Significance
Microtubules are polymers of αβ-tubulin that play important roles in the cell. Regulation of their dynamics is critical for function and includes the posttranslational modification of tubulin. While most of tubulin modifications reside in the flexible C-terminal tail of tubulin, acetylation of α-tubulin on K40 is localized to the inside of the microtubule, within the so-called αK40 loop. Using high-resolution cryo-EM maps of acetylated and deacetylated microtubules, in conjunction with molecular-dynamics methods, we found that acetylation restricts the range of motion of the αK40 loop. In the deacetylated state, the loop extends deeper into the microtubule lumen and samples a greater number of conformations that we propose increase its accessibility to the acetylase and likely influence lateral contacts.
Keywords: cryo-EM, MD, tubulin modifications, microtubule, acetylation
Abstract
Acetylation of K40 in α-tubulin is the sole posttranslational modification to mark the luminal surface of microtubules. It is still controversial whether its relationship with microtubule stabilization is correlative or causative. We have obtained high-resolution cryo-electron microscopy (cryo-EM) reconstructions of pure samples of αTAT1-acetylated and SIRT2-deacetylated microtubules to visualize the structural consequences of this modification and reveal its potential for influencing the larger assembly properties of microtubules. We modeled the conformational ensembles of the unmodified and acetylated states by using the experimental cryo-EM density as a structural restraint in molecular dynamics simulations. We found that acetylation alters the conformational landscape of the flexible loop that contains αK40. Modification of αK40 reduces the disorder of the loop and restricts the states that it samples. We propose that the change in conformational sampling that we describe, at a location very close to the lateral contacts site, is likely to affect microtubule stability and function.
Microtubules (MTs) are essential cytoskeletal polymers important for cell shape and motility and critical for cell division. They are built of αβ-tubulin heterodimers that self-assemble head-to-tail into protofilaments (PFs). PFs associate in parallel to form a polar, hollow tube (2), with the most common number of PFs being 13, but with variations present depending on species or on in vitro polymerization conditions. Lateral contacts between PFs involve key residues in the so-called M-loop (between S7 and H9) in one tubulin monomer and the H2–H3 loop and β-hairpin structure in the H1′–S2 loop of the other tubulin subunit across the lateral interface. These lateral contacts are homotypic (α–α and β–β contacts), except at the MT “seam,” where the contacts are heterotypic (α–β and β–α contacts) (3). MTs undergo dynamic instability, the stochastic switching between growing and shrinking states (4, 5). These dynamics are highly regulated in vivo by multiple mechanisms that affect tubulin and its interaction with a large number of regulatory factors.
One mechanism that cells can use to manipulate MT structure and function involves the posttranslational modification (PTM) of tubulin subunits. Through the spatial–temporal regulation of proteins by the covalent attachment of additional chemical groups, proteolytic cleavage or intein splicing, PTMs can play important roles in controlling the stability and function of MTs (6). Most of tubulin PTMs alter residues within the highly flexible C-terminal tail of tubulin that extends from the surface of the MT and contributes to the binding of microtubule-associated proteins (MAPs) (7, 8). These PTMs include detyrosination, Δ2-tubulin generation, polyglutamylation, and polyglycylation (9). However, acetylation of α-tubulin on K40 stands out as the main tubulin PTM that localizes to the inside of the MT, within a loop of residues P37 to D47, often referred to as the αK40 loop. This modification is carried out by α-tubulin acetyltransferase αTAT1 and removed by the NAD+-dependent deacetylase SIRT2 and by HDAC6 (7, 10). Despite some interesting functional studies concerning this modification, structural insights into the effect of this “hidden PTM” on MT properties are still missing.
Shortly after its discovery over 30 y ago (11), acetylation of αK40 was found to mark stable, long-lived (t1/2 > 2 h) MT subpopulations, including the axonemes of cilia and flagella or the marginal bands of platelets (7, 10), and to protect MTs from mild treatments with depolymerizing drugs, such as colchicine (12) and nocodazole (13). Multiple studies have suggested that reduced levels of αK40 acetylation cause axonal transport defects associated with Huntington’s disease, Charcot–Marie–Tooth disease, amyotrophic lateral sclerosis, and Parkinson’s disease (14–17). These defects can be reversed by restoring αK40 acetylation levels (18). On the other hand, elevated levels of αK40 acetylation promote cell–cell aggregation, migration, and tumor reattachment in multiple aggressive, metastatic breast cancer cell lines (19, 20). While these studies imply that tubulin acetylation has an effect on axonal growth and transport, a mechanistic model is still missing.
Whether acetylated MTs are stable because they are acetylated or whether stable structures are better at acquiring this modification remains a point of contention. For example, a previous study showed that acetylation did not affect tubulin polymerization kinetics in vitro (21). However, this study was confounded by two factors: (i) MTs acetylated by flagellar extract were compared with native brain tubulin, which is ∼30% acetylated, and (ii) only a single round of polymerization/depolymerization was performed after in vitro acetylation, which is insufficient to remove αTAT1 or other MAPs. Thus, the results of that study may be limited by the purity and preparation of the sample. Our previous structural work also found no significant differences between 30% acetylated and 90% deacetylated MTs at a resolution of ∼9 Å, particularly at the modification site, the αK40 residue within the αK40 loop, which was invisible in both cases due to the intrinsic disorder and/or to the remaining heterogeneity of the loop (i.e., that study may have been limited by the low purity of the samples) (10). More recent in vitro studies, using pure samples of 96% acetylated and 99% deacetylated MTs, argue that αK40 acetylation induces a structural change that improves the flexibility and resilience of MTs (22, 23). These studies find that acetylated MTs maintain their flexural rigidity, or persistence length, after repeated rounds of mechanical stress, while deacetylated MTs show a 50% decrease in rigidity and are 26% more likely to suffer from complete breakage events (22, 23).
