Significance
Although the role of adjuvant and antigen molecules in the generation of adaptive immune responses is well known, the development of vaccines remains an outstanding challenge, particularly for cancer immunotherapy. We investigated a set of spherical nucleic acids (SNAs) in which the adjuvant, antigen, and overall architecture of the nanostructure are conserved but are differentiated in a key feature: the position and conjugation chemistry of the peptide antigen. Comparison of these compositionally equivalent yet structurally distinct SNAs led to our identification of a structure with superior performance as a therapeutic vaccine across several animal models and a mechanistic basis for the improvement in antitumor immune responses; these results indicate the potential for success in the rational development of SNA vaccines.
Keywords: spherical nucleic acids, cancer vaccinology, structural design, nanotechnology, immunotherapy
Abstract
In the case of cancer immunotherapy, nanostructures are attractive because they can carry all of the necessary components of a vaccine, including both antigen and adjuvant. Herein, we explore how spherical nucleic acids (SNAs), an emerging class of nanotherapeutic materials, can be used to deliver peptide antigens and nucleic acid adjuvants to raise immune responses that kill cancer cells, reduce (or eliminate) tumor growth, and extend life in three established mouse tumor models. Three SNA structures that are compositionally nearly identical but structurally different markedly vary in their abilities to cross-prime antigen-specific CD8+ T cells and raise subsequent antitumor immune responses. Importantly, the most effective structure is the one that exhibits synchronization of maximum antigen presentation and costimulatory marker expression. In the human papillomavirus-associated TC-1 model, vaccination with this structure improved overall survival, induced the complete elimination of tumors from 30% of the mice, and conferred curative protection from tumor rechallenges, consistent with immunological memory not otherwise achievable. The antitumor effect of SNA vaccination is dependent on the method of antigen incorporation within the SNA structure, underscoring the modularity of this class of nanostructures and the potential for the deliberate design of new vaccines, thereby defining a type of rational cancer vaccinology.
Fighting cancer through immunotherapy, by engaging and steering a patient’s immune system to attack cancer cells, is a powerful therapeutic approach (1–5). In particular, the success of adoptive cell transfer strategies and checkpoint inhibitors (targeting PD-1, PD-L1, and CTLA4), especially for treating melanoma and lung cancer, has revealed the power of unlocking the immune system to attack tumors (6–8). Indeed, a dramatic response to checkpoint inhibitors in a subset of patients with advanced cancer has been documented. In addition to such approaches, injectable vaccines are particularly attractive because, in principle, they do not involve cell harvesting, and thereby provide a convenient, safe, and low-cost way to boost a patient’s immune system (9, 10).
A major challenge in the development of vaccines is the design and selection of the vehicle for delivering adjuvant and antigen molecules (1). In principle, as with any therapeutic agent, the structure could have a significant influence on safety, efficacy, and potency (11–15). In the case of vaccines, the way in which multiple molecular components are formulated could have a major influence on the biodistribution and delivery to cells of the immune system, and on the activation of immunostimulatory pathways that ultimately lead to the priming and expansion of antigen-specific T cells (16, 17). Nanoparticle vaccines, in particular, provide a way to enhance the delivery of immunostimulatory molecules to the immune system through benefits in biodistribution and codelivery of adjuvant and antigen to immune cells (18). Importantly, vaccine designs that use nanostructures, functionalized with both adjuvant and antigen molecules, have shown the ability to enhance the activation of antigen-presenting cells (APCs) and priming of antigen-specific cytotoxic T lymphocytes (CTLs) over that of mixtures of adjuvant and antigen molecules (19). These developments underscore the need for vaccine design strategies that can effectively address multiple and specific types of immune system cells and activate corresponding pathways (e.g., antigen presentation, costimulatory molecular expression). Furthermore, Gu and Mooney (20) and Koshy and Mooney (21) have hypothesized that the timing of activation and intracellular processing of vaccine components may also be crucial to creating the most active vaccines, and Irvine and coworkers (22) have shown the importance of the temporal programming of dendritic cell (DC) activation by adjusting the order and dose of cytokine injections. In addition, Rincon-Restrepo et al. (23) have shown the effects of nanoparticle size and structure on the intracellular distribution of protein antigens delivered by vaccine particles. Designing vaccines that can control the intracellular spatial distribution of immunostimulatory molecules and the timing of activation of these pathways may be promising for optimizing the induction of antitumor immune responses. Exploiting this opportunity and, in the process, perhaps increasing the success of clinical trials based upon vaccine candidate architectures, however, requires a structural scaffold and modularity that enable the systematic study of the variables that can influence vaccine performance, while conserving other features of the vaccine formulation (e.g., selection, amounts, stoichiometric ratio of antigen and adjuvant).
Spherical nucleic acids (SNAs) are clinically used nanoparticle conjugates consisting of densely packed, highly oriented, therapeutic oligonucleotides (immune-modulatory, antisense, and siRNA gene regulatory) surrounding a nanoparticle core (24–27). SNAs, unlike their linear cousins, possess the ability to enter cells without the need for auxiliary transfection reagents. In 2015, our laboratory reported a class of immunostimulatory SNAs (IS-SNAs) designed to activate the TLR9 pathway and concomitantly deliver a surrogate antigen for the treatment of mouse lymphoma (28). What remains unclear in the design of SNAs as cancer vaccines, however, is how differences in the chemical linkages between the nanoparticle core, oligonucleotide, and peptide can influence and provide ways to improve antigen-specific immune responses. Although prior work has looked at a range of compositionally different constructs and has seen promising activity against simple mixtures of adjuvant and antigen, no study has systematically looked at the consequences of systematically controlling the presentation of antigen and adjuvant at fixed compositional amounts (28). Since IS-SNAs are well-defined nanostructures generated from chemically synthesized and purified molecular components (liposomal cores, chemically functionalized oligonucleotides, and peptides), they enable the systematic study of vaccine structure–activity relationships, and allow one to rationally and iteratively design vaccines with optimum immunostimulatory function.
Herein, we describe a comparison of three SNA structures clearly differentiated in the chemistry of antigen incorporation, and evaluate the ability of these structures to induce antigen-specific immune responses in several mouse models of cancer. We chose these designs to evaluate the importance of SNA structure on their ability to (i) codeliver antigen and adjuvant to individual APCs (and not just populations of APCs); (ii) control the kinetics of release of adjuvant and antigen from the SNA, as well as the timing of antigen presentation and DC activation; and (iii) lead to intracellular processing of peptide antigen for effective presentation by the MHC-I pathway (cross-presentation). These functions are essential for generating an antigen-specific immune response and performing as vaccines. Orchestrating the codelivery and timing of immunostimulatory pathways may lead to successful induction of antigen-specific CTLs, while poor coordination of these events (e.g., induction of costimulatory markers but not of antigen presentation or of antigen presentation without costimulatory markers) could lead to T cell fatigue or anergy.
