Significance
Jasmonates are indispensable for plant growth, as well as for adaptation to biotic and abiotic stresses. The synthesis of jasmonates is initiated in the chloroplast, where the precursor, 12-oxophytodienoic acid (OPDA) is derived from plastid lipids. Despite the fact that much information has been gained on the enzymes involved in the biosynthesis and signaling pathways of jasmonates, the export of OPDA from plastids remains enigmatic. In the present study, we showed that JASSY, a protein localized to the outer chloroplast envelope, facilitates export of OPDA from the chloroplast. Loss-of-function mutants lead to abolished jasmonate accumulation, which in turn results in increased susceptibility to cold treatment, as well as pathogen attack.
Keywords: jasmonate, plant hormones, chloroplast, membrane pore, cold acclimation
Abstract
Jasmonates are vital plant hormones that not only act in the stress response to biotic and abiotic influences, such as wounding, pathogen attack, and cold acclimation, but also drive developmental processes in cooperation with other plant hormones. The biogenesis of jasmonates starts in the chloroplast, where several enzymatic steps produce the jasmonate precursor 12-oxophytodienoic acid (OPDA) from α-linolenic acid. OPDA in turn is exported into the cytosol for further conversion into active jasmonates, which subsequently induces the expression of multiple genes in the nucleus. Despite its obvious importance, the export of OPDA across the chloroplast membranes has remained elusive. In this study, we characterized a protein residing in the chloroplast outer membrane, JASSY, which has proven indispensable for the export of OPDA from the chloroplast. We provide evidence that JASSY has channel-like properties and propose that it thereby facilitates OPDA transport. Consequently, a lack of JASSY in Arabidopsis leads to a deficiency in accumulation of jasmonic acids, which results in impaired expression of jasmonate target genes on exposure to various stresses. This results in plants that are more susceptible to pathogen attack and also exhibit defects in cold acclimation.
Jasmonates (JAs) play an important role in various cellular responses, including the reaction to biotic and abiotic stresses as well as the formation of reproductive organs. JA biosynthesis has been investigated in some detail, and thus the enzymes involved are well understood even with respect to mechanisms and regulation (1, 2). JAs are derived from α-linolenic acid (α-LeA), which is released by phospholipase 1 from galactolipids in the chloroplast thylakoid membrane (3). As an initial step, α-LeA is oxygenated by a lipoxygenase (LOX). Among six LOXs in Arabidopsis, LOX2 is thought to drive the bulk of JA formation during the first 2 h after initiation, for example, by wounding (4, 5). Subsequently, 13-allene oxide synthase (AOS) and 13-allene oxide cyclase introduce dioxygen and cyclize the fatty acid molecules to 12-oxophytodienoic acid (OPDA) (6, 7). Of note, OPDA is also found esterified to galactolipids (8, 9). However, it has not been entirely clarified whether esterified oxylipins are produced from free fatty acid intermediates or whether synthesis can occur on fatty acids while they are esterified into lipids (10).
In any case, OPDA is exported from the chloroplast, by as-yet unknown components. After import of OPDA into peroxisomes, possibly by an ATP-binding cassette (ABC) transporter, it is reduced by the peroxisomal OPDA reductase (OPR3) (11–13). These cyclic intermediates are then processed by the peroxisomal fatty acid β-oxidation machinery, producing JA. JA is in turn exported to the cytosol, where JAR1 forms the bioactive compound (+)-7-iso-jasmonoyl-l-isoleucine (JA-Ile) (14).
With identification of the JASMONATE-ZIM-DOMAIN (JAZ) proteins, the mechanistic role of JA-Ile in gene activation has been elucidated. While endogenous levels of JA-Ile are low, the JAZ proteins bind to various transcription factors, thereby repressing their activity (15). MYC2, a basic helix-loop-helix transcription factor, was the first transcription factor identified in this process and was found to regulate the expression of multiple genes that aid the reaction to various stresses, such as insects/herbivores and wounding (16). On a rise in endogenous JA-Ile levels due to an abiotic or a biotic stimulus, JAZ is bound by an F-box protein, CORONATINE INSENSITIVE 1 (COI1), wherein JA-Ile acts as a molecular glue. COI1 in turn is part of the ubiquitin proteasome degradation machinery and forms a Skp1/Cullin/F-box (SCFCOI1) complex, which has E3 ubiquitin ligase activity. This complex formation thus results in the ubiquitination and degradation of JAZ, thereby allowing the transcription factors to bind to their targets and activate gene expression (17).
Not only do JAs function in biotic stress response, but several studies also have identified JAZ as a repressor of the transcription factors INDUCER OF CBF EXPRESSION 1 and 2 (ICE1 and ICE2), which activate the C-repeat binding factor (CBF) pathway. Consequently, several genes responsible for cold and freezing tolerance are activated (18–20).
Recently, a protein of as-yet unknown function was identified in a proteomics study of chloroplast outer envelopes (OEs). Intriguingly, this protein was found to be coexpressed with a number of genes involved in the JA response (21). This protein, which we termed JASSY, contains a steroidogenic acute regulatory protein-related lipid transfer (START) domain, suggesting a function in the binding and/or transport of hydrophobic molecules. Loss of functional JASSY results in JA deficiency and thus influences cold acclimation as well as pathogen susceptibility in Arabidopsis mutants lacking JASSY. In this study, we investigated the role of JASSY in the export of OPDA from chloroplasts.
Results
JASSY Is Localized to the Chloroplast OE.
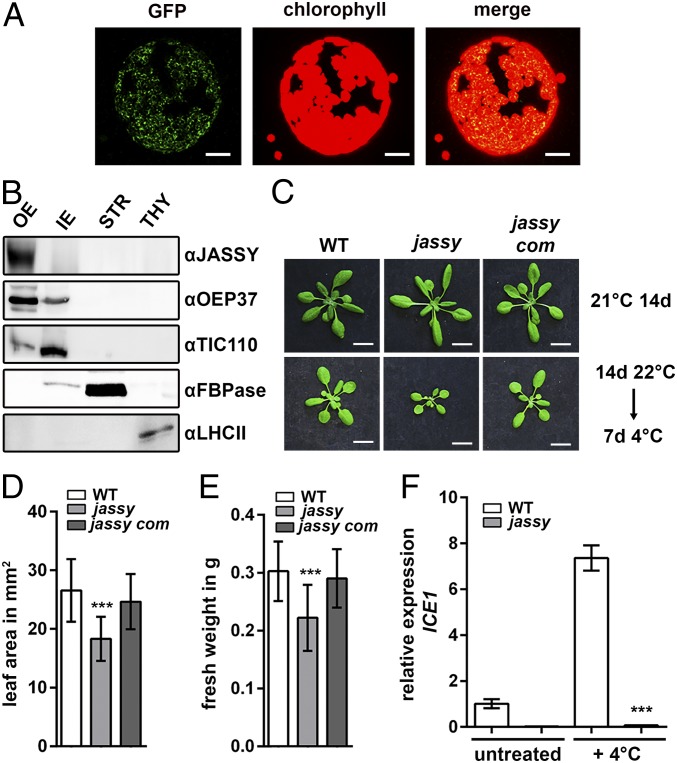
Chloroplasts contribute to several cellular metabolic pathways by performing crucial enzymatic reactions. Since they are surrounded by two envelope membranes, multiple channels and transporters are required within these membranes to facilitate the exchange of metabolites. Due to recent advances in analysis of subfractionated membranes using proteomics approaches, a number of as-yet uncharacterized proteins potentially residing in the chloroplast envelope membranes have been identified. Among these is a novel potential OE protein, here termed JASSY (At1g70480) (21). We started by investigating the subcellular localization of JASSY via the expression of a GFP fusion protein, as well as chloroplast fractionation and immunologic detection analyses. As a first step, we generated a C-terminal GFP fusion construct, which was used for the transfection of tobacco leaves via Agrobacterium. The fusion protein showed a clear chloroplast localization, indicating that JASSY is targeted to the chloroplast (Fig. 1A).
