomic DNA was sonicated into 100- to 300-bp fragments, which were end-repaired and ligated with adenine at their 3′ ends. After ligation with Illumina DNA adaptors, unmethylated cytosine residues were converted to uracils using the Qiagen EpiTect Bisulfite Kit.
For data analysis, clean reads were mapped to the TAIR 10 genome using Bismark, allowing up to two mismatches. DMR identification was performed as described previously (70). In brief, the DNA methylation level in every 200-bp window at 50-bp intervals was compared between WT and mutant seedlings using Fisher’s exact test with P ≤ 0.05. FDRs were estimated using the Benjamini–Hochberg correction of calculated Fisher’s P values. Windows with seven or more differentially methylated cytosines (defined as C with P < 0.01 in Fisher’s exact test) and a >1.5-fold change in DNA methylation levels were retained and combined if the gap size was no more than 100 bp to generate DMRs. Finally, DMR length was adjusted to start at the first mC and end at the last mC.
Metabolite Measurement.
Ten-day-old Arabidopsis seedlings grown on plates containing MS medium were used to measure acetyl-CoA, butyryl-CoA, and hexanoyl-CoA levels. Sample preparation and procedures for measuring metabolites were described previously (71). The experiments were performed with three biological replicates.
Data Availability.
Sequence data referred to in this article can be found in the GenBank/EMBL databases under the following accession numbers: AT2G36490 for ROS1, AT3G14980 for ROS4/IDM1, AT5G66750 for DDM1, AT4G11130 for RDR2, AT3G51840 for ACX4, AT3G06860 for MFP2, AT2G33150 for KAT2, AT1G64230 for UBC28, and AT3G18780 for ACTIN2. Primary datasets for the whole-genome bisulfite sequences of Col-0, ros1-4, acx4-4, acx4-1, mfp2-2, and kat2-3 mutant plants have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE98214), as have histone acetylation ChIP-seq data (accession no. GSE98214). Whole-genome bisulfite sequencing data of C24 WT, ros1-1, and ros4 plants were obtained from the GEO database (accession no. SRP042060) (23).
Supplementary Material
Acknowledgments
This study was supported by the Natural Science Foundation of China (Grant 31330041).
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequence data referred to in this article have been deposited in the GenBank/EMBL database (accession nos. AT2G36490 for ROS1, AT3G14980 for ROS4/IDM1, AT5G66750 for DDM1, AT4G11130 for RDR2, AT3G51840 for ACX4, AT3G06860 for MFP2, AT2G33150 for KAT2, AT1G64230 for UBC28, and AT3G18780 for ACTIN2). Primary datasets for the whole-genome bisulfite sequences of Col-0, ros1-4, acx4-4, acx4-1, mfp2-2, and kat2-3 mutant plants have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE98214). Histone acetylation ChIP-seq data also have been deposited in the GEO database (accession no. GSE98214). Whole-genome bisulfite sequencing data of C24 WT, ros1-1, and ros4 plants were obtained from the GEO database (accession no. SRP042060).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904143116/-/DCSupplemental.
