Significance
Here we have directly visualized conformational changes in the 5′UTR of the HIV-1 genome using single-molecule fluorescence techniques. We find that the monomeric 5′UTR can spontaneously transition between two conformations, which have distinct intramolecular base pairing. One of the observed conformations is competent for dimerization with a second 5′UTR molecule. Our results are consistent with a model in which dimerization initiates by way of localized intermolecular kissing-loop base pairing, which is promoted by tRNA primer annealing. The intermolecular interface then extends, giving rise to the putative extended dimer, which is stabilized by HIV-1 NC. Thus, the 5′UTR is intrinsically dynamic, and both viral and host factors play a role in modulating the RNA conformation and dynamics.
Keywords: HIV-1, 5′ untranslated region, RNA conformational dynamics, single-molecule FRET, in-gel FRET
Abstract
The highly conserved 5′ untranslated region (5′UTR) of the HIV-1 RNA genome is central to the regulation of virus replication. NMR and biochemical experiments support a model in which the 5′UTR can transition between at least two conformational states. In one state the genome remains a monomer, as the palindromic dimerization initiation site (DIS) is sequestered via base pairing to upstream sequences. In the second state, the DIS is exposed, and the genome is competent for kissing loop dimerization and packaging into assembling virions where an extended dimer is formed. According to this model the conformation of the 5′UTR determines the fate of the genome. In this work, the dynamics of this proposed conformational switch and the factors that regulate it were probed using multiple single-molecule and in-gel ensemble FRET assays. Our results show that the HIV-1 5′UTR intrinsically samples conformations that are stabilized by both viral and host factor binding. Annealing of tRNALys3, the primer for initiation of reverse transcription, can promote the kissing dimer but not the extended dimer. In contrast, HIV-1 nucleocapsid (NC) promotes formation of the extended dimer in both the absence and presence of tRNALys3. Our data are consistent with an ordered series of events that involves primer annealing, genome dimerization, and virion assembly.
The 5′ untranslated region (5′UTR) of the HIV-1 RNA genome is a structured and highly conserved sequence that regulates diverse processes during the viral replication cycle, including RNA splicing, translation initiation, genome dimerization and packaging into assembling virions, and initiation of reverse transcription (1). HIV-1 assembly occurs at the plasma membrane of infected cells where the viral Gag and Gag-Pol polyproteins accumulate. The nucleocapsid (NC) domain of Gag contains a pair of zinc-knuckle motifs that select the HIV-1 genome during virion assembly by way of specific interactions with the packaging signal (Psi) located in the 5′UTR (2, 3). Packaging of two copies of the genome is required to form an infectious virion (4). Mutations that decrease dimerization of the genome reduce the packaging of genome into virions (5, 6). Genome dimerization is proposed to be regulated by a conformational switch in the 5′UTR that exposes the dimerization initiation site (DIS) located within Psi (7) (Fig. 1A). In the dimerization-incompetent monomeric form of the 5′UTR, the U5 region pairs with and sequesters the DIS. Alternatively, the U5 region can pair with a stem loop containing the AUG Gag start codon, exposing the DIS and forming the dimerization-competent conformation (Fig. 1A). The ability to adopt complex and dynamic structures allows individual RNA molecules to regulate distinct processes during virus replication. In this way, the HIV-1 5′UTR can switch between multiple roles by changing its conformation (2, 3). A variety of biochemical and structural studies suggest that the 5′UTR is on a structural tipping point, able to readily adopt different conformations with the potential to regulate function (8–10). Following initiation of dimerization by intermolecular kissing loop base-pairing between the palindromic DIS sequences from two genome molecules, further structural rearrangements have been proposed that lead to more extensive intermolecular base-pairing, forming the extended dimer conformation of the 5′UTR (Fig. 1A) (5, 11).
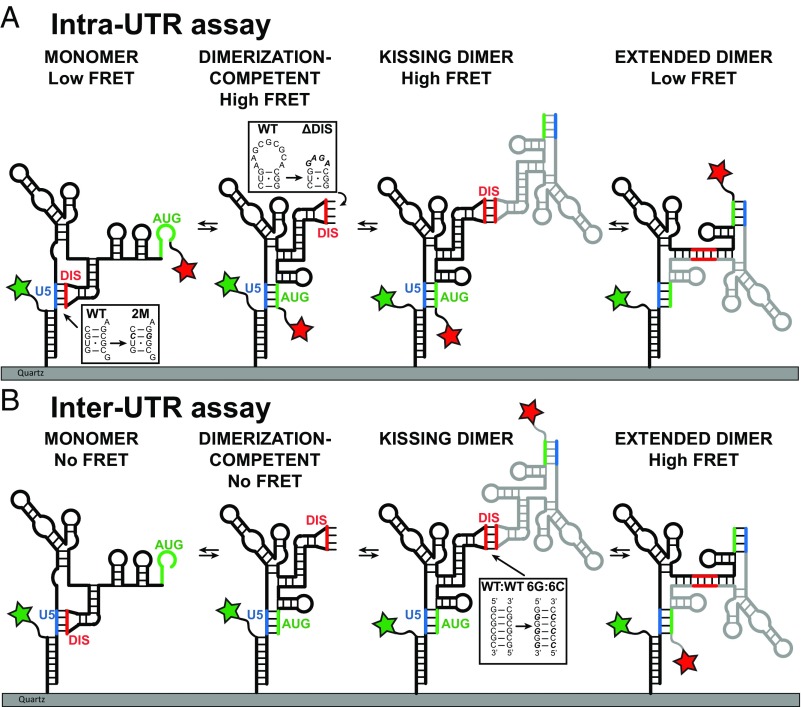
Fig. 1.
Experimental design of smFRET imaging assays to report on the conformation and dimerization of the HIV-1 5′UTR. (A) Secondary structural model of surface-immobilized HIV-1 5′ UTR construct. The Intra-UTR assay reports on the conformation of the immobilized FH238 molecule. The donor fluorophore (green star) is attached to FH.Oligo, which is annealed to FH238, and the acceptor fluorophore (red start) is attached directly to the 3′ end of FH238. Insets indicate the locations and sequences of the 2M and ΔDIS mutations. (B) The Inter-UTR assay reports on dimerization and the conformation of the dimerized complex. The donor fluorophore (green star) is attached to FH.Oligo, which is annealed to FH238, and the acceptor fluorophore is attached to the 3′ end of the free 238 molecule. Inset indicates the location and sequence of 6G and 6C mutations. In both assays, known and putative conformations were predicted to yield the indicated FRET states.
Host tRNALys3 serves as the primer for initiation of reverse transcription by annealing to the HIV-1 genome at the primer binding site (PBS) within the 5′UTR. Human tRNALys3 is known to be selectively incorporated into the virus, along with the other major isoacceptor, tRNALys1,2 (12, 13). The PBS is located just upstream of the DIS, but the timing of tRNALys3 annealing with respect to genome dimerization remains unknown. NMR studies support the presence of at least two mutually exclusive 5′UTR conformational states (3), and in-gel structure-probing of the WT 5′UTR revealed a mixed population with structural differences between monomer and dimer, consistent with a structural switch (14). However, these studies did not address the dynamics of the conformational change or probe possible triggers such as tRNA annealing or viral factor binding. Cell-based assays showed that genomic RNA (gRNA) lacking the PBS sequence is packaged into virions 60% less than WT gRNA, consistent with a possible role for tRNA annealing in gRNA packaging (15). Indeed, annealing of antisense oligonucleotides complementary to the PBS (cPBS18) and other regions of the 5′UTR promoted the DIS-exposed conformer (15–17). Annealing of tRNALys3 was also reported to stimulate dimerization, albeit not as efficiently as cPBS18 (15).
Here we sought to elucidate the putative conformational dynamics of the HIV-1 5′UTR and probe the role of host and viral factors in modulating those dynamics. To this end, we developed single-molecule Förster resonance energy transfer (smFRET) imaging assays, coupled with ensemble in-gel FRET measurements that report explicitly on the conformational changes in the 5′UTR that relate to dimerization. We find that the monomeric 5′UTR spontaneously interconverts between the dimerization-competent and dimerization-incompetent forms, which is mediated by alternative base pairing between U5 and DIS or AUG. Our observations are also consistent with formation of an extended dimer that forms following an initial kissing interaction between the DIS of two 5′UTR molecules, and which is facilitated by NC. We show that tRNALys3 annealing to the PBS occurs on the monomeric 5′UTR and that tRNA annealing can promote the kissing dimer but not the extended dimer. The kissing dimer is then converted to the extended dimer by NC. This suggests an ordered series of events that involves primer annealing, genome dimerization, and virion assembly.
Results
smFRET Imaging Demonstrates That the 5′UTR Spontaneously Transitions Between Multiple Conformational States.
