Significance
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a disease which afflicts approximately 2 million people in the United States and many more around the globe. A combination of factors might trigger ME/CFS, and there is currently no well-established blood-based biomarker to diagnose it. Taking advantage of advancements in micro/nanofabrication, direct electrical detection of cellular and molecular properties, microfluidics, and artificial intelligence techniques, we developed a nanoelectronics blood-based assay that can potentially establish a diagnostic biomarker and a drug-screening platform for ME/CFS. Given the significance of this assay, we envision it has the potential to be widely employed in research laboratories and clinics in the future as an aid to physicians as well as to our colleagues in the ME/CFS research community.
Keywords: myalgic encephalomyelitis/chronic fatigue syndrome, diagnostic biomarker, nanoelectronics biosensor, artificial intelligence, machine learning
Abstract
There is not currently a well-established, if any, biological test to diagnose myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). The molecular aberrations observed in numerous studies of ME/CFS blood cells offer the opportunity to develop a diagnostic assay from blood samples. Here we developed a nanoelectronics assay designed as an ultrasensitive assay capable of directly measuring biomolecular interactions in real time, at low cost, and in a multiplex format. To pursue the goal of developing a reliable biomarker for ME/CFS and to demonstrate the utility of our platform for point-of-care diagnostics, we validated the array by testing patients with moderate to severe ME/CFS patients and healthy controls. The ME/CFS samples’ response to the hyperosmotic stressor observed as a unique characteristic of the impedance pattern and dramatically different from the response observed among the control samples. We believe the observed robust impedance modulation difference of the samples in response to hyperosmotic stress can potentially provide us with a unique indicator of ME/CFS. Moreover, using supervised machine learning algorithms, we developed a classifier for ME/CFS patients capable of identifying new patients, required for a robust diagnostic tool.
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a disease that affects at least 2 million Americans and millions more globally (1–3). Several studies have found that this disease may be triggered by a combination of factors such as major life stressors, infection (viral: ENV, HHV-6, HHV-7, stomach viruses, cytomegalovirus, and bacterial infections), toxin exposure, immunodeficiency, nutritional deficiencies, genetic susceptibility, and several others (1, 4, 5). There is currently no single biomarker to diagnose ME/CFS (1, 6). As a result, diagnosing ME/CFS patients is a lengthy and costly process, which constitutes a fundamental impediment to patient care. This lag in diagnosis also causes barriers to research, complicating patient recruitment and the handling of heterogeneous samples of patients with only marginally similar conditions. However, in one of these very recent studies (1), patients with ME/CFS showed abnormalities in 20 out of 63 biochemical pathways, suggesting such metabolic features as potential biomarkers. All the while, ME/CFS patients experience one of the lowest quality of life illnesses, with an unadjusted EuroQol 5-dimensional 3-level (EQ-5D-3L) mean of 0.47 compared with a mean of 0.69 recorded for lung cancer (7, 8). This clearly indicates the central importance of identifying a reliable biomarker for ME/CFS. Researchers have investigated a panoply of potential biomarkers, many of which would indicate improper immune function and signs of autoimmunity (5, 9), for example, differences in cytokine profiles; natural killer cells; 5-HT autoimmune activity; and the responsiveness of T cells (4, 5, 9–19). The immune system is a typical focal point for ME/CFS research supported by the observation that CFS is often preceded by a viral infection and has many long-term flu-like symptoms (4, 5, 20). Various researchers have even hypothesized that the source of the illness is viral in nature (19, 21). There are other subsets of symptoms beyond the flu-like symptoms and fatigue, such as muscle joint pain; unrefreshing sleep; and high sensitivity to light, sound, odor, taste, touch, and vibration. Many of the patients usually suffer from different types of paresthesias, such as tingling and numbness in different parts of the body. Other symptoms include postural orthostatic tachycardia syndrome; light-headedness; gastrointestinal symptoms such as nausea and abdominal pain; headaches of a new type, pattern, or severity; autonomic and endocrine symptoms such as poor temperature regulation; cold or heat intolerance; and recurrent sore throats. In some instances, researchers have linked cytokine profiles and inflammation to the severity of ME/CFS in patients (4). One study examined cytokine profiles postexertion to explore potential differences in ME/CFS and sedentary controls (2). Others have focused on finding physiological anomalies by looking at exercise intolerance and cardiac impairment (2, 22). Molecular aberrations have also been observed in numerous studies of ME/CFS blood cells (1, 4). In addition, several studies have shown that inducing a biological stressor on peripheral blood mononuclear cells (PBMCs) in the form of hyperosmotic stress forces the cells to consume ATP, a key metabolite, which is hypothesized to be deficient in ME/CFS patients (1, 23, 24).
To explore the aforementioned ATP hypothesis and to study the biology of the disease and its molecular aberrations, we applied a nanoelectronic array, called nanoneedle bioarray, to monitor blood cells’ electrical responses to an induced stressor. The nanoneedle bioarray is a versatile, ultrasensitive, and high-throughput nanoelectronic device developed to detect molecular and cellular interactions and their electrical properties in real time. The array directly measures the impedance modulations resulting from cellular and/or molecular interactions. We developed this ultrasensitive, cost-effective assay by exploiting recent advancements in nanofabrication and microfluidics. We posit that this assay can potentially offer an outstanding biomarker to rapidly and inexpensively diagnose ME/CFS with high accuracy. In addition, this assay may offer a remarkable opportunity for the discovery of new treatments for this debilitating disease. In this study, we used ME/CFS PBMCs as the model of study. PBMCs are generally the cells of choice in many fields of research since they are well characterized and can be easily isolated (25) (SI Appendix). We also applied different machine learning algorithms to make our platform a more robust and precise potential diagnostic tool for ME/CFS.
Results and Discussion
Initial Motivation.
The cardinal feature of ME/CFS is considered to be a worsening of symptoms postexertion known as postexertional malaise (20). To mimic this condition on a cellular level, we introduced a stressor into the patient’s clinical samples, composed of isolated PBMCs incubated in their own plasma. We initially hypothesized that stressing PBMCs (in a high-salt environment) would result in extensive consumption and potential depletion of ATP, a high-energy metabolite. Salt stress, and, more generally, hyperosmotic stress, is a commonly applied stress model in studies on various types of cells such as plant, yeast, bacteria, mice, and human (26–32). To stimulate hyperosmotic stress in our study, we increased the plasma’s NaCl concentration to 200 mmol/L. NaCl is a typical stimulator of the osmotic response, which acts in essentially the same way as many other agents, such as sugars (33). However, as shown in Fig. 1C, the promising experimental findings produced by the use of our assay led us to the conclusion that the significantly different impedance response of the hyperosmotic-stressed PBMCs we identified constitutes a reliable method of differentiating CFS patients from healthy controls, which is the main focus of this work.