References
- 1.Qian W, et al. (2012) A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336:1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutendra G, et al. (2014) A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158:84–97. [DOI] [PubMed] [Google Scholar]
- 3.Tariq M, Paszkowski J (2004) DNA and histone methylation in plants. Trends Genet 20:244–251, and erratum (2005) 21:36. [DOI] [PubMed] [Google Scholar]
- 4.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M (2002) Histone acetylation regulates the time of replication origin firing. Mol Cell 10:1223–1233. [DOI] [PubMed] [Google Scholar]
- 5.Shen Y, Wei W, Zhou DX (2015) Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci 20:614–621. [DOI] [PubMed] [Google Scholar]
- 6.Sivanand S, Viney I, Wellen KE (2018) Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends Biochem Sci 43:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong BW, et al. (2017) The role of fatty acid β-oxidation in lymphangiogenesis. Nature 542:49–54. [DOI] [PubMed] [Google Scholar]
- 8.Nagaraj R, et al. (2017) Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 168:210–223.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, et al. (2017) Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat Plants 3:814–824. [DOI] [PubMed] [Google Scholar]
- 10.Fatland BL, et al. (2002) Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol 130:740–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He XJ, Chen T, Zhu JK (2011) Regulation and function of DNA methylation in plants and animals. Cell Res 21:442–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Lang Z, Zhu JK (2018) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19:489–506. [DOI] [PubMed] [Google Scholar]
- 14.Wu SC, Zhang Y (2010) Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol 11:607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gehring M, et al. (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong Z, et al. (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111:803–814. [DOI] [PubMed] [Google Scholar]
- 17.Parrilla-Doblas JT, Ponferrada-Marín MI, Roldán-Arjona T, Ariza RR (2013) Early steps of active DNA demethylation initiated by ROS1 glycosylase require three putative helix-invading residues. Nucleic Acids Res 41:8654–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, et al. (2012) Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci USA 109:11425–11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan CG, et al. (2017) A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res 27:226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang Z, et al. (2015) The methyl-CpG-binding protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol Cell 57:971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian W, et al. (2014) Regulation of active DNA demethylation by an α-crystallin domain protein in Arabidopsis. Mol Cell 55:361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, et al. (2015) Methyl-CpG-binding domain protein MBD7 is required for active DNA demethylation in Arabidopsis. Plant Physiol 167:905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, et al. (2014) REPRESSOR OF SILENCING5 encodes a member of the small heat shock protein family and is required for DNA demethylation in Arabidopsis. Plant Cell 26:2660–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan CG, et al. (2015) MET18 connects the cytosolic iron-sulfur cluster assembly pathway to active DNA demethylation in Arabidopsis. PLoS Genet 11:e1005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong Z. (2019) A SUVH-DNAJ/SDJ protein complex activates the expression of promoter-methylated genes in Arabidopsis. J Integr Plant Biol 61:90–92. [DOI] [PubMed] [Google Scholar]
- 26.Harris CJ, et al. (2018) A DNA methylation reader complex that enhances gene transcription. Science 362:1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao X, et al. (2019) A group of SUVH methyl-DNA binding proteins regulate expression of the DNA demethylase ROS1 in Arabidopsis. J Integr Plant Biol 61:110–119. [DOI] [PubMed] [Google Scholar]
- 28.Zhao QQ, Lin RN, Li L, Chen S, He XJ (2019) A methylated-DNA-binding complex required for plant development mediates transcriptional activation of promoter methylated genes. J Integr Plant Biol 61:120–139. [DOI] [PubMed] [Google Scholar]
- 29.Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J 41:859–874. [DOI] [PubMed] [Google Scholar]
- 30.Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51:1126–1136. [DOI] [PubMed] [Google Scholar]
- 31.Li J, et al. (2015) A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27:908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi H, et al. (1999) A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J Biol Chem 274:12715–12721. [DOI] [PubMed] [Google Scholar]
- 33.Lei M, et al. (2015) Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA 112:3553–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kindl H. (1993) Fatty acid degradation in plant peroxisomes: Function and biosynthesis of the enzymes involved. Biochimie 75:225–230. [DOI] [PubMed] [Google Scholar]
- 35.Pan R, Liu J, Hu J (December 21, 2018) Peroxisomes in plant reproduction and seed-related development. J Integr Plant Biol 10.1111/jipb.12765. [DOI] [PubMed] [Google Scholar]
- 36.Shockey JM, Fulda MS, Browse JA (2002) Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129:1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41:156–181. [DOI] [PubMed] [Google Scholar]