In designing these three SNAs, our aim was to conserve composition (i.e., TLR9-agonist oligonucleotide, peptide antigen, nanoparticle core) but to vary the position and conjugation chemistry of the peptide antigen. Each of the three SNA structures consists of a unilamellar liposome core (40–45 nm in diameter, dioleoyl-sn-glycero-3-phosphocholine) that presents and orients TLR9 agonist oligonucleotides (3′-cholesterol–functionalized and “1826” CpG sequence specific for the activation of murine TLR9) at the surface. The three SNA architectures (E, A, and H) we examined varied in the position and conjugation chemistry of the peptide antigen in the following ways: (i) soluble antigen encapsulated within the liposome core (“encapsulated” model, E), (ii) antigen located at the surfaces of SNAs by chemical conjugation to oligonucleotides (functionalized at the 3′-terminus with cholesterol groups) adsorbed to the liposome surface (“anchored” model, A), and (iii) antigen located at the surfaces of SNAs by chemical conjugation of the antigen to oligonucleotides hybridized to CpG oligonucleotides adsorbed to the liposome surface (“hybridized” model, H). For antigens chemically conjugated to oligonucleotides, we used a biochemically labile linker for the traceless release of antigen, as previously reported by our group (29). For each of the three SNA structures, we used three different peptide antigens to evaluate immune responses in vitro and in vivo: OVA1 (C-SIINFEKL), melanoma-derived antigen gp100 (C-KVPRNQDWL), and human papillomavirus-16 oncoprotein E6 antigen (VYDFAFRDLC). We evaluated the influence of these structural variations on the uptake, codelivery of CpG and antigen, intracellular trafficking and retention of antigen, kinetics of activation and antigen presentation, induction of antigen-specific CD8+ T cell responses, and, ultimately, in vivo antitumor efficacy. We also compared these activities with those of “unformulated” vaccines: mixtures of soluble TLR9-agonist and peptide antigen, without any chemical conjugation.
Results
Design and Synthesis of SNAs with Variation in Antigen Incorporation.
Our approach to generating well-differentiated SNA structures E, A, and H takes advantage of the modular nature and chemical synthesis of SNAs (Fig. 1A). Each of the molecular components of these SNAs was synthesized and purified (chemically functionalized oligonucleotides, peptides, and liposomes) and incorporated into the liposomal SNA structure through the initial formation of liposomes, followed by the adsorption of the adjuvant to their surface via hydrophobic anchoring groups (cholesterol). For SNA E, antigen was loaded into the core during the liposome formation process. For SNA A, a peptide–oligonucleotide–3′-cholesterol conjugate was coadsorbed to the liposomes along with 3′-cholesterol–functionalized CpG. For SNA H, a peptide-oligonucleotide conjugate, with a nucleotide sequence complementary to CpG, was hybridized with CpG oligonucleotides before adsorption to the liposomes (SI Appendix, Table S1). Details of the synthetic procedures and the characterization of the physical properties and chemical composition of the SNAs are available in SI Appendix (SI Appendix, Fig. S1 A–D). To compare SNAs that differ in structure, but not in composition, we prepared E, A, and H SNAs that were similar in the stoichiometry of CpG and antigen to liposome (75 molecules of each per liposomal structure with an average diameter of 55–60 nm, including the oligonucleotide shell) (SI Appendix, Fig. S1 E and F). We synthesized SNAs E, A, and H with different antigens (OVA-1, gp100, and E6), and subsequently compared their immunostimulatory properties and explored their performance as therapeutic vaccines in clinically relevant mouse tumor models (SI Appendix, Table S2).
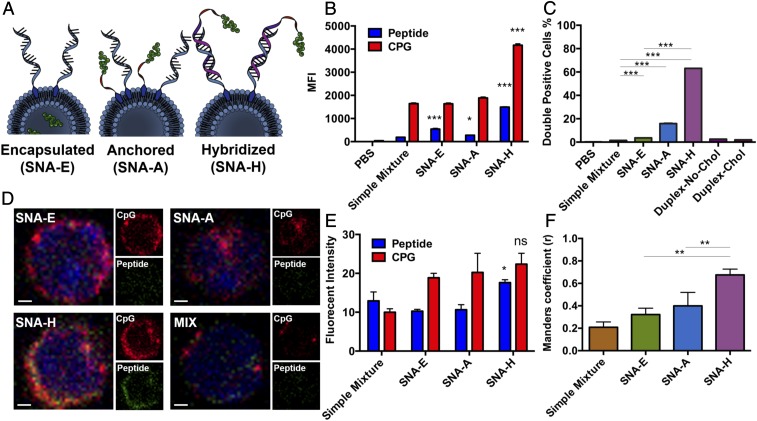
Fig. 1.
Evaluation of the dependence of CpG and antigen codelivery on SNA structure. (A) Scheme of three designs of SNA E, SNA A, and SNA H. (B) Uptake of CpG (Cy5) and OVA1 (TMR) by BMDCs in vitro, measured by flow cytometry. MFI, medium fluorescence intensity. (C) Fraction of cells showing high levels of both CpG and OVA1, recovered from the DLNs of mice (n = 3) 2 h following s.c. injection with reagents as indicated, as determined by flow cytometry. Values are an average of three replicates. Chol, cholesterol. (D) Images of cells recovered from DLNs from mice 4 h following immunization by s.c. injection, visualized by confocal microscopy. OVA1 peptide labeled with TMR is shown in green, and CpG labeled with Cy5 is shown in red. (Scale bars, 1 μm.) (E) Fluorescence intensity for OVA1 peptide and CpG of the images. (F) Subcellular colocalization of peptide and CpG was quantified by Manders coefficient (values of r > 0.6 indicate strong colocalization). Data presented as mean ± SEM (B, C, E, and F). ***P < 0.001; **P < 0.01; *P < 0.05. ns, not significant.
Codelivery of Immunostimulatory Oligonucleotides and Peptide Antigens to DCs.