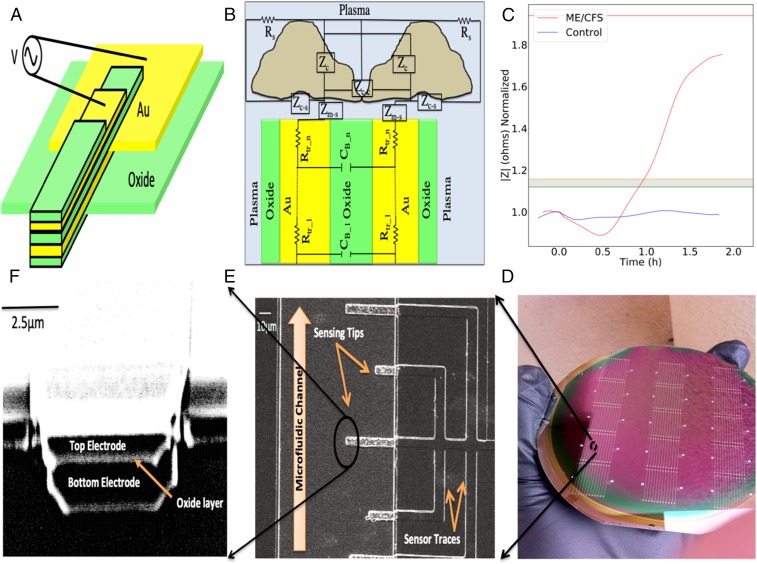
Fig. 1.
(A) Schematic of a single nanoelectronic sensor (not to scale). (B) Circuit model of a sensor−solution interface, where Zm-s is media−sensor surface interactions, Zc-c is cell−cell interactions, Zc-s is cell−sensor surface adhesion, Zc is a cell impedance (membrane capacitance Cm, and cytoplasm conductivity of the cells, σcp), and Rs is resistance of the solution. (C) The experimentally obtained impedance versus time curves illustrating the electrical response of hyperosmotic-stressed samples of a bed-bound ME/CFS patient and a healthy control in real time. A gray region is defined experimentally and its top and bottom borders are shown with orange and green lines, respectively (for further details see Trial Population and Statistical Analysis). (D) Array of nanoneedle sensors fabricated on a 4-in wafer. (E and F) SEM images of a nanoelectronic sensor tips, (E) top view and (F) from the microfluidics channel side.
Features of Assay and Theory of How Assay Operates.
Our assay is designed as an ultrasensitive assay capable of directly measuring biomolecular interactions in real time, at low cost, and in a multiplex format. The array processing and multiplexing is an important feature to enable high-throughput sensing and monitoring tasks on a large scale. Fabrication of an array of thousands of sensors in parallel in a microfluidic channel integrated with their individual on-chip amplifiers and read-out systems can provide a single, portable platform that features a variety of clinical applications, including the detection of various biomarkers in parallel for improved early-stage disease diagnosis. The way in which the assay operates is theorized as follows: The assay detects any impedance modulation due to the presence and/or interactions of biomolecules of interest at the active sensing region of the sensors. Impedance (Z), briefly, is the ratio of applied voltage (V) to the induced current (A). It has two components: resistance, the in-phase component (Zre), and reactance, the out-of-phase component (Zim). With respect to an electrochemical system, the resistance component is commonly dominated by a change in local conductivity (e.g., change of ion concentration in osmolytes) (34), while the reactance component is commonly dominated by a change in local relative dielectric permittivity (34, 35) (e.g., local replacement of buffer molecules with biomolecules) (for further details, see SI Appendix). Using our sensors, we recorded and calculated three parameters of in-phase impedance (Zre), out-of-phase impedance (Zim), and impedance magnitudes to further enhance the precision and accuracy of the sensors. Utility of the assay for label-free detection of proteins and nucleic acids has previously been demonstrated (34–40). In this study, we applied the assay to objectively and continually quantify the effect of hyperosmotic stress on patients’ clinical samples. For each experiment, each of which lasted approximately 3 h, the sensors collected ∼40,000 data points with a sampling frequency of 5 Hz.
Configuration and Microfabrication of Assay.
The nanoelectronic assay structure consists of two conductive layers with an insulating layer in between. There are two additional protective oxide layers above and underneath the sensors (Fig. 1A). The nanometer-sized sensing region of the sensor consists of a 30-nm-thin oxide layer sandwiched between two 100-nm-thin gold layers. The top protective oxide layer is intended to prevent the exposure of the top conductive electrodes to the solutions. There is a thermally grown oxide layer underneath the bottom electrodes to electrically insulate the sensors from the substrate. Each sensor’s width is ∼3 μm to 5 μm. A scanning electron microscopy (SEM) image of a single sensor is shown in Fig. 1 E and F. The sensors are designed in 3D format to improve the molecules-to-sensors hit rate and to help diffusion occur in multiple directions. The sensors were fabricated on 4-in silicon wafers (Fig. 1D) following several optical photolithography, deposition, and etching procedures. (Details of the fabrication process can be found in Materials and Methods.)
Real-Time Impedance Measurements.
