Significance
Cholestatic itch is a debilitating symptom that affects hundreds of thousands of patients every year, drastically decreasing patient quality of life. Additional therapeutic targets are needed, as current therapies are ineffective for many patients. In recent years, the Mas-related G protein-coupled receptor (MRGPR) family of receptors has been linked to itch. Here, we determine that MRGPRX4 is specifically activated by numerous bile acids (BAs) up-regulated in cholestatic patient sera. To test the relevance of these findings in vivo, we generated humanized mice expressing MRGPRX4 in sensory neurons. MRGPRX4+ humanized mice display higher levels of itch in response to injection of BAs and in a mouse model of cholestasis, indicating that MRGPRX4 could play a role in cholestatic itch.
Keywords: cholestasis, itch, pruritus, MRGPRX4, bile acids
Abstract
Patients suffering from cholestasis, the slowing or stoppage of bile flow, commonly report experiencing an intense, chronic itch. Numerous pruritogens are up-regulated in cholestatic patient sera, including bile acids (BAs). Acute injection of BAs results in itch in both mice and humans, and BA-modulating therapy is effective in controlling patient itch. Here, we present evidence that human sensory neuron-expressed Mas-related G protein-coupled receptor X4 (MRGPRX4), an orphan member of the Mrgpr family of GPCRs, is a BA receptor. Using Ca2+ imaging, we determined that pathophysiologically relevant levels of numerous BAs activated MRGPRX4. No mouse Mrgpr orthologs were activated by BAs. To assess the in vivo relevance of BA activation of MRGPRX4, we generated a humanized mouse with targeted expression of MRGPRX4 in itch-encoding sensory neurons. BAs activated MRGPRX4+ sensory neurons at higher levels compared with WT neurons. Compared with control animals, MRGPRX4+ mice scratched more upon acute injection of BAs and in a model of cholestatic itch. Overall, these data suggest that targeting MRGPRX4 is a promising strategy for alleviating cholestatic itch.
In the United States, hundreds of thousands of patients report experiencing a severe, generalized pruritus (itch) due to cholestasis, or the slowing or stoppage of bile flow. Cholestatic itch is often nonresponsive to standard pharmacological treatments (1–3) and instead requires physically removing the causative obstruction (such as gallstones), draining the bile, or transplanting the liver to alleviate itch (1, 4, 5). Because these procedures are often curative, the responsible pruritogens are hypothesized to originate from the liver and bile. Numerous candidate pruritogens are present in bile and up-regulated in cholestatic patients, including opioids, lysophosphatidic acid, bilirubin, and bile acids (BAs). Therapies targeting these mechanisms, such as opioid antagonists, rifampicin, and BA-binding resins like cholestyramine, are frontline therapy for cholestatic itch (6–9).
BAs are amphipathic cholesterol derivatives synthesized in liver. BAs have previously defined roles as detergents, hormones, and antiinflammatory agents (10–13). Primary BA synthesis aids in biliary secretion of phospholipids, drugs, and toxins. In liver, BAs are conjugated with taurine or glycine for biliary secretion. Secreted BAs can be reabsorbed in the ileum and retrogradely transported to liver where they inhibit synthesis in an inhibitory feedback loop. In cholestatic patients, BAs accumulate in both serum and tissue because bile flow is abrogated.
BAs are pruritic as injecting BAs in humans evokes itch (14, 15). In mice, TGR5, a G protein-coupled receptor (GPCR) with micromolar affinity toward BAs (16, 17), is expressed in itch sensory neurons (10). TGR5−/− mice scratched less in response to injection of multiple activating BAs, indicating that the receptor mediates a component of acute BA-associated itch (10). However, recent clinical trials have found that TGR5-specific agonists do not elicit pruritus, suggesting that activating TGR5 alone is not sufficient to evoke itch in humans (18). Accordingly, BA-induced itch is likely not TGR5 dependent in humans, a scenario different from mice.
Cholestatic pruritus is classified as nonhistaminergic itch. Cholestatic patients do not exhibit classic signs of histamine release, such as erythema or swelling (3). Moreover, antihistamines are ineffective in treating cholestatic pruritus, with only a few patients reporting clinical improvement in their symptoms (3, 19). The Mas-related family of G protein-coupled receptors (MRGPRs) are GPCRs that mediate nonhistaminergic itch. In mice, MRGPRs are expressed in small-diameter sensory neurons that code exclusively for itch and not pain (20). Over the years, increasingly more members of the MRGPR family have been identified as receptors for known pruritogens (21).
In mice, there are 27 expressed Mrgprs, only a few of which have known ligands (22). In humans, there are eight expressed Mrgprs (MRGPRX1–X4 and -D–G). MRGPRX1 and MRGPRD have been implicated in nonhistaminergic itch as receptors for pruritic compounds. MRGPRX1, -X3, -X4, -D, and -E are expressed in human dorsal root ganglia (DRG) and trigeminal ganglia (TG), while MRGPRX2 has been detected in human mast cells (23–26). Recently, we have identified murine receptor Mrgpra1 and human receptor MRGPRX4 as receptors for bilirubin that may mediate a component of cholestatic itch (27). Another published MRGPRX4 agonist, nateglinide, is a clinically approved type 2 diabetes drug that causes itch as a prominent side effect (28, 29), further implicating MRGPRX4 as an itch receptor. In this study, we screened major bile constituents against a panel of both mouse and human MRGPRs. From this screen, we determined that numerous BAs specifically activated MRGPRX4. Of all mouse and human receptors tested, BAs only activated MRGPRX4. Using newly generated humanized mice in which we express MRGPRX4 selectively in itch-sensing neurons, we demonstrated that MRGPRX4 contributed to the BA-induced component of cholestatic itch.