To probe the conformational changes of the 5′UTR related to genome dimerization we established single-molecule and bulk FRET imaging assays focused on a 238-nucleotide (nt) RNA segment (238.WT), which contains the entire 5′UTR except for the transactivation response element (TAR) and PolyA stem loops, which are dispensable for 5′UTR dimerization and packaging into assembling virions (18). Our approach required site-specific attachment of donor and acceptor fluorophores to the 5′UTR construct (Materials and Methods). We extended the 5′ end of the 5′UTR construct with a FRET handle (FH) sequence, forming FH238.WT, to facilitate annealing to a cDNA handle (FH.Oligo), which was fluorescently labeled at its 5′ end. Annealing a 3′-biotinylated DNA handle to the FH238.WT construct allowed tethering of the construct to a streptavidin-coated quartz surface for time-resolved imaging of donor and acceptor fluorescence via total internal reflection fluorescence (TIRF) microscopy, as required for smFRET imaging (Fig. 1A). The second fluorophore was chemically linked to the 3′ end of the 5′UTR through standard means (19). Native gel analyses confirmed that the 5′ extension to FH238.WT, annealing of the FH.Oligo, and the attachment of a fluorophore to the 3′ end did not impair the dimerization or native gel migration patterns of the RNA constructs (Fig. 2A and SI Appendix, Fig. S1 A and B).
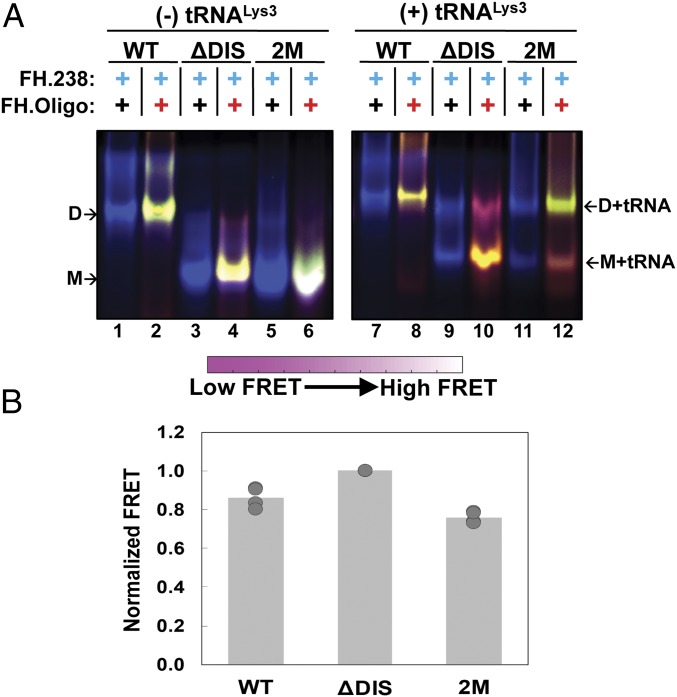
Fig. 2.
In-gel Inter-UTR FRET assay and effect of tRNALys,3 heat annealing. (A) In-gel FRET of 5′UTR constructs as indicated above each lane. Lanes 1–6 do not contain tRNALys3, while lanes 7–12 contain tRNALys3-bound 5′UTR. The locations of the monomer (M), tRNALys3-bound monomer (M + tRNA), dimer (D), and tRNALys3-bound dimer (D + tRNA) bands are indicated. In all panels, labeling schemes are indicated above each lane (black +, unlabeled RNA; blue +, donor-labeled RNA; red +, acceptor-labeled RNA). False color overlays of individual fluorescent images are shown, as described in Materials and Methods. The color bars indicate how the range of colors corresponds qualitatively to in-gel FRET efficiency. All gels were run in the presence of 10 mM Mg2+ in the running buffer and gel matrix. The data shown are from a single gel where the gap represents lanes that were removed as they are unrelated to this experiment. (B) Quantification of in-gel FRET shown in A. The major species in lanes 2, 4, and 6 were quantified as described in Materials and Methods and normalized to FH238.ΔDIS, which was set to a FRET efficiency of 1.0. Results are the average of four independent experiments. In each case, data from the four trials are shown with the mean value indicated by the height of the bar.
Based on previously published models, we predicted that the dimer-competent monomer conformation with the DIS exposed would generate a high-FRET state, while the dimer-incompetent conformation in which the DIS is sequestered through base pairing with U5 would generate a low-FRET state (5, 20–22) (Fig. 1A). To test this prediction, in addition to FH238.WT, which is capable of dimerizing, two monomeric mutant RNA constructs were designed. In one construct the DIS loop was replaced with a GAGA tetraloop (FH238.ΔDIS). This construct cannot form the U5:DIS interaction and is expected to stably adopt the DIS-exposed conformer (22). A second construct (FH238.2M) contains two point mutations, G108C in U5 and C258G in DIS, that eliminate two base-pairing interactions in the palindromic DIS but still allow the U5:DIS interaction to occur with a similar stability as WT, through the compensatory mutation in the U5 region. The G108C mutation is also predicted to destabilize the U5:AUG interaction by changing a G:U base pair to a C:U base pair. Native PAGE analysis confirmed that both mutant constructs were monomeric, as expected (Fig. 2A and SI Appendix, Fig. S1 A and B).
To evaluate the conformation of the monomeric 5′UTR constructs we then performed in-gel FRET. FH238.WT displayed a FRET efficiency that was slightly higher than that of the 2M construct, consistent with the 2M construct favoring the dimer-incompetent conformation (Fig. 2B). However, the monomeric ΔDIS construct displayed 25–30% higher FRET than the WT or 2M constructs. This result is consistent with a shift in the conformational equilibrium toward the DIS-exposed high-FRET state for FH238.ΔDIS.
We next asked whether the 5′UTR constructs could transition between the DIS-exposed and DIS-sequestered conformations. To this end, we applied smFRET imaging to the surface-immobilized FH238.WT, FH238.ΔDIS, and FH238.2M constructs in the presence of 10 mM MgCl2 (Fig. 1A, Intra-UTR assay). The majority (66%) of the smFRET trajectories acquired for the FH238.WT construct indicated only a stable low- (0.11 ± 0.04) or high-FRET state (0.96 ± 0.05) (Fig. 3A), with no transitions in FRET observed. Owing to the slow timescale of dynamics, only 28% of the traces indicated transitions between the high- and low-FRET states. Six percent of the traces exhibited intermediate-FRET states; this subpopulation was not responsive to the experimental variables investigated here. The FH238.ΔDIS construct showed increased occupancy in the high-FRET state compared with FH238.WT (Fig. 3B), consistent with in-gel FRET results and the promotion of the DIS-exposed conformation by the ΔDIS construct. Conversely, the 2M mutant showed slightly increased occupancy in the low-FRET state and resulted in a histogram that was more similar to WT than to the ΔDIS construct (Fig. 3C), consistent with the in-gel FRET results. This is consistent with the 2M mutations destabilizing the U5:AUG interaction but not affecting the stability of U5-DIS base-pairing. These data indicate that the low-FRET state reflects the DIS-sequestered, dimerization-incompetent conformation, and the high-FRET state reflects the DIS-exposed, dimerization-competent conformation.
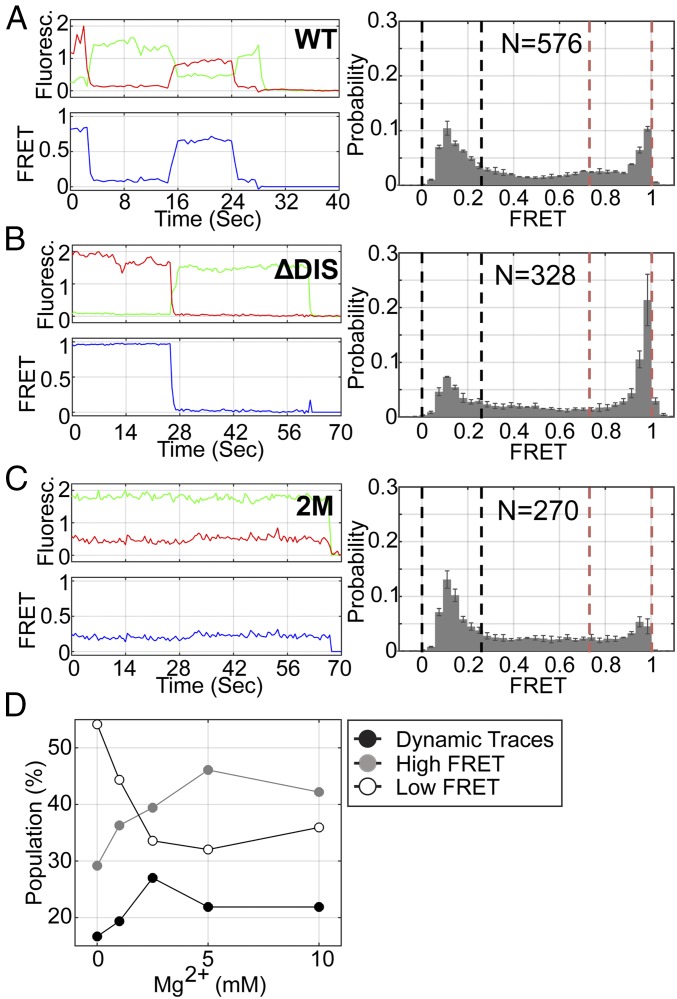
Fig. 3.