To demonstrate the clinical utility of the assay presented here for disease diagnostic applications, we monitored the electrical response of hyperosmotic-stressed samples of a bed-bound ME/CFS patient and a healthy control in real time. Prepared samples (Materials and Methods and SI Appendix) consisted of PBMCs (200 cells per μL) incubated in their plasma and prepared within 5 h of use. Fig. 1C illustrates the experimentally obtained impedance versus time curves of these experiments. Before the electrochemical impedance spectroscopy (Vrms = 250 mV at f = 15 KHz) (Materials and Methods), all sensors were subjected to an extensive cleaning procedure to eliminate any potential for contamination (Materials and Methods). For each experiment, there was a waiting time of ∼20 min for the impedance to reach a baseline value. The baseline value is defined as <2% impedance fluctuation. After reaching the baseline, we introduced a small volume (∼6 μL) of hyperosmotic stressor to the samples. Testing of the sample from the healthy control showed a transient decrease in impedance signal after raising the plasma’s NaCl concentration to 200 mmol/L. The signal gradually returned to a value near the baseline signal (0.88% ± 0.2, 2.69% ± 0.2, and 1.17% ± 0.28 for |Z|, Zre, and Zimg, respectively) generated in the unstressed state and did not change with time when tested across ∼3 h. Electrical impedance data were continually collected until the impedance reached a plateau (<0 slope for >500 s). The same steps were followed for the ME/CFS patient samples. Similarly, this sample established a baseline impedance value that did not change in the absence of the stressor. The hypertonic stressor added to this sample also resulted in a transient decrease in measured impedance, comparable to the healthy control’s sample, which gradually returned to the baseline after about 40 min. However, the increase in impedance was followed by a marked excursion above the initial baseline value by 74.92% ± 0.69, 301.67% ± 3.55, and 64.73% ± 0.62, for |Z|, Zre, and Zimg, respectively, figures that are significantly greater than the values observed for the healthy control. As shown in Fig. 1C, the ME/CFS samples’ response to the stressor is a unique characteristic of the impedance pattern and is dramatically different from the response observed among the control samples. We believe the observed robust impedance modulation difference of the samples in response to hyperosmotic stress (ME/CFS versus healthy) can potentially provide us with a unique indicator of ME/CFS. One should also note that, for both experiments, the baseline point served as a reference point for data normalization purposes. It allowed us to eliminate possible critical variations such as human error, possible dissimilarity of the sensors due to the complicated microfabrication process, and possible blood sample alterations due to the lengthy sample preparation process.
Trial Population and Statistical Analysis.
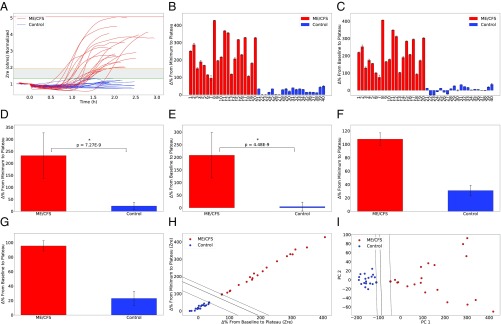
To pursue the goal of developing a reliable biomarker for ME/CFS and to demonstrate the utility of our platform for point-of-care diagnostics, we further validated the array by testing patients with moderate to severe ME/CFS. A total of 40 patients and healthy controls were prospectively included. All of the ME/CFS patients included in this study had previously been diagnosed by a physician in line with the Canadian Consensus Criteria (CCC) (20). These experiments were approved by Stanford IRB, and written consent was obtained following Stanford IRB-40146 before any testing or analysis began. Of the 20 ME/CFS patients studied, 5 were severely ill and 15 were moderately affected by their condition. Of the 20 healthy controls studied, 5 were age- and gender-matched to 5 of our ME/CFS patients (with both sets of samples collected simultaneously), 10 were selected at random and recruited by us, and 5 were anonymous donors collected by the Stanford Blood Center. The recruited healthy controls (excluding the blood center samples) were not diagnosed with ME/CFS or related diseases and had no blood relatives diagnosed with ME/CFS. Patient and control samples were handled exactly the same in all preprocessing steps. For each experiment, we analyzed and recorded all three parameters of in-phase impedance (Zre), out-of-phase impedance (Zim), and the impedance magnitudes (Materials and Methods). Impedance signal excursion above the initial baseline value for ME/CFS patients ranged from 75.61% ± 12.69 to 406.2% ± 1.32, 12.46% ± 0.13 to 94.98% ± 0.92, and 7.42% ± 1.45 to 81.49% ± 0.88 for Zre, |Z|, and Zimg, respectively. The ranges for healthy controls were −27.33% ± 0.56 to 34.7% ± 5.18, −14.89% ± 0.32 to 18.02% ± 0.41, and −16.44% ± 0.22 to 18.38% ± 0.44 for Zre, |Z|, and Zimg, respectively. Additionally, minimum-to-plateau impedance signal change for ME/CFS patients ranged from 95.79% ± 12.8 to 427.29% ± 1.57, 14.13% ± 1.24 to 109.25% ± 0.95, and 13.36% ± 0.99 to 94.99% ± 0.9 for Zre, |Z|, and Zimg, respectively. Experimentally calculated values of these signals for healthy controls are 0.05% ± 0.53 to 49.15% ± 4.64, 1.15% ± 0.17 to 19.69% ± 0.46, and 0.16% ± 0.71 to 19.29% ± 0.74 for Zre, |Z|, and Zimg, respectively (Fig. 2 B and C). According to our analyses, the in-phase impedance (Zre) from the baseline to plateau showed the greatest separability (P = 4.48E-9), while out-of-phase impedance (Zim) and magnitude (|Z|) signals were also significantly separable (P = 2.73E-5 and P = 1.12E-5, respectively). Supplementary to analyzing the percentage change to the plateau from the baseline, the percentage change to the plateau from the minimum also showed strong separability, as shown in Fig. 2 D and E. Similarly, in-phase impedance showed the greatest separability (P = 7.27E-9), while Zim and |Z| signals were also significantly separable (P = 5.06E-5 and P = 2.67E-5, respectively). Furthermore, other parameters such as slopes and the time taken to reach both the minimum and plateau were also analyzed. Most notably, the maximum positive slope showed the greatest separability (P = 1.19E-7, P = 1.67E-5, and P = 2.22E-5 for Zre, |Z|, and Zimg, respectively), although it was not as significant as the percentage change from the minimum to the plateau (P = 7.27E-9 for Zre) and from the baseline to the plateau (P = 4.48E-9 for Zre). Fig. 2A shows the experimentally obtained impedance versus time curves of all ME/CFS and healthy control samples. To generate each plot, about ∼40,000 data points per experiment were collected. Fig. 2A also shows an experimentally defined gray region. According to our experimental results, the ME/CFS signals that lay in the top range of this gray region belonged to the moderately affected ME/CFS patients, while severely ill patients all showed signals far above the gray region. Considering these findings, we think there might be a correlation between the level of disease severity and signal strength. Additionally, the healthy controls that lay in the bottom range of the gray region had a familial link to fibromyalgia. Also, the repeatability and reproducibility of the assay were validated for both ME/CFS patients and healthy controls (Fig. 2 F and G). Repeating the experiments for the healthy control showed the baseline-to-plateau impedance signal varied at 7.08% ± 0.23, 18.95% ± 0.69, and 5.04% ± 0.36 for |Z|, Zre, and Zimg, respectively, while variations for the minimum-to-plateau signals were 5.96% ± 0.35, 16.23% ± 0.65, and 4.02% ± 0.47 for |Z|, Zre, and Zimg, respectively. For the ME/CFS sample, variations for |Z|, Zre, and Zimg were 1.66% ± 0.42, 14.09% ± 2.71, and 1.35% ± 0.41, and 1.68% ± 0.41, 17.71% ± 2.62, and 1.29% ± 0.41 for the baseline-to-plateau and for the minimum-to-plateau signals, respectively.