We compared the ability of E, A, and H SNA structures to enter DCs and deliver both CpG oligonucleotides and peptide antigens to individual DCs. The delivery of both types of molecules and the induction of signaling for the parallel pathways of antigen presentation and costimulatory marker expression are essential steps for activating APCs and further priming antigen-specific T cells. Upon treatment of bone marrow-derived DCs (BMDCs) with each SNA structure functionalized with CpG (labeled with Cy5) and OVA1 antigen [labeled with tetra-methyl-rhodamine (TMR)] and analysis of cellular uptake, we found significant advantages for SNA H in the uptake of both CpG and antigen (Fig. 1B). To investigate these effects in vivo, we injected mice s.c. with the same set of SNAs. Extraction of the draining lymph node (DLN) after 2 h and analysis of the CD11c+ DCs by flow cytometry showed a wide range in the fraction of cells containing high levels of both CpG and OVA1. The fraction of DCs with high levels of uptake for both CpG and OVA1 depended on SNA structure and followed the order of E < A < H. Indeed, SNA H remarkably led to >60% of a DC population showing codelivered adjuvant and antigen, far greater than that for SNAs E and A (Fig. 1C). In contrast, for mixtures of CpG and OVA1 (no coupling between the components), the fraction of DCs showing codelivery was negligible (<1.5%). The comparison of results for SNA H and dsDNA conjugated to OVA1 that is not formulated into an SNA structure (<2% codelivery) establishes the critical influence of SNA structure in achieving high levels of codelivered oligonucleotide and peptide. Indeed, the observation that SNA H is generated from dsDNA and liposomes, yet shows significantly higher uptake and codelivery of CpG and antigen than dsDNA, suggests that these components enter presumably as intact SNA structures. Taken together, these data show that the dependence of the codelivery of CpG and antigen on SNA structure and the superiority of SNA H are amplified in vivo. The structural features of SNA H that drive the enhancement of codelivery are (i) the linkage of antigen to CpG by chemical conjugation and nucleic acid hybridization and (ii) the enhancement of cellular uptake of oligonucleotides by the SNA architecture. SNA H is not susceptible to erosion in codelivery through the mechanisms likely responsible for the separation of antigen and CpG in SNAs E and A (i.e., leakage of peptide through liposome membranes, desorption of antigen-functionalized oligonucleotides from liposomes) (30).
We analyzed the codelivery of adjuvant and antigen molecules by SNAs by imaging (by confocal microscopy) the DCs extracted from mice immunized by SNAs with Cy5-labeled CpG and TMR-labeled OVA. The images show comparable levels of CpG delivered by each SNA structure, but higher levels of OVA1 codelivered by SNA H than those codelivered by SNAs A and E (Figs. 1 D and E and SI Appendix, Fig. S1G). Manders coefficient values (Fig. 1F) showed a decreasing r score for SNAs H (r = 0.68), A (r = 0.40), and E (r = 0.32), indicating that the highest levels of subcellular colocalization of CpG and OVA1 are accomplished by SNA H at an early time point (4 h after vaccination), when intracellular processing of antigen is at an early stage.
Trafficking of Peptide Antigens Within DCs, Delivered by Different SNA Structures.
We compared the uptake, trafficking, and retention of peptide antigens delivered by SNAs E, A, and H. Upon treatment of BMDCs with SNA structures formulated with OVA1 labeled with Cy5 for 2 h, the cells were washed and incubated in fresh medium and monitored by confocal fluorescence microscopy over a further 24-h period. The presence of OVA1 in the late endosomes (LEs) and endoplasmic reticulum (ER) was determined by colocalization of Cy5 (red) and fluorescent markers (green) for the LEs and ER, respectively, in confocal microscope images (Fig. 2 A and B). We found clear trends that differentiate the SNA structures in the uptake of OVA1 (at the earliest time points of 2 and 4 h) and in the retention of OVA1 at the late time points. The order in overall delivery of OVA1 is H > A > E at the early time point of 2 h. At 24 h, only SNA H enabled substantial retention of peptide within the cells (57% of the maximum levels observed at 2 h). Both SNAs E and A, however, showed a rapid decline in the presence of peptide (<8% of maximum levels observed at 2 h) (Fig. 2C). The subsequent analysis of subcellular distribution of OVA1 indicated that this effect was driven by the sustained retention of OVA1 delivered by SNA H in the endosome (Fig. 2D) and ER (Fig. 2E), the site of MHC-1 peptide loading, through the 24-h period following SNA treatment. The higher uptake of peptide antigen delivered by SNA H, followed by retention at substantial levels of these peptides in the endocytic pathway and ER for a 24-h period, is dependent on the structure of SNA H, and provides a major advantage in generating longer windows of time for efficient cross-priming of antigen-specific T cells by DCs.
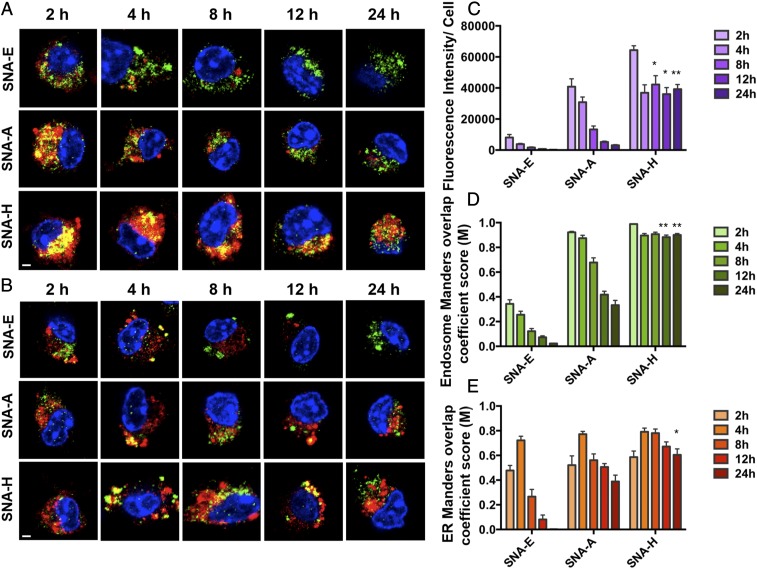
Fig. 2.
Evaluation of the time-dependent intracellular fate of antigens delivered by the three SNA structures by confocal microscopy. Images of OVA1 peptide (Cy5, red) colocalized with the LE (A; Alexa-568, green, Rab9) or ER (B; Alexa-568, green, PDI) delivered by SNA E, SNA A, and SNA H are shown. (Scale bars, 2 μm.) (C) Peptide fluorescent intensity per cell over time. (D) Manders overlap coefficient (M) representing the fraction of endosomes in which the Rab9 signal is colocalized with Cy5. (E) M representing the fraction of the ER in which the PDI signal is colocalized with Cy5. SNA H has a major advantage over SNA A and SNA E in the temporal release of antigen, by way of increased retention of peptide within the endosomes of BMDCs throughout the 24-h period. All analysis values are an average of 10–15 randomly selected images. Data presented as mean ± SEM (C–E). ***P < 0.001; **P < 0.01; *P < 0.05.
Activation of DCs and Cross-Priming of T Cells by SNAs.