Since the αK40 residue is less than 15 Å away from the lateral interface between PFs, a possible model for the molecular mechanism of acetylation is that it alters inter-PF interactions by promoting a conformation of the αK40 loop that confers flexural rigidity, thus increasing its resistance to mechanical stress—a phenomenon called PF sliding (22, 23). Molecular dynamics (MD) simulations have suggested a model where αK40 forms a stabilizing salt bridge with αE55 within the core of the α-tubulin monomer that in turn stabilizes αH283 within the M-loop of its neighboring α-tubulin monomer (24). Another study proposed that αK40 acetylation may specify 15-PF MTs, which are known to be 35% stiffer than 13-PF MTs and more effective at forming MT bundles (25).
Given the uncertainties remaining concerning the effect of αK40 acetylation on MTs, we decided to characterize the conformational properties of the αK40 loop in the acetylated and deacetylated MTs that could have an effect on MT structure and properties. To that end, we produced near atomic-resolution cryo-EM maps of 96% acetylated (Ac96) and 99% deacetylated (Ac0) MTs. By improving sample purity, we were able to visualize more density for the αK40 loop in the acetylated state. Using MD methods to fit the cryo-EM map, we found that acetylation shifts the conformational landscape of the αK40 loop by restricting the range of motion of the loop. In contrast, in the Ac0 state, the αK40 loop extends deeper into the lumen of the MT, and samples a greater number of conformations. These motions are likely to increase the accessibility of the loop to αTAT1, in agreement with the hypothesis that αTAT1 acts by accessing the MT lumen (26), and likely influence lateral contacts, in agreement with the causative effect of acetylation on the mechanical properties of MTs (22).
Results and Discussion
High-Resolution Cryo-EM Reconstructions of Pure Acetylated (Ac96) and Deacetylated (Ac0) MTs.
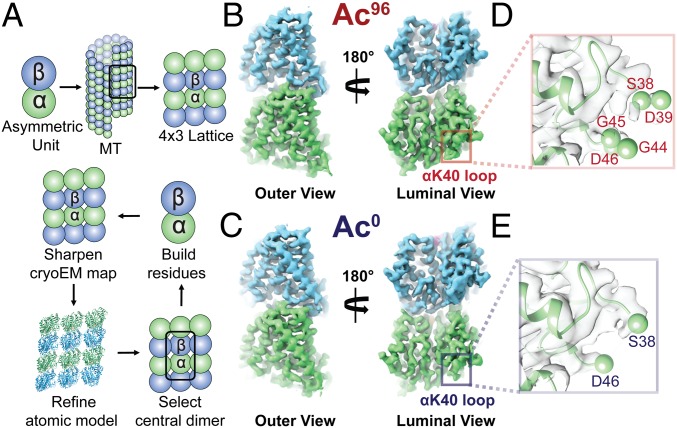
Using recent biochemical schemes designed to enrich for specific acetylation states (22), we generated Ac96 and Ac0 MTs for use in our cryo-EM studies. We prepared cryo-EM samples as previously described (3, 27) of Ac96 and Ac0 MTs in the presence of end-binding protein 3 (EB3). EB3 served as a fiducial marker of the dimer that facilitated alignment of MT segments during image processing (28). SI Appendix, Table S1 summarizes the data collection, refinement, and validation statistics for each high-resolution map we visualized (SI Appendix, Figs. S1 and S2). Using the symmetrized MT reconstruction, which takes advantage of the pseudohelical symmetry present in the MT, we extracted a 4 × 3 array of dimers for further B-factor sharpening, refinement (29), and model building (Fig. 1A). This array includes all possible lateral and longitudinal nonseam contacts for the central dimer, which was later extracted for model building and map analysis (Fig. 1 B and C).
Fig. 1.
High-resolution maps of 96% acetylated (Ac96) and <1% acetylated (Ac0) MTs. (A) Schematic of the model-building and refinement process in PHENIX. We sharpened a representative 4 × 3 lattice, refined the corresponding atomic structure (3JAR) into our map, and extracted out the central dimer to build additional residues into the αK40 loop. We performed this process iteratively for both the Ac96 and Ac0. The structure of the Ac96 (B) and Ac0 (C) αβ-tubulin heterodimers, respectively, are shown from the outer and luminal views with close-ups of αK40 loop in each state (D and E) low-pass filtered to 3.7 Å.
The αK40 loop has been poorly resolved in previous EM reconstructions, and existing models contain a gap between residues Pro37 and Asp48 (SI Appendix, Fig. S3A) (26). While the loop has been resolved in a number of X-ray crystallographic structures, the conformations stabilized in the crystal lattice are likely consequences of the presence of calcium and/or crystal contacts (SI Appendix, Fig. S3B). For our symmetrized maps, we were able to build residues S38–D39 and G44–D47 into the Ac96 state and S38 and D46–D47 into the Ac0 state (Fig. 1 D and E). Qualitatively, the maps suggest that the αK40 loop is slightly more ordered in the Ac96 state, with the protrusion of density following Pro37 extending away from or toward Asp48 in the Ac96 or Ac0 states, respectively. However, it is likely that multiple conformations of the loop, perhaps as a function of each loop’s individual position around a helical turn, are averaged together and result in the low signal-to-noise levels we observe in the map.