Results
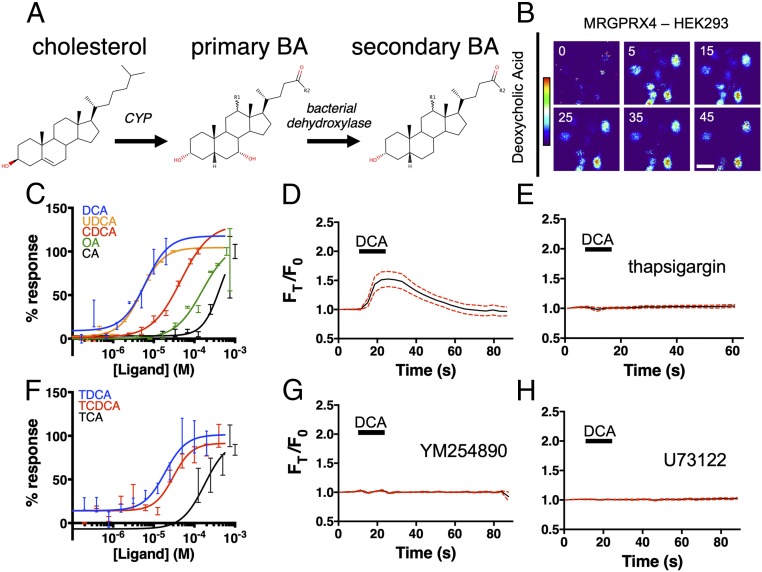
In liver hepatocytes, cholesterol is metabolized via cytochrome P450 (CYP) to primary BAs (Fig. 1A). Primary BAs are subsequently metabolized to other BAs, after which they are conjugated to either glycine or taurine for more efficient biliary secretion. In the intestine, bacteria metabolize BAs to form secondary BAs, which can return to the bloodstream and liver through enterohepatic circulation (Fig. 1A). We recently identified MRGPRX4 as a bilirubin receptor that may contribute to cholestatic pruritus (27). We performed a counter screen of MRGPRX4 and Mrgpra1 against additional bile constituents using G⍺15-expressing HEK cells. G⍺15 is a permissive Gα protein capable of coupling GPCRs to intracellular calcium stores via phospholipase C (PLC) (30). As measured by intracellular Ca2+ changes, numerous BAs, both primary and secondary, activated MRGPRX4 (Fig. 1 B and C). We tested BA activity against 12 closely related mouse Mrgprs (SI Appendix, Fig. S1A) as well as sequence-similar human receptors, MRGPRX1–X3. No mouse receptors, including the previously identified mouse ortholog Mrgpra1 or sequence-similar human receptors, were activated by BAs (SI Appendix, Fig. S1 B–E). Based on this result, we concluded that BAs specifically activated MRGPRX4 among the Mrgpr family members.
Fig. 1.
BAs activate MRGPRX4, a human sensory neuron-expressed GPCR. (A) The molecular stereostructures of primary and secondary BAs and their metabolic precursor, cholesterol. Major structural differences are highlighted in red. R1 denotes the potential presence of an α-hydroxyl substituent in some BA derivatives. R2 denotes additional substituents that vary among BAs. (B–H) Ca2+ imaging of HEK293 cells stably expressing MRGPRX4. (B) Representative Fura-2 fluorescence heat map images of HEK cells showing changes in intracellular [Ca2+] induced by DCA (10 µM). (Scale bar, 20 µm.) (C and F) Concentration–Ca2+ response curves of BAs against MRGPRX4. Data are a representative experiment of three independent replicates performed in triplicate, depicted as mean ± SEM (C) Concentration–Ca2+ response curves of unconjugated BAs. OA, oleic acid. (F) Concentration–Ca2+ response curves of tauro-conjugated BAs. (D, E, G, and H) Ca2+ imaging traces. A total of 10 µM DCA was added where indicated by black bars. Mean ± 95% CI depicted. n = 15. (E, G, and H) Cells were preincubated with either 10 µM sarco/endoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin (E), 10 µM of the Gαq inhibitor YM254890 (G), or 10 µM PLC inhibitor U73122 (H) for 30 min before imaging.
Ca2+ imaging of MRGPRX4-expressing HEK293 cells displayed robust Ca2+ flux in response to 10 μM deoxycholic acid (DCA) (Fig. 1 B and D). Thapsigargin, a noncompetitive inhibitor of the sarcoendoplasmic reticulum Ca2+ ATPase, completely inhibited BA-associated Ca2+ influx (Fig. 1E), indicating that MRGPRX4-induced Ca2+ signal was dependent on intracellular Ca2+ stores. Previous studies have identified MRGPRX4 as being Gαq linked (27, 29). In line with this, DCA-associated Ca2+ flux appears to be similarly Gαq linked, since both the Gαq inhibitor, YM254890, and the PLC inhibitor, U73122, inhibited Ca2+ signal (Fig. 1 G and H).
Of the BAs tested, DCA and ursodeoxycholic acid (UDCA) displayed the highest potency in terms of MRGPRX4-associated Ca2+ signal (Fig. 1C and SI Appendix, Fig. S1F). Both DCA and UDCA had EC50s of ∼5 μM, similar to their activity toward previously published receptors like TGR5 (17). Unlike TGR5, however, MRGPRX4 exhibited a larger range of potency for various BAs. For example, cholic acid (CA) displayed much lower potency against MRGPRX4 (EC50 ∼430 μM), well outside physiologic range. In contrast, the EC50 of CA toward TGR5 is ∼10 μM (17).
Primary bile acids can be conjugated to either glycine or taurine to form water-soluble bile salts. We tested three tauro-conjugated bile acids, tauro-DCA (TDCA), tauro-CDCA (TCDCA), and tauro-cholic acid (TCA) against MRGPRX4. All conjugated bile acids were capable of activating MRGPRX4 at concentrations similar to the respective unconjugated BA (Fig. 1F and SI Appendix, Fig. S1G).