Fig. 1.
JASSY is localized to the OE of chloroplasts and is important for cold tolerance. (A) Transient expression of JASSY GFP fusion proteins in tobacco. Protoplasts were isolated after the transfection, and fluorescence was observed at 488 nm. (Scale bar: 5 µm.) (B) Pea chloroplasts were fractionated into OEs, IEs, stroma (STR), and thylakoids (THY). Fractions were separated by SDS/PAGE and subjected to immunoblotting with antisera against JASSY, OEP37, TIC110, FBPase, and LHCII. (C) WT, jassy, and jassy com were grown under standard conditions (Upper) or shifted to 4 °C for 7 d after growth for 14 d under standard conditions (Lower). (Scale bar: 1 cm.) (D) The leaf areas of WT, jassy, and jassy com after 7 d of cold treatment (C, Lower) was determined in square millimeters. The t test was used to indicated significance between WT and jassy. n >20. (E) Fresh weights of WT, jassy, and jassy com after 7 d of cold treatment (C, Lower) was measured in grams. The t test was used to indicate significance between WT and jassy. n >20. (F) Relative mRNA expression levels of ICE1 in WT and jassy monitored by qPCR in untreated plants and after 24 h at 4 °C; the t test indicated significance. Similar results were obtained in three biological replicates.
Interestingly, the GFP expression resulted in a ring-shaped signal, again indicating that JASSY might be associated with the chloroplast envelope. To investigate the sublocalization, we fractionated pea chloroplasts into OEs, inner membrane (IEs), thylakoids, and stroma. By doing so, we were able to locate JASSY exclusively in the OE fraction (Fig. 1B). Antisera against the OE protein 37 (OEP37, OE), the translocon of the chloroplast IE 110 (TIC110, IE), the fructose-1,6-bisphosphatase (FBPase, stroma) and the light-harvesting complex II (LHCII, thylakoids) served as markers to show the purity of the isolated chloroplast fractions (Fig. 1B).
To further analyze the localization, we performed an in vitro import assay using radiolabeled JASSY and isolated chloroplasts. We did not observe a shift in size of the translation product after the import reaction, indicating that the protein is not processed in the chloroplast stroma (SI Appendix, Fig. S1A). To determine whether JASSY is merely attached to the chloroplast surface or is indeed efficiently imported, chloroplasts were incubated with thermolysin after the import reaction. The radiolabeled JASSY protein was resistant to this protease treatment (SI Appendix, Fig. S1A). As a control, the ferredoxin-NADP+ reductase (FNR), which contains a transit peptide and is targeted to the stroma, was treated in the same manner. In this case, the unprocessed translation product was fully digested, whereas the mature protein was protected from the peptidase (SI Appendix, Fig. S1A). Both experiments were performed in parallel, showing that thermolysin treatment was efficient.
Since JASSY does not contain any predicted hydrophobic α-helical transmembrane domains, we treated pea OEs with several chaotropic reagents to analyze the mode of membrane interaction (SI Appendix, Fig. S1B). After treatment with 1 M NaCl, 6 M urea, 0.1 M Na2CO3, and 2 M NaBr, JASSY remained entirely in the insoluble fraction after centrifugation in all cases. Only treatment with 0.1 NaOH released a portion of the JASSY protein from the membrane. As a control, membranes were incubated with 1% SDS, which resulted in total solubilization of JASSY. The integral, β-barrel protein Toc75 served as control and, as expected, was released to the supernatant only after treatment with 1% SDS (SI Appendix, Fig. S1B). Therefore, we conclude that JASSY is stably inserted into the OE.
Loss of JASSY Decreases Cold Tolerance and Increases Susceptibility to Pathogen Attack.
A homozygous T-DNA insertion mutant of JASSY was isolated, in which the expression of JASSY was entirely abolished as shown on both protein and mRNA levels (SI Appendix, Fig. S1 C and D). Isolated chloroplasts from Arabidopsis wild type (WT) and mutant were fractionated into envelopes (mixed fractions containing IEs and OEs), stroma, and thylakoids. The fractions were subjected to SDS/PAGE, and immunoblotting was performed with an antiserum raised against the recombinant Arabidopsis JASSY protein. A band at the expected size of 36.3 kDa was again detected in the envelope fraction, which was absent in the mutant. The antibody recognized two additional bands in the stromal fraction; however, these were also present in the jassy knockout mutant and thus are assumed to represent cross-reactions of the antiserum (SI Appendix, Fig. S1E). Treatment with antisera against the translocon of chloroplast OE 75 (Toc75, OE), FBPase, and LHCII served as markers to show the purity of the isolated chloroplast fractions (SI Appendix, Fig. S1E).
Under normal growth conditions, jassy does not show an altered phenotype compared with the WT (Fig. 1C). However, when 14-d-old plants grown under standard long-day conditions were transferred to 4 °C for 7 d, a significant reduction in growth was observed (Fig. 1C). This was monitored by measurements of the leaf area as well as of the total fresh weight, both of which were significantly reduced in the mutant after cold treatment (Fig. 1 D and E). To ensure that the phenotype was caused by disruption of JASSY, we complemented the mutant by expressing the JASSY cDNA. Several independent complemented lines were obtained in which the cold phenotype was fully rescued. The phenotype of a representative line, jassy com, is shown in Fig. 1C, along with full recovery of the leaf area and weight in Fig. 1 D and E. Gene expression was likewise restored in jassy com, as shown by RT-PCR (SI Appendix, Fig. S1D).
Since coexpression data obtained from ATTED-II (atted.jp) suggested that JASSY is coexpressed with genes involved in JA metabolism, and JA-deficient mutants are known to be susceptible to cold stress (19, 22), we analyzed expression of the transcription factor ICE1 by quantitative PCR (qPCR). ICE1 is activated by JA and plays an important role in cold acclimation by inducing the expression of CBF3. CFBs in turn activate downstream targets mediating the cold acclimation response. Intriguingly, expression levels of ICE1 were below the limit of detection in the jassy mutant, whereas in the WT, ICE1 mRNA expression was up-regulated almost eightfold after 24 h of cold treatment (Fig. 1F).