The 5′UTR transitions between monomeric and dimerization-competent conformations. (A) (Left) Fluorescence (green, donor; red, acceptor) and FRET (blue) trajectories acquired from an individual FH238.WT molecule. (Right) Population FRET histogram indicating that FH238.WT occupies a mixture of high- and low-FRET states. Black and red dashed lines indicate the upper and lower bound of low- and high-FRET SD, respectively. The number of FRET trajectories compiled into the histogram is indicated (N). The same data for the (B) FH238.ΔDIS and (C) FH238.2M constructs. (D) Fraction of the population of FH238.WT molecules displaying stable high-FRET (gray), stable low-FRET (open), and dynamic (black) traces as a function of MgCl2 concentration.
To provide further evidence that the high-FRET state corresponds to the DIS-exposed conformation, we varied the Mg2+ concentration since high Mg2+ is known to promote dimerization in gel-shift, calorimetry, and NMR experiments (20, 21, 23, 24). The majority of the smFRET trajectories acquired from individual FH238.WT molecules (ca. 54%) in the absence of Mg2+ indicated a predominant low-FRET state consistent with our expectations of the DIS-sequestered conformation (Fig. 3D and SI Appendix, Fig. S2A). Approximately 29% indicated the stable high-FRET state, and 17% indicated transitions between the two states. As expected, increasing the Mg2+ concentration led to stabilization of the high-FRET state and an increase in the number of molecules displaying only high FRET (Fig. 3D and SI Appendix, Fig. S2A). Kinetic analysis of the traces demonstrating dynamics indicated that the lifetime of the low-FRET state decreased from 16 s in the absence of Mg2+ to 12 s at 10 mM Mg2+ (SI Appendix, Fig. S2B). Interestingly, the lifetime of the high-FRET state was insensitive to Mg2+. Taken together, these data are consistent with the monomeric 5′UTR spontaneously transitioning between DIS-sequestered and DIS-exposed conformations. Furthermore, consistent with previous reports (5), these dynamics stem from sequestration of the DIS through labile base-pairing with U5. The presence of Mg2+ shifts the dynamic equilibrium in favor of the DIS-exposed conformation with maximum dynamics observed near physiological concentrations of 2.5 mM Mg2+ (Fig. 3D).
FRET Assays Are Consistent with Different Dimerization States of the 5′UTR.
Given our observation of reversible exchange of the monomeric 5′UTR between DIS-exposed and DIS-sequestered conformations, we next asked if dimerization stabilizes the DIS-exposed conformation or induces further rearrangements. To this end, we incubated the labeled FH238.WT with unlabeled 238.WT that lacked the handle-annealing sequence in the presence of 10 mM Mg2+, followed by evaluation by smFRET imaging. The unlabeled 5′UTR without the handle sequence is incapable of binding to the surface and acts as a dimerization partner for the surface-bound labeled 5′UTR (Fig. 1A, Intra-UTR assay). Surprisingly, we observed a predominant low-FRET state, indistinguishable from the state observed for the DIS-sequestered, dimerization-incompetent monomer (Fig. 4A, Left). However, analysis by native gel electrophoresis indicated that the 5′UTR had dimerized (Fig. 2A). Secondary structural models suggest that formation of an extended dimer might generate a low-FRET state in the current assay (Fig. 1A). We cannot rule out the possibility that other dimeric conformations also exist that contribute to the low-FRET state in this assay.
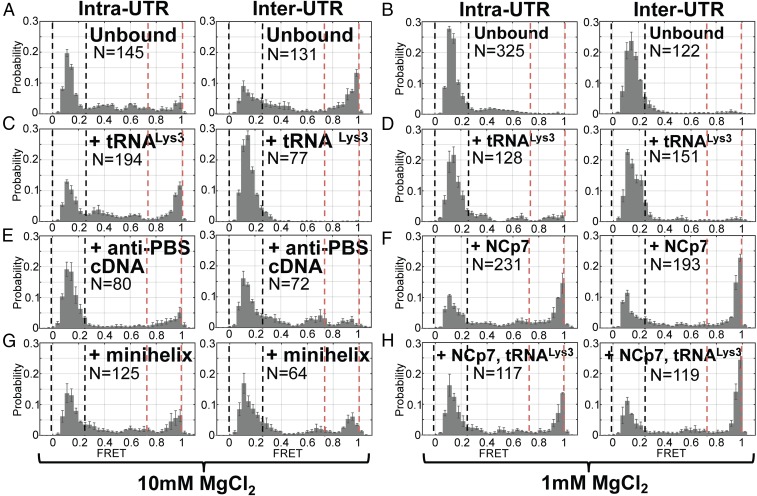
Fig. 4.
After tRNALys3 annealing, NCp7 is required for transition to the extended dimer conformation. Population FRET histograms from FH238.WT dimerized with 238.WT and imaged in 10 mM (A, C, E, and G) or 1 mM (B, D, F, and H) MgCl2 using either the Intra-UTR (Left) or Inter-UTR FRET (Right) assay (Fig. 1), as indicated above for each panel set. Black and red dashed lines indicate the upper and lower bound of low- and high-FRET SD, respectively. Before imaging in both MgCl2 concentrations, the FW238.WT complex was annealed to tRNALys3 (C and D), the 18-nt anti-PBS cDNA (E), or the tRNALys3-derived acceptor-TΨC minihelix (G). Alternatively, the FH238.WT complex was incubated with NCp7 (F) or NCp7 and tRNALys3 (H), followed by removal of NCp7 with proteinase K digestion before imaging (Materials and Methods). The number of FRET trajectories compiled into each histogram is indicated (N).
To further interrogate the conformation of the dimerized 5′UTR, we designed a second smFRET assay that we anticipated would distinguish between the extended dimer and additional dimer conformations. We first annealed an unlabeled FH238.WT to a labeled FH.Oligo. We then allowed this complex to dimerize with a labeled 238.WT that lacked the handle annealing sequence (Fig. 1B, Inter-UTR assay). In this assay, FRET was only observed when a surface-bound unlabeled FH238.WT interacts with a labeled 238.WT; this assay is insensitive to conformational changes of the monomer. With this assay, we predicted that the extended dimer conformation would bring the fluorophores into close proximity, giving rise to a high-FRET state, while other dimer conformations would yield lower-FRET states. Consistent with this expectation, when FH238.WT:238.WT dimers were formed in the presence of 10 mM Mg2+ the smFRET trajectories indicated a predominant high-FRET state, with significant occupancy in a low-FRET state (Fig. 4A, Right). When dimerization was performed at 1 mM Mg2+, only the low-FRET dimer species was detected (Fig. 4B). These data are consistent with the dimeric 5′UTR molecules adopting either extended dimer or alternative dimer conformations, which may include the kissing dimer. According to this interpretation, elevated Mg2+ stabilizes the high-FRET extended dimer conformation. These data further suggest that the predominant low-FRET state observed in the Intra-UTR assay reflects, at least in part, formation of the extended dimer.
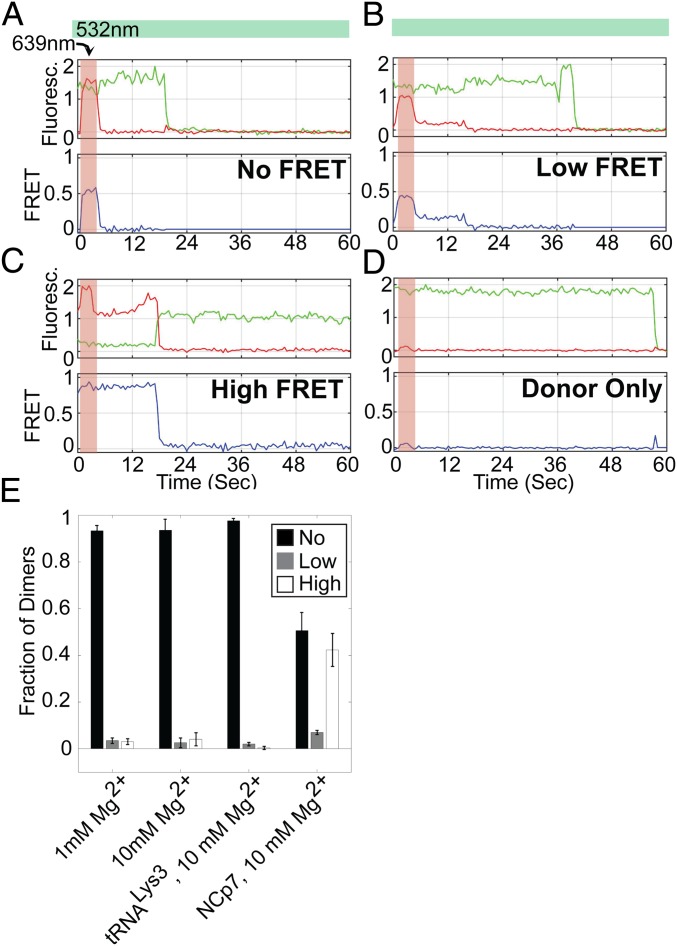
We next sought to determine whether all dimeric 5′UTR species had been detected in the Inter-UTR assay or whether a subpopulation might not be generating a detectable FRET signal due to the interfluorophore distance remaining too large. To this end, we conducted a variation of the Inter-UTR assay in which the acceptor fluorophore was transiently excited directly by a laser in addition to excitation of the donor fluorophore. This provided a direct means of detecting the presence of both fluorophores in individual 5′UTR dimers, even in the absence of a detectable FRET signal. We thus determined the number of 5′UTR dimers that yielded high-, low-, and 0-FRET states (Fig. 5 A–D). Surprisingly, regardless of whether dimers were formed in the presence of 1 or 10 mM Mg2+, the vast majority generated 0 FRET (Fig. 5E). Low- and high-FRET trajectories represented only 8.4–9.5% of the total population of dimers. Taken together, these data demonstrate that under the current conditions, dimerized 5′UTR can adopt multiple conformations, with the putative extended dimer reflecting only a small subpopulation and the remainder corresponding to alternative conformations, which likely include the kissing dimer. In a small subset of the observed molecules, transitions into and out of the extended dimer were seen, indicating that the 5′UTR is intrinsically capable of undergoing these conformational changes without interaction with a protein or RNA factor (SI Appendix, Fig. S3).