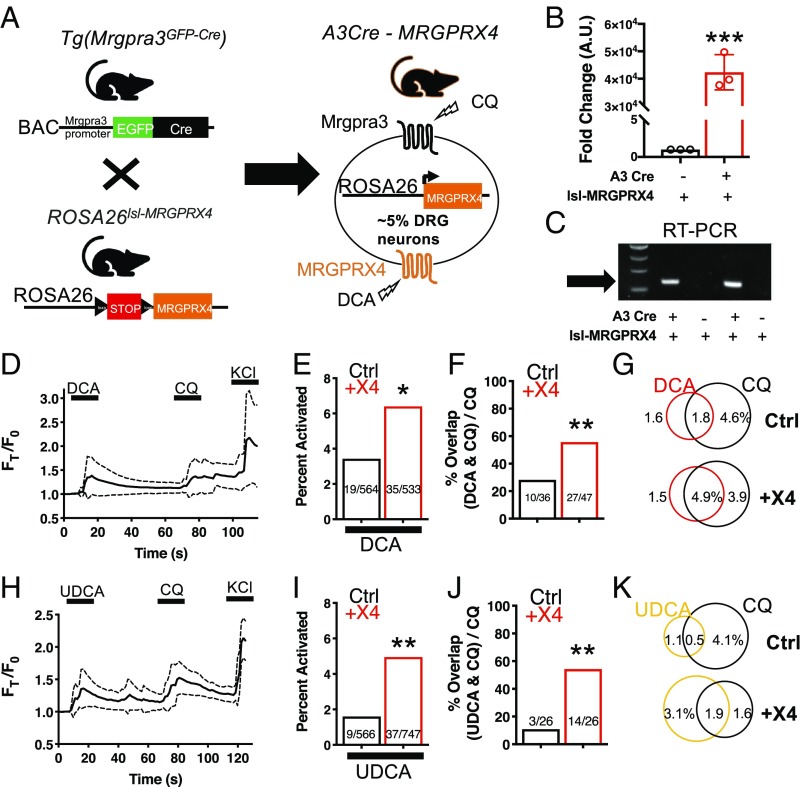
MRGPRX4 is primarily expressed in sensory neurons of the DRG. To test whether BAs interact with MRGPRX4 in neurons, we generated humanized mice expressing MRGPRX4. We crossed Tg(Mrgpra3-GFP-Cre) (A3-Cre) animals with newly generated ROSA26loxSTOPloxl-MRGPRX4 (lsl-MRGPRX4) animals (Fig. 2A). A3-Cre animals express Cre recombinase under control of the Mrgpra3 promoter, which labels ∼5% of the DRG sensory neurons that specifically encode for itch (20). In lsl-MRGPRX4 animals, expression of MRGPRX4 is under Cre-loxp control. Progeny that inherit both A3-Cre and lsl-MRGPRX4 (+X4) would be expected to express MRGPRX4 in mouse Mrgpra3-containing itch neurons (Fig. 2A). Cre-null but transgene-positive mice (Cre-, lsl-MRGPRX4) were studied as comparative controls.
Fig. 2.
The +X4 humanized mouse sensory neurons are more readily activated by BAs. (A) Diagram showing mating strategy to generate humanized (+X4) mice. Mrgpra3-Cre expression is restricted to mouse DRG. Approximately 5% of DRG would be expected to express receptor. qPCR (B and C) RT-PCR of RNA collected from both control (A3-Cre negative) and +X4 (A3-Cre positive) mice expressing Rosa26lsl-MRGPRX4. (B) Mean ± 95% CI depicted. n = 3 representative samples. ***P < 0.001, Student’s t test. (C) Arrow indicates the expected band based on primer design. Four independent samples collected from different mice are depicted. (D and H) Ca2+ imaging traces of Ctrl and +X4 DRG neurons. Mean ± 95% CI depicted. n = 10 neurons. After a 10-s baseline, either 10 µM DCA (D) or 100 µM UDCA (H) was applied where indicated by black bars. After 1 min, 1 mM CQ was added. (E and I) Percent activation of Ctrl and +X4 DRG neurons from either 10 µM DCA (E) or 100 µM UDCA (I). (F and J) Percent overlap of CQ-activated Ctrl and +X4 DRG neurons (∼5% of total) activated by both DCA and CQ (F) and both UDCA and CQ (J). (E, F, I, and J) *P < 0.05; **P < 0.01; χ2 test. A neuron was considered to be activated if ∆F > 0.2 for at least 30 s. (G and K) Venn diagrams of DCA- (G), UDCA- (K), and CQ-activated neurons as a percent of total DRG neurons.
The +X4 animals expressing A3-Cre and lsl-MRGPRX4 have high levels of MRGPRX4 mRNA in sensory neurons as assayed by both qPCR and RT-PCR (Fig. 2 B and C). Based on previously determined potencies, we selected DCA and UDCA for further experimentation. As assayed by Ca2+ imaging, DCA activated a subpopulation of small-diameter (∼20 μm) +X4 DRG neurons (Fig. 2D and SI Appendix, Fig. S2 A and B). At the tested concentration, TGR5 receptor present in both control (Ctrl) and +X4 neurons would be activated. Indeed, DCA activated 3.4% of control neurons, likely due to activation of TGR5 (Fig. 2E). Approximately 25% of control neurons activated by DCA were also activated by chloroquine (CQ), a ligand for Mrgpra3 (Fig. 2 F and G). Based on the genetic strategy employed for +X4 generation, we hypothesized that DCA would activate a higher percentage of Mrgpra3, CQ-sensitive, sensory neurons in +X4 animals. DCA activated 6.4% of +X4 sensory neurons (Fig. 2E). In addition, DCA activated significantly more CQ-sensitive, Mrgpra3-expressing, neurons in +X4 DRG (Fig. 2 F and G). Based on these data, we concluded that +X4 humanized sensory neurons expressed functional MRGPRX4 receptor that could be activated by DCA.