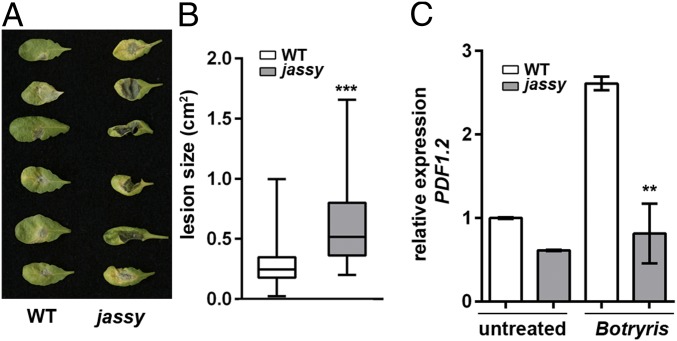
Apart from its role in response to abiotic stresses, JA is an important player in the reaction to biotic stresses, such as defense against pathogen attack. Therefore, we treated WT and mutant plants with the Arabidopsis pathogen Botrytis cinerea for 2 d. Whereas the WT showed only mild infection, the leaves of jassy exhibited large lesions (Fig. 2A). The lesion size was quantified, thus verifying the observed phenotype (Fig. 2B). Plant defensin 1.2 (PDF1.2) is a well-characterized marker gene specifically induced by JA on pathogen treatment or wounding (23). Therefore, we tested the expression of PDF1.2 before and after pathogen treatment to determine whether the observed pathogen susceptibility is due to a defect in the JA signaling pathway. Indeed, whereas PDF1.2 expression was enhanced in the WT on pathogen treatment, no changes in expression level were observed in jassy (Fig. 2C).
Fig. 2.
Loss of JASSY results in advanced susceptibility to B. cinerea in Arabidopsis. (A) Disease severity in WT and jassy plants at 2 d after B. cinerea treatment. (B) Botrytis-induced lesion size on leaves as shown in A was determined in square centimeters. The t test was used to indicated significance. n = 100. (C) Relative mRNA expression levels of PDF1.2 in WT and jassy plants was monitored by qPCR in untreated and B. cinerea-treated plants. The t test was used to indicate significance. n = 3. Similar results were obtained in three biological replicates.
The Expression of JA-Responsive Genes Is Not Activated in the jassy Mutant on Wounding.
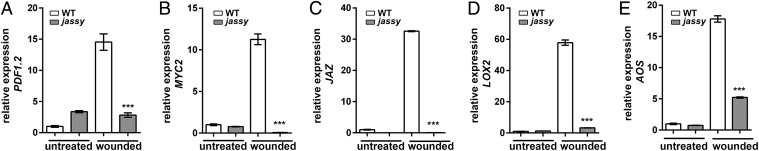
To further investigate whether the loss of JASSY induces a defect in JA signaling, we analyzed the expression of several JA-regulated genes in response to wounding (Fig. 3). Similar to the reaction following pathogen attack, no increase in PDF1.2 expression was observed in jassy at 90 min after wounding, in contrast to WT, which showed a 14-fold higher expression (Fig. 3A). A similar result was obtained for MYC2, a basic helix-loop-helix transcription factor that is a well-described master regulator of the JA signaling pathway (24) (Fig. 3B). Moreover, JAZ repressor proteins are known to be up-regulated on wounding. A 30-fold up-regulation was observed in the WT, whereas no reaction was observed on the expression level in the jassy mutant (Fig. 3C). Not only transcription factors and defense genes are activated by JAZ degradation, but also expression of the JA biosynthesis enzymes is induced in a positive feedback loop by JA after wounding. In line with our previous results, we found that LOX2 as well as AOS were lacking such induction in the jassy mutant (Fig. 3 D and E). The reduced expression was completely restored in the complemented mutant line; the expression of PDF1.2 is shown as a representative example (SI Appendix, Fig. S1F). Therefore, JASSY seems to be crucial for the initiation of JA-induced signaling pathways.
Fig. 3.
mRNA expression levels of JA-related genes. Expression levels of the indicated transcripts were determined in untreated plants and at 90 min after wounding by qPCR. (A) PDF1.2. (B) MYC2. (C) JAZ. (D) LOX2. (E) AOS. Similar results were obtained in three biological replicates in all cases. The t test was used to indicate significance for all datasets. n = 3.
Lack of JASSY Prevents JA Accumulation.
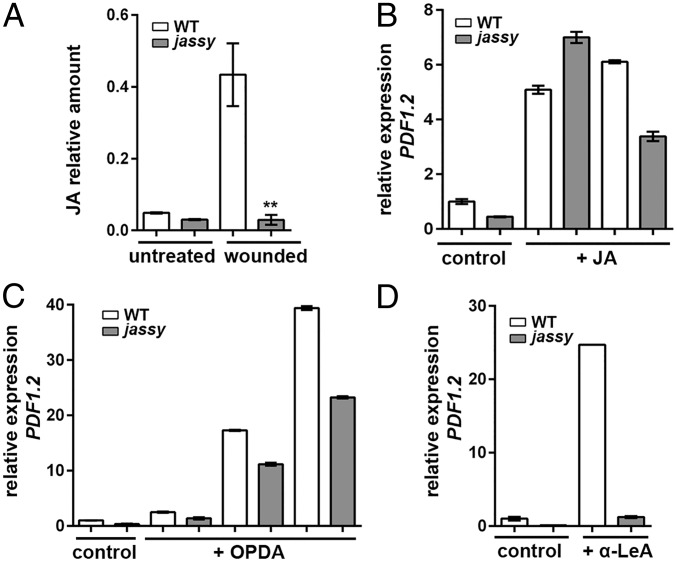
Since we observed no activation of the JA-responsive pathway on a transcriptional level, we aimed to determine whether a lack of JASSY leads to a general defect in the accumulation of JA. Since JA is barely detectable under standard conditions, JA levels were measured before and 90 min after wounding. Strikingly, in jassy, no increase in basal levels upon wounding was observed, whereas in the WT, a clear accumulation of JA was detected (Fig. 4A).
Fig. 4.
Measurement of JA levels in WT and jassy and treatment with JA-Ile, OPDA, and α-LeA. (A) The relative amount of JA was determined in untreated WT and jassy plants as well as at 90 min after wounding. The t test was used to indicate significance. n = 3. (B) WT and jassy mutants were sprayed with 0.4% ethanol (control) or with JA-Ile. The mRNA expression of PDF1.2 was determined at 30 and 120 min after the treatment. Similar results were obtained in three biological replicates. (C) WT and jassy mutants were sprayed with 0.3% ethanol (control) or with OPDA, and the mRNA expression of PDF1.2 was determined at 90, 120, and 180 min after the treatment. (D) WT and jassy mutants were sprayed with 0.3% ethanol (control) or with α-LeA, and the mRNA expression of PDF1.2 was determined at 30 min after the treatment. Similar results were obtained in three biological replicates.
To strengthen the hypothesis that indeed the lack of JA was responsible for the observed defects in gene expression, we treated WT and jassy mutants externally by spraying plants with JA-Ile. Strikingly, the expression of PDF1.2 could be fully recovered in jassy after 30 min (Fig. 4B). While expression in the WT was induced even further at 120 min after the JA-Ile treatment, expression levels were slightly decreased in the mutant. The observed induction clearly demonstrates that JASSY functions not in JA-induced gene expression per se, but rather in a step related to the biosynthesis of JA. The lowered transcript abundance of PDF1.2 at 120 min after the JA treatment likewise indicates that the positive feedback loop in response to JA accumulation, which results in an increased de novo synthesis of JA, is hampered in jassy.