Conformational Differences Across MT States Are Confirmed by Nonsymmetrized Reconstructions.
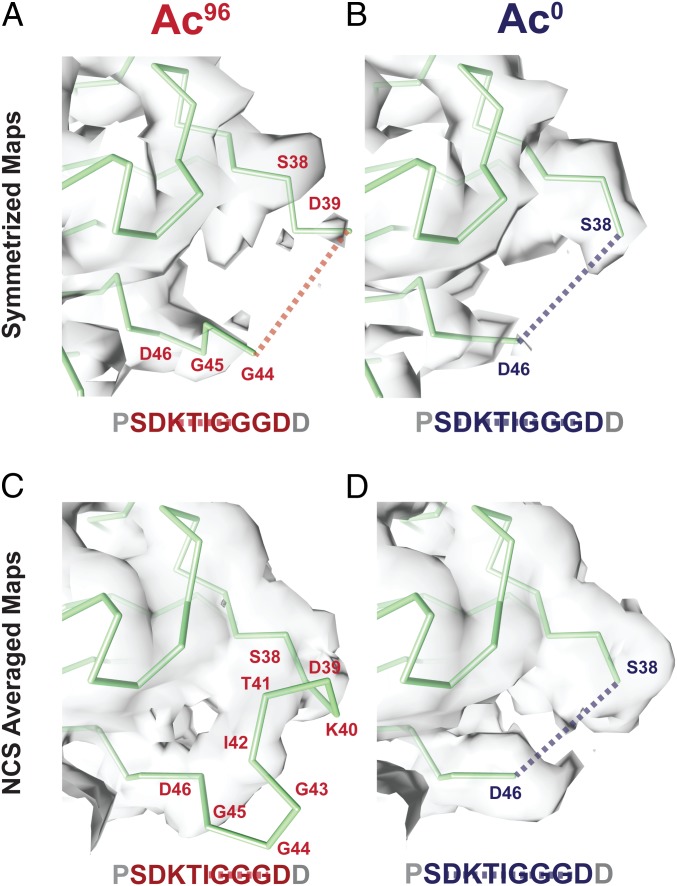
We considered the possibility that the symmetrizing procedure used to improve signal and resolution in our image analysis was averaging different αK40 loop conformation within different PFs and thus interfering with our interpretation of the loop structure in the two states. To test the hypothesis, we analyzed the nonsymmetrized maps calculated with C1 symmetry for the Ac96 and Ac0 states. We extracted a full turn of 13 adjacent dimers. This full-turn map revealed additional density extending out further along the loop in the Ac96 state compared with the symmetrized maps filtered to the same resolution (4 Å) (Fig. 2). Furthermore, the density for the loop observed at the seam was distinct from that at the nonseam contacts. To maximize the interpretability of the subunits making nonseam contacts, we used noncrystallographic symmetry (NCS) averaging as an alternative method to increase the signal-to-noise levels in the maps. This procedure improved the density for nonglycine backbone atoms in the αK40 loop in the Ac96 state, allowing us to trace an initial Cα backbone for this region, while in the Ac0 state the loop remained unmodelable (Fig. 2 C and D). This interpretation agrees with the qualitative difference in the density, which indicate less disorder for the Ac96 state than Ac0 state, of the traditionally symmetrized and C1 maps.
Fig. 2.
Symmetrized and NCS-averaged C1 maps of Ac96 and Ac0 MTs reveal the αK40 loop is more ordered in the Ac96 state. Close-up views of the αK40 loop (P37–D47) in the (A) Ac96 and (B) Ac0 states in the symmetrized maps low-pass filtered to 4 Å and the (C) Ac96 and (D) Ac0 states in the NCS-averaged C1 maps low-pass filtered to 4 Å. The dotted lines indicate missing residues.
This NCS averaging method had multiple advantages over the traditional averaging technique for pseudohelical processing implemented in FREALIGN (27). First, the model coordinates used for the averaging are based on the matrix of α-tubulin monomers along a full turn rather than the single α-tubulin monomer. Second, in the FREALIGN averaging approach, the signal from the dimers at the seam are down-weighted, whereas NCS averaging allows us to separate the signal from the seam, and thus to deconvolute the signal from the nonseam locations. Third, this procedure also acts to low-pass filter the map to 4 Å (the high-resolution limit of the C1 map; SI Appendix, Fig. S6), which should suppress noise from the more disordered parts of the map, including alternative conformations of the αK40 loop. Using this NCS-based approach, we were able to resolve density and build a model for three additional residues, the acetylated K40, T41, and I42. These residues pack toward the globular domain of α-tubulin, consistent with the favorability of burying these relatively hydrophobic residues in the Ac96 state. Despite observing only very weak density, we have modeled the glycine-rich region that extends into the lumen as a tight turn, which we note is only possible due to the expanded Ramachandran space accessible to glycine residues (Fig. 2C). In contrast, and despite better global resolution, we did not observe any density consistent with a stable conformation of the loop in the Ac0 map. Based on this result, which is consistent across the NCS-averaged and traditionally symmetrized maps, we did not build any additional residues into the Ac0 density (Fig. 2D).
Ensemble Modeling of the Loop in Each State Using Density-Restrained MD.