TGR5 has been previously identified as being expressed in Mrgpra3+ itch neurons (10). To better disambiguate MRGPRX4 and TGR5 activity, we selected UDCA as a ligand for additional experimentation. All BAs tested against MRGPRX4 are reported to activate TGR5 (31). However, UDCA, the second most potent activator of MRGPRX4, is a significantly less potent TGR5 agonist (17). Based on these data, we predicted that lower concentrations of UDCA would activate more +X4 neurons than control neurons. Indeed, UDCA activated a similar percentage of +X4 DRG neurons compared with DCA but activated a significantly smaller percentage of control neurons (Fig. 2 H and I). Additionally, UDCA activated only 10% of CQ-sensitive control neurons while activating 54% of +X4 neurons (Fig. 2 J and K).
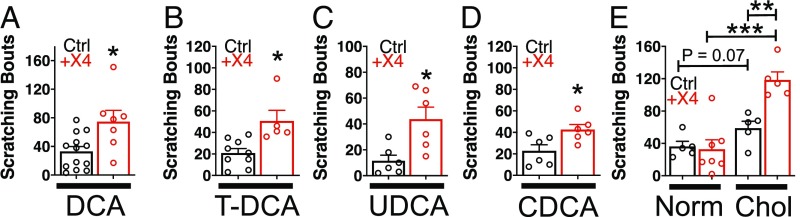
To test the in vivo relevance of bile acid agonism of MRGPRX4, we injected bile acids into control and +X4 animals and assessed acute itch. As before, the genetic strategy of MRGPRX4 humanization results in +X4 animals expressing MRGPRX4 receptor in Mrgpra3+ itch neurons. Therefore, we hypothesized that a true, in vivo ligand–MRGPRX4 interaction would activate Mrgpra3+ neurons and produce itch behavior. Compared with control, +X4 animals scratched significantly more in response to injection of pathophysiologic concentrations of DCA, UDCA, CDCA, and TDCA at the nape (Fig. 3 A–D). This increase was specific to BAs, as histamine and chloroquine, two canonical pruritogens, stimulated similar levels of itch in control and +X4 animals (SI Appendix, Fig. S3 A and B). Additionally, injection of cholestatic patient plasma, containing BAs and numerous other pruritogens (SI Appendix, Fig. S3C), elicited greater itch in +X4 animals compared with control animals (Fig. 3D). This difference between Ctrl and +X4 animals was not observed upon injection of control plasma (Fig. 3D). Compared with normal plasma, patient plasma elicited greater itch in control animals (Fig. 3D). This could result from numerous pruritogens acting at established itch receptors in mice, such as bilirubin at Mrgpra1 or BAs at TGR5.
Fig. 3.
Mice expressing MRGPRX4 scratch more after injection with unconjugated and conjugated BAs. (A) Scratching bouts associated with injection of DCA. A total of 50 µL of 1 mM DCA was injected into the nape of Ctrl and +X4 mice. Ctrl n = 13; +X4 n = 6. (B) Scratching bouts associated with injection of TDCA. A total of 50 µL of 1 mM TDCA was injected into the nape of Ctrl and +X4 mice. Ctrl n = 9; +X4 n = 5. (C) Scratching bouts associated with injection of UDCA. A total of 50 µL of 2 mM UDCA was injected into the nape of Ctrl and +X4 mice. Ctrl n = 6; +X4 n = 6. (D) Scratching bouts associated with injection of CDCA. A total of 50 µL of 2 mM CDCA was injected into the nape of Ctrl and +X4 mice. Ctrl n = 6; +X4 n = 6. (E) Scratching bouts associated with injection of human plasma from either normal or cholestatic patients (Chol) into Ctrl and +X4 mice. A total of 50 µL of plasma was injected into the nape of mice. For normal plasma: Ctrl n = 5; +X4 n = 7. For Chol plasma: Ctrl n = 5; +X4 n = 5. (A–D) Mean ± SEM depicted. Each open circle represents an individual mouse. (A–D) *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t test.
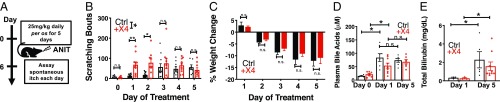
Having established that unconjugated and conjugated BAs elicited greater acute itch in +X4 animals, we sought to determine whether this observation remained true in a mouse model of cholestatic itch. To induce cholestasis, we administered α-napthyl isothiocyanate (ANIT) to mice. ANIT is a well-established model that elicits intrahepatic cholestasis in mice (27, 32). We treated control and +X4 animals with 25 mg/kg ANIT daily for 5 d while assessing spontaneous itch and weight 24 h after each treatment (Fig. 4A). Over the course of the experiment, cholestatic itch in +X4 animals was significantly different from control animals (Fig. 4B). The majority of this difference was observed at early timepoints. Specifically, +X4 animals scratched significantly more than baseline just 24 h after a single dose of ANIT (Fig. 4B). Subsequent treatment did not significantly increase pruritus. In control animals, itch was not significantly elevated until the third day of treatment (Fig. 4B). After day 3, control mice did not scratch more with subsequent treatments. During days 3–5 of treatment, itch between control and +X4 animals was indistinguishable (Fig. 4B). ANIT does not appear to directly activate MRGPRX4, since ANIT treatment did not elicit itch in either WT or +X4 animals immediately after treatment (SI Appendix, Fig. S4A). Control and +X4 animals displayed no differences in weight change from ANIT treatment. Both groups lost significant weight over the 5-d treatment course (Fig. 4C). Control and +X4 animals had similar degrees of cholestatic injury after either 1 or 5 d of ANIT treatment, as assessed by plasma levels of BAs, total bilirubin, albumin, aspartate aminotransferase (AST), alkaline phosphatase (ALP), and alanine aminotransferase (ALT) (Fig. 4 D and E and SI Appendix, Fig. S4 B–E).
Fig. 4.