Antigen-specific T cell responses depend upon the interaction between activated DCs and T cells; the quality of this interaction, and of the subsequent T cell response, is dependent upon the concerted presentation of antigen and expression of costimulatory markers by DCs upon vaccination (22). We therefore compared the kinetics of the parallel pathways of presentation of SNA-delivered OVA1 and the expression of the costimulatory marker CD86 in BMDCs. Following the treatment of BMDCs with SNAs for 1 h (2.5 μM in OVA1 and CpG) and subsequent washing to remove SNAs from the cell culture medium, cells were resuspended and incubated in fresh medium for up to 48 h. Although the maximum expression of CD86 took place ∼24 h after treatment for all three SNA structures (Fig. 3A), notably, the time at which OVA1 presentation was maximized was different among the SNAs (∼16 h for SNA E, ∼20 h for SNA A, and ∼24 h for SNA H; Fig. 3A). A major consequence of the slower kinetics of antigen presentation induced by SNAs A and H (compared with SNA E), due to the processing and dissociation of OVA1 from these SNA structures, was greater overlap in time where DCs present both antigen and costimulatory markers. Importantly, the kinetic data for SNA H showed synchronization of maximized antigen presentation and costimulatory marker expression (Fig. 3A). Taken together with the superior ability of SNA H to codeliver CpG and peptide to DCs, these data suggest that SNA H may be ideal for the priming of antigen-specific T cells.
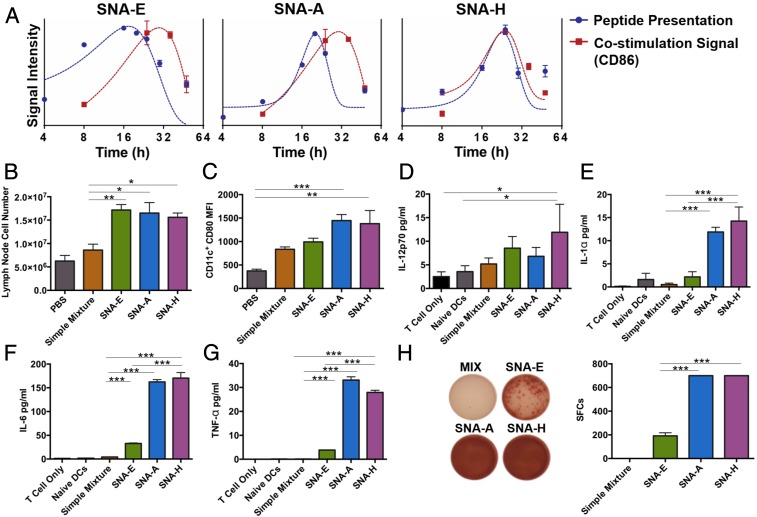
Fig. 3.
Kinetics of DC activation with SNAs. (A) Kinetics of antigen (OVA1) presentation and expression of costimulation marker (CD86) by BMDCs upon treatment with SNAs, determined by flow cytometry. Data points are the average of three measurements of gating, and curves obtained by fitting the data to Gaussian functions are to guide the eye. (B) Number of DLN cells from mice (n = 3) 16 h following immunization by s.c. injection with reagents as indicated. (C) Expression of costimulatory marker CD80 by DLN DCs collected from immunized mice above. (D–G) DCs isolated from immunized mice above were cocultured with purified OT1 CD8+ T cells for 48 h. Secretion of IL-12p70, IL-1α, IL-6, or TNF-α in the culture supernatant was determined by ELISA. (H) Presence of IFN-γ–secreting CD8+ T cells was measured by ELISpot assay [representative images (Left) and counts from three replicate measurements (Right)]. Data presented as mean ± SEM (B–H). ***P < 0.001; **P < 0.01; *P < 0.05.
Immunization by s.c. injection of SNAs resulted in DC activation and antigen presentation in vivo. In all three SNA designs, the DLNs of immunized C57BL/6 mice swelled and showed increased cellularity (16 h following immunization), compared with those of mice immunized with a mixture of CpG and OVA1 (Fig. 3B). CD80 expression on CD11c+ DCs in DLNs was higher for SNAs A and H than for SNA E or a mixture of CpG and OVA1 (Fig. 3C), while expression levels of CD86 and CD40 were comparable across all treatment groups (SI Appendix, Fig. S2 A and B).
We next examined the ability of DCs activated by SNAs in vivo to cross-prime CD8+ T cells. DCs from the DLNs were harvested from immunized mice and cocultured with OT1 CD8+ T cells for 2 d ex vivo. The secretion of proinflammatory cytokines (IL-12p70, IL-1α, IL-6, and TNF-α) was highly dependent on SNA structure. Although each SNA structure (E, A, and H) led to greater levels of cytokine secretion than for mixtures of CpG and OVA1 (Fig. 3 D–G), SNAs H and A were superior to SNA E in stimulating the secretion of IL-1α, IL-6, and TNF-α by OVA1-specific T cells. In addition, we used an enzyme-linked immune absorbent spot (ELISpot) assay to examine the number of IFN-γ–secreting T cells generated by coculturing with DCs from immunized mice. The DCs extracted from SNA H- and SNA A-immunized mice showed a greater ability to induce IFN-γ production from OT1 CD8+ T cells, compared with those extracted from SNA E-immunized mice (Fig. 3H). Importantly, vaccination with oligonucleotides conjugated to OVA1 not formulated as SNAs had a negligible effect on non-antigen–specific DC activation (SI Appendix, Fig. S2 C–E). These observations demonstrate that differences in SNA structure ultimately lead to substantial differences in the quality of antigen-specific T cell responses.
Antigen-Specific CTL Responses Generated by Vaccination with SNAs.
We evaluated the quality of antigen-specific CTL responses induced by the vaccination of immunocompetent mice (C57BL/6) by SNAs E, A, and H and, for comparison, mixtures of CpG and antigen. We performed a comparison of SNA structures for three different antigens (31, 32): OVA1 (Fig. 4 A–D and I and SI Appendix, Fig. S3A), E6 (Fig. 4 E–H and J), and gp100 (SI Appendix, Fig. S3B). We found that the influence of SNA structure on raising antigen-specific T cells is not limited to OVA or restricted by the selection of antigen. The data in Fig. 4 show that SNA structures were superior to mixtures of CpG and peptide antigen at generating cytotoxic and memory phenotypes in antigen-specific CD8+ T cells in vivo through the incorporation of OVA1 (Fig. 4 A and B) and E6 (Fig. 4 E and F). The effector function of antigen-specific CD8+ T cells raised in immunized mice, as measured by IFN-γ secretion via both ELISpot assay and flow cytometry, was significantly increased for mice vaccinated with SNAs A and H, for both OVA1 and E6 (Fig. 4 C, D, G, and H). The comparable performance of SNAs A and H in raising antigen-specific T cell responses is an outcome far downstream of vaccination and the uptake of SNAs by APCs. The advantage of SNA H over SNA A in codelivery of CpG and antigen (as observed in Fig. 1) acts only as part of a mechanism for enhancing antigen-specific T cell responses. Other factors (e.g., kinetic profiles for processing of CpG and antigen, intracellular trafficking of CpG and antigen, sensitivity to antigen presentation levels for T cell priming) in which SNAs H and A are similar may contribute to the overall similarity in SNAs H and A in downstream T cell function. Vaccination with mixtures of CpG and peptide yielded negligible numbers of IFN-γ–secreting T cells, as did vaccination with SNA E for E6 (Fig. 4 G and H).