For regions that exhibit a high degree of disorder, like the αK40 loop, a single, static structure is a poor description of the native state. Ensemble models can help to elucidate how populations of conformations change upon perturbations, such as PTMs (30, 31). To derive an ensemble of conformations representing the Ac96 and Ac0 states, we used the atomic structure built into the Ac96 map as the starting model to initiate metainference-based MD simulations, which augment a standard force field with a term representing the density derived from the cryo-EM map (32). In contrast to molecular dynamics and flexible fitting (MDFF) and other refinement methods that seek to converge on a single structure (33), this method models a structural ensemble by maximizing the collective agreement between simulated and experimental maps, and accounts for noise using a Bayesian approach (34). Initiating simulations for both the Ac96 and Ac0 states from starting models that differ only in the acetyl group and distinct input experimental density maps allowed us to test whether acetylation restricts the motion of the loop, trapping it in a tighter ensemble of conformations.
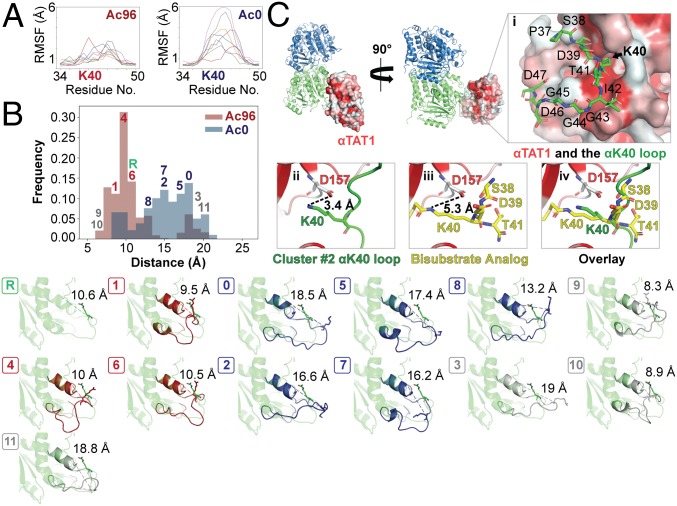
To analyze the conformational dynamics of the loop, we analyzed the root-mean-square fluctuations of residues 36–48 within replicas for each simulation. This analysis shows that the αK40 loop fluctuations are more restricted in the Ac96 state than in the Ac0 state (Fig. 3A). Next, we analyzed the distribution of conformations adopted by the loop by analyzing the distance between K40 and the globular domain of α-tubulin (represented by L26) and by clustering together the snapshots from all replicas of both simulations based on the root-mean-square deviations (RMSDs) of residues 36–48. Similar to the starting reference model, where the distance is 10.6 Å, Ac96 is enriched in conformations that pack close to the globular domain of the α-tubulin core (Fig. 3B). These conformations, exemplified by clusters 1, 4, and 6, position the acetylated K40 to interact with residues along H1. In contrast, the Ac0 state favors conformations that extend toward the MT lumen, as exemplified by clusters 0, 2, 5, 7, and 8 (Fig. 3B). Clusters 3, 9, 10, and 11, labeled in gray, had equal numbers of frames enriched in Ac96 and Ac0 and sampled rare (<5%) extreme states on both the exposed and packed ends of the conformational spectrum (Fig. 3B).
Fig. 3.
Acetylation restricts the motion and alters the conformational ensemble of the αK40 loop. (A) Per-residue root-mean-square fluctuation (RMSF) analyses were determined over the course of 12 ns for residues 34–50 the C1 maps using GROMACs in PLUMED and graphed using the MDAnalysis. The different colored lines refer to the eight different replicas. (B) Ensemble modeling of the loop across Ac96 and Ac0 states using density restrained MD. Frames were classified into 1 of 11 clusters by conformation. Clusters either had a greater number of Ac96 frames (red), Ac0 frames (blue), or an equal number of frames from both states (gray). The reference is shown in green. The unique conformations of each of the 12 clusters (0–11) are shown below. (C) αTAT1 cocrystalized with a bisubstrate analog consisting of α-tubulin residues 38–41 (PDB ID code 4PK3) is shown bound to αβ-tubulin sampling the αK40 loop of cluster 2 from the metainference ensemble. αTAT1 is colored by hydrophobicity, where hydrophobic regions are red. (C, i) After backbone alignment of the surrounding residues of the bisubstrate analog, αK40 from cluster 2 fits directly into the catalytic groove, highlighted by the black arrow. (C, ii–iv) Similar to the bisubstrate analog (shown in yellow), αK40 from the cluster #2 loop (shown in green) is positioned very close to αTAT1:D157, which stabilizes the enzyme–tubulin interface.
These computational results are consistent with the visual analysis of the density for both the NCS and traditionally symmetrized maps, which indicated that the loop is more ordered after acetylation. The residual disorder identified by the simulations using the Ac96 map may be important for deacetylation by SIRT2. On the other hand, the increased flexibility we observe for the Ac0 state suggests a potential mechanism by which αTAT1 could acetylate K40. Previous proposals argued that acetylation can occur from the outside (35) or inside of the lumen (26). However, to catalyze the modification, a flexible region within αTAT1 would have to extend ∼25 Å through a MT wall fenestration between four tubulin dimers to reach αK40, or the MT would have to undergo a major structural rearrangement in the lattice to allow αTAT1 to enter the lumen. Previous work demonstrated that the αTAT1 active site and its MT recognition surface is concave and could not stretch through the lumen (26). Our findings support the idea that αTAT1 modifies the loop from within the lumen of the MT because the deacetylated loop samples extended structures that could be accessible to αTAT1 and because the structural rearrangement caused by acetylation is small and local to the αK40 loop.