The +X4 humanized mice display increased cholestatic itch concordant with serum BA elevation. (A) Experimental flowchart of cholestatic model. ANIT was given daily per os for 5 d. After 24-h treatment, spontaneous itch was assessed for 1 h. (B) Spontaneous scratching bouts from ANIT-treated animals. Itch was assessed for 1 h. Scratching bouts from Ctrl and +X4 were significantly different. P < 0.05 by ANOVA. For day 0: Ctrl n = 5, +X4 n = 7; for day 1: Ctrl n = 14, +X4 n = 11; for day 2: Ctrl n = 13, +X4 n = 9; for day 3: Ctrl n = 11, +X4 n = 9; for day 4: Ctrl n = 10, +X4 n = 9; and for day 5: Ctrl n = 7, +X4 n = 9. (C) Percent weight change from baseline of Ctrl and +X4 mice during ANIT treatment. For day 1: Ctrl n = 5, +X4 n = 7; for all other days: Ctrl n = 4, +X4 n = 6. (D) Plasma BA levels from Ctrl and +X4 mice at indicated days of ANIT treatment. For day 0: Ctrl n = 4, +X4 n = 4; for day 1: Ctrl n = 6, +X4 n = 9; for day 5: Ctrl n = 6, +X4 n = 7. (E) Total plasma bilirubin levels at indicated days of treatment. For day 1: Ctrl n = 3, +X4 n = 3; for day 5: Ctrl n = 5, +X4 n = 9. (B–E) Mean ± SEM depicted. Circles represent individual mice. n.s., not significant; *P < 0.05; **P < 0.01; Student’s t test.
Based on the observed time course of itch behavior, we hypothesized that humanized +X4 mice scratched more at earlier timepoints due to a ligand of MRGPRX4, such as BAs, being elevated during those periods. Indeed, at day 1, ANIT treatment resulted in significant elevation of plasma BAs, similar to levels observed on day 5. Importantly, plasma bilirubin, albumin, and liver enzyme levels were not elevated at day 1 (Fig. 4E and SI Appendix, Fig. S4 B–E). This distinction suggested that the increase in itch on days 1–2 in +X4 animals resulted from an increase in BAs and not bilirubin, another ligand of MRGPRX4. We hypothesized that +X4 cholestatic itch is similarly elevated compared with control at later timepoints (days 3–5) due to signaling from the intact bilirubin itch receptor Mrgpra1 in Ctrl animals and other potential pruritogen–receptor interactions.
Discussion
In this study, we demonstrate that numerous unconjugated and conjugated BAs are ligands for MRGPRX4, a human sensory neuron-expressed GPCR. BAs activated humanized (+X4) mouse sensory neurons at higher percentages, indicating that BAs are capable of interacting with neuronally expressed receptor. Humanized mice scratched more in response to both acute injection of BAs and a chronic model of cholestatic itch. Cholestatic itch in humanized mice rose in conjunction with plasma BA concentration. Our results strongly support that MRGPRX4 is a receptor for BAs. Given the receptor’s expression pattern in humans, it is possible that MRGPRX4 mediates a BA component of cholestatic itch.
Despite being structurally similar, BAs displayed a wide range of affinity for MRGPRX4. CA, the least potent BA, has 3α, 7α, and 12α hydroxyl groups. In contrast, the higher potency BAs, DCA and UDCA, have only a 3α hydroxyl and a 12α or 7β hydroxyl group, respectively. Based on this, we hypothesize that BA potency for MRGPRX4 is structurally determined by hydroxyl groups at the 7 and 12 position carbons, and their effects on hydrogen bonding, sterics, and water solubility. Cholesterol has no detectable activity toward MRGPRX4 and is most clearly structurally different from BAs in that cholesterol exhibits more planarity among its four rings due to a double bond between carbons 5 and 6.
UDCA, one of the more potent determined agonists of MRGPRX4, has been shown to reduce itch in a significant number of patients suffering from pruritus associated with intrahepatic cholestasis of pregnancy and primary biliary cholangitis (33–35). Our data suggest that UDCA could promote itch via MRGPRX4. There are many possible explanations for this apparent discrepancy. Notably, UDCA may act as an antipruritic agent by decreasing hepatocellular injury, inflammation, and fibrosis (33–35). UDCA treatment also does not appear to increase total serum BA levels despite being a BA. Rather, while serum UDCA levels increase, UDCA either decreases or leaves total serum BAs unchanged. The molecular mechanisms by which UDCA improves cholestatic injury could result from a combination of effects, such as improving hepatocellular function, immunosuppressive effects through unknown mechanisms, and improvements in BA flow through the digestive system. Improvement of cholestasis in the absence of total BA serum level elevation would be expected to improve pruritus and is consistent with our data.
The role of BAs in eliciting cholestatic itch in patients is complex. Cholestyramine, a BA chelating resin, aids in BA secretion and is frontline therapy for cholestatic pruritus. In a double-blind trial, cholestyramine was shown to be effective for treating itch. However, more recently, colesevalam, a BA-binding resin many times more effective than cholestyramine, displayed contradictory trial results (36). This discrepancy in the effectiveness of serum BA-modulating therapy is buttressed by clinical observations that serum BA levels do not correlate with pruritus. Not all patients with pruritus have elevated serum BAs, and not all patients with elevated serum BAs exhibit itch. For example, serum BA levels are maximally elevated during liver failure, but these patients often report little to no itch.