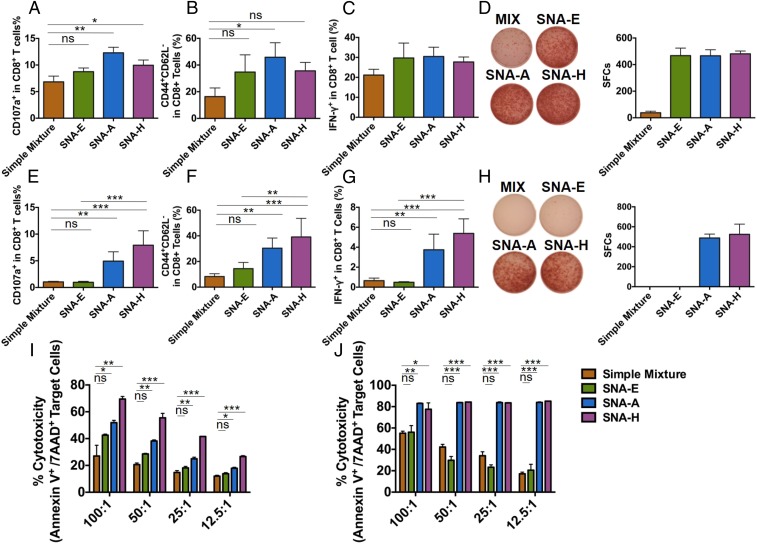
Fig. 4.
Antigen-specific CTL responses induced by SNA vaccination. C57BL/6 mice (n = 3) were immunized by three s.c. injections of SNAs or a mixture of OVA1 antigen (A–D and I) or E6 antigen (E–H and J) on days 0, 14, and 28. One week later, splenic T cells were analyzed by flow cytometry. Percentages of CD8+ T cells that were positive for CD107a (marker for cytotoxic activity) (A and E), for CD44+CD62L− (effector memory phenotype) (B and F), and for IFN-γ (C and G) are shown. The presence of IFN-γ–secreting splenic CD8+ T cells from immunized mice above was measured by ELISpot assay 48 h after restimulation ex vivo with OVA1 (D) or E6 antigen (H) [representative images (Left) and counts from three replicate measurements (Right)]. A comparison of OVA1-specific (I) or E6-specific (J) cytotoxicity induced by different SNAs is shown. Purified splenic CD8+ T cells from immunized mice above were cocultured with corresponding target tumor cells at the indicated ratios for 24 h, and tumor cell apoptosis was measured using annexin V and 7-aminoactinomycin D (7-AAD) staining by flow cytometry. Data are presented as mean ± SEM. ***P < 0.001; **P < 0.01; *P < 0.05. ns, not significant.
For T cells raised by SNAs formulated with OVA1, SNA H led to the greatest efficacy in killing target cells (EG.7-OVA) in a dose-dependent fashion (Fig. 4I). Furthermore, the killing of target cells showed a clear dependence on SNA structure, following the order of H > A > E > mixture of CpG and OVA1. For the targeted killing of TC-1 cells, vaccinations with SNAs H and A with E6 led to comparable CTL performances that were far superior to that induced by SNA E or a mixture of CpG and E6. These data indicate that the structure of SNA H, by way of the advantages in its interaction with DCs, ultimately leads to superior antigen-specific T cell responses in vivo. The effect of SNA structure on CTL activity was, however, more emphatic for E6 than for OVA1. Whether the differences observed between these two antigen systems are driven primarily by the intrinsic immunogenicity of the E6 and OVA1 antigens, or by the influence of the peptide antigens on the properties of SNAs, warrants further investigation. Taken together, these experiments indicate the potentially broad applicability of SNA structures, particularly SNA H, in raising immune responses to different tumor-specific antigens and, ultimately, their use in cancer immunotherapy.
SNA Structure-Dependent Antitumor Immune Responses.
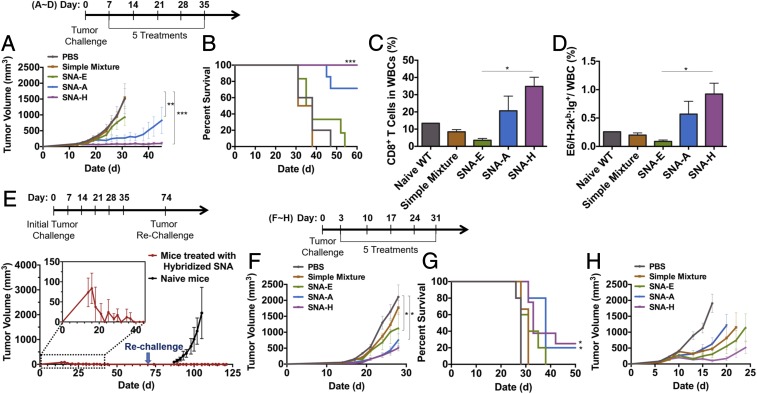
To evaluate SNA structures as potential immunotherapeutic agents for cancer, we tested three well-established tumor-bearing mouse models with SNAs. TC-1 tumors were generated by s.c. implantation of TC-1 cells in the flanks of C57BL/6 mice and then allowing them to grow to ∼50 mm3 before treatment with SNA structures E, A, and H, each formulated with the E6 antigen (seven to 10 mice per group). Additional groups for untreated mice and treatment with a mixture of CpG and E6 peptide served as control and reference groups, respectively. Treatment consisted of an initial vaccination followed by four boosts, with 7 d between each boost (Fig. 5A, scheme). Treatment with SNA H strikingly led to tumor regression and survival of 100% of the animals in the group through 60 d (Fig. 5 A and B). In contrast, treatment with mixtures of CpG and E6 or SNA E failed to deliver significant improvements in tumor burden or survival over the untreated group, suggesting that the antitumor efficacy of SNAs is highly dependent upon the SNA structure. Within the SNA H treatment group, 30% of the animals were in a tumor-free condition until 72 d. These tumor-free mice were subsequently rechallenged (on day 72) with an inoculation of fresh TC-1 cells into the flank opposing the initial tumor site, but were not given any additional therapy. These mice rejected the implanted TC-1 cells, while tumor growth was aggressive in a reference group (naive mice that had received no prior vaccination) (Fig. 5E). This observation suggests that the immunological memory generated by treatment with SNA H leads to long-term tumor protection. The growth of TC-1 tumors was also significantly inhibited by treatment with SNA A (Fig. 5A); 70% of the animals treated with SNA A survived through 60 d.