Fig. 2.
Trial population and statistical analysis. (A) The experimentally obtained impedance versus time curves of 40 ME/CFS and healthy control samples used in this study with an experimentally defined gray region. Top and bottom borders of the gray region are shown with orange and green lines, respectively. To generate each plot, about ∼40,000 data points per experiment were collected. (B−E) Analyzed percentage change of (B) minimum-to-plateau and (C) baseline-to-plateau impedance signals, in which both showed a strong separability, (D) with P = 7.27E-9 for minimum-to-plateau and (E) P = 4.48E-9 for baseline-to-plateau impedance signals. (F and G) Repeatability and reproducibility validation of the assay for both ME/CFS patients and healthy controls. (H) Primary ME/CFS classifier created by applying supervised SVM machine learning algorithm to our experimental datasets. (I) Perfect linearly separable dataset in PCA space after performing PCA on a data matrix comprising six features of impedance change signals from the baseline and minimum to the plateau for all three components of impedance (|Z|, Zre, and Zim). *P < 1e-8.
Sample Selection.
To adapt this assay for use as a potential diagnostic tool for ME/CFS, we optimized the assay using several different clinical samples. We tested patients’ PBMCs in their whole-blood, serum, and plasma samples to find the most reliable and distinguishable indicators. According to our experimental data, both serum and plasma samples showed a unique, reliable, and repeatable pattern, while such a pattern was not observed for the whole-blood samples. As a result, platform validations and optimization studies were performed for the plasma samples. Furthermore, we explored a number of different preparation and storage methods to minimize the need for freshly drawn blood, and to provide the possibility of using patient samples collected around the globe. We conducted the assay with samples kept at 4 °C, room temperature, and in a 37 °C storage cabinet, as well as those frozen in −20 °C freezers and in liquid nitrogen (at 200 cells per μL). We concluded that plasma used within 5 h of preparation at 200 cells per μL gives the most reliable and reproducible results. The other most successful techniques tested were 24 h storage at room temperature and liquid nitrogen freezing for 1 wk, both of which preserved the pattern of fresh samples, although the response was slightly attenuated.
Discussion.
Although the initial hypothesis of this study was that the ATP consumption rate differs in ME/CFS patients’ blood cells compared with healthy control blood cells, our experimental results on 40 ME/CFS individual patients and healthy controls revealed that ME/CFS hyperosmotic-stressed samples (PBMCs in plasma) display a unique characteristic in their impedance pattern, and one that is significantly different than what was observed in the controls. Our work thus far indicates that the impedance signature itself can potentially represent a distinctive indicator of ME/CFS patients versus controls and can potentially establish a diagnostic metric for the disease. Furthermore, it can also provide insights into the biology of this complex syndrome. Previous studies have also sought to find a differentiating attribute of ME/CFS by inducing exertional malaise (41). While the exact mechanisms behind the discovered differences remain unclear and further analysis is needed to pinpoint their precise source, we envision that a number of mechanisms may be triggered when plasma samples containing PBMCs are exposed to a stressor. To this end, frequency-resolved impedance readings (10 Hz to 1 MHz) at different time points were subjected to a mathematical model developed by Giaever and Keese (42) to understand the exciting circuit model representing the sensor−solution interface components. The impedance results are integrations of the impedance that are attributed to the media−sensor surface interactions (Zm-s), cell−cell interactions (Zc-c), cell−sensor surface adhesion (Zc-s), impedance of cells (membrane capacitance Cm, cytoplasm conductivity of the cells, σcp), resistance of the solution (Rs), and other components (e.g., proteins, exosomes, and lipids) in plasma. The assumption here is that the current flows radially into the space between the cell’s ventral surface and the substrate and then escapes between the cells. The current density is assumed to be consistent in the y direction, and the cells are disk-shaped objects with membrane surfaces and filled with a conducting electrolyte. Moreover, to identify the mechanisms and components involved at the cellular and molecular levels, we have started to investigate the plasma components (e.g., proteins, exosomes, and lipids) and individual cell types (e.g., T cells) separately. This investigation forms part of on-going studies that require further investigation before mechanisms may be suggested with a good degree of certainty.