Based on these data, we hypothesize that cholestatic itch is multifactorial, with numerous pruritogens present in bile that likely activate a variety of neuronal receptors. It is also possible that a single pruritogen may activate multiple receptors or that multiple bile components activate the same receptor. For example, we recently identified that MRGPRX4 is activated by bilirubin at pathophysiologic concentrations observed in cholestasis (27). With this is mind, our data support a model whereby both BAs and bilirubin contribute to cholestatic itch via agonism at the same receptor, MRGPRX4. In this model, even without taking additional pruritogen–receptor interactions into account, serum levels of a single marker would not be expected to perfectly correlate with itch. Therefore, to properly interpret correlations relying on serum metabolite values, multiple components would likely need to be considered in relation to itch. Additional hypotheses for this lack of correlation include possible desensitization and internalization of MRGPRX4 during late-stage liver failure and/or populational polymorphisms in MRGPRX4 sequence that alter receptor affinity. Further research is required to address these possibilities. However, despite these open questions, our data suggest that MRGPRX4 may be a potential therapeutic target for cholestatic itch.
Methods
Animal Care and Use.
All experiments were performed in accordance with protocols approved by the Animal Care and Use Committee at The Johns Hopkins University School of Medicine. All mice used were 6- to 12-wk-old males and females (20–30 g) that had been generated on a 129svj/C57BL6J mixed background. There were no significant differences in itch between male and female mice.
Human Plasma Isolation.
Plasma from patients suffering from cholestasis was isolated under a protocol approved by the Institutional Review Board (IRB) at The Johns Hopkins University School of Medicine (study no. IRB00154650). Formal informed consent was obtained for this study. Control plasma was isolated from donors who did not exhibit kidney or liver disease, had no complaints of itch, and were free from any detectable viral infection (hepatitis C and B viruses and HIV) (control study no. NA_00013177, the Johns Hopkins Department of Dermatology Patient Database and Tissue Bank). Whole blood was collected into PAXgene tubes (PreAnalytiX 761115) and centrifuged for 5 min at 300 × g. Plasma was collected, aliquoted, and stored at −20 °C until experimentation. All samples were deidentified for this study.
Molecules and Preparation.
The following molecules were used (Sigma): DCA, chenodeoxycholic acid (CDCA), CA, UDCA, oleic acid, taurocholic acid, TDCA, taurochenodeoxycholic acid, ANIT, CQ, thapsigargin, FMRF peptide, BAM8-22 peptide, and histamine. PAMP9-20 was custom synthesized by Genscript. U73122 was obtained from Santa Cruz Biotechnology, YM-254890 from Wako Chemicals, and Fluo 4-AM and Fura 2-AM were from Molecular Probes.
Bile acids were freshly prepared before each experiment according to solubility and manufacturer’s recommendations. When applicable, bile acids were prepared in aqueous buffer, otherwise in either DMSO or 0.1N NaOH. Final concentration of DMSO in all applicable tested solutions was <0.5%. ANIT and cyclosporin A were dissolved in olive oil and freshly prepared as needed. All other drugs were prepared as 100 μL–1,000 μL aliquots and stored at −20 °C before thawing at 4 °C. Freeze/thaw cycles were avoided whenever possible.
Calcium Imaging and Analysis.
Behavioral assays were performed as previously described (27). Briefly, cells were imaged in calcium imaging buffer (CIB) (10 mM Hepes, 1.2 mM NaHCO3, 130 mM NaCl, 3 mM KCl, 2.5 mM CaCl2, 0.6 mM MgCl2, 20 mM glucose, and 20 mM sucrose at pH 7.4 and 290–300 mOsm). To monitor changes in intracellular [Ca2+], cells were loaded with either Fura 2-AM (HEK293 cells) or Fluo 4-AM (DRG neurons) for 30 min in the dark at 37 °C in CIB just before imaging. With Fura 2-AM, emission at 510 nm was monitored from excitation at both 340 nm and 380 nm. With Fluo 4-AM, emission at 520 nm was monitored from excitation at 488 nm.
HEK293 cells.
HEK293 cells stably expressing the murine G protein α-subunit Gα15, a unique Gα protein that nonselectively couples a large variety of GPCRs to PLC, and one MRGPR of interest were plated on poly-D-lysine–coated coverslips. After 12–24 h, cells were loaded with the Fura 2-AM. Unless otherwise specified, compounds were perfused into the imaging chamber for ∼15 s after a baseline period was established. Response was monitored at 5-s intervals for an additional 60 s. HEK293 cell lines stably expressing MRGPRs of interest were previously generated (27). MRGPRs tested include murine receptors: Mrgpra1, Mrgpra10, Mrgpra2, Mrgpra14, Mrgpra3, Mrgpra12, Mrgpra16, Mrgpra19, Mrgpra4, Mrgprc11, Mrgprb5, Mrgprb4, and human receptors: MRGPRX1, MRGPRX2, MRGPRX3, and MRGPRX4.
DRG neurons.
Unless otherwise noted, DRG neurons were imaged for 20 s to establish a baseline before compounds were added. At the end of every imaging trial, 50 mM KCl was added as a positive control. Cells were identified as responding if the intracellular [Ca2+] rose by either 30% compared with baseline or 50% compared with the [Ca2+]i change assayed during addition of 50 mM KCl. Cells included in calculating percentages displayed at least a 50% increase in [Ca2+]i compared with baseline upon addition of KCl. Damaged, detached, high-baseline, and motion-activated cells were excluded from analysis.
EC50 determinations.
HEK293 cells stably expressing Gα15 and MRGPRX4 were seeded in poly-D-lysine–coated 96-well plates at 25,000 cells per well and incubated overnight. Cells were loaded with FLIPR Calcium 5 dye (Molecular Devices) according to the manufacturer’s instructions in Hanks’ balanced salt solution (HBSS) with 20 mM Hepes, pH 7.4. BAs were freshly dissolved and diluted into HBSS with 20 mM Hepes buffer. Compounds were added and wells were imaged with a Flexstation 3 (Molecular Devices). EC50 values were determined from dose–responses performed in triplicate, repeated two to four times by subtracting minimum signal from maximum signal during the test period.
Generation of +X4 Humanized Mice.