Fig. 5.
SNA structures determine the antitumor efficacy of SNA vaccination. Seven days after tumor implantation, TC-1 tumor-bearing C57BL/6 mice (n = 7–10) were treated with PBS, SNA E, SNA A, SNA H, or a mixture of CpG and E6. (A) Tumor growth curves for each treatment group. (B) Survival of tumor-bearing mice shown in Kaplan–Meier curves. (C) Percentage of WBCs on day 40 that are CD8+ T cells. (D) Percentage of WBCs on day 40 that are E6-specific CD8+ T cells, as determined by staining T cells with E6 dimer. (E) Design for tumor rechallenge experiment. The memory effect and sustained rejection of tumor rechallenge in SNA H-treated mice that had rejected the initial TC-1 tumor implantation and were tumor-free at least until day 72 (red line) and, as a control group (black line), the growth of tumors in naive C57BL/6 mice upon inoculation with TC-1 cells are shown. Tumor growth (F) and Kaplan–Meier survival curves (G) of LLC1-OVA–bearing C57BL/6 mice treated with SNA E, SNA A, SNA H, or a mixture of CpG and OVA1 are shown. (H) Tumor growth curve of EG.7-OVA–bearing C57BL/6 mice treated with SNA E, SNA A, SNA H, or a mixture of CpG and OVA1. ***P < 0.001; **P < 0.01; *P < 0.05. Statistical significance for survival analysis in B and G was calculated by the log-rank test.
The efficacy of SNA H and SNA E in tumor inhibition and survival was consistent with the tumor antigen-specific CD8+ T cell responses raised by these vaccines. The percentages of overall CD8+ T cells and E6-specific CD8+ T cells within WBCs were highest for peripheral blood sampled (on day 40) from animals treated with SNAs H and A (34.8% and 20.7%, respectively, for CD8+ T cells; 0.9% and 0.6%, respectively, for E6-specific CD8+ T cells). These percentages were significantly lower for the other treatment groups (3.5% and 8.4%, respectively, for CD8+ T cells in the SNA E- and PBS-treated groups; 0.1% and 0.2%, respectively, for E6-specific CD8+ T cells) (Fig. 5 C and D).
We also found that the quality of antitumor immune responses in mice bearing LLC1-OVA tumors and E.G7-OVA tumors was highly SNA structure-dependent. Treatment with SNAs H and A functionalized with OVA peptide resulted in the best outcomes in tumor growth inhibition and animal survival; 80% of animals in these groups survived through day 31, a time point at which 100% of the animals had perished in groups of animals that were untreated or treated with a mixture of CpG and OVA (Fig. 5 F and G). The use of SNAs in prophylactic vaccination was capable of delaying LLC1-OVA tumor initiation and growth. Animals were vaccinated 21 d and 7 d (primary injection and boost, respectively) before implantation of LLC1-OVA cells. Each SNA structure was superior to a mixture of CpG and OVA peptide in delaying the initiation of tumor growth and prolonging survival (SI Appendix, Fig. S4 A–D). Prophylactic vaccination with SNA H led to the best outcomes, resulting in a 15-d delay in tumor initiation, longer than that observed for vaccination with SNAs A (13 d) or E (11 d) (SI Appendix, Fig. S4C). With E.G7-OVA tumor treatment, we used the same dosing and treatment plan used in the treatment of mouse models of LLC1-OVA tumors. Treatment with SNA H functionalized with OVA peptide resulted in the best outcomes in tumor growth inhibition (Fig. 5H), while SNAs E and A led to outcomes comparable to those for mixtures of antigen and CpG.
Our examination of the effects of SNA structure on the three different tumor models revealed that treatment with SNA H leads to the best outcomes in tumor burden and animal survival. Treatment with SNA A leads to significantly better outcomes than those for SNA E or mixtures of CpG and antigen; in the LLC1-OVA model, treatments with SNAs A and H led to comparable outcomes, while the E.G7-OVA model revealed the best outcomes for SNA H. These results also showed differences in the efficacy of SNA vaccination and the dependence on SNA structure between the TC-1 (E6), LLC1 (OVA), and E.G7 (OVA) models, particularly in the elimination of TC-1 tumors upon treatment with SNA H. These differences are likely due to the immunogenicity of the antigens used (E6 and OVA1) and the aggressiveness of the cells used to generate the tumor models. These tumor models have been used to illustrate the antitumor activity of vaccines using other materials (e.g., polymer-based delivery of antigen and adjuvant). Our study of SNAs in this study, however, shows efficacy using structures composed of US Food and Drug Administration-approved classes of materials (i.e., liposomes, oligonucleotides) and provides a potential way to avoid the chronic liver toxicity that may arise from the use of polymeric materials (33–35).
Discussion
This study of compositionally equivalent yet structurally distinct SNAs has determined that differences in SNA structure can lead to major improvements in raising cellular immune responses and outcomes in antitumor immunotherapy. A key lesson from this study is that even within a single class of materials, the way in which adjuvant molecules and tumor-associated antigens are structured within a vaccine can profoundly influence the activation of immune responses. Numerous comparisons of uptake and intracellular trafficking (Figs. 1 and 2), DC activation (Fig. 3), T cell activation (Fig. 4), and therapeutic outcomes in vivo (Fig. 5) showed the inability of mixtures of CpG and peptide antigen to boost effective immune responses, while consistently resulting in the ability of SNA structures to invoke responses in a manner clearly dependent upon how the SNA structures incorporate antigen and adjuvant molecules (H > A > E). These differences are emphatic in the interaction of SNAs with DCs, by controlling the codelivery of CpG and peptide, tuning the subcellular trafficking and retention of peptides within individual cells, and synchronizing the kinetics of processing of CpG and antigen; these differences ultimately drive the quality of the effector function of antigen-specific killing of tumor cells in vivo and range from essentially ineffective to curative.