In terms of the mechanisms involved, one of the first candidates to be considered is Na/K ATPase, which is present in the plasma membrane of all cells. It is responsible for pumping sodium and potassium ions across the cell membrane using an active transport mechanism that requires the consumption of ATP. The normal intracellular concentration of PBMCs is around 10 mM, while that of plasma is about 135 mM. The reverse is true for potassium (around 140 mM intracellular and 5 mM extracellular). Upon the increase in extracellular sodium ion concentration, passage of some additional Na ions into cells may occur by diffusion, depending on the permeability of the PBMCs to this ion. As for the active transport of the ions by Na/K ATPase, the pumping of three Na ions to the outside of the cell and two K ions to the inside takes place in each cycle of operation, thereby sustaining the large excess of Na ions outside and high concentration of K ions inside required for maintaining cell potential. Accordingly, increasing sodium concentration in plasma while keeping its original low potassium level unchanged would be in line with the main function of the membrane enzyme in preserving the concentration gradients of Na and K ions across PBMC plasma membranes. While an increase in Na ion concentration per se does not appear to be antagonistic to the normal functioning of Na/K ATPase, it may act as a factor causing osmotic stress. In such a situation, some specific mechanisms may be triggered as a response to the new, unfavorable situation to achieve intracellular water homeostasis (43). Indeed, a number of reports in the literature indicate that the addition of salts or other osmotic agents to the cell environment may induce the production of inflammatory cytokines (20, 39–44). These studies indicate that the development of inflammation as a consequence of osmotic stress may be a general phenomenon affecting PBMCs (44, 45) and a number of other cell types (46–50). Both lower (20 mM to 41 mM) (48) and higher (100 mM to 135 mM) (47) concentrations of NaCl than the amount tested in the present study (65 mM) have been investigated. In a study on the effect of osmotic stress on human bronchial epithelial cells, IL-8 production was stimulated by additional NaCl (ranging from 50 mM to 150 mM) in a time- and dose-dependent manner (50). These findings confirm the ability of cells to alter gene expression in response to changes in the osmotic environment (51). These mechanisms may contribute to altering media−sensor surface interactions (Zm-s), cell−cell interactions (Zc-c), cell−sensor surface adhesion (Zc-s), impedance of cells (membrane capacitance Cm, cytoplasm conductivity of the cells σcp), resistance of the solution (Rs), and other plasma components (e.g., proteins, exosomes, and lipids), which can modulate the value of the impedance signals measured by our sensors. A number of other mechanisms have been reported as being involved in the effect of high salt concentration on cells other than PBMCs, which may play a role relevant to the present study. These include a size change induced by an increase in osmotic pressure, and accumulation of glycerol and amino acids (27), which might contribute to altering the impedance of cells (membrane capacitance Cm, cytoplasm conductivity of the cells, σcp), cell−cell interactions (Zc-c), and cell−sensor surface adhesion (Zc-s), which can modulate the value of the impedance signals measured by the sensors here. However, the simultaneous standard live microscopy imaging of heterogeneous population of the PBMCs at different critical points of the experiments was not able to distinguish a significant visually detectable cell size difference between ME/CFS patient cells and healthy control cells. Nonetheless, high-resolution imaging (e.g., transmission electron microscopy) of specific cell types at different critical points of the experiments might help us to better understand the possible contribution of this mechanism to our results. Additionally, phospholipid synthesis and expansion of endoplasmic reticulum membrane (52) can also contribute to altering the impedance of cells (membrane capacitance Cm, and cytoplasm conductivity of the cells σcp), which can modulate the value of the impedance signals measured by the sensors. A change in the composition of the plasma membrane (28) is another possible mechanism that can contribute to altering the impedance of cells (membrane capacitance Cm), cell−cell interactions (Zc-c), and cell−sensor surface adhesion (Zc-s), all of which can modulate the value of the impedance signals measured by the sensors. Moreover, the reinforcing of the membrane by increasing the concentration of ubiquinone-8 can also contribute to altering the impedance of cells (membrane capacitance Cm), cell−cell interactions (Zc-c), and cell−sensor surface adhesion (Zc-s), and to modulating the value of the impedance signals measured by our sensors. Although all of the above are possible contributing mechanisms, further experiments are required to understand the precise contributing mechanisms behind the observed differences, and whether they are specific to ME/CFS or whether the response might be found among other similar diseases.
Additionally, our very attractive preliminary results (not included in this paper) involving severely ill ME/CFS patients and healthy controls indicated a clear difference in the behavior of PBMCs among ME/CFS patients when placed in the patients’ own plasma compared with when they were incubated in healthy plasma. These observations were one of the motivations behind experimenting with the components of ME/CFS patients’ plasma at the molecular level, in addition to studying cells. The observed impedance patterns may be due to the presence or absence of plasma-associated factors. Interestingly, cytokine production by PBMCs (53) and human retina pigment epithelium cells (48) treated with lipopolysaccharide have been found to be enhanced by hyperosmotic stress, a finding that may be related to the present study. It is also tempting to speculate that these reports and our own findings are relevant to some recent studies indicating an important inflammatory component in ME/CFS (4, 9, 54).
Moreover, to create a classifier for ME/CFS patients capable of identifying new patients, required for a robust diagnostic tool, we developed a trained kernel Support Vector Machine (SVM), a supervised machine-learning algorithm, using our experimental data. To classify new patients based on whether they fall to the right of the decision boundary, we initially selected the two features with the largest significance: change from the baseline to the plateau and change from the minimum to the plateau for the in-phase components of the impedance. Using these features, a cubic polynomial kernel SVM was able to classify the two populations, although the two features are highly correlated, as shown in Fig. 2H. To further improve the robustness of the classifier by decreasing the correlation between the two axes, a principal component analysis (PCA) was performed on a data matrix comprising six features: the change in impedance from the baseline and minimum to the plateau for all three components of impedance (|Z|, Zre, and Zim). The results (n = 2) yielded a seamless dataset that is linearly separable in PCA space (Fig. 2I).
Next, we aim to perform further experiments to understand the specific mechanisms contributing to the observed results, and to test the performance of the assay on other similar condition diseases. Additionally, we are working on adapting the technology to a platform capable of preclinical testing of drugs and therapies on cells from ME/CFS patients, leading toward development of a portable, handheld, and easy-to-use platform that can be operated by researchers and clinicians at any skill level.
In summary, there is currently no well-established blood-based biomarker to diagnose ME/CFS, which afflicts ∼2 million people in the United States and many more around the globe. The molecular aberrations observed in numerous studies of ME/CFS blood cells offer the opportunity to develop a diagnostic assay for blood samples. Taking advantage of recent advancements in micro/nanofabrication, direct electrical detection of cellular and molecular properties, and microfluidics, we developed an ultrasensitive and cost-effective nanoelectronic assay capable of continual monitoring of cellular and molecular events in real time from a very small sample volume (∼50 μL). By studying ME/CFS at the molecular level, we demonstrate the clinical utility of the assay for disease diagnostic applications. According to our experimental results, ME/CFS blood cells display a unique characteristic in their impedance pattern when subjected to hyperosmotic stress that is significantly different from the control (P < 4.5E-9, n = 40). To make our platform a more robust diagnostic tool, potentially capable of accurately identifying new patients, we developed a precise ME/CFS classifier by applying a supervised SVM machine learning algorithm to our experimental datasets. In addition, we performed PCA on a data matrix of important features, which established a seamless linearly separable dataset in PCA space. This technology is a blood-based impedance biomarker that can potentially establish a diagnostic biomarker and a drug-screening platform for ME/CFS in conjunction with preexisting evaluation measures such as the CCC. This is a low-cost, rapid, miniaturized, minimally invasive, and highly sensitive assay. Given the significance of this assay and its reliability, we envision it has the potential to be widely employed in other research laboratories and clinics in the near future as an aid to physicians as well as to our colleagues in the ME/CFS research community.