Fig. 5.
Direct excitation of both fluorophores in the Inter-UTR FRET assay verifies presence of No-FRET FH238 dimers. Dimerized FH238.WT complexes were imaged under direct excitation of the donor fluorophore (532 nm) throughout the observation window (indicated by the green bar) in addition to transient excitation of the acceptor fluorophore (639 nm; indicated by the red bar). Example fluorescence (donor, green; acceptor, red) and FRET (blue) traces of (A) a no-FRET dimer, (B) a low-FRET dimer, and (C) a high-FRET dimer. (D) Also shown is an example of a complex lacking the acceptor fluorophore, which indicates minimal increase in background fluorescence due to the 639-nm excitation. (E) Quantification of no-FRET (black), low-FRET (white), and high-FRET (gray) dimers under the different Mg2+ concentrations and in the presence of the indicated factors.
Native in-gel FRET was performed to further assess whether the kissing or extended dimer was preferentially formed during 5′UTR dimerization. To prevent homodimer formation between RNAs with identical fluorophores from convoluting the ensemble FRET measurements, we analyzed the dimerization of 238-nt 5′UTR constructs containing two complementary nonpalindromic DIS sequences. A 5′-extended FH238 containing a 6G DIS (FH238.6G) was incubated in the presence or absence of a nonhandled 238 containing a 6C DIS (238.6C) (Fig. 1B, boxed). Neither construct formed significant amounts of homodimer, but heterodimers readily formed (SI Appendix, Fig. S1C). Additionally, when both fluorophores were placed on the FH238.6G (analogous to the Intra-UTR assay), a high-FRET state was observed, consistent with the formation of a DIS-exposed monomer (SI Appendix, Fig. S1C, lane 3). Using this same labeling scheme, when the FH238.6G dimerized with 238.6C a high-FRET dimer was observed (SI Appendix, Fig. S1C, lane 7). In contrast, when labeling and dimerization were configured as in the Inter-UTR assay, a low-FRET dimer was observed (SI Appendix, Fig. S1C, lane 6). These results are consistent with the smFRET data and suggest that the predominant form of the 5′UTR dimer in the absence of other factors is not the extended dimer and likely includes the kissing dimer as well as other dimer conformations.
Annealing of tRNALys3 Reduces Formation of the Extended Dimer.
Previous experiments suggested that annealing of an anti-PBS DNA oligonucleotide or tRNALys3 to the 5′UTR increases dimerization (15). We therefore applied our FRET assays to determine whether annealing of an anti-PBS cDNA, a tRNALys3 acceptor-TΨC minihelix (25), or full-length tRNALys3 to the FH238.WT affected the conformational equilibrium described above. The FH238.WT was dimerized in the presence of 10 mM Mg2+ and one of the three complementary sequences, followed by imaging using the Intra- and Inter-UTR assays. In the Intra-UTR assay, tRNALys3 annealing led to an increase in the high-FRET state compared with unbound FH238.WT (Fig. 4C, Left), suggesting promotion of the DIS-exposed conformation and the kissing dimer. In the Inter-UTR assay, we observed no high FRET, suggesting that tRNALys3 annealing had inhibited transition to the extended dimer. Transient direct excitation of the acceptor confirmed again that most dimers existed in the 0-FRET conformation (Fig. 5E). Interestingly, the anti-PBS DNA oligonucleotide and tRNALys3 minihelix did not strongly promote the DIS-exposed conformation in the Intra-UTR assay but permitted limited occupancy in the high-FRET state in the Inter-UTR assay, which was not seen with tRNALys3 (Fig. 4 E and G). The unique behavior of the full-length tRNA suggests that interactions between tRNALys3 and the 5′UTR outside of the 18-nt PBS region are responsible for modulating the 5′UTR conformation.
Ensemble in-gel FRET assays were also performed in the presence of tRNALys3. Changes in FRET efficiency due to heat annealing of tRNALys3 to FH238.WT, FH238.ΔDIS, and FH238.2M were not quantified due to the limitations of the experiment; every molecule is doubly labeled preventing differentiation of dimeric FRET states in this case. Interestingly, FH238.2M RNA formed significant amounts of dimer only when annealed to tRNALys3 (Fig. 2A, lane 12). These data suggest that tRNA annealing promotes a DIS-exposed conformation and formation of the kissing dimer, even with a mutated DIS, but appears to prevent formation of the extended dimer.
Incubation with NCp7 Increases FRET States Consistent with Extended Dimer.
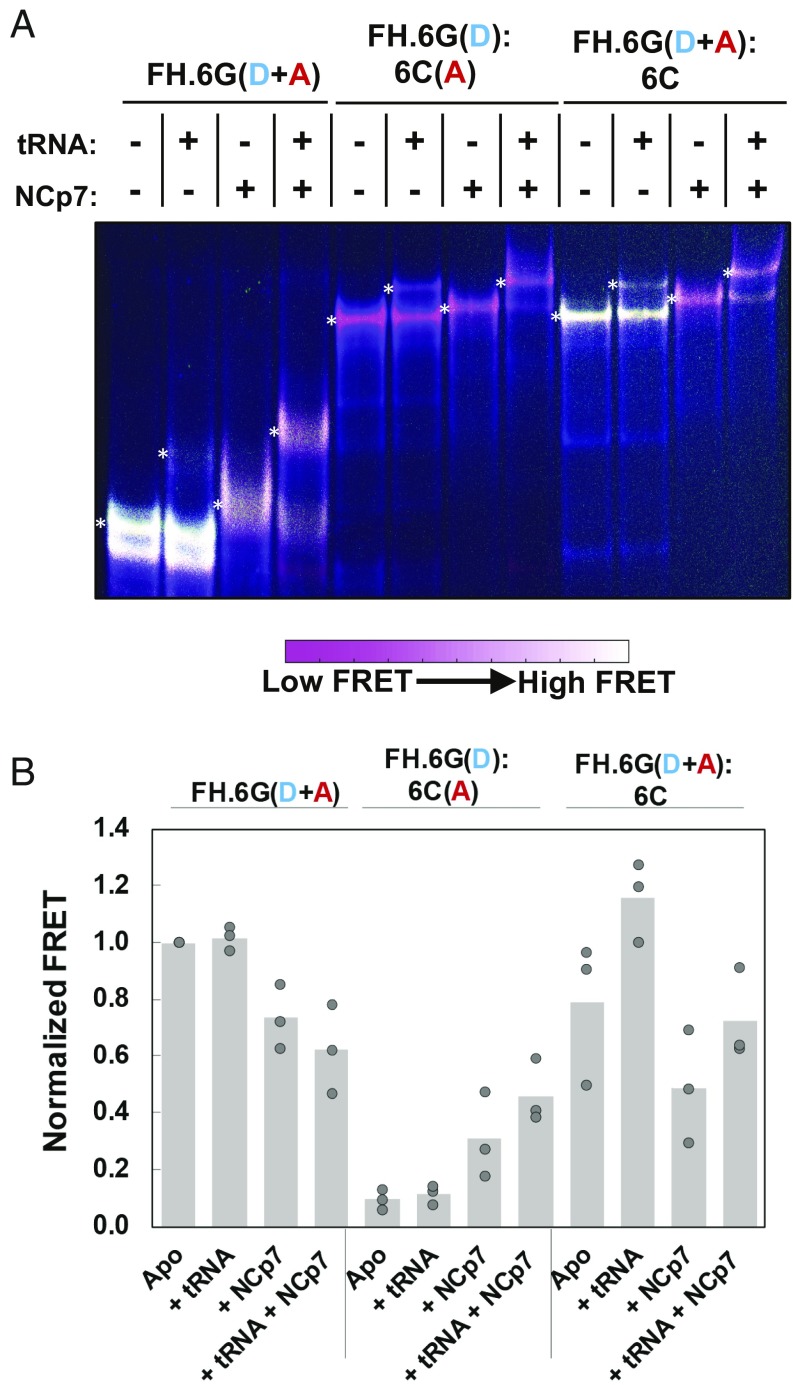
It is well established that the NC domain of Gag facilitates selective incorporation of a dimerized genome into assembling virions (26–29). However, the precise nature of the NCp7-bound 5′UTR dimer is not known. Previous fluorescence and NMR data suggest that NCp7 facilitates extension of the intermolecular base pairing following formation of the kissing dimer (30). We therefore used our FRET assays to better characterize the conformation of the 5′UTR following binding by NCp7. To this end, we applied our smFRET assays to probe the conformation of the 5′UTR that had been dimerized in the presence of NCp7. These experiments were only conducted at 1 mM Mg2+ since high Mg2+ inhibits NCp7 annealing activity (31). The Intra-UTR assay showed that dimerizing FH238.WT in the presence of NCp7 led to a mixture of low and high FRET, with significantly more high FRET than seen in the absence of NCp7 (Fig. 4F, Left). In-gel FRET analysis of FH238.6G and 6C 5′UTRs incubated with NCp7 confirmed that 5′UTR dimerization occurs efficiently in the presence of NCp7, suggesting that the smFRET signal represents a mixture of the extended dimer and alternate dimer conformations (Fig. 6A, compare lanes 7 and 11). Likewise, the Inter-UTR smFRET assay indicated a mixture of high and low FRET, further evidence of a mixture of dimer conformations (Fig. 4F, Right). Both monomeric 5′UTR and dimeric 5′UTR labeled according to the Intra-UTR assay experienced a decrease in FRET when incubated with NCp7 (Fig. 6A, compare lanes 1 and 3 and lanes 9 and 11, respectively), while dimeric 5′UTR labeled according to the Inter-UTR assay experienced a slight increase (Fig. 6A, compare lanes 5 and 7). These results are quantified in Fig. 6B. Transient direct excitation of the acceptor fluorophore in the Inter-UTR assay indicated that in contrast to experiments in the absence of NCp7, a significant proportion of the dimers generated a high-FRET state, and the proportion of molecules that demonstrated 0 FRET was reduced (Fig. 5E). These data support the hypothesis that NCp7 facilitates transition to the extended dimer, which is not formed in the absence of NCp7 at 1 mM Mg2+ (Fig. 4B).
Fig. 6.