Vaccine immunotherapy for cancer has shown limited success clinically, despite vaccines that have exploited the knowledge and availability of adjuvants, tumor-associated antigens, and our understanding of APC–T cell interactions (36). The lack of outstanding clinical responses to date may be due to the shortcomings of previously examined vaccines in their ability to codeliver the molecular components of vaccines to APCs and to control the timing of multiple pathways required to generate high-quality cellular immune responses, as well as to the absence of these considerations in efforts to develop vaccines. The modularity of SNAs in our study of vaccine designs here has enabled the identification of SNA H as superior among SNAs nearly identical in composition but different in structure, and the mechanistic basis for its higher performance. This structure-based approach using SNAs, and analysis of mechanisms that correlate improvements in APC activation and T cell priming to improvements to therapeutic outcomes in vivo, defines a rational design approach that allows one to identify functional components and systematically optimize vaccine effectiveness, increasing their chances of being translated to the clinic. This is especially true for the SNA system, where the core adjuvant has been successfully taken into the clinic and is now being used in patients who have cancer. Therefore, given the scalability and clinical relevance of SNAs, this work defines a promising route to creating effective vaccines, potentially for many conditions. The improvements in vaccine performance demonstrated in multiple tumor models under both preventive and therapeutic settings suggest the promise of rationally designed SNAs in immunotherapy for cancer. In addition, the combination of SNA vaccines and immune checkpoint inhibitors may provide a route for further potential improvement in therapeutic outcomes, given the suppressive mechanisms engaged by the tumor microenvironment.
Materials and Methods
SNA Synthesis.
DNA oligonucleotide synthesis, antigen conjugation, and liposome nanoparticle functionalization and characterization are discussed in SI Appendix (SI Appendix, Fig. S1).
In Vitro SNA Administration and Analysis.
SNAs were added to the culture medium for BMDCs at varying concentrations (100 nM–2.5 μM). To measure SNA uptake, we incubated SNAs with BMDCs at 100 nM in both antigen and adjuvant for 30 min. Cells were stained with markers for DCs (CD11c, catalog no. 117308; Biolegend) and fixed (4% paraformaldehyde) before analysis by flow cytometry. Gating parameters in the analysis of flow cytometry data were established from cells that served as negative controls, which were treated with PBS or medium only. To analyze antigen presentation and costimulatory molecule expression, SNAs were added to BMDCs for 1 h [the antigen and adjuvant concentration was fixed at 2.5 μM (for both)] and were subsequently removed (time points below) by washing the cells and adding fresh media. At designated time points (Fig. 3A) from the start of the experiment to 48 h, cells were stained with fluorescent antibodies for CD11c, OVA1-H-2kb, and CD86, and cell populations were characterized by values for positive gating. Values from three repetitions of the kinetics experiment were used to fit curves with Gaussian functions. To study the priming and proliferation of T cells in vitro, SNAs functionalized with OVA1 or gp100 were added to the whole splenocytes of OT1 mice or pmel mice, respectively. T cell proliferation was measured by flow cytometry 72 h after treatment by analysis of the dilution of proliferation dye efluor 450 (SI Appendix, Fig. S3 A and B).
Analysis of Intracellular Distribution of SNAs by Confocal Microscopy.
To visualize SNA uptake and intracellular trafficking, we plated purified BMDCs [selected using a kit from Stem Cell (catalog no. 18556)] on chamber slides and treated them with SNAs for 2 h (2.5 μM in peptide and CpG). Cells were then incubated with fresh medium for up to 24 h. Cells were then fixed (4% paraformaldehyde) and permeabilized (0.5% Triton in PBS) before staining with organelle markers (LE: Rab9, Abcam ab179815; ER: PDI, Abcam ab31811) and DAPI (no. 1351303edu; Bio-Rad). Images of cells were collected by a Zeiss LSM 800 microscope using the same parameters for image acquisition for each sample (e.g., laser power, master gain, offset). For statistical analysis of images, 10–15 separate images were randomly selected, and the determination of Manders overlap coefficient (M) used the reconstruction of 10 Z-stack images of each cell. Detailed description of Manders colocalization (r) and M calculations are described in ref. 37.
In Vivo T Cell Memory Response Analysis.
For in vivo T cell memory response analyses, C57BL/6 mice were vaccinated (on days 0, 14, and 28 with simple mixtures or SNA structures composed of 6 nmol of peptide and 6 nmol of CpG), and the spleens were collected on day 35. We used flow cytometry and ELISpot assays to measure IFN-γ expression level from the CD8+ T cells of vaccinated mice. Memory phenotype markers (CD62L and CD44) and T cell degranulation markers (CD107a) were also measured by flow cytometry. Antigen-specific T cells were expanded ex vivo by taking splenocytes from vaccinated mice and coculturing them with γ-irradiated, peptide-pulsed, naive splenocytes for 5 d. Upon selection of CD8+ T cells from these cocultures, the cytotoxicity of these cells was determined by coculturing the T cells with target cells overnight. The apoptosis of target cells was analyzed by annexin V and 7-aminoactinomycin D staining through flow cytometry.
IFN-γ ELISpot Assay.
The ELISpot assay was performed as described previously using the mouse IFN-γ ELISpot Ready-SET-Go! Kit (no. 15531137; eBioscience) (38, 39). Briefly, 2 × 105 splenocytes from each vaccinated mouse were incubated with or without OVA1 or E6 peptide at 37 °C for 48 h. The IFN-γ spots were analyzed by an ImmunoSpot Microanalyzer (Cellular Technology Limited).
Flow Cytometry.
Antibodies were purchased from Biolegend. Surface staining, annexin V staining, and intracellular IFN-γ staining were performed as published previously (40). Samples were acquired on a BD LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo software.
For the analysis of codelivery of OVA1 and CpG to DCs, “high” levels of uptake were defined by identifying and counting double-positive cells and comparing that population with cells only treated with PBS (used as the gate in the analysis of flow cytometry data).
Tumor Inoculation and in Vivo Efficacy Studies.
All animals were used under an approved protocol of the Institutional Animal Care and Use Committee of Northwestern University. To implant tumors, TC-1 (2 × 105), LLC1-OVA (2 × 105), or E.G7-OVA (5 × 105) cells were injected s.c. into the right flank of C57BL/6 mice (∼8–12 wk old). To assess therapeutic efficacy, we vaccinated with SNAs or simple mixtures of peptide and CpG by s.c. injection (doses were 6 nmol of peptide and 6 nmol of CpG) once per week, starting on day 3 (LLC1-OVA and E.G7-OVA) or day 7 (TC-1), when the tumors are palpable. Tumors were measured by calipers every other day. Animal health was monitored by body weight and activity measurement.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health (NIH) under Award U54CA199091. It was also supported by the Prostate Cancer Foundation and the Movember Foundation under Award 17CHAL08, the IDP Sherman Fairchild Foundation through the Robert H. Lurie Comprehensive Cancer Center, and the Vannevar Bush Faculty Fellowship program sponsored by the Basic Research Office of the Assistant Secretary of Defense for Research and Engineering and funded by the Office of Naval Research through Grant N00014-15-1-0043. S.W. and G.Y. were supported by fellowships associated with the Chemistry of Life Processes Predoctoral Training Program T32GM105538 at Northwestern University. Z.H. was supported in part by the Northwestern University Graduate School Cluster in Biotechnology, Systems, and Synthetic Biology, which is affiliated with the Biotechnology Training Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902805116/-/DCSupplemental.