Considering that JASSY localizes to the chloroplast OE, it is reasonable to assume that it is likely involved in the export of OPDA. In this case, external treatment with OPDA is likewise expected to restore the transcription of JA-responsive genes. Thus, OPDA was subjected to WT and jassy plants, and expression levels of PDF1.2 were determined at 90, 120, and 180 min after the treatment. Indeed, it was again observed that the expression of PDF1.2 was induced in jassy, almost to WT levels (Fig. 4C). In contrast, treatment solely with α-LeA resulted in PDF1.2 expression in the WT, but not in the mutant (Fig. 4D). Taken together, these data strongly suggest that JASSY mediates OPDA export from the chloroplast.
JASSY Binds to OPDA and Functions as a Membrane Channel.
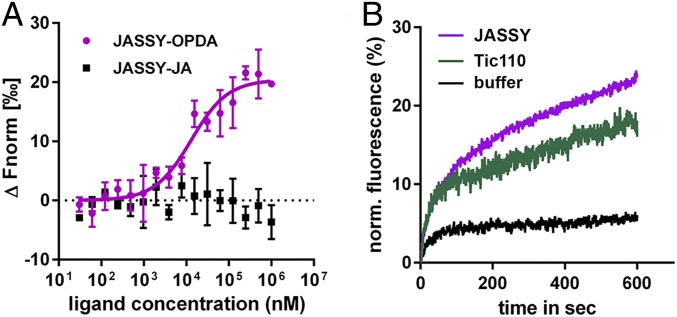
Since JASSY belongs to the Bet v1-like superfamily, which comprises domains known to function in binding hydrophobic components, we analyzed a potential direct interaction between OPDA and recombinant soluble JASSY protein purified from Escherichia coli using microscale thermophoresis (MST). Binding occurred, even though relatively high concentrations of OPDA had to be applied, resulting in a KD of approximately 1 mM (SI Appendix, Fig. S2A). Moreover, no binding curve could be fitted for the tested interaction between JASSY and JA (SI Appendix, Fig. S2B). To analyze whether the affinity increased in a membrane environment, the recombinant JASSY protein was incorporated into liposomes. Indeed, MST measurements with JASSY proteoliposomes and OPDA revealed a much stronger binding event with a KD of 12.8 ± 3.6 µM (Fig. 5A). Again, no binding occurred between JASSY liposomes and JA (Fig. 5A).
Fig. 5.
JASSY binds to OPDA and forms a pore in proteoliposomes. (A) Recombinant JASSY protein was reconstituted into liposomes, and MST measurements were performed with OPDA and JA. A binding curve could be fitted only with OPDA and a KD value of 12.7 ± 3.6 µM was calculated. n = 3. (B) Liposome leakage assay. JASSY or TIC110 protein was added to liposomes, and fluorescence was recorded every millisecond (excitation at 494 nm and emission at 515 nm). Initial fluorescence was set to 0, and total liposome dequenching resulting from 0.5% Triton X-100 was set to 100%. 1× PBS buffer, pH 7.4 was added to the liposomes as a negative control.
To elucidate whether JASSY has the capacity to form a membrane pore, we performed a liposome leakage assay. To this end, carboxyfluorescein-containing liposomes were generated, into which the recombinant JASSY protein was incorporated. During the reconstitution, fluorescence emission of the carboxyfluorescein dye was recorded. Indeed, fluorescence increased continuously over time, indicating release of the carboxyfluorescein dye from the liposomes. The IE protein TIC110 behaved in a similar manner and served as a positive control. In contrast, adding buffer to the liposomes did not result in leakage of carboxyfluorescein (Fig. 5B).
Since the liposome leakage assay indeed suggested that JASSY is able to form membrane pores, we used the planar lipid bilayer technique for an electrophysiological characterization to investigate this observation further. This technique has been used to study pore formation, activity dynamics, and substrate sensitivity of various other membrane channels, including protein translocases, metabolite transporters, pore-forming toxins, and ion channels (25–29) at single-molecule resolution. The technique uses ion conductivity as an activity readout for pore formation. To this end, we inserted JASSY into preformed liposomes via detergent-mediated reconstitution.
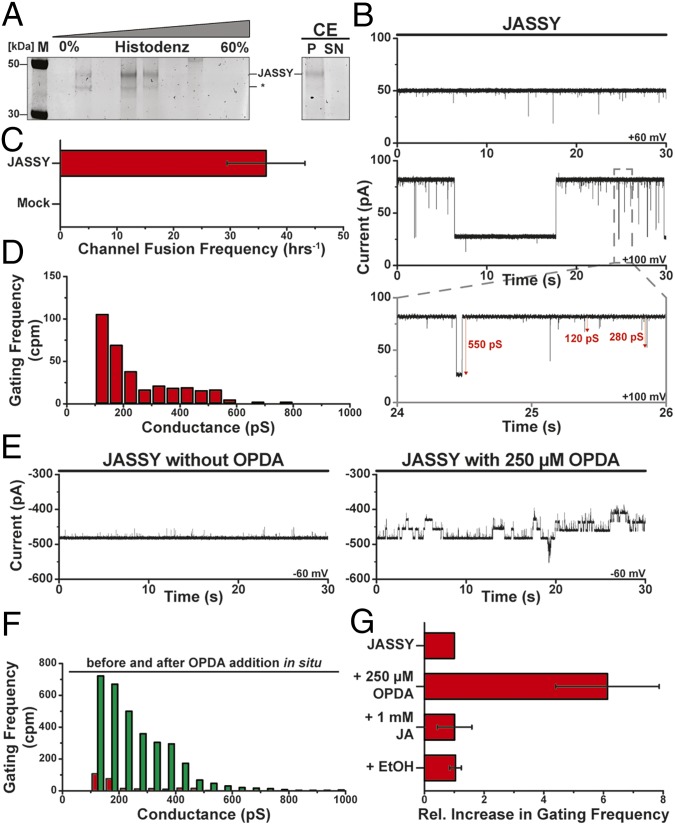
To assess incorporation success, we analyzed comigration of the protein with liposomes in a density flotation assay and found the JASSY-containing floating proteoliposomes. We confirmed that incorporated JASSY was resistant to carbonate extraction and as such behaved as an integral membrane protein (Fig. 6A). In the next step, JASSY-containing vesicles and identically treated liposomes with a purified sample of an empty plasmid mock expression were separately fused with planar lipid bilayers and subjected to high-resolution electrophysiological characterizations. While JASSY-containing proteoliposomes readily fused with bilayers and showed stable channel activity (Fig. 6 A and B), the mock sample did not lead to channel insertion even after prolonged incubation of several hours (Fig. 6C). JASSY channels exhibited frequent voltage-dependent gating with dynamic conductance states between 100 and 600 pS at 250 mM KCl (Fig. 6D). A reversion potential of 31.4 ± 2.0 mV at a 12.5-fold KCl gradient was measured; thus, the channels displayed a mild preference for potassium over chloride (SI Appendix, Fig. S3). It will be interesting to see in future studies how exactly OPDA is passing the channel and if different routes for different charged molecules exist, as has been suggested for other relatively wide pores (30, 31).
Fig. 6.