Materials and Methods
Fabrication.
We fabricated the arrays using the following protocol: First, 200 nm of SiO2 was thermally grown on a silicon wafer to insulate the substrate from the other layers. This process was done in a high-temperature atmospheric furnace to grow silicon dioxide (SiO2) on silicon wafers (∼3 h at 1,100 °C). In subsequent steps, the bottom conductive electrodes were patterned and deposited through optical photolithography, and metal deposition processes, followed by lift-off steps. To do that, first, standard silicon wafers were prebaked on a hot plate at 200 °C for ∼2 h. Then the manual resist spinning step was performed by applying 10 drops of hexamethyldisilizane to the wafers as an adhesive layer. This was followed by the coating of the wafers with MaP 1215 photoresists. The wafers were then transferred to a contact aligner system to perform precision mask-to-wafer alignment followed by near-UV photoresist exposure (∼3 s). Exposed photoresists were developed, and the patterned wafers were transferred to an evaporation system to deposit the bottom metallic electrodes. In the evaporation system, first, 3 nm of Cr was deposited as an adhesive layer, which was followed by the deposit of 100 nm of gold to create the sensors’ bottom conductive electrodes. The wafers then went through a metal lift-off process (∼30 min in Acetone) to remove the remaining photoresists and form the final configuration of the bottom electrodes. The next step was deposition of 30 nm of silicon dioxide (sensing region) using plasma-assisted atomic layer deposition technique. This is a high-quality, conformal, uniform, pinhole- and particle-free oxide film to minimize the electrical shorting effect and maximize the sensors’ yield. This was followed by the fabrication of the top conductive electrodes. A similar procedure as for the bottom electrodes was followed to fabricate the top electrodes. These conductive electrodes were then coated with a protective oxide layer (SiO2). These protective layers were deposited using a plasma-enhanced chemical vapor deposition system. Several etching steps followed by a lithography step were performed to form channels underneath the sensors. Then, the oxide from electrical measuring pads, called bonding pads, was removed. To achieve this, the patterned wafers went through a wet etching process (6:1 Buffered Oxide Etch) to expose these bonding pads. The final step was further cleaning of the sensing tips and forming them with sharp edges in the channel. This step was achieved using the focused ion beam etching process.
Statistical Analysis.
The critical points we used for analysis of the impedance versus time curves were baseline, minimum, and plateau levels; minimum and maximum slope; and the time taken to reach the minimum and plateau. The baseline, defined earlier, functioned as a reference point to normalize any process variations. The baseline was calculated by averaging the impedance for 200 s starting at 250 s before the addition of the stressor. The real-time impedance was then normalized by the baseline yielding a value of 1 at the baseline. To determine the remaining features, when salt is introduced, we split the data into two sections to remove the large drop in impedance associated with the introduction of salt. The first section contained the baseline and the initial drop, while the second contained the sample’s response to the addition of salt. A high-order polynomial (d = 14) was fit to the second section and used to estimate the rest of the critical points in closed form. The minimum was the point at which the fitted curve is at its lowest. The plateau was the point after the minimum where the impedance is at its largest in the fitted portion. To calculate the minimum and maximum slopes, the fitted polynomial was differentiated. The minimum slope was determined as the minimum value of the differentiated curve between the start of the fitted section and the time of the minimum. Similarly, the maximum slope was determined as the maximum value of the differentiated curve between the time of the minimum, and the time of the plateau. The uncertainty in the measurement of the baseline minimum and plateau was estimated as the SD of the raw data from 60 s before the critical point to 60 s after. All P values were two-tailed and calculated using Welch’s t test.
Electrochemical Impedance Spectroscopy.
To perform electrochemical impedance spectroscopy, a Versa STAT3 potentiostat (Princeton Instruments) was used. An operating voltage of 250 mV RMS was established. We chose to measure impedance at a frequency of 15 KHz, as this frequency was shown to be the optimal working frequency of the sensors (34). The sampling rate was five samples for all experiments. All measurements were performed at room temperature.
Nanosensors’ Preexperiment Cleaning.
To eliminate the potential for sensor contamination, we conducted an extensive sensor cleaning process before each experiment. The cleaning procedure consisted of two main steps: (i) series of liquid washes to remove macroscopic particles, (ii) followed by ultraviolet ozone (UVO) cleaning to eliminate organic contaminants. The liquid wash was performed by dipping a Micro CleanFoam TX757B swab (Texwipe) in 2% SDS and then using the swab to remove any oils or fats deposited on the wafer, by gently rubbing the swab on the silicon surface. The wafer was then rinsed with an excess of deionized (DI) water to wash off the SDS residue. An acetone rinse was then performed using a new foam swab to dislodge any remaining particles. A wash of isopropanol was then performed to remove the residue left by the acetone. The final wash was performed using DI water, and the sensors were then dried with compressed air. Next, the sensors were sanitized using a UVO cleaner (Model 42; Jetlight) for 30 min to remove organic contamination and hydrophilize the SiO2 surface. The full cleaning procedure (liquid wash steps followed by the UVO cleaning step) was repeated twice more (three times in total) to fully eliminate any contamination, residue, and organic contamination.
Sample Preparation.
For each subject, blood was collected in an 8-mL sodium citrate CPT tube and a 6-mL lithium heparin tube. Following the protocol explained in SI Appendix, PBMCs were isolated from the whole-blood plasma, and the cells were adjusted to a concentration of 200 cells per μL and delivered for the experiments.
Experimental Setup.
To make electrical connections to the sensors’ small (50 μm × 50 μm) measuring pads, an S-1160 probe station (Signatone) was used. A biocompatible silicone wells FlexWell Incubation Chamber (Grace Bio-Labs) was cut and placed over a sensor as a microfluidic well to contain the sample. For each experiment, 50 μL of the prepared sample (SI Appendix) was injected into the microfluidic wells, and real-time electrical impedance measurements were then recorded.