To test this idea, we superposed the backbone of the bisubstrate analog consisting of α-tubulin residues 38–41 from the αTAT1 cocrystal structure (PDB ID code 4PK3) with representatives of the clusters from the metainference ensemble. Not surprisingly, most arrangements generated severe clashes between αTAT1 and the globular domain of α-tubulin (SI Appendix, Fig. S4). However, the model from cluster 2 had only a single severe clash (between αTAT1:R74 and α-tubulin:G57) and displayed reasonable surface complementarity with αTAT1, positioning αK40 directly into the catalytic groove (Fig. 3C, i). Here, αK40 remains within a close proximity of αTAT1:D157 (Fig. 3C, ii–iv), which coordinates an essential hydrogen-bond network at the enzyme–tubulin interface and when mutated decreases acetylation by ∼96% and ∼92% for free tubulin and within assembled MTs, respectively (26). Notably, although cluster 2 was not the most extended conformation, it was highly enriched in the Ac0 ensemble. This finding bolsters the model that acetylation occurs from inside the lumen (26) and suggests that deacetylated α-tubulin samples the conformations that are competent for catalysis by αTAT1 with relatively little energetic cost.
Acetylation Induces a Local Structural Rearrangement of the αK40 Loop That Promotes Stability by Weakening Lateral Contacts.
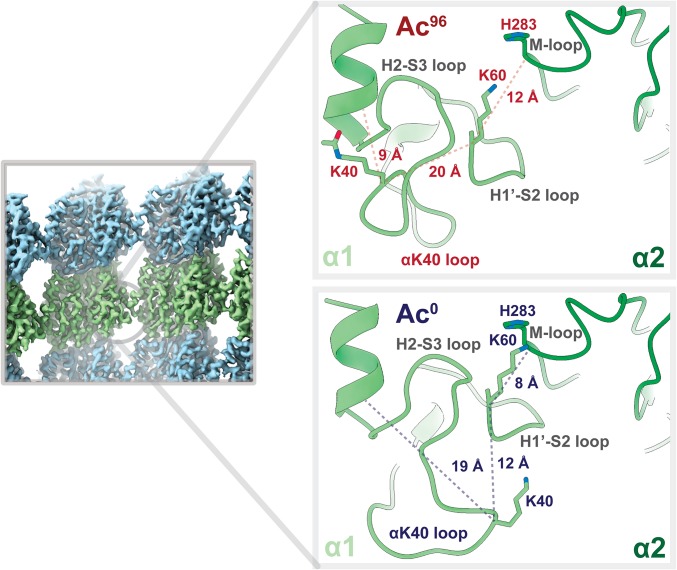
Collectively our structural and MD results show that acetylation restricts the motion of the αK40 loop. These results led us to hypothesize that the change in the structural ensemble of the αK40 loop upon acetylation, while subtle and local, may affect lateral contacts. These local changes may disrupt the small lateral interface between α-tubulin subunits. The origin of this effect may be highly distributed, as we do not visualize any stable interactions between the Ac0 state of the loop and the globular domain. However, upon acetylation, the structural ensemble becomes more restricted and the potential for the loop to strengthen any of these interactions between monomers is lost. For example, in many of the extended conformations favored by the Ac0 state, K40 in a α1-monomer is close to the M-loop of the neighboring α2-monomer and may buttress the H1′–S2 loop, providing support for the vital α1K60:α2H283 lateral interaction (Fig. 4). In contrast, when K40 is acetylated, it packs ∼10 Å closer to the globular domain of the α1-monomer, reducing the potential for intermonomer interactions (Fig. 4).
Fig. 4.
Acetylation may weaken lateral interactions. Close-up view of the lateral contacts between two α-tubulin monomers at a nonseam location (α1, light green; α2, dark green). K40 in α1 of the Ac0 state is 8 Å closer to the M-loop of α2 and appears to buttress the H1′–S2 loop, providing support for the vital α1K60–α2H283 lateral interaction. By contrast, that support is lost in the Ac96 state because the acetylated K40 now packs much closer to the hydrophobic, inner core.
We tested whether K40 acetylation alters the electrostatic interaction energy and the hydrogen-bonding network at the lateral interface by estimating the electrostatic interactions. We used the Debye–Hückel (DH) formula to calculate interaction energies for each conformation sampled in the two metainference ensembles and compared the resulting distributions (36–38). We found that acetylation does indeed weaken lateral interactions (SI Appendix, Fig. S5). Additionally, the Ac0 ensemble contains conformations with strong DH interaction energies that do not exist in the Ac96 ensemble (SI Appendix, Fig. S5). While the effects of acetylation are subtle, the local effects at the lateral contacts site may have an additive effect that stabilizes the MT lattice. This idea is consistent with previous work that argues that the weakening of lateral interactions is a protective mechanism to prevent preexisting lattice defects from spreading into large areas of damage under repeated stress—a mechanism that could be exploited by cancer cells (22, 23).