JASSY forms an OPDA-sensitive membrane channel. (A) JASSY proteoliposomes were subjected to density gradient flotation and subsequent sodium carbonate extraction (CE). *The JASSY degradation band. (B) Current flux through channels formed by JASSY was recorded at various indicated membrane potentials. Conductance changes of selected gating events are indicated. (C) Frequency of channel appearance due to proteoliposome fusion with the lipid bilayer was determined for both JASSY- and mock-expressed incorporations (n = 3, 2 h per experiment); error bars represent SEM. (D) Gating event frequency was calculated from current recording sets as shown in B. n = 6. (E) Current trace of multiple JASSY channels was recorded before (Left) and after (Right) in situ incubation with OPDA. (F) Frequency of gating events before (red) and after (green) incubation of JASSY with OPDA calculated from three independent experiments, as shown in D. (G) Average increase in gating frequency of events >100 pS was quantified for OPDA, JA, and ethanol. n = 3. Error bars represent SEM.
As we have shown that JASSY binds to OPDA, we probed possible substrate specific alterations of OPDA on the channel properties. Ethanol-solubilized OPDA was added to preinserted channels in situ, and channel characteristics before and after OPDA addition were evaluated. OPDA addition led to significantly increased gating frequencies of incorporated JASSY channels (Fig. 6E). Gating frequency analysis suggested that all conductance states were enhanced similarly, with an additional increased abundance of a high conductance state above 600 pS (Fig. 6F). While OPDA addition led to a sixfold increase in gating frequency on average, no significant change in gating frequency or any other determined channel characteristics was observed after addition of ethanol or JA (Fig. 6G).
Discussion
Whereas the enzymatic steps involved in JA biosynthesis have been investigated and studied in detail for decades now, only slow progress has been made concerning the transport of JAs and its precursors. Recently, JAT1 was identified as a transporter for JA-Ile into the nucleus (14). As to OPDA, the ABC transporter COMATOSE (CTS) has been suggested to be at least partially involved in OPDA import into peroxisomes. CTS is thought to transport a wide range of peroxisomal fatty acids or acyl-CoA–coupled fatty acids. Levels of JAs are reduced in the mutants but are not absent; thus, it is expected that other pathways for OPDA entry into peroxisomes exist in parallel (12, 32, 33). However, so far nothing is known about the export of OPDA from the chloroplast. In this study, we provide evidence that JASSY is a novel OE protein that fulfils this function.
Due to the defect in OPDA export, JA accumulation in Arabidopsis leaves is impaired entirely in the mutant, leading to disturbed responses to wounding, pathogen attack, and cold acclimation. Not only did we observe these effects on a phenotypical level, but we could also show that gene expression in the respective JA-responsive signaling cascades was not induced in mutants lacking JASSY. Experiments showing that gene expression could be reactivated with JA-Ile as well as OPDA spraying clearly demonstrate that JASSY functions upstream of JA perception in the nucleus as well as upstream of OPDA to JA conversion in the cytosol and peroxisome.
Over the last few years, a number of JA-related mutants have been analyzed to help elucidate the JA biosynthesis pathway as well as the role of JA during stress response and development. For example, the fad3/fad7/fad8 mutant is deficient in JA synthesis due to lack of α-LeA, the JA precursor in the chloroplast (34, 35). Moreover, this mutant, just like many other mutants involved in JA biosynthesis or perception, such as dad1 (36), aos (37), opr3 (38), and coi (39), are male sterile. Interestingly, we did not observe problems with either male or female fertility in jassy; however, McConn and Browse showed that a leaky allele of the fad3/fad7/fad8 triple mutant, which accumulates traces of α-LeA, is fertile (40). Since we also observed a small amount of JA in jassy, this might be sufficient for flower development and seed production. Alternatively, the fertility could be explained by the fact that the Arabidopsis genome harbors a second gene—At1g23560; JASSY-2—with 46% identity on the protein sequence level to JASSY. However, JASSY-2 is solely transcribed in flower buds, in contrast to JASSY, which is ubiquitously expressed in all developmental stages (www.bar.utoronto.ca). Therefore, JASSY-2 might take over the function of JASSY in flowers and play a role in flower maturation.
JASSY belongs to the Bet v1-like superfamily, a large protein family containing proteins that despite their low sequence similarity share a similar 3D structure (41). The common structure is characterized by a β-α2-β6-α fold, forming a U-shaped incomplete β-barrel wrapped around a long α-helix, thus providing a large hydrophobic binding cavity. The Bet v1 family is divided into 11 subfamilies, including the pathogenesis-related protein 10 (PR10), START domain proteins, and oligoketide/cyclase/dehydrases, in the latter of which JASSY shows the highest similarity to that described previously (41). Moreover, a domain of unknown function (DUF220) is predicted in JASSY, partially overlapping with the START domain. The hydrophobic cavity of most of the Bet v1 proteins functions in binding ligands, such as lipids, sterols, or secondary metabolites (41). In line with this, we observed that JASSY is able to bind OPDA, an interaction likely mediated by this conserved domain and likely important for the transport process.
Moreover, we found that JASSY is not only able to permeabilize membranes, but also forms a voltage-gated channel in planar lipid bilayers. These channels are substrate sensitive and show an OPDA-dependent activation, implicating a mode of transport of JA precursors across the OEs through JASSY. Although the relatively high OPDA concentrations applied in these experiments might not reflect the overall OPDA concentration in chloroplasts, local concentrations of OPDA in the vicinity of JASSY may be significantly elevated in vivo. Moreover, regulation of the channel activity remains to be elucidated. Acylation has been observed to play roles in fatty acid export from the chloroplast as well as OPDA import into peroxisomes and might be used to ensure unidirectional transport of OPDA (42, 43).
Where the function of JAs has been well established in all higher land plants, the presence of JAs in lower land plants and algae is still controversial (44). JAs have been detected in a broad range of bryophytes, but not in the moss Physcomitrella patens (45, 46). Nevertheless, OPDA can be detected in the latter and seems to play a role in fertility of the moss (47). Interestingly, JASSY is conserved among land plants, bryophytes, and green algae, supporting the concept that OPDA functions as a signaling molecule in Physcomitrella as well as the idea that JA may also trigger responses in other mosses, as well as algae (SI Appendix, Fig. S4).
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana WT Columbia ecotype and the mutant were grown on soil under long-day conditions (16-h light/8-h dark, 22 °C, 120 µE m−2 s−1), unless indicated otherwise. Homozygous jassy mutants were isolated and confirmed by genotyping PCR. The WT allele was amplified with At1g70480 LP (Ex5 rev) and At1g70480 RP (Ex3 for), the mutant allele with LB2 and At1g70480 RP (Ex3 for). RT-PCR was used to confirm the absence of the transcript in the mutant Oligonucleotide sequences are listed in SI Appendix, Table S1. The jassy mutant (SAIL_860179) was obtained from the European Arabidopsis Stock Centre. For complementation, the coding sequence was cloned into the vector pAUL11 (48). The construct was introduced into Agrobacterium tumefaciens strain GV3101, and jassy was transformed by floral dip (49).
For cold treatment, plants were first grown under long-day conditions for 14 d and then transferred to 4 °C, 16-h light/8-h dark, 22 °C, 120 µE m−2 s−1 for the indicated time periods. Wounding of Arabidopsis leaves was performed as follows. Leaves of 4-wk-old plants were wounded by cutting with sharp razor blades. Each leaf was wounded three times on each occasion. No leaf was wounded more than once. Wounded plants were covered and harvested leaves were immediately stored in liquid nitrogen. JA, OPDA, and α-LeA for feeding experiments were obtained from Cayman Chemical. Plants were sprayed with either 200 µM JA-Ile, 50 µM OPDA or 300 µM of α-LA. Untreated plants were sprayed with 0.3% ethanol as a control. Plants were harvested for RNA isolation at the indicated time points.