In-gel FRET assay and effect of NCp7-mediated tRNALys,3 annealing. (A) Native 6% polyacrylamide gel electrophoresis of FH238.6G (FH.6G) and 238.6C (6C) HIV-1 5′UTR constructs. The complexes were either labeled according to Inter-UTR FRET (lanes 1–4 and 9–12) or Intra-UTR FRET (lanes 5–8). The gel was run in the presence of 1 mM Mg2+ in the running buffer and gel matrix. The presence (+) or absence (−) of both tRNALys,3 and NCp7 is shown above each lane. The locations of the monomer (M), tRNALys,3-bound monomer (M + tRNA), dimer (D), and tRNALys,3-bound dimer (D + tRNA) bands are indicated. The color bar indicates how the range of colors corresponds qualitatively to in-gel FRET efficiency. (B) Quantification of in-gel FRET shown in A, with the slowest migrating band quantified for each lane (indicated by a white asterisk to the left of the quantified band). The FRET signal is normalized to high-FRET monomeric FH238.6G in lane 1. In each case, data from the three trials are shown with the mean value indicated by the height of the bar.
We next asked if tRNALys3 annealing at 1 mM Mg2+ in the presence of NCp7 also led to dimerization and, if so, whether the extended dimer is readily formed. Both smFRET assays yielded results that were nearly identical to that observed with NCp7 in the absence of tRNALys3, indicating both the extended dimer and alternative dimer conformations (Fig. 4 F and H). In-gel FRET analysis indicated that tRNALys3 annealing to the dimer was observed in both the absence and presence of NCp7, but the extent of annealing was greater in the presence of NC (Fig. 6A, compare lanes 6 and 8 and lanes 10 and 12). NCp7-mediated annealing of tRNALys3 did not significantly alter FRET intensities compared with samples incubated with NCp7 in the absence of tRNALys3 (Fig. 6B). Thus, while tRNALys3 promotes a transition to the DIS-exposed conformation, NCp7 is required to efficiently transition to the extended dimer.
Discussion
The structure of the HIV-1 5′UTR has been extensively investigated. However, characterization of the 5′UTR structure is challenging due to the intrinsic heterogeneity of the RNA, which plays many regulatory roles during the viral lifecycle. Chemical and enzymatic probing has therefore yielded numerous models of the HIV-1 5′UTR secondary structure (28). In addition to monomeric states, various dimeric states have also been proposed to form, further increasing the complexity of structural possibilities. As dimeric gRNA is found within infectious HIV-1 virions, Gag-mediated gRNA packaging and virion assembly is dependent upon formation of a monomeric 5′UTR conformer wherein the DIS is exposed to potentiate gRNA dimerization through initial kissing dimer formation. Moreover, before reverse transcription initiation, tRNALys3 must be annealed to the PBS within the HIV-1 5′UTR. Although the exact timing of this event in the viral lifecycle is still poorly understood, it is well established that the nucleic acid chaperone activity of the NC domain of Gag facilitates tRNALys3 annealing (32–35) and that tRNA is found annealed to gRNA in HIV-1 protease-negative immature virions (36). Due to the close proximity between known NC binding sites, the PBS, and the DIS, which are all located in the 5′UTR, we hypothesized that NC and/or host tRNALys3 binding may serve to modulate the conformation and dynamics of the 5′UTR, shifting the equilibrium to a DIS-exposed state that is competent to dimerize.
Previous studies showing that the HIV-1 5′UTR can adopt multiple conformational states have not directly addressed the conformational dynamics of the full 5′UTR RNA or tested whether factor binding affects these dynamics. Previous smFRET analyses have monitored conformational changes in an ∼100-nt RNA template mimicking the PBS domain but lacking the Psi domain (37, 38). One study was conducted in the absence or presence of heat-annealed or HIV-1 NC-annealed tRNALys3 and concluded that NC reduced the dynamics of the tRNA-annealed complex and facilitates formation of an interaction between a previously identified primer activation signal (PAS) in the viral RNA and a complementary region in the tRNA TΨC arm (37, 39). A more recent study measured the effect of HIV-1 reverse transcriptase (RT) binding to the tRNA-annealed complex (38). This study concluded that the conformationally heterogeneous complex was stabilized upon RT binding. Here we have probed 5′UTR dynamics at the single-molecule level using a dimerization-competent viral RNA construct containing both the PBS and Psi domains. In the absence of bound factors, we observed transitions between the DIS-exposed and DIS-sequestered forms of the monomeric HIV-1 5′UTR. These results suggest that a model wherein full-length HIV-1 transcripts exist as a single pool of RNA that is capable of transitioning between alternative conformational and functional states is plausible. An alternative model has recently been proposed, wherein distinct populations of full-length HIV-1 RNA transcripts, presumably with different propensities to adopt different conformations, are destined for different fates depending on the transcription start site (23, 40). Indeed, separate gRNA pools have been observed in other retroviruses, such as MLV. Our constructs lack the 5′-TAR/polyA domains and do not directly address this alternative model (41, 42).
HIV-1 gRNA dimerization is a multistep process that regulates genome encapsidation, a critical step in infectious viral particle production (11, 43). Although gRNA is initially selected by Gag in the cytoplasm, recent studies suggest that dimerization occurs at the plasma membrane (44). Moreover, differences in gRNA dimer structure in protease-deficient viruses relative to mature virions suggest that the processed NC protein likely plays a role in formation of the mature gRNA extended dimer (45, 46). Whereas Gag performs the initial tRNALys3/PBS annealing reaction, mature NC is also required for final tRNALys3 primer placement and remodeling of the RNA complex (47, 48). Our results indicate that in the absence of other factors, dimeric states of the 5′UTR can form, but only a relatively small percentage of the RNAs are present as extended dimers. We have shown that NCp7 promotes formation of the extended dimer under physiological Mg2+ concentration, a condition under which the extended dimer does not readily form in the absence of NC. This is consistent with the role of NC as a nucleic acid chaperone, enabling transition of the 5′UTR to the stable extended dimer. Furthermore, tRNALys3 annealing to the PBS stabilizes the DIS-exposed form of the 5′UTR, allowing kissing loop dimerization but preventing extended dimer formation. Although the precise timing of tRNA primer annealing to gRNA is not known in any retroviral system, these results are consistent with a model wherein primer annealing occurs just before or concomitant with dimerization.