- 38.Preisig-Müller R, Gühnemann-Schäfer K, Kindl H (1994) Domains of the tetrafunctional protein acting in glyoxysomal fatty acid beta-oxidation: Demonstration of epimerase and isomerase activities on a peptide lacking hydratase activity. J Biol Chem 269:20475–20481. [PubMed] [Google Scholar]
- 39.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G (2015) Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab 21:805–821. [DOI] [PubMed] [Google Scholar]
- 40.Rylott EL, et al. (2006) The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal beta-oxidation is essential for seedling establishment. Plant J 45:930–941. [DOI] [PubMed] [Google Scholar]
- 41.Richmond TA, Bleecker AB (1999) A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11:1911–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castillo MC, León J (2008) Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J Exp Bot 59:2171–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Germain V, et al. (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28:1–12. [DOI] [PubMed] [Google Scholar]
- 44.Wiszniewski AAG, Bussell JD, Long RL, Smith SM (2014) Knockout of the two evolutionarily conserved peroxisomal 3-ketoacyl-CoA thiolases in Arabidopsis recapitulates the abnormal inflorescence meristem 1 phenotype. J Exp Bot 65:6723–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang ZP, et al. (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, et al. (2016) The second subunit of DNA polymerase delta is required for genomic stability and epigenetic regulation. Plant Physiol 171:1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steimer A, et al. (2000) Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12:1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wasternack C, Song S (2017) Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J Exp Bot 68:1303–1321. [DOI] [PubMed] [Google Scholar]
- 49.Bussell JD, Reichelt M, Wiszniewski AAG, Gershenzon J, Smith SM (2014) Peroxisomal ATP-binding cassette transporter COMATOSE and the multifunctional protein abnormal INFLORESCENCE MERISTEM are required for the production of benzoylated metabolites in Arabidopsis seeds. Plant Physiol 164:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fatland BL, Nikolau BJ, Wurtele ES (2005) Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell 17:182–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai L, Sutter BM, Li B, Tu BP (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Öst A, Pospisilik JA (2015) Epigenetic modulation of metabolic decisions. Curr Opin Cell Biol 33:88–94. [DOI] [PubMed] [Google Scholar]
- 53.Jacob Y, et al. (2010) Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Earley KW, et al. (2010) Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24:1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donohoe DR, et al. (2012) The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell 48:612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao X, et al. (2016) Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat Commun 7:11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eastmond PJ, Graham IA (2000) The multifunctional protein AtMFP2 is co-ordinately expressed with other genes of fatty acid beta-oxidation during seed germination in Arabidopsis thaliana (L.) Heynh. Biochem Soc Trans 28:95–99. [DOI] [PubMed] [Google Scholar]
- 58.Reumann S, Ma C, Lemke S, Babujee L (2004) AraPerox: A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol 136:2587–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keating ST, El-Osta A (2015) Epigenetics and metabolism. Circ Res 116:715–736. [DOI] [PubMed] [Google Scholar]
- 60.Meng J, et al. (2018) METHIONINE ADENOSYLTRANSFERASE4 mediates DNA and histone methylation. Plant Physiol 177:652–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carrer A, Wellen KE (2015) Metabolism and epigenetics: A link cancer cells exploit. Curr Opin Biotechnol 34:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia R, et al. (2006) ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 18:85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3:1018–1025. [DOI] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, et al. (2008) Model-based analysis of ChIP-seq (MACS). Genome Biol 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson JT, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zang C, et al. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics 25:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Q, et al. (2010) DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22:2336–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, et al. (2013) DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA 110:8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Purves RW, Ambrose SJ, Clark SM, Stout JM, Page JE (2015) Separation of isomeric short-chain acyl-CoAs in plant matrices using ultra-performance liquid chromatography coupled with tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 980:1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data referred to in this article can be found in the GenBank/EMBL databases under the following accession numbers: AT2G36490 for ROS1, AT3G14980 for ROS4/IDM1, AT5G66750 for DDM1, AT4G11130 for RDR2, AT3G51840 for ACX4, AT3G06860 for MFP2, AT2G33150 for KAT2, AT1G64230 for UBC28, and AT3G18780 for ACTIN2. Primary datasets for the whole-genome bisulfite sequences of Col-0, ros1-4, acx4-4, acx4-1, mfp2-2, and kat2-3 mutant plants have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE98214), as have histone acetylation ChIP-seq data (accession no. GSE98214). Whole-genome bisulfite sequencing data of C24 WT, ros1-1, and ros4 plants were obtained from the GEO database (accession no. SRP042060) (23).