Pea plants (Pisum sativum L., cv. Arvica) were grown under a 12-h light (120 μmol m−2 s−1) and 12-h dark regimen at 21 °C. N. benthamiana was grown in soil under greenhouse conditions.
Pathogen Assay.
Four-week-old plants grown under short day conditions (8 h light/16 h dark, 22 °C, 120 µE m−2 s−1) were inoculated by applying one drop of 20 μL B. cinerea spore suspension with 2 × 105 spores per milliliter to each leaf (50). The extent of disease was analyzed by quantifying lesion size using ImageJ software.
Agrobacterium-Mediated Transient Expression of Fluorescent Proteins in Tobacco.
Four- to 6-wk-old Nicotiana benthamiana leaves were infiltrated with Agrobacterium for transient expression of gene constructs. The A. tumefaciens strain Agl1 was transformed with the respective gene constructs, and infiltration was performed as described previously (51). Protoplasts were prepared essentially as described previously (52), except cell walls were digested for 90 min at 40 rpm in 1% cellulase R10 and 0.3% macerase R10 after vacuum infiltration. Fluorescence was observed with a Leica TCS SP5 confocal laser scanning microscope at 20 °C.
Quantitative Real-Time RT-PCR Analysis.
RNA was extracted from untreated and treated plants as indicated using the Qiagen RNeasy Plant Kit. After quantification of the RNA and digestion of DNA with TURBO DNA-free kit (Life Technologies), first-strand cDNA was synthesized using M-MLV reverse transcriptase (Promega) from 1 µg of RNA. qPCR was performed in a Bio-Rad CFX96 real-time PCR detection system with SYBR Green Real-Time PCR Master Mix (Roche). Expression levels were normalized to the expression of ACTIN2 (AT3g18780). Gene-specific oligonucleotides are listed in SI Appendix, Table S1.
Chloroplast Isolation, Subfractionation, and in Vitro Import.
Pea chloroplast isolation for in vitro protein import and subfractionation was performed as described previously (53, 54). Isolation and subfractionation of chloroplasts from 4-wk-old Arabidopsis plants was also performed as described previously (55). Templates for in vitro transcription were cloned into pF3A (FNR) and pSP65 (JASSY) (Promega), and in vitro transcription was performed with SP6 polymerase (Fermentas). In vitro translation in reticulocyte lysate (Promega) was performed according to the manufacturer’s instructions.
For in vitro import chloroplasts equivalent to 10 μg chlorophyll and in vitro translation product were suspended in a total volume of 100 μL of import mix buffer [250 mM methionine, 250 mM cysteine, 5% BSA, 100 mM ATP, 1 M calcium gluconate, 1 M NaHCO3, and 1× HMS (50 mM Hepes, 3 mL MgSO4, and 0.3 M sorbitol)]. The reaction was incubated at 25 °C for 15 min. Chloroplasts were reisolated with a 300-μL Percoll cushion (330 mM sorbitol, 1 M Hepes/KOH, 40% Percoll) and centrifugation at 7,000 × g for 5 min at 4 °C. The pellet was washed twice in washing buffer I (330 mM sorbitol, 1 M Hepes, and 1 M MgCl2). The pellet was resuspended in 100 μL of wash buffer II (330 mM sorbitol, 50 mM Hepes, and 0.5 mM CaCl2) that contained 1 mg/mL thermolysin where indicated, followed by incubation on ice for 20 min. Then 0.5 M EDTA was added to stop the reaction. Chloroplasts were washed once in washing buffer III (330 mM sorbitol, 50 mM Hepes, and 5 mM EDTA). The pellet was resuspended in 20 µL of protein. Chloroplasts were separated by SDS/PAGE and analyzed by autoradiography.
Antisera.
For production of antisera in rabbits, full-length JASSY was expressed in LB medium in Rosetta 2(DE3) cells (Novagen), and JASSY was purified from inclusion bodies with Ni-NTA affinity chromatography (GE Healthcare). Antisera were generated by Pineda. Other antisera used included Tic110 (56), FBPase (57), OEP37 (58), Toc75 (59), and LHCII (60). Proteins were separated by SDS/PAGE, and immunodetection was performed and with enhanced chemiluminescence as described previously (61).
JA Measurements.
JA was extracted from frozen leaf material before and after wounding as described by Glauser and Wolfender (48). In brief, 200 mg fresh weight frozen material was ground in a 2-mL reaction tube using a ball mill (TissueLyser II; Qiagen) at 30 Hz for 1 min. For extraction, 1.5 mL of precooled isopropanol was added, along with 0.5% (vol/vol) formic acid and 5 ng mL−1 chloramphenicol as an internal standard. After extensive mixing to fully resuspend the tissue, the samples were mixed using the ball mill at 30 Hz for 4 min, then centrifuged at 16,400 rpm (Centrifuge 5417R; Eppendorf). The supernatant was transferred to a new 2-mL reaction tube and dried using a vacuum concentrator (Concentrator 5301; Eppendorf). Dry pellets were resuspended in 85% methanol using sonication (Sonifier B12; Branson Sonic Power). The solution was centrifuged at 16,400 rpm (Centrifuge 5417R; Eppendorf) for 90 s, after which 1 mL of the supernatant was loaded onto a C18 solid-phase extraction (SPE) column (Sep-Pak Vac 1 cc 50 mg; Waters). SPE columns were equilibrated previously using 1 mL of 100% and 1 mL of 85% (vol/vol) methanol. After SPE separation, the total flow trough was collected, followed by a 1-mL 85% (vol/vol) methanol washing step. The flow-through and the methanol wash were combined and dried overnight using the vacuum concentration. Dry samples were filled with argon and stored at −80 °C until further measurement.
Liquid chromatography-mass spectrometry (LC-MS) analysis was done using a Dionex UltiMate 3000 ultra-high-performance liquid chromatograph (Thermo Fisher Scientific) along with an Impact II ultra-high-resolution Qq time-of-flight mass spectrometer (Bruker). The dry samples were resolved in 100 µL of 85% (vol/vol) methanol. For separation, 20 µL was injected onto a C18 reversed phase column (Ultra Aqueous C18, 3 µm, 100 × 2.1 mm; Restek) with a 400-µL min−1 flow at 30 °C. The solvents used were water (A) and acetonitrile (B), both including 0.1% (vol/vol) formic acid. The 30-min gradient started at 5% B for 2 min, followed by a ramp to 95% B within 20 min. After a 3-min washing step at 95% B, the gradient turned back to 5% B within 1 min and remained there for a 4-min equilibration.
For MS detection, an electrospray ionization source was used in positive mode at 4 kV capillary and 0.5 kV endplate voltage. Nitrogen was the dry gas at 8 L min−1, 8 bar, and 200 °C. The Impact II mass spectra were recorded in MS mode from 50 to 1,300 m/z with 40,000 resolution, 1 Hz scan speed, and 0.3 ppm mass accuracy. Compounds were annotated in a targeted approach using specific mass (m/z) at retention time and the isotopic pattern. All data were acquired with otofControl 4.0 and HyStar 3.2 (Bruker). Data evaluation was performed with DataAnalysis 5.1, ProfileAnalysis 2.3, and MetaboScape 1.0 (Bruker). Additional data evaluation was done with Microsoft Excel. BioSolve or Sigma-Aldrich supplied all solvents and standards in LC-MS grade.