Supplementary Material
Acknowledgments
We thank Dr. Laurel Diane Crosby, Professor Craig Heller, and Dr. Bruce Schaar for useful comments, discussions, and support. This work was supported by National Institutes of Health Grant P01 HG000205 and Open Medicine Foundation.
Footnotes
Conflict of interest statement: R.W.D. is Director of the Scientific Advisory Board of the Open Medicine Foundation.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901274116/-/DCSupplemental.
References
- 1.Naviaux RK, et al. Metabolic features of chronic fatigue syndrome. Proc Natl Acad Sci USA. 2016;113:E5472–E5480. doi: 10.1073/pnas.1607571113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moneghetti KJ, et al. Value of circulating cytokine profiling during submaximal exercise testing in myalgic encephalomyelitis/chronic fatigue syndrome. Sci Rep. 2018;8:2779. doi: 10.1038/s41598-018-20941-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prins JB, van der Meer JWM, Bleijenberg G. Chronic fatigue syndrome. Lancet. 2006;367:346–355. doi: 10.1016/S0140-6736(06)68073-2. [DOI] [PubMed] [Google Scholar]
- 4.Montoya JG, et al. Cytokine signature associated with disease severity in chronic fatigue syndrome patients. Proc Natl Acad Sci USA. 2017;114:E7150–E7158. doi: 10.1073/pnas.1710519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorusso L, et al. Immunological aspects of chronic fatigue syndrome. Autoimmun Rev. 2009;8:287–291. doi: 10.1016/j.autrev.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Devanur LD, Kerr JR. Chronic fatigue syndrome. J Clin Virol. 2006;37:139–150. doi: 10.1016/j.jcv.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Falk Hvidberg M, Brinth LS, Olesen AV, Petersen KD, Ehlers L. The health-related quality of life for patients with myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) PLoS One. 2015;10:e0132421. doi: 10.1371/journal.pone.0132421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brea J. 2016 What happens when you have a disease doctors can’t diagnose. Available at https://me-pedia.org/wiki/What_happens_when_you_have_a_disease_doctors_can%27t_diagnose_-_TED_Talk_(2016). Accessed April 16, 2019.
- 9.Maes M, et al. In myalgic encephalomyelitis/chronic fatigue syndrome, increased autoimmune activity against 5-HT is associated with immuno-inflammatory pathways and bacterial translocation. J Affect Disord. 2013;150:223–230. doi: 10.1016/j.jad.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 10.Jason LA, Corradi K, Torres-Harding S, Taylor RR, King C. Chronic fatigue syndrome: The need for subtypes. Neuropsychol Rev. 2005;15:29–58. doi: 10.1007/s11065-005-3588-2. [DOI] [PubMed] [Google Scholar]
- 11.Aoki T, Miyakoshi H, Usuda Y, Herberman RB. Low NK syndrome and its relationship to chronic fatigue syndrome. Clin Immunol Immunopathol. 1993;69:253–265. doi: 10.1006/clin.1993.1178. [DOI] [PubMed] [Google Scholar]
- 12.Fang H, et al. Gene expression profile exploration of a large dataset on chronic fatigue syndrome. Pharmacogenomics. 2006;7:429–440. doi: 10.2217/14622416.7.3.429. [DOI] [PubMed] [Google Scholar]
- 13.Caligiuri M, et al. Phenotypic and functional deficiency of natural killer cells in patients with chronic fatigue syndrome. J Immunol. 1987;139:3306–3313. [PubMed] [Google Scholar]
- 14.Maher KJ, Klimas NG, Fletcher MA. Chronic fatigue syndrome is associated with diminished intracellular perforin. Clin Exp Immunol. 2005;142:505–511. doi: 10.1111/j.1365-2249.2005.02935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natelson BH, Haghighi MH, Ponzio NM. Evidence for the presence of immune dysfunction in chronic fatigue syndrome. Clin Diagn Lab Immunol. 2002;9:747–752. doi: 10.1128/CDLI.9.4.747-752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlo-Stella N, et al. A first study of cytokine genomic polymorphisms in CFS: Positive association of TNF-857 and IFNgamma 874 rare alleles. Clin Exp Rheumatol. 2006;24:179–182. [PubMed] [Google Scholar]
- 17.Tomoda A, et al. Cytokine production and modulation: Comparison of patients with chronic fatigue syndrome and normal controls. Psychiatry Res. 2005;134:101–104. doi: 10.1016/j.psychres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Robertson MJ, et al. Lymphocyte subset differences in patients with chronic fatigue syndrome, multiple sclerosis and major depression. Clin Exp Immunol. 2005;141:326–332. doi: 10.1111/j.1365-2249.2005.02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy G, Spence V, Underwood C, Belch JJF. Increased neutrophil apoptosis in chronic fatigue syndrome. J Clin Pathol. 2004;57:891–893. doi: 10.1136/jcp.2003.015511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carruthers BM, et al. Myalgic encephalomyelitis/chronic fatigue syndrome: Clinical working case definition, diagnostic and treatment protocols. J Chronic Fatigue Syndr. 2003;11:7–115. [Google Scholar]
- 21.Grinde B. Is chronic fatigue syndrome caused by a rare brain infection of a common, normally benign virus? Med Hypotheses. 2008;71:270–274. doi: 10.1016/j.mehy.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Hollingsworth KG, Hodgson T, Macgowan GA, Blamire AM, Newton JL. Impaired cardiac function in chronic fatigue syndrome measured using magnetic resonance cardiac tagging. J Intern Med. 2012;271:264–270. doi: 10.1111/j.1365-2796.2011.02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booth NE, Myhill S, McLaren-Howard J. Mitochondrial dysfunction and the pathophysiology of myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) Int J Clin Exp Med. 2012;5:208–220. [PMC free article] [PubMed] [Google Scholar]
- 24.Gerwyn M, Maes M. Mechanisms explaining muscle fatigue and muscle pain in patients with myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS): A review of recent findings. Curr Rheumatol Rep. 2017;19:1. doi: 10.1007/s11926-017-0628-x. [DOI] [PubMed] [Google Scholar]
- 25.Delves PJ, Martin SJ, Burton DR, Roitt IM. Roitt’s Essential Immunology. 12th Ed Wiley-Blackwell; Hoboken, NJ: 2011. [Google Scholar]