In conclusion, this integrative approach combines the structural insight of cryo-EM with the sampling efficiency of MD to investigate how PTMs can transform a conformational ensemble (39, 40). Our high-resolution maps serve as a blueprint for the scale of conformational change and relevant degrees of freedom that the αK40 loop can sample. We show that acetylation of tubulin induces electrostatic perturbations that restrict the motion of the αK40 loop, weakening lateral contacts. The sum of many weak reduced lateral contacts reduces the inter-PF interactions. The increased flexibility provided by fewer inter-PF interactions provides a mechanism by which αTAT1 can locally fine-tune the load-bearing capacity and mechanical resistance to stress of MTs (22, 41). Deacetylation increases lateral contacts, which renders MTs inflexible, brittle, and highly susceptible to breakage under stress (22, 23). The pattern of increased lateral interactions between subunits leading to mesoscale instability may be conserved in other polymeric systems (42–44). Therefore, αK40 acetylation may function as an evolutionarily conserved “electrostatic switch” to regulate MT stability (39, 40). Cancer cells may exploit this subtle form of regulation to promote cell adhesion, invasive migration, and other markers of aggressive metastatic behavior (19).
Materials and Methods
Sample Preparation for Cryo-EM.
Porcine brain tubulin was purified as previously described (45) and reconstituted to 10 mg/mL in BRB80 buffer [80 mM Pipes, pH 6.9, 1 mM ethylene glycol tetraacetic acid (EGTA), 1 mM MgCl2] with 10% (vol/vol) glycerol, 1 mM GTP, and 1 mM DTT, and flash-frozen in 10-μL aliquots until needed. The acetylated and deacetylated MTs (15 µM) were copolymerized with EB3 (25 µM), at 37 °C for ∼15 min in the presence of 10% Nonidet P-40, 1 mM DTT, and BRB80 buffer. The EB3-decorated MTs were added to glow-discharged C-flat holey carbon grids (CF-1.2/1.3–4C, 400 mesh, copper; Protochips) inside a Vitrobot (FEI) set at 37 °C and 85% humidity before plunge-freezing in ethane slush and liquid nitrogen, respectively, as previously described (3).
Cryo-EM.
Micrographs were collected using a Titan Krios microscope (Thermo Fisher Scientific) operated at an accelerating voltage of 300 kV. All cryo-EM images were recorded on a K2 Summit direct electron detector (Gatan), at a nominal magnification of 22,500×, corresponding to a calibrated pixel size of 1.07 Å. The camera was operated in superresolution mode, with a dose rate of ∼2 e− per pixel per s on the detector. We used a total exposure time of 4 s, corresponding to a total dose of 25 electrons/Å2 on the specimen. The data were collected semiautomatically using the SerialEM software suite (46).
Image Processing.
Stacks of dose-fractionated image frames were aligned using the UCSF MotionCor2 software (47). MT segments were manually selected from the drift-corrected images (acetylated dataset, 205 images; deacetylated MT dataset, 476 images) using the APPION image processing suite (48). We estimated the CTF using CTFFIND4 (49) and converted the segments to 84% overlapping boxes (512 × 512 pixels) for particle extraction. The remaining nonoverlapping region is set to 80 Å and corresponds to the tubulin dimer repeat (asymmetric unit). Consequently, there are ∼13 unique tubulin dimers per MT particle. To determine the initial global alignment parameters and PF number for each MT particle, raw particles were compared with 2D projections of low-pass filtered MT models (∼20 Å, 4° coarse angular step size) with 12, 13, 14, and 15 PFs (50) using the multireference alignment (MSA) feature of EMAN1 (51). Finally, 13-PF MT particles (acetylated dataset, 20,256 particles; deacetylated MT dataset, 29,396) were refined in FREALIGN, version 9.11 (52, 53), using pseudohelical symmetry to account for the presence of the seam. To verify the location of the seam, we used the 40-Å shift approach to categorize MTs based on their azimuthal angle, as previously described (28).
Atomic Model Building and Coordinate Refinement.
COOT (54) was used to build the missing polypeptides of the αK40 loop in α-tubulin, using the available PDB 3JAR as a starting model. Successively, all ab initio atomic models were iteratively refined with phenix.real_space_refine into EM maps sharpened with phenix.autosharpen (29, 55). For visual comparisons between states, potential density thresholds were interactively adjusted in Coot to maximize isocontour similarity around backbone atoms distant from the αK40 loop. For Figs. 1 and 2, all densities are represented in Chimera at a threshold of 1.1.
MD Simulations.
Code for map preparation, simulation execution, and analysis is available at https://github.com/fraser-lab/plumed_em_md.
To prepare the cryo-EM maps, we fitted the maps with a Gaussian mixture model (GMM) by applying a divide-and-conquer approach (34), using generate_gmm.py and convert_GMM2PLUMED.sh. Cross-correlations to the experimental maps were greater than 0.99. All simulations were performed with GROMACS 2016.5 (36) and the PLUMED-ISDB module (56) of the PLUMED library (57) using the Charmm36-jul2017 forcefield (58) with patches for acetylated lysine (aly) (59) and the TIP3P water model. For the deacetylated simulations, the same starting model was used with a manual edit of the PDB to eliminate the acetylation (with all hydrogens replaced by GROMACS during model preparation). The initial model was minimized then equilibrated for 2 ns, using prep_plumed.py. MD simulations were performed on a metainference ensemble of eight replicas for an aggregate simulation time of 96 ns for each acetylation state, using prep_plumed2.py and prep_plumed3.py. Contributions of negative scatterers (atoms OD1 and OD2 of Asp residues; OE1 and OE2 of Glu) were excluded from contributing to the predicted maps during the simulation. This modification effectively eliminates the contribution of these side chains to the agreement between density maps, in keeping with the nonexistent density of negatively charged side chains in EM maps, while allowing them to contribute to the simulation through the energy function. Clustering and convergence analyses (32) were performed and analyzed using MDAnalysis (60).