Protein Expression and Purification of Recombinant JASSY.
The coding sequence of JASSY was cloned into pET51b+. Soluble JASSY protein used for MST, electrophysiology, and carboxyfluorescein assays was expressed overnight at 18 °C in E. coli in Rosetta 2(DE3) cells, and the protein was purified with Ni-NTA and Strep-Tactin (IBA Lifesciences).
Generation of Liposomes for MST and Carboxyfluorescein Assay.
Here 20 mg of phosphatidylcholine lipids (Larodan Fine Chemicals) was were washed with chloroform/methanol (1:1) and dried under N2. For MST analysis, dried liposomes were resuspended in an MST-suitable buffer (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, and 0.05% Tween-20) to a concentration of 20 mg/mL. Lipids were subjected five freeze/thaw cycles, and the mixed lamellar liposomes were incubated with JASSY (1 mg/mL) at a 1:1 ratio for 1.5 h at 4 °C under agitation.
For the carboxyfluorescein assay, the lipids were resuspended in 1× PBS, pH 7.4 to a final concentration of 20 mg/mL, and 20 mM of carboxyfluorescein was added to 20 mg/mL PC lipids. Subsequently, lipids were subjected to five freeze/thaw cycles, and the mixed lamellar liposomes were extruded through a membrane with 200-nm pore size to generate unilamellar vesicles. To remove nonencapsulated dye, the liposomes were dialyzed against 1× PBS buffer, pH 7.4 overnight at 4 °C.
Microscale Thermophoresis.
Soluble JASSY protein was diluted in MST buffer (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, and 0.05% Tween-20) to 200 nM. JASSY proteoliposomes were diluted to 2.4 µM in the same buffer. Tris-NTA dyes were diluted in PBST buffer (137 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 pH 7.4, and 0.05% Tween-20) to a final concentration of 100 nM. Then 100 µL of protein was mixed with 100 µL of each dye separately, and the reaction mixtures were then incubated for 30 min at room temperature in the dark. Final concentrations of pure JASSY protein and the JASSY proteoliposomes were 50 nM and 600 nM, respectively. The concentrations of OPDA and JA varied between 30 nM and 1 mM. Reactions were incubated for 10 min at room temperature and then loaded into standard Monolith NT.115 glass capillaries (NanoTemper).
Carboxyfluorescein Assay.
For fluorescence measurements, 5 μL of liposomes were mixed with 995 μL of 1× PBS pH 7.4 to generate a suitable fluorescent signal. After the addition of purified proteins, fluorescence was recorded every millisecond for 300 or 600 s with a PerkinElmer LS55 fluorescence spectrometer with an excitation wavelength of 494 nm and an emission wavelength of 515 nm.
Electrophysiology and Liposome Preparation.
To prepare the samples for electrophysiological characterization, l-a-phosphatidylcholine and l-a-phosphatidylethanolamine (both from Avanti Polar Lipids) were mixed at an 80:20 molar ratio, dried under nitrogen, then desiccated under vacuum. The dried lipids were fully resuspended in liposomes buffer (150 mM NaCl and 50 mM Tris/HCl, pH 7.5) to 5 mg/mL and subjected to seven freeze/thaw cycles before being extruded through a 200-nm filter. The liposomes and purified recombinant JASSY, or the identical treated mock expression using an empty plasmid, were solubilized with 80 mM of the dialyzable detergent MEGA-9 (Glycon) separately for 15 min at room temperature. Both parts were pooled and incubated for another 30 min at room temperature with final lipid and protein concentrations of 1.5 mg/mL and 0.2 mg/mL, respectively. The mixture was dialyzed in a 3.5-kDa cutoff dialysis tube against 5 L of liposome buffer to remove detergent, first for 2 h at room temperature and then overnight at 4 °C. The success of incorporation was monitored by density gradient flotation assay and sodium carbonate extraction as described previously (62).
The electrophysiological experiments were performed using the planar lipid bilayer technique as described previously (63). In brief, liposomes with incorporated JASSY were added below the lipid bilayer to allow osmotically driven fusion in the cis chamber (250 mM KCl and 10 mM Mops/Tris, pH 7), while the trans chamber contained a low salt buffer (20 mM KCl and 10 mM Mops/Tris, pH 7). After insertion of a channel into the bilayer, buffers in both chambers (cis and trans) were perfused to symmetrical standard buffer conditions using 20× chamber volumes. For determination of the reversal potential, asymmetric buffer conditions as used for liposome fusion were applied. Electrical recordings were performed using Ag/AgCl electrodes in a 2 mM KCl agar bridge, connected to a GeneClamp 500B amplifier via a CV-5–1GU headstage (Molecular Devices), with the cis electrode connected to the ground. The signal was digitized by a Digidata 1440a A/D converter and recorded with AxoScope 10.3 and Clampex 10.3 software (Molecular Devices). Automated data analysis was performed as described previously (30). To determine substrate effects, OPDA in ethanol, with corresponding amounts of ethanol or JA in ethanol were added to both sides of bilayer-incorporated JASSY channels. The buffer in both chambers was circulated by stirring for 2 min after addition, then left alone for another 2 min before recording.
Computational Analyses.
Sequences were obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). Trees were generated by using CLC Main Workbench software (Qiagen). Alignments were generated using the algorithm provided by CLC Main Workbench (developed by Qiagen). Graphs and statistical analyses were generated using GraphPad Prism version 6.0 (www.graphpad.com).
Supplementary Material
Acknowledgments
We thank Tamara Hechtl for excellent technical assistance and Annabel Mechela for help with Arabidopsis envelope preparations. This work was supported by the Chinese Scholarship Program (L.G.) and the German Research Foundation (SFB-TR 175 Projects B05 and B06 to J.S. and S.S.; SFB 1190 Project P12 to M.M.; and FOR1905 Project P01 to N.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900482116/-/DCSupplemental.