Interactions between tRNALys3 and the 5′UTR outside of the 18-nt PBS have previously been observed (49–51), and smFRET data with truncated primer constructs suggest these interactions may serve to block formation of the extended dimer conformation of the 5′UTR in the absence of NC (Fig. 4 E and G). Surprisingly, tRNALys3 annealing even induced dimerization of the 2M construct (Fig. 2A), which contains only a 4-nt palindromic loop sequence and is observed exclusively as a monomer in the absence of tRNA (Figs. 2A and 3A). Previous DIS mutagenesis and evolution studies have suggested that 4-nt palindromes are maintained in the infectious HIV-1 population (52). It is also possible that contacts outside of the DIS can act as secondary dimerization sites, as previous cell-based studies have shown that the DIS stem loop is dispensable for production of infectious virions (53, 54). Indeed, we also observed that tRNALys3 annealing slightly promoted dimerization of the ΔDIS construct, although not to the extent of the 2M construct (Fig. 2A). Our data suggest that in these cases, tRNALys3 annealing to the PBS is likely a prerequisite for efficient gRNA dimerization, stabilizing the kissing dimer to ensure budding virions contain two copies of gRNA, as well as a tRNA primer for initiation of reverse transcription.
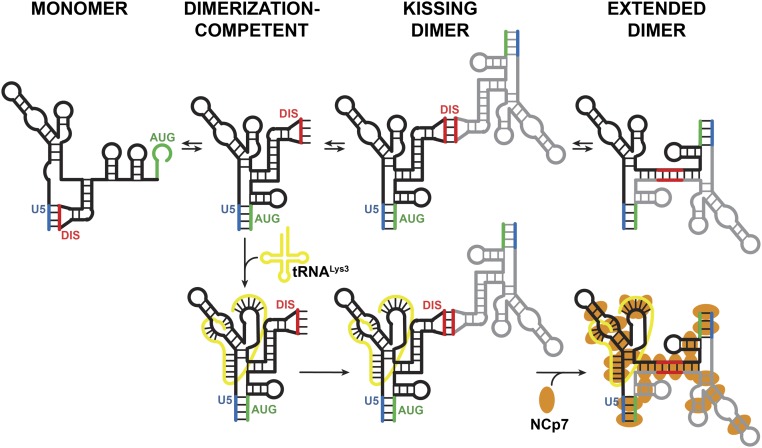
A recent cryo-EM study of tRNALys3 annealed to a 101-nt gRNA PBS construct in the presence of reverse transcriptase showed that tRNALys3 adopts an extended helical conformation in the initiation complex (55). Our results show that the full tRNA is required for strongly promoting the DIS-exposed conformer and annealing of shorter oligonucleotides does not induce these same effects. These data support an important functional role for the additional tRNA interactions in the viral lifecycle. Taken together with previous data, this work allows us to propose a final model in which tRNA primer annealing stabilizes the DIS-exposed conformation of the 5′UTR, facilitating kissing dimer formation (Fig. 7). We speculate that this kissing dimer is then recognized by Gag for packaging to ensure infectious particles contain both a dimeric genome and a tRNA primer. After viral maturation, NCp7 remodels the 5′UTR:tRNA complex to form an extended dimer and an annealed tRNA state that is optimal for initiation of reverse transcription (56).
Fig. 7.
Model of 5′UTR conformation and dimerization and the effects of tRNALys3 and NCp7. The 2M and ΔDIS mutations in the 5′UTR promote the dimerization-incompetent and dimerization-competent conformations, respectively. The WT 5′UTR exists in equilibrium between both conformations. tRNALys3 promotes the DIS-exposed monomer and kissing dimer but inhibits the formation of the extended dimer. NC enriches for the extended dimer conformation, irrespective of tRNALys3 binding. Elevated Mg2+ promotes formation of the extended dimer in the absence of other factors.
This smFRET imaging approach utilized here has been able to distinguish between a number of HIV-1 5′UTR conformations of biological interest. This system is able to not just capture static states of the 5′UTR but also monitor transitions between states. Using this system, we have shown that the 5′UTR readily transitions between conformations on the timescale of minutes. This approach is also amenable to the addition of host and viral factors, which will facilitate future studies of the potential role of these factors in remodeling the structure of the 5′UTR in other steps of the lifecycle, such as initiation of reverse transcription and Gag/Gag-Pol translation.
Materials and Methods
Design of FRET Constructs.
To position one fluorescent dye at the base of the U5 stem, 238-nt RNA constructs consisting of nt 106–343 of the HIV-1 NL4-3 5′UTR were extended by an 18-nt “handle” at the 5′ end to allow annealing to a complementary 5′ fluorophore-labeled DNA. Random 5′ extension sequences were generated in silico and screened for secondary structure formation using mFold (57). The 5′ extension sequence (5′ GGUUGGAGGUUAUGGAGCA 3′), which lacked any predicted interactions with the 5′UTR, was used for all constructs. The complementary 5′ fluorophore-labeled DNA oligonucleotide was designed with a 5′-T overhang (5′ TTTTGCTCCATAACCTCCAACC 3′) to ensure rotational mobility of the dye after annealing. A mutant construct wherein the DIS sequence was mutated to a GAGA tetraloop (ΔDIS) was used as a DIS-exposed control, while a mutant construct containing the G108C and C258G point mutations was used as a DIS-sequestered control (2M). Additionally, DIS → 6G and DIS → 6C mutant constructs were used to facilitate some of the in-gel FRET assays.
RNA Production.
DNA templates for in vitro transcription of FH238.WT were amplified by PCR using pHIV-1_356_WT plasmid (SI Appendix, Table S1). For handled constructs, the forward primer contained the T7 promoter and the engineered FRET handle binding sequence, while the reverse primer ended at the 3′ end of the 5′UTR (SI Appendix, Table S2). Handle-free RNA was produced using only the T7 promoter in the forward primer. The DNA templates encoding ΔDIS and 2M constructs were prepared by site-directed mutagenesis as described previously (58) and amplified as for the WT sequence. The DNAs encoding DIS → 6G and DIS → 6C constructs were generated from plasmids p6G-Mini-MSL and p6C-Mini-BSL (43), a gift of Dr. Wei-Shau Hu, NIH, Frederick, MD, by PCR amplification as described above. All primers and plasmids used are listed in SI Appendix, Tables S1 and S2. RNA was produced by T7 expression using either T7 RNA polymerase purified in-house as described (59) or a T7 RiboMAX Express Large-Scale RNA Production System (Promega Corporation). All RNA constructs were purified by HPLC purification over a Xbridge SEC column (Waters Corporation) or by elution from an 8% denaturing polyacrylamide gel using the crush and soak method (60). RNAs were periodically checked for purity on an 8% polyacrylamide/6M urea denaturing gel run at 200 V for 2 h and stained with ethidium bromide.
Human tRNALys3 was generated by T7 RNA polymerase transcription from a tRNALys3-encoding pUC19 plasmid linearized by FOK1 and transcribed as described above.
Fluorescent Labeling of 5′UTR RNA.
5′UTR RNA used in FRET studies was labeled by periodate oxidation of the 3′ end as described (19). RNA used in smFRET experiments was coupled to hydrazide-conjugated LD650 (Lumidyne Technologies). Briefly, coupling was performed by mixing 6 nmol RNA with 114 µL 25 mM NaIO4 and 114 µL 1 M NaOAc, pH 5.2, and incubating for 2 h on ice protected from light. Following precipitation of the RNA with isopropanol, the RNA pellet was resuspended in 60 µL 1 M NaOAc, pH 5.2, 27 µL 1 M NaCl, and 33 µL water. The resuspended RNA was then mixed with 60 µL 10 mM LD650-hydrazide. The total volume was increased to 600 µL with water and tumbled at room temperature for 2 h protected from light. Labeled RNA was purified by HPLC using an Xbridge SEC column (Waters Corporation).
RNA used in ensemble in-gel FRET studies was labeled by periodate oxidation of the 3′ end as described (19). Hydrazide-linked Alexa-488 or Alexa-647 was then coupled to the 3′ aldehyde. Briefly, 1 nmol of RNA was mixed with 1.3 µL 400 mM NaIO4 and 6.7 µL 3 M NaOAc (pH 5.2) in a total volume of 200 µL and incubated at room temperature for 2 h protected from light. Following ethanol precipitation, the RNA pellet was resuspended in water and mixed with 3.3 µL 3 M NaOAc (pH 5.2) and 10 µL 10 mM hydrazide-coupled dye to a total volume of 100 µL. The mixture was then tumbled overnight at 4 °C. Labeled RNA was purified by ethanol precipitation followed by elution over an Illustra Microspin G-25 column (GE Healthcare) using the manufacturer’s protocol and a final ethanol precipitation.
Fluorescent Labeling of DNA Handle.