- 26.Dixit AK, Jayabaskaran C. Phospholipid mediated activation of calcium dependent protein kinase 1 (CaCDPK1) from chickpea: A new paradigm of regulation. PLoS One. 2012;7:e51591. doi: 10.1371/journal.pone.0051591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreishcheva EN, et al. Adaptation to salt stress in a salt-tolerant strain of the yeast Yarrowia lipolytica. Biochemistry (Mosc) 1999;64:1061–1067. [PubMed] [Google Scholar]
- 28.Sévin DC, Sauer U. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat Chem Biol. 2014;10:266–272. doi: 10.1038/nchembio.1437. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Oh J, Sung GH. Regulation of MAP kinase Hog1 by calmodulin during hyperosmotic stress. Biochim Biophys Acta. 2016;1863:2551–2559. doi: 10.1016/j.bbamcr.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farabaugh KT, et al. Protein kinase R mediates the inflammatory response induced by hyperosmotic stress. Mol Cell Biol. 2017;37:e00521-16. doi: 10.1128/MCB.00521-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brocker C, Thompson DC, Vasiliou V. The role of hyperosmotic stress in inflammation and disease. Biomol Concepts. 2012;3:345–364. doi: 10.1515/bmc-2012-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esfandyarpour R, et al. Label-free electronic probing of nucleic acids and proteins at the nanoscale using the nanoneedle biosensor. Biomicrofluidics. 2013;7:044114. doi: 10.1063/1.4817771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esfandyarpour R, Yang L, Koochak Z, Harris JS, Davis RW. Nanoelectronic three-dimensional (3D) nanotip sensing array for real-time, sensitive, label-free sequence specific detection of nucleic acids. Biomed Microdevices. 2016;18:7. doi: 10.1007/s10544-016-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esfandyarpour R, Esfandyarpour H, Javanmard M, Harris JS, Davis RW. Microneedle biosensor: A method for direct label-free real time protein detection. Sens Actuators B Chem. 2013;177:848–855. doi: 10.1016/j.snb.2012.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esfandyarpour R, Esfandyarpour H, Harris JS, Davis RW. Simulation and fabrication of a new novel 3D injectable biosensor for high throughput genomics and proteomics in a lab-on-a-chip device. Nanotechnology. 2013;24:465301. doi: 10.1088/0957-4484/24/46/465301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esfandyarpour R, Javanmard M, Koochak Z, Harris JS, Davis RW. Nanoelectronic impedance detection of target cells. Biotechnol Bioeng. 2014;111:1161–1169. doi: 10.1002/bit.25171. [DOI] [PubMed] [Google Scholar]
- 39.Esfandyarpour R, Esfandyarpour H, Javanmard M, Harris JS, Davis RW. Electrical detection of protein biomarkers using nanoneedle biosensors. MRS Proc. 2012;1414:mrsf11-1414-hh04-04. [Google Scholar]
- 40.Esfandyarpour R, Javanmard M, Koochak Z, Esfandyarpour H, Harris JS, Davis RW. Thin film nanoelectronic probe for protein detection. MRS Proc. 2013;1572:mrss13-1572-ss01-10. [Google Scholar]
- 41.Keller BA, Pryor JL, Giloteaux L. Inability of myalgic encephalomyelitis/chronic fatigue syndrome patients to reproduce VO2peak indicates functional impairment. J Transl Med. 2014;12:104. doi: 10.1186/1479-5876-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giaever I, Keese CR. Micromotion of mammalian cells measured electrically. Proc Natl Acad Sci USA. 1991;88:7896–7900. doi: 10.1073/pnas.88.17.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho SN. Intracellular water homeostasis and the mammalian cellular osmotic stress response. J Cell Physiol. 2006;206:9–15. doi: 10.1002/jcp.20445. [DOI] [PubMed] [Google Scholar]
- 44.Shapiro L, Dinarello CA. Osmotic regulation of cytokine synthesis in vitro. Proc Natl Acad Sci USA. 1995;92:12230–12234. doi: 10.1073/pnas.92.26.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro L, Dinarello CA. Hyperosmotic stress as a stimulant for proinflammatory cytokine production. Exp Cell Res. 1997;231:354–362. doi: 10.1006/excr.1997.3476. [DOI] [PubMed] [Google Scholar]
- 46.Li DQ, et al. JNK and ERK MAP kinases mediate induction of IL-1beta, TNF-alpha and IL-8 following hyperosmolar stress in human limbal epithelial cells. Exp Eye Res. 2006;82:588–596. doi: 10.1016/j.exer.2005.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimoto S, et al. Hyperosmolarity-induced interleukin-8 expression in human bronchial epithelial cells through p38 mitogen-activated protein kinase. Am J Respir Crit Care Med. 1999;159:634–640. doi: 10.1164/ajrccm.159.2.9712090. [DOI] [PubMed] [Google Scholar]
- 48.Hubert A, Cauliez B, Chedeville A, Husson A, Lavoinne A. Osmotic stress, a proinflammatory signal in Caco-2 cells. Biochimie. 2004;86:533–541. doi: 10.1016/j.biochi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Zhang D, et al. High-salt enhances the inflammatory response by retina pigment epithelium cells following lipopolysaccharide stimulation. Mediators Inflamm. 2015;2015:197521. doi: 10.1155/2015/197521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loitsch SM, et al. Reactive oxygen intermediates are involved in IL-8 production induced by hyperosmotic stress in human bronchial epithelial cells. Biochem Biophys Res Commun. 2000;276:571–578. doi: 10.1006/bbrc.2000.3504. [DOI] [PubMed] [Google Scholar]
- 51.Finan JD, Guilak F. The effects of osmotic stress on the structure and function of the cell nucleus. J Cell Biochem. 2010;109:460–467. doi: 10.1002/jcb.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Otto NM, et al. Hyperosmotic stress enhances cytokine production and decreases phagocytosis in vitro. Crit Care. 2008;12:R107. doi: 10.1186/cc6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stringer EA, et al. Daily cytokine fluctuations, driven by leptin, are associated with fatigue severity in chronic fatigue syndrome: Evidence of inflammatory pathology. J Transl Med. 2013;11:93. doi: 10.1186/1479-5876-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.