Clustering of the two metainference ensembles was carried out using the Gromos method (61) after aggregating the trajectories of the Ac0 and Ac96 simulations. A subset of every 10 frames were selected from the entire aggregated trajectory. The distance between two conformations was defined as the RMSDs calculated on the Cα atoms of the αK40 loop. The clustering cutoff was equal to 2.5 Å, with similar results obtained with other cutoffs. The population of each cluster in the Ac0 and Ac96 ensembles was then recalculated from the two separate trajectories.
Changes in the electrostatic interaction energies at the lateral contacts were determined using the using the DH formula:
where is the vacuum’s dielectric constant; is the dielectric constant of the solvent; and are the charges of the ith and jth atoms, respectively; is the distance between these two atoms; and is the DH parameter (37) defined in terms of the temperature and the ionic strength of the solution .
The DH energy is calculated between the following two groups of atoms, denoted as A and B in the formula above: (i) all atoms in residue range 30–60 of chain A (α1-subunit) and (ii) all atoms in residue range 200–380 of chain E (α2-subunit) in PDBs 6O2S and 602T. Residues not included in this range do not significantly contribute to the DH interaction energy between adjacent α-subunits. Parameters used in the calculation of the DH energy are as follows: temperature (T = 300 K), dielectric constant of solvent ( = 80; water at room temperature), and ionic strength ( = 0.3–1 M).
Accession Numbers.
All electron density maps have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb/ (EMDB ID codes EMD-0612, EMD-0613, EMD-0614, and EMD-0615). Atomic models have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 602Q, 602R, 602S, and 602T).
Supplementary Material
Acknowledgments
We thank P. Grob and J. Fang for cryo-EM data collection support; A. Chintangal and P. Tobias for computational support; T. H. D. Nguyen, B. Greber, E. Kellogg, B. LaFrance, S. Howes, and S. Pöpsel for supportive discussions; and A. Roll-Mecak for helpful comments based on our preprint. We also acknowledge the Berkeley Bay Area Cryo-EM Facility and additional scientific resources at University of California, Berkeley. J.S.F. was funded by the University of California, San Francisco–University of California, Berkeley Sackler Faculty Exchange Program and National Institute of General Medical Sciences (NIGMS) Grant R01-GM123159. This work was funded through NIGMS Grants R35-GM127018 and P01-GM063210 (to E.N.) and the National Science Foundation Grant 2016222703 and the National Academy of Sciences National Research Council Ford Foundation Grant (to L.E.-W.). E.N. is a Howard Hughes Medical Investigator.
Footnotes
Conflict of interest statement: E.N. and C.J. were coauthors in the 2016 review article “Microtubules: 50 Years on from the discovery of tubulin” [Borisy G, et al. (1)].
Data deposition: All electron density maps have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb/ (EMDB ID accession nos. EMD-0612–EMD-0615). Atomic models have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 602Q–602T). Code for map preparation, simulation execution, and analysis is available on GitHub at https://github.com/fraser-lab/plumed_em_md.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900441116/-/DCSupplemental.
References
- 1.Borisy G, et al. (2016) Microtubules: 50 years on from the discovery of tubulin. Nat Rev Mol Cell Biol 17:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nogales E, Whittaker M, Milligan RA, Downing KH (1999) High-resolution model of the microtubule. Cell 96:79–88. [DOI] [PubMed] [Google Scholar]
- 3.Zhang R, Alushin GM, Brown A, Nogales E (2015) Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell 162:849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchison TJ. (1993) Localization of an exchangeable GTP binding site at the plus end of microtubules. Science 261:1044–1047. [DOI] [PubMed] [Google Scholar]
- 5.Mitchison T, Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312:237–242. [DOI] [PubMed] [Google Scholar]
- 6.Walsh G, Jefferis R (2006) Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol 24:1241–1252. [DOI] [PubMed] [Google Scholar]
- 7.Magiera MM, Singh P, Gadadhar S, Janke C (2018) Tubulin posttranslational modifications and emerging links to human disease. Cell 173:1323–1327. [DOI] [PubMed] [Google Scholar]
- 8.Janke C, Montagnac G (2017) Causes and consequences of microtubule acetylation. Curr Biol 27:R1287–R1292. [DOI] [PubMed] [Google Scholar]
- 9.Janke C, Bulinski JC (2011) Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat Rev Mol Cell Biol 12:773–786. [DOI] [PubMed] [Google Scholar]
- 10.Howes SC, Alushin GM, Shida T, Nachury MV, Nogales E (2014) Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol Biol Cell 25:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeDizet M, Piperno G (1987) Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc Natl Acad Sci USA 84:5720–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeDizet M, Piperno G (1986) Cytoplasmic microtubules containing acetylated α-tubulin in Chlamydomonas reinhardtii: Spatial arrangement and properties. J Cell Biol 103:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Brabander MJ, Van de Veire RML, Aerts FEM, Borgers M, Janssen PA (1976) The effects of methyl (5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl) carbamate, (R 17934; NSC 238159), a new synthetic antitumoral drug interfering with microtubules, on mammalian cells cultured in vitro. Cancer Res 36:905–916. [PubMed] [Google Scholar]
- 14.Dompierre JP, et al. (2007) Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci 27:3571–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.d’Ydewalle C, et al. (2011) HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot–Marie–Tooth disease. Nat Med 17:968–974. [DOI] [PubMed] [Google Scholar]