References
- 1.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T (2014) Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat Rev Cancer 14:135–146. [DOI] [PubMed] [Google Scholar]
- 2.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD (2016) The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol 13:273–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irvine DJ, Swartz MA, Szeto GL (2013) Engineering synthetic vaccines using cues from natural immunity. Nat Mater 12:978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, et al. (2008) Nanoparticles in medicine: Therapeutic applications and developments. Clin Pharmacol Ther 83:761–769. [DOI] [PubMed] [Google Scholar]
- 5.Ashley CE, et al. (2011) The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat Mater 10:389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulley JL, et al. (2015) Avelumab (MSB0010718C), an anti-PD-L1 antibody, in advanced NSCLC patients: A phase 1b, open-label expansion trial in patients progressing after platinum-based chemotherapy. J Clin Oncol 33(Suppl 15):8034. [Google Scholar]
- 7.Patnaik A, et al. (2015) Phase 1 study of pembrolizumab (pembro; MK-3475) plus ipilimumab (IPI) as second-line therapy for advanced non-small cell lung cancer (NSCLC): KEYNOTE-021 cohort D. J Clin Oncol 33(Suppl 15):8011. [Google Scholar]
- 8.Weber JS, et al. (2015) Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): A randomised, controlled, open-label, phase 3 trial. Lancet Oncol 16:375–384. [DOI] [PubMed] [Google Scholar]
- 9.Boutros C, et al. (2016) Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat Rev Clin Oncol 13:473–486. [DOI] [PubMed] [Google Scholar]
- 10.Naidoo J, et al. (2015) Toxicities of the anti-PD-1 and anti-PD-L1 immune checkpoint antibodies. Ann Oncol 26:2375–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langer R, Tirrell DA (2004) Designing materials for biology and medicine. Nature 428:487–492. [DOI] [PubMed] [Google Scholar]
- 12.Peer D, et al. (2007) Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol 2:751–760. [DOI] [PubMed] [Google Scholar]
- 13.Kanasty R, Dorkin JR, Vegas A, Anderson D (2013) Delivery materials for siRNA therapeutics. Nat Mater 12:967–977. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Zhao W, Tan W (2008) Bioconjugated silica nanoparticles: Development and applications. Nano Res 1:99–115. [Google Scholar]
- 15.Sutton D, Nasongkla N, Blanco E, Gao J (2007) Functionalized micellar systems for cancer targeted drug delivery. Pharm Res 24:1029–1046. [DOI] [PubMed] [Google Scholar]
- 16.Frey S, Castro A, Arsiwala A, Kane RS (2018) Bionanotechnology for vaccine design. Curr Opin Biotechnol 52:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irvine DJ, Hanson MC, Rakhra K, Tokatlian T (2015) Synthetic nanoparticles for vaccines and immunotherapy. Chem Rev 115:11109–11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couvreur P. (2013) Nanoparticles in drug delivery: Past, present and future. Adv Drug Deliv Rev 65:21–23. [DOI] [PubMed] [Google Scholar]
- 19.Kemp JA, Shim MS, Heo CY, Kwon YJ (2016) “Combo” nanomedicine: Co-delivery of multi-modal therapeutics for efficient, targeted, and safe cancer therapy. Adv Drug Deliv Rev 98:3–18. [DOI] [PubMed] [Google Scholar]
- 20.Gu L, Mooney DJ (2016) Biomaterials and emerging anticancer therapeutics: Engineering the microenvironment. Nat Rev Cancer 16:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshy ST, Mooney DJ (2016) Biomaterials for enhancing anti-cancer immunity. Curr Opin Biotechnol 40:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzeng A, et al. (2016) Temporally programmed CD8α+ DC activation enhances combination cancer immunotherapy. Cell Rep 17:2503–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rincon-Restrepo M, et al. (2017) Vaccine nanocarriers: Coupling intracellular pathways and cellular biodistribution to control CD4 vs CD8 T cell responses. Biomaterials 132:48–58. [DOI] [PubMed] [Google Scholar]
- 24.Rosi NL, et al. (2006) Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 312:1027–1030. [DOI] [PubMed] [Google Scholar]
- 25.Zheng D, et al. (2012) Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci USA 109:11975–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapadia CH, Melamed JR, Day ES (2018) Spherical nucleic acid nanoparticles: Therapeutic potential. BioDrugs 32:297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen SA, et al. (2013) Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med 5:209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radovic-Moreno AF, et al. (2015) Immunomodulatory spherical nucleic acids. Proc Natl Acad Sci USA 112:3892–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skakuj K, et al. (2018) Conjugation chemistry-dependent T-cell activation with spherical nucleic acids. J Am Chem Soc 140:1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meckes B, Banga RJ, Nguyen ST, Mirkin CA (2018) Enhancing the stability and immunomodulatory activity of liposomal spherical nucleic acids through lipid-tail DNA modifications. Small 14:1702909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo M, et al. (2017) A STING-activating nanovaccine for cancer immunotherapy. Nat Nanotechnol 12:648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakker ABH. (1996) Melanocyte lineage-specific antigen gp100 in T cell-mediated immunotherapy of melanoma. PhD dissertation (Radboud University, Nijmegen, The Netherlands).
- 33.Li WA, Mooney DJ (2013) Materials based tumor immunotherapy vaccines. Curr Opin Immunol 25:238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, et al. (2010) Polymer- and lipid-based nanoparticle therapeutics for the treatment of liver diseases. Nano Today 5:296–312. [Google Scholar]
- 35.Nel A, Xia T, Mädler L, Li N (2006) Toxic potential of materials at the nanolevel. Science 311:622–627. [DOI] [PubMed] [Google Scholar]
- 36.Nel AE, et al. (2015) Where are we heading in nanotechnology environmental health and safety and materials characterization? ACS Nano 9:5627–5630. [DOI] [PubMed] [Google Scholar]
- 37.Manders EM, et al. (1992) Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci 103:857–862. [DOI] [PubMed] [Google Scholar]
- 38.Qin L, et al. (2016) Exogenous IL-33 overcomes T cell tolerance in murine acute myeloid leukemia