References
- 1.Wasternack C. (2007) Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100:681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wasternack C, Hause B (2013) Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaller A, Stintzi A (2009) Enzymes in jasmonate biosynthesis: Structure, function, regulation. Phytochemistry 70:1532–1538. [DOI] [PubMed] [Google Scholar]
- 4.Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92:8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53:275–297. [DOI] [PubMed] [Google Scholar]
- 6.Song WC, Brash AR (1991) Purification of an allene oxide synthase and identification of the enzyme as a cytochrome P-450. Science 253:781–784. [DOI] [PubMed] [Google Scholar]
- 7.Hamberg M, Fahlstadius P (1990) Allene oxide cyclase: A new enzyme in plant lipid metabolism. Arch Biochem Biophys 276:518–526. [DOI] [PubMed] [Google Scholar]
- 8.Stelmach BA, et al. (2001) A novel class of oxylipins, sn1-O-(12-oxophytodienoyl)-sn2-O-(hexadecatrienoyl)-monogalactosyl diglyceride, from Arabidopsis thaliana. J Biol Chem 276:12832–12838. [DOI] [PubMed] [Google Scholar]
- 9.Hisamatsu Y, Goto N, Sekiguchi M, Hasegawa K, Shigemori H (2005) Oxylipins arabidopsides C and D from Arabidopsis thaliana. J Nat Prod 68:600–603. [DOI] [PubMed] [Google Scholar]
- 10.Genva D, et al. (2018) New insights into the biosynthesis of esterified oxylipins and their involvement in plant defense and developmental mechanisms. Phytochem Rev 18:343–358. [Google Scholar]
- 11.Nyathi Y, et al. (2010) The Arabidopsis peroxisomal ABC transporter, comatose, complements the Saccharomyces cerevisiae pxa1 pxa2Delta mutant for metabolism of long-chain fatty acids and exhibits fatty acyl-CoA–stimulated ATPase activity. J Biol Chem 285:29892–29902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theodoulou FL, et al. (2005) Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants: Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol 137:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye ZW, et al. (2016) Arabidopsis acyl-CoA–binding protein ACBP6 localizes in the phloem and affects jasmonate composition. Plant Mol Biol 92:717–730. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, et al. (2017) Transporter-mediated nuclear entry of jasmonoyl-isoleucine is essential for jasmonate signaling. Mol Plant 10:695–708. [DOI] [PubMed] [Google Scholar]
- 15.Chung HS, et al. (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146:952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16:1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wasternack C, Song S (2017) Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J Exp Bot 68:1303–1321. [DOI] [PubMed] [Google Scholar]
- 18.Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451. [DOI] [PubMed] [Google Scholar]
- 19.Sharma M, Laxmi A (2016) Jasmonates: Emerging players in controlling temperature stress tolerance. Front Plant Sci 6:1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, et al. (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J Exp Bot 68:1361–1369. [DOI] [PubMed] [Google Scholar]
- 21.Simm S, et al. (2013) Defining the core proteome of the chloroplast envelope membranes. Front Plant Sci 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25:2907–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521. [DOI] [PubMed] [Google Scholar]
- 24.Kazan K, Manners JM (2013) MYC2: The master in action. Mol Plant 6:686–703. [DOI] [PubMed] [Google Scholar]
- 25.Meinecke M, Bartsch P, Wagner R (2016) Peroxisomal protein import pores. Biochim Biophys Acta 1863:821–827. [DOI] [PubMed] [Google Scholar]
- 26.Krüger V, et al. (2012) The mitochondrial oxidase assembly protein1 (Oxa1) insertase forms a membrane pore in lipid bilayers. J Biol Chem 287:33314–33326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domańska G, et al. (2010) Helicobacter pylori VacA toxin/subunit p34: Targeting of an anion channel to the inner mitochondrial membrane. PLoS Pathog 10.1371/journal.ppat.1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colombini M. (1979) A candidate for the permeability pathway of the outer mitochondrial membrane. Nature 279:643–645. [DOI] [PubMed] [Google Scholar]
- 29.Schrempf H, et al. (1995) A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J 14:5170–5178.
- 30.Denkert N, et al. (2017) Cation selectivity of the presequence translocase channel Tim23 is crucial for efficient protein import. eLife 6:e28324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Im W, Roux B (2002) Ion permeation and selectivity of OmpF porin: A theoretical study based on molecular dynamics, Brownian dynamics, and continuum electrodiffusion theory. J Mol Biol 27:851–869. [DOI] [PubMed] [Google Scholar]
- 32.Footitt S, et al. (2007) The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol 144:1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bussell JD, Reichelt M, Wiszniewski AA, Gershenzon J, Smith SM (2014) Peroxisomal ATP-binding cassette transporter COMATOSE and the multifunctional protein abnormal INFLORESCENCE MERISTEM are required for the production of benzoylated metabolites in Arabidopsis seeds. Plant Physiol 164:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Routaboul JM, Fischer SF, Browse J (2000) Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol 124:1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijayan P, Browse J (2002) Photoinhibition in mutants of Arabidopsis deficient in thylakoid unsaturation. Plant Physiol 129:876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13:2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JH, et al. (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31:1–12. [DOI] [PubMed] [Google Scholar]
- 38.Sanders PM, et al. (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12:1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6:751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radauer C, Lackner P, Breiteneder H (2008) The Bet v 1 fold: An ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol 8:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo AJ, Ohlrogge JB, Pollard M (2004) On the export of fatty acids from the chloroplast. J Biol Chem 279:16101–16110. [DOI] [PubMed] [Google Scholar]
- 43.Kienow L, et al. (2008) Jasmonates meet fatty acids: Functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J Exp Bot 59:403–419. [DOI] [PubMed] [Google Scholar]
- 44.Han GZ. (2017) Evolution of jasmonate biosynthesis and signaling mechanisms. J Exp Bot 68:1323–1331. [DOI] [PubMed] [Google Scholar]
- 45.Záveská Drábková L, Dobrev PI, Motyka V (2015) Phytohormone profiling across the bryophytes. PLoS One 10:e0125411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponce De León I, et al. (2012) Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol Plant Pathol 13:960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stumpe M, et al. (2010) The moss Physcomitrella patens contains cyclopentenones but no jasmonates: Mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol 188:740–749. [DOI] [PubMed] [Google Scholar]
- 48.Glauser G, Wolfender JL (2013) A non-targeted approach for extended liquid chromatography-mass spectrometry profiling of free and esterified jasmonates after wounding. Methods Mol Biol 1011:123–134. [DOI] [PubMed] [Google Scholar]
- 49.Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. [DOI] [PubMed] [Google Scholar]
- 50.Weiberg A, et al. (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweiger R, Müller NC, Schmitt MJ, Soll J, Schwenkert S (2012) AtTPR7 is a chaperone-docking protein of the Sec translocon in Arabidopsis. J Cell Sci 125:5196–5207. [DOI] [PubMed] [Google Scholar]
- 52.Koop HU, et al. (1996) Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta 199:193–201. [DOI] [PubMed] [Google Scholar]
- 53.Waegemann K, Soll J (1995) Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol 50:255–267. [DOI] [PubMed] [Google Scholar]
- 54.Waegemann K, Soll J (1991) Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J 1:149–158. [Google Scholar]
- 55.Gerdes L, et al. (2006) A second thylakoid membrane-localized Alb3/OxaI/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. J Biol Chem 281:16632–16642. [DOI] [PubMed] [Google Scholar]
- 56.Lübeck J, Soll J, Akita M, Nielsen E, Keegstra K (1996) Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J 15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- 57.Benz JP, et al. (2009) Arabidopsis Tic62 and ferredoxin-NADP(H) oxidoreductase form light-regulated complexes that are integrated into the chloroplast redox poise. Plant Cell 21:3965–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goetze TA, Philippar K, Ilkavets I, Soll J, Wagner R (2006) OEP37 is a new member of the chloroplast outer membrane ion channels. J Biol Chem 281:17989–17998. [DOI] [PubMed] [Google Scholar]
- 59.Seedorf M, Soll J (1995) Copper chloride, an inhibitor of protein import into chloroplasts. FEBS Lett 367:19–22. [DOI] [PubMed] [Google Scholar]
- 60.Duy D, et al. (2007) PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 19:986–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamberti G, Gügel IL, Meurer J, Soll J, Schwenkert S (2011) The cytosolic kinases STY8, STY17, and STY46 are involved in chloroplast differentiation in Arabidopsis. Plant Physiol 157:70–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarasenko D, et al. (2017) The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology. J Cell Biol 216:889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Montilla-Martinez M, et al. (2015) Distinct pores for peroxisomal import of PTS1 and PTS2 proteins. Cell Rep 13:2126–2134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.