The +X4 humanized mice were generated by crossing Tg(Mrgpra3-Cre) mice with newly generated ROSA26lslMRGPRX4 mice. Tg(Mrgpra3-Cre) mice were generated as previously described (20). A BAC clone containing the entire Mrgpra3 gene was modified to express EGFP-Cre and inserted using homologous recombination. ROSA26lslMRGPRX4 mice were generated by homologous recombination of an MRGPRX4 cDNA construct under loxp-STOP-loxp control to the ROSA26 locus.
DRG Dissociation and Culture.
DRG from all spinal levels were collected in cold DH10 media [90% DMEM/F-12, 10% FBS, penicillin (100 units/mL), and streptomycin (100 μg/mL)]. DRG were digested with a dispase (5 mg/mL)/collagenase type I (1 mg/mL) enzyme mixture at 37 °C for 45 min. After trituration, cells were spun at 300 × g and resuspended in DH10 before being plated on glass coverslips coated with poly-D-lysine (0.5 mg/mL) and laminin (10 μg/mL; Invitrogen) (if used for calcium imaging). Before imaging, DRG were cultured with DH10 supplemented with 50 ng/mL NGF at 37 °C overnight.
RT-PCR and qPCR.
RNA was purified from dissected DRG with a Qiagen RNEasy kit according to the manufacturer’s suggestions before first-strand cDNA generation using a SuperScript III kit (Invitrogen) according to the manufacturer’s instructions. A negative control reaction was included where SuperScript III reverse transcriptase was replaced by water. For RT-PCR, 25 μL PCR reactions were run with 12.5 μL RedTaq ReadyMix (Sigma), 0.5 μL dimethyl sulphoxide (DMSO), 0.25 μL each of 50 µM forward and reverse primer, 10 μL water, and 2 μL mixture from cDNA or negative control. Primers utilized for MRGPRX4 were forward, 5′-GCCTGAGTATGCTGAGCG and reverse, 5′-CTAAACAGCAGGGACAGGC. The PCR protocol utilized was 4 min at 95 °C, 30 s at 55 °C, 40 s at 72 °C, and 25 s at 95 °C (with the prior three steps repeated 35 times), and a final 4-min step at 72 °C. For qPCR, TaqMan primers for human MRGPRX4 and murine β-actin were ordered from Thermo Fisher Scientific. Reactions consisted of 7.5 μL EagleTaq Universal master mix with ROX (Sigma), 5 μL of RNase-free water, 0.5 μL primers, and 2 μL cDNA. Samples were run in triplicate using an Applied Biosystems QuantStudio 6 Flex Real-Time PCR system. The ΔΔCt method was used to quantify relative amounts of cDNA for MRGPRX4 normalized to β-actin.
Behavioral Studies.
Behavioral assays were performed as previously described (27), with a few relevant adjustments. All applicable behavioral tests were performed and analyzed with the experimenter blind to genotype. All behavior tests between Ctrl and +X4 humanized mice were performed with littermates. Briefly, all behavior experiments were performed between 8 AM and 12 PM. On the day before the experiment, animals were placed in the test chamber for 30 min before being subjected to a series of three mock injections with 5-min break periods in between. On the day of the experiment, animals were first allowed to acclimatize to the test chamber for 10 min before injection. Pruritic compounds were s.c. injected into the nape of the neck (50 μL volume), and scratching behavior was observed for 30 min. A bout of scratching was defined as a continuous scratching movement directed by either hindpaw to the injection site. Scratching behavior was quantified by counting the number of scratching bouts within the 30-min observation period. For plasma itch studies, plasma from three cholestatic patients and three control patients were separately pooled and utilized for experimentation. After injection, animals were observed for 60 min.
Mouse Models of Cholestasis and Sample Collection.
ANIT (Sigma) was solubilized in olive oil (Sigma). Animals were dosed with 25 mg/kg ANIT per os daily for 5 d. One day before treatment, animals were acclimatized for itch behavior. Each day, 24 h subsequent to treatment, animals were placed in test chambers and videotaped for 1 h before being weighed. The number of scratching bouts were counted and binned in 5-min intervals during this period. When behavior assessment concluded, animals were administered pentobarbital (50 mg/kg, i.p.) and blood was collected via cardiac puncture and placed into heparinized tubes (BD Biosciences). After centrifugation, plasma was collected, aliquoted, and stored at −80 °C until analysis. BA levels were assessed by a fluorometric kit from Cell Biolabs. All other reported plasma substances were determined by the Johns Hopkins Animal Pathology Lab.
Data Analysis.
Data were analyzed as previously described (27). Group data were expressed as mean ± SEM unless otherwise noted. Two-tailed unpaired Student’s t tests, two-way ANOVA, and χ2 tests were used to determine significance in statistical comparisons. Differences were considered significant at P < 0.05. Statistical power analysis was used to justify sample size, and variance was determined to be similar among all treatment groups as determined by F test. No samples or animals subjected to successful procedures and/or treatments were excluded from analysis. All behavior experiments were designed in a blocked manner with consideration for both genotype and treatment.
Supplementary Material
Acknowledgments
We thank Dr. James Hamilton and Dr. Shawn Kwatra for assisting with the IRB-approved collection of patient plasma samples and Dr. Caiying Guo and the Gene Targeting and Transgenics Resources team at Janelia Research Campus for assistance in generating the ROSA26-lslMRGPRX4 mice. This work was supported by NIH Grants R01NS054791 (to X.D.) and MH18501 (to S.H.S.).
Footnotes
Conflict of interest statement: J.M. is a consultant for Escient Pharmaceuticals, a company focused on developing small molecule inhibitors for MRGPRs. X.D. is a co-founder of Escient Pharmaceuticals.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903316116/-/DCSupplemental.