For smFRET studies, 5′-amino modifier C6-TTTTGCTCCATAACCTCCAACC-3′ DNA handle (FH.Oligo; Integrated DNA Technologies) was labeled by coupling with NHS-linked LD550 (Lumidyne Technologies). Briefly, 90 µL of 250 µM handle was mixed with 90 nmol LD550 in 90 µL DMSO and 150 µL Na2CO3 0.1 M, pH 8.7, and incubated at room temperature for 2 h. Labeled DNA handle was purified by HPLC purification over an Xbridge SEC column (Waters Corporation). For in-gel FRET studies, HPLC-purified 5′ Alexa-488- or Alexa-647-labeled FH.Oligos (SI Appendix, Table S3) were purchased from IDT. The dried pellet was resuspended in 200 mM KCl for long-term storage at −20 °C.
Annealing and Dimerization of 5′UTR Constructs.
For native gel experiments, the FH.Oligo was annealed to the 5′ end of the handled RNA by incubating 1.0 µM FH.Oligo and 1.0 µM 5′UTR RNA in 50 mM Hepes, pH 7.5, at 80 °C for 2 min followed by incubation at 65 °C for 4 min. MgCl2 was then added to a final concentration of 10 mM, and samples were incubated at 37 °C for 6 min and then cooled on ice for 30 min. For tRNALys3 heat-annealing experiments, tRNALys3 was added at the desired ratio (either 0:1 or 2:1 tRNA:5′UTR) before incubation at 80 °C. For NC-mediated annealing experiments, tRNALys3 was folded separately from the 5′UTR RNA using the same conditions as described above and added to samples at the desired ratio after incubation on ice.
For smFRET experiments, FH.Oligo was annealed to the 5′ end of the 5′UTR-derived constructs by heating 0.1 µM FH.Oligo and 0.1 µM FH238 construct to 80 °C for 2 min, heating at 65 °C for 4 min, followed by heating at 37 °C for 6 min, and then cooling on ice for 30 min. Excess unannealed handle was then removed by passing over an Amicon Ultra 30K spin column (Merck Millipore Ltd.).
The 5′UTR constructs were dimerized in 50 mM Hepes by heating 0.1 µM FH238 with 0.1 µM 238, 0.1 µM labeled FH.Oligo, and 0.2 µM tRNALys3, if required, to 80 °C for 2 min followed by heating at 65 °C for 4 min. NCp7 (obtained as a synthetic peptide from GenScript Corporation or New England Peptide) and MgCl2 are then added, if required, and incubated at 37 °C for 6 min. Samples were then cooled on ice for 30 min. Excess unannealed handle was then removed by passing over an Amicon Ultra 30K spin column (Merck Millipore Ltd.). To remove NCp7, 0.2 µL of 20 µg/µL Proteinase K (Thermo Scientific) was added to 40 µL of 0.1 µM RNA dimerization reaction and incubated at 37 °C for 30 min.
In-Gel FRET Assays.
For in-gel experiments performed with the FRET constructs alone or with tRNALys3, RNA/DNA were folded as described above in the presence or absence of tRNALys3 and diluted to a final 5′UTR concentration of 200 nM in FRET buffer [50 mM Hepes (pH 7.5), 10 mM MgCl2, and 50 mM NaCl], followed by incubation at room temperature for 30 min. For in-gel FRET experiments with NCp7, FRET complexes and tRNALys3 were folded separately as described above, added together in a 1:1 ratio, and diluted to a final 5′UTR concentration of 200 nM in NCp7 buffer [50 mM Hepes (pH 7.5), 1.0 mM MgCl2, 75 mM KCl, 200 µM Tris(2-carboxyethyl)phosphine]. NCp7 or an equal volume of NCp7 buffer as a no-NC control was then added to a final ratio of 6 nt:1NCp7 followed by incubation at 37 °C for 30 min. NCp7 was removed by adding Proteinase K (Thermo Scientific) to a final concentration of 1.0 µg/µL followed by incubation at 37 °C for 30 min. Samples were electrophoresed on 4.5 or 6% native polyacrylamide gels containing TBM [Tris-Borate-MgCl2 containing 1.0 mM MgCl2 (for experiments involving NCp7) or 10 mM MgCl2 (for all other experiments)] in both the gel and running buffer. Gels were run at 120V for 10–16 h at 4 °C. RNA was visualized using a Typhoon FLA 9500 phosphorimager (GE Healthcare Life Sciences). Gels were scanned three times in succession to image donor emission due to direct excitation (Ex. = 473 nm, BPB1 Filter [520–540 nm]), acceptor emission due to FRET (Ex. = 473 nm, LPR Filter [>665 nm]), and acceptor emission due to direct excitation (Ex. = 635 nm, LPR Filter [>665 nm]). Gel images were processed and quantified using ImageQuant TL software (GE Healthcare Life Sciences). Fluorescent intensities were calculated via densitometry. Apparent FRET (Fapp) values were calculated according to Eq. 1 (61), where IF is the FRET intensity, IB is the background intensity, ν is the excitation wavelength (1 = 473 nm, 2 = 635 nm), and ν′ is the emission wavelength (665 nm). For each gel, Fapp values were normalized to high FRET 238.ΔDIS or FH.6G monomer species, which were set to a value of 1.0.
| [1] |
TIRF Microscopy.
smFRET imaging was performed on a customized prism-based TIRF microscope as previously described (62). The surface-bound nucleic acids were illuminated by the evanescent field generated by total internal reflection of a 532-nm Genesis MX solid-state laser (Coherent) at a power of ∼95 mW, measured at the prism. Direct excitation of the acceptor fluorophore was performed by total internal reflection of a 639-nm Genesis MX solid-state laser (Coherent) at a power of ∼120 mW, measured at the prism. In Fig. 5, the 639-nm laser was pulsed for 2 s at the beginning of acquisition; the 532-nm laser illumination was present throughout acquisition. In all cases, movies were collected at 2 frames/s using custom-built LabView acquisition software (National Instruments) or Micro-Manager (Open Imaging).
Experiments were performed in Imaging Buffer containing 50 mM Tris, pH 7.5, 50 mM Hepes, pH 7.5, 50 mM NaCl, and 1 mM MgCl2 or 10 mM MgCl2. In the application of the Intra-UTR assay to dimerized 5′UTR RNA, imaging was performed in the presence of 0.1 μM handle-free FH238.WT RNA. For the investigation of the effect of tRNALys3, anti-PBS cDNA, or Minihelix on 5′UTR conformation, imaging was performed in the presence of 0.2 μM of each RNA or DNA species. Bulk measurements indicated fluorescence anisotropy consistent with freely tumbling fluorophores, with the exception of measurements done in presence of NCp7, in which case a modest increase in anisotropy was noted (SI Appendix, Table S4). As a result, before smFRET imaging, NCp7 was removed by proteinase K digest as described above. Where noted, 0.01 𝜇M labeled 238.WT 5′UTR was included in imaging buffer for the Inter-UTR smFRET experiments. Also contained in the imaging buffer was a mixture of triplet-state quenchers [1 mM trolox, 1 mM cyclooctatetraene, 1 mM nitrobenzyl alcohol (63)] and an enzymatic system for removal of molecular oxygen, which included 2 mM protocatechuic acid and 8 nM protocatechuate 3,4-deoxygenase (64). All smFRET experiments were conducted at room temperature.
smFRET Analysis.
smFRET data were processed and analyzed using the SPARTAN software package implemented in MATLAB (65). smFRET trajectories were automatically identified according to several criteria: (i) donor and acceptor fluorescence trajectories both displayed a single photobleaching event, which is indicative of a single fluorophore pair; (ii) FRET was detectable for minimally five frames before photobleaching of either fluorophore; (iii) the correlation coefficient of donor and acceptor fluorescence traces was less than −0.2; and (iv) the signal-to-noise ratio of the total fluorescence, defined as the ratio of the magnitude of the photobleaching event to the variance of the background signal, was greater than 8. Trajectories that passed these criteria where then verified manually by visual inspection. Kinetic analysis of the smFRET trajectories was performed with hidden Markov modeling. Specifically, the individual trajectories were fit to a model containing a high- and low-FRET state using the segmental k-means algorithm (66). The high- and low-FRET dwell times were compiled into histograms and fit to exponential functions (A exp –t/τ, where τ is the average dwell time) using a least-squares fitting algorithm. All smFRET data analysis and figure preparation was conducted in MATLAB.
smFRET data collected with the transient acceptor excitation pulse were initially processed as described in above. Dimers were identified as traces indicating both donor and acceptor fluorescence during the 640-nm laser pulse. Low-FRET and high-FRET traces were identified by the FRET value observed after conclusion of the 639-nm laser pulse. The no-FRET population was identified as traces that indicated both fluorophores during the 639-nm laser pulse and only donor fluorescence after conclusion of the pulse.