- 16.Kim JY, et al. (2016) HDAC6 inhibitors rescued the defective axonal mitochondrial movement in motor neurons derived from the induced pluripotent stem cells of peripheral neuropathy patients with HSPB1 mutation. Stem Cells Int 2016:9475981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, et al. (2012) MEC-17 deficiency leads to reduced α-tubulin acetylation and impaired migration of cortical neurons. J Neurosci 32:12673–12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godena VK, et al. (2014) Increasing microtubule acetylation rescues axonal transport and locomotor deficits caused by LRRK2 Roc-COR domain mutations. Nat Commun 5:5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boggs AE, et al. (2015) α-Tubulin acetylation elevated in metastatic and basal-like breast cancer cells promotes microtentacle formation, adhesion, and invasive migration. Cancer Res 75:203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Martile M, Del Bufalo D, Trisciuoglio D (2016) The multifaceted role of lysine acetylation in cancer: Prognostic biomarker and therapeutic target. Oncotarget 7:55789–55810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maruta H, Greer K, Rosenbaum JL (1986) The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J Cell Biol 103:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portran D, Schaedel L, Xu Z, Théry M, Nachury MV (2017) Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat Cell Biol 19:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z, et al. (2017) Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cueva JG, Hsin J, Huang KC, Goodman MB (2012) Posttranslational acetylation of α-tubulin constrains protofilament number in native microtubules. Curr Biol 22:1066–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaaban S, Brouhard GJ (2017) A microtubule bestiary: Structural diversity in tubulin polymers. Mol Biol Cell 28:2924–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szyk A, et al. (2014) Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 157:1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alushin GM, et al. (2014) High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 157:1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R, Nogales E (2015) A new protocol to accurately determine microtubule lattice seam location. J Struct Biol 192:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonomi M, Vendruscolo M (2018) Determination of protein structural ensembles using cryo-electron microscopy. Curr Opin Struct Biol 56:37–45. [DOI] [PubMed] [Google Scholar]
- 31.Vahidi S, et al. (2018) Reversible inhibition of the ClpP protease via an N-terminal conformational switch. Proc Natl Acad Sci USA 115:E6447–E6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonomi M, Pellarin R, Vendruscolo M (2018) Simultaneous determination of protein structure and dynamics using cryo-electron microscopy. Biophys J 114:1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singharoy A, et al. (2016) Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. eLife 5:e16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanot S, et al. (2017) Multi-scale Bayesian modeling of cryo-electron microscopy density maps. bioRxiv:10.1101/113951. Preprint, posted March 4, 2017.
- 35.Yajima H, et al. (2012) Conformational changes in tubulin in GMPCPP and GDP-taxol microtubules observed by cryoelectron microscopy. J Cell Biol 198:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447. [DOI] [PubMed] [Google Scholar]
- 37.Do TN, Carloni P, Varani G, Bussi G (2013) RNA/peptide binding driven by electrostatics—insight from bidirectional pulling simulations. J Chem Theory Comput 9:1720–1730. [DOI] [PubMed] [Google Scholar]
- 38.Theillet F-X, et al. (2014) Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem Rev 114:6661–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narayanan A, Jacobson MP (2009) Computational studies of protein regulation by post-translational phosphorylation. Curr Opin Struct Biol 19:156–163. [DOI] [PubMed] [Google Scholar]
- 40.Beltrao P, Bork P, Krogan NJ, van Noort V (2013) Evolution and functional cross-talk of protein post-translational modifications. Mol Syst Biol 9:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gittes F, Mickey B, Nettleton J, Howard J (1993) Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol 120:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eun YJ, Kapoor M, Hussain S, Garner EC (2015) Bacterial filament systems: Toward understanding their emergent behavior and cellular functions. J Biol Chem 290:17181–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Castilla A, et al. (2017) Structure of the calcium-dependent type 2 secretion pseudopilus. Nat Microbiol 2:1686–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krupka M, et al. (2017) Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat Commun 8:15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castoldi M, Popov AV (2003) Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif 32:83–88. [DOI] [PubMed] [Google Scholar]
- 46.Mastronarde DN. (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. [DOI] [PubMed] [Google Scholar]
- 47.Zheng SQ, et al. (2017) MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lander GC, et al. (2009) Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A, Grigorieff N (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egelman EH. (2007) The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J Struct Biol 157:83–94. [DOI] [PubMed] [Google Scholar]
- 51.Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. [DOI] [PubMed] [Google Scholar]
- 52.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N (2013) Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol 183:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grigorieff N. (2007) FREALIGN: High-resolution refinement of single particle structures. J Struct Biol 157:117–125. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger TC, et al. (2018) Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr D Struct Biol 74:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonomi M, Camilloni C (2017) Integrative structural and dynamical biology with PLUMED-ISDB. Bioinformatics 33:3999–4000. [DOI] [PubMed] [Google Scholar]
- 57.Tribello GA, Bonomi M, Branduardi D, Camilloni C, Bussi G (2014) PLUMED 2: New feathers for an old bird. Comput Phys Commun 185:604–613. [Google Scholar]
- 58.Huang J, MacKerell AD Jr (2013) CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J Comput Chem 34:2135–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J, et al. (2017) CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat Methods 14:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O (2011) MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J Comput Chem 32:2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daura X, et al. (1999) Peptide folding: When simulation meets experiment. Angew Chem Int Ed 38:236–240. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.