References
- 1.Hegade VS, et al. (2016) The safety and efficacy of nasobiliary drainage in the treatment of refractory cholestatic pruritus: A multicentre European study. Aliment Pharmacol Ther 43:294–302. [DOI] [PubMed] [Google Scholar]
- 2.Beuers U, Kremer AE, Bolier R, Elferink RP (2014) Pruritus in cholestasis: Facts and fiction. Hepatology 60:399–407. [DOI] [PubMed] [Google Scholar]
- 3.Bergasa NV. (2014) Pruritus of cholestasis. Itch: Mechanisms and Treatment, Frontiers in Neuroscience, eds Carstens E, Akiyama T (CRC Press, Boca Raton, FL).
- 4.Hofmann AF, Huet PM (2006) Nasobiliary drainage for cholestatic pruritus. Hepatology 43:1170–1171. [DOI] [PubMed] [Google Scholar]
- 5.Appleby VJ, Hutchinson JM, Davies MH (2015) Safety and efficacy of long-term nasobiliary drainage to treat intractable pruritus in cholestatic liver disease. Frontline Gastroenterol 6:252–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergasa NV, et al. (1995) Effects of naloxone infusions in patients with the pruritus of cholestasis. A double-blind, randomized, controlled trial. Ann Intern Med 123:161–167. [DOI] [PubMed] [Google Scholar]
- 7.Bergasa NV, et al. (1992) A controlled trial of naloxone infusions for the pruritus of chronic cholestasis. Gastroenterology 102:544–549. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann AF. (2002) Rifampicin and treatment of cholestatic pruritus. Gut 51:756–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Airede AK, Weerasinghe HD (1996) Rifampicin and the relief of pruritus of hepatic cholestatic origin. Acta Paediatr 85:887–888. [DOI] [PubMed] [Google Scholar]
- 10.Alemi F, et al. (2013) The TGR5 receptor mediates bile acid-induced itch and analgesia. J Clin Invest 123:1513–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alemi F, et al. (2013) The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 144:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieu T, Jayaweera G, Bunnett NW (2014) GPBA: A GPCR for bile acids and an emerging therapeutic target for disorders of digestion and sensation. Br J Pharmacol 171:1156–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K (2011) The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol 54:1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varadi DP. (1974) Pruritus induced by crude bile and purified bile acids. Experimental production of pruritus in human skin. Arch Dermatol 109:678–681. [PubMed] [Google Scholar]
- 15.Kirby J, Heaton KW, Burton JL (1974) Pruritic effect of bile salts. BMJ 4:693–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama T, et al. (2002) Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun 298:714–719. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata Y, et al. (2003) A G protein-coupled receptor responsive to bile acids. J Biol Chem 278:9435–9440. [DOI] [PubMed] [Google Scholar]
- 18.Hodge RJ, et al. ; SB-756050 Project Team (2013) Safety, pharmacokinetics, and pharmacodynamic effects of a selective TGR5 agonist, SB-756050, in type 2 diabetes. Clin Pharmacol Drug Dev 2:213–222. [DOI] [PubMed] [Google Scholar]
- 19.Bergasa NV. (2008) Pruritus in primary biliary cirrhosis: Pathogenesis and therapy. Clin Liver Dis 12:385–406, x. [DOI] [PubMed] [Google Scholar]
- 20.Han L, et al. (2013) A subpopulation of nociceptors specifically linked to itch. Nat Neurosci 16:174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meixiong J, Dong X (2017) Mas-related G protein-coupled receptors and the biology of itch sensation. Annu Rev Genet 51:103–121. [DOI] [PubMed] [Google Scholar]
- 22.McNeil B, Dong X (2012) Peripheral mechanisms of itch. Neurosci Bull 28:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lembo PM, et al. (2002) Proenkephalin A gene products activate a new family of sensory neuron–Specific GPCRs. Nat Neurosci 5:201–209. [DOI] [PubMed] [Google Scholar]
- 24.McNeil BD, et al. (2015) Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature 519:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flegel C, et al. (2015) RNA-seq analysis of human trigeminal and dorsal root ganglia with a focus on chemoreceptors. PLoS One 10:e0128951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goswami SC, et al. (2014) Itch-associated peptides: RNA-seq and bioinformatic analysis of natriuretic precursor peptide B and gastrin releasing peptide in dorsal root and trigeminal ganglia, and the spinal cord. Mol Pain 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meixiong J, et al. (2019) Identification of a bilirubin receptor that may mediate a component of cholestatic itch. eLife 8:e44116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozlitina J, et al. (2019) An African-specific haplotype in MRGPRX4 is associated with menthol cigarette smoking. PLoS Genet 15:e1007916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lansu K, et al. (2017) In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat Chem Biol 13:529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Offermanns S, Simon MI (1995) G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J Biol Chem 270:15175–15180. [DOI] [PubMed] [Google Scholar]
- 31.Sato H, et al. (2008) Novel potent and selective bile acid derivatives as TGR5 agonists: Biological screening, structure-activity relationships, and molecular modeling studies. J Med Chem 51:1831–1841. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka Y, Aleksunes LM, Cui YJ, Klaassen CD (2009) ANIT-induced intrahepatic cholestasis alters hepatobiliary transporter expression via Nrf2-dependent and independent signaling. Toxicol Sci 108:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuzaki Y, et al. (1990) Improvement of biliary enzyme levels and itching as a result of long-term administration of ursodeoxycholic acid in primary biliary cirrhosis. Am J Gastroenterol 85:15–23. [PubMed] [Google Scholar]
- 34.de Caestecker JS, Jazrawi RP, Petroni ML, Northfield TC (1991) Ursodeoxycholic acid in chronic liver disease. Gut 32:1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glantz A, et al. (2008) Intrahepatic cholestasis of pregnancy: Amelioration of pruritus by UDCA is associated with decreased progesterone disulphates in urine. Hepatology 47:544–551. [DOI] [PubMed] [Google Scholar]
- 36.Kuiper EM, et al. (2010) The potent bile acid sequestrant colesevelam is not effective in cholestatic pruritus: Results of a double-blind, randomized, placebo-controlled trial. Hepatology 52:1334–1340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.