Supplementary Material
Acknowledgments
We thank Dr. Wei-Shau Hu (NIH Frederick) for plasmids encoding the 6G/6C 5′UTR constructs and Robert Blakemore for helpful discussion. This work was supported by NIH Grants R01 GM113887, GM065056, and U54 GM103368 (to K.M.-F.); T32 GM118291 (to J.P.K.); and R21 AI136711-01 (to J.B.M.) and by the Collaborative Development Programs of the Pittsburgh Center for HIV-Protein Interactions (P50 GM082251-12) and the Center for HIV-1 RNA Studies (U54 GM103297-07) (to J.B.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902271116/-/DCSupplemental.
References
- 1.Bieniasz P, Telesnitsky A (2018) Multiple, switchable protein:RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu Rev Virol 5:165–183. [DOI] [PubMed] [Google Scholar]
- 2.van Bel N, Das AT, Cornelissen M, Abbink TEM, Berkhout B (2014) A short sequence motif in the 5′ leader of the HIV-1 genome modulates extended RNA dimer formation and virus replication. J Biol Chem 289:35061–35074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu K, et al. (2011) NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed EO. (2015) HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell RS, Liang C, Wainberg MA (2004) Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology 1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B (2004) A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J Virol 78:10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keane SC, et al. (2016) NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc Natl Acad Sci USA 113:13033–13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huthoff H, Berkhout B (2001) Two alternating structures of the HIV-1 leader RNA. RNA 7:143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooms M, Verhoef K, Southern E, Huthoff H, Berkhout B (2004) Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res 32:819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vrolijk MM, Ooms M, Harwig A, Das AT, Berkhout B (2008) Destabilization of the TAR hairpin affects the structure and function of the HIV-1 leader RNA. Nucleic Acids Res 36:4352–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois N, Marquet R, Paillart J-C, Bernacchi S (2018) Retroviral RNA dimerization: From structure to functions. Front Microbiol 9:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang M, et al. (1993) Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol 67:3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavon-Eternod M, Wei M, Pan T, Kleiman L (2010) Profiling non-lysyl tRNAs in HIV-1. RNA 16:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenyon JC, Prestwood LJ, Le Grice SFJ, Lever AML (2013) In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res 41:e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seif E, Niu M, Kleiman L (2013) Annealing to sequences within the primer binding site loop promotes an HIV-1 RNA conformation favoring RNA dimerization and packaging. RNA 19:1384–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkhout B, et al. (2002) In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J Biol Chem 277:19967–19975. [DOI] [PubMed] [Google Scholar]
- 17.Stephenson JD, et al. (2013) Three-dimensional RNA structure of the major HIV-1 packaging signal region. Structure 21:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heng X, et al. (2012) Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol 417:224–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinaldi AJ, Suddala KC, Walter NG (2014) Native Purification and labeling of RNA for single molecule fluorescence studies. RNA-RNA Interactions, Methods in Molecular Biology (Springer, New York), pp 63–95.
- 20.Keane SC, et al. (2015) RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348:917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huthoff H, Berkhout B (2002) Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry 41:10439–10445. [DOI] [PubMed] [Google Scholar]
- 22.Stephenson JD, Kenyon JC, Symmons MF, Lever AML (2016) Characterizing 3D RNA structure by single molecule FRET. Methods 103:57–67. [DOI] [PubMed] [Google Scholar]
- 23.Kharytonchyk S, et al. (2016) Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci USA 113:13378–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Bel N, Ghabri A, Das AT, Berkhout B (2015) The HIV-1 leader RNA is exquisitely sensitive to structural changes. Virology 483:236–252. [DOI] [PubMed] [Google Scholar]
- 25.Stello T, Hong M, Musier-Forsyth K (1999) Efficient aminoacylation of tRNA(Lys,3) by human lysyl-tRNA synthetase is dependent on covalent continuity between the acceptor stem and the anticodon domain. Nucleic Acids Res 27:4823–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkowitz R, Fisher J, Goff SP (1996) RNA packaging. Curr Top Microbiol Immunol 214:177–218. [DOI] [PubMed] [Google Scholar]
- 27.D’Souza V, Summers MF (2005) How retroviruses select their genomes. Nat Rev Microbiol 3:643–655. [DOI] [PubMed] [Google Scholar]
- 28.Lu K, Heng X, Summers MF (2011) Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol 410:609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuzembayeva M, Dilley K, Sardo L, Hu W-S (2014) Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aduri R, Briggs KT, Gorelick RJ, Marino JP (2013) Molecular determinants of HIV-1 NCp7 chaperone activity in maturation of the HIV-1 dimerization initiation site. Nucleic Acids Res 41:2565–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vo M-N, Barany G, Rouzina I, Musier-Forsyth K (2009) Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Implications for reverse transcription. J Mol Biol 386:773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones CP, Saadatmand J, Kleiman L, Musier-Forsyth K (2013) Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. RNA 19:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng YX, et al. (1999) The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: Possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol 73:4251–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo F, Saadatmand J, Niu M, Kleiman L (2009) Roles of Gag and NCp7 in facilitating tRNA(Lys)(3) Annealing to viral RNA in human immunodeficiency virus type 1. J Virol 83:8099–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roldan A, Warren OU, Russell RS, Liang C, Wainberg MA (2005) A HIV-1 minimal gag protein is superior to nucleocapsid at in vitro annealing and exhibits multimerization-induced inhibition of reverse transcription. J Biol Chem 280:17488–17496. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, et al. (1997) Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J Virol 71:726–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beerens N, et al. (2013) Role of the primer activation signal in tRNA annealing onto the HIV-1 genome studied by single-molecule FRET microscopy. RNA 19:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coey AT, et al. (2018) Dynamic interplay of RNA and protein in the human immunodeficiency virus-1 reverse transcription initiation complex. J Mol Biol 430:5137–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beerens N, Groot F, Berkhout B (2001) Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J Biol Chem 276:31247–31256. [DOI] [PubMed] [Google Scholar]
- 40.Masuda T, et al. (2015) Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep 5:17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin JG, Grimley PM, Ramseur JM, Berezesky IK (1974) Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol 14:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin JG, Rosenak MJ (1976) Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: Evidence for persistence of viral messenger RNA. Proc Natl Acad Sci USA 73:1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolaitchik OA, et al. (2013) Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog 9:e1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, et al. (2016) HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc Natl Acad Sci USA 113:E201–E208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu W, et al. (2006) Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J Virol 80:1242–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jalalirad M, Laughrea M (2010) Formation of immature and mature genomic RNA dimers in wild-type and protease-inactive HIV-1: Differential roles of the Gag polyprotein, nucleocapsid proteins NCp15, NCp9, NCp7, and the dimerization initiation site. Virology 407:225–236. [DOI] [PubMed] [Google Scholar]
- 47.Seif E, Niu M, Kleiman L (2015) In virio SHAPE analysis of tRNA(Lys3) annealing to HIV-1 genomic RNA in wild type and protease-deficient virus. Retrovirology 12:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleiman L, Jones CP, Musier-Forsyth K (2010) Formation of the tRNALys packaging complex in HIV-1. FEBS Lett 584:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huthoff H, Bugala K, Barciszewski J, Berkhout B (2003) On the importance of the primer activation signal for initiation of tRNA(lys3)-primed reverse transcription of the HIV-1 RNA genome. Nucleic Acids Res 31:5186–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldschmidt V, Ehresmann C, Ehresmann B, Marquet R (2003) Does the HIV-1 primer activation signal interact with tRNA3(Lys) during the initiation of reverse transcription? Nucleic Acids Res 31:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beerens N, Berkhout B (2002) The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virol 76:2329–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hussein ITM, et al. (2010) Delineation of the preferences and requirements of the human immunodeficiency virus type 1 dimerization initiation signal by using an in vivo cell-based selection approach. J Virol 84:6866–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berkhout B, van Wamel JL (1996) Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol 70:6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill MK, et al. (2003) The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J Virol 77:8329–8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen KP, et al. (2018) Architecture of an HIV-1 reverse transcriptase initiation complex. Nature 557:118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu W, Gorelick RJ, Rein A (1994) Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol 68:5013–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones CP, Cantara WA, Olson ED, Musier-Forsyth K (2014) Small-angle X-ray scattering-derived structure of the HIV-1 5′ UTR reveals 3D tRNA mimicry. Proc Natl Acad Sci USA 111:3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellinger T, Ehricht R (2018) Single-step purification of T7 RNA polymerase with a 6-histidine tag. Biotechniques 24:718–720. [DOI] [PubMed] [Google Scholar]
- 60.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clegg RM, Murchie AI, Lilley DM (1994) The solution structure of the four-way DNA junction at low-salt conditions: A fluorescence resonance energy transfer analysis. Biophys J 66:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das DK, et al. (2018) Direct visualization of the conformational dynamics of single influenza hemagglutinin trimers. Cell 174:926–937.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dave R, Terry DS, Munro JB, Blanchard SC (2009) Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J 96:2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aitken CE, Marshall RA, Puglisi JD (2008) An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J 94:1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juette MF, et al. (2016) Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods 13:341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin F. (2004) Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J 86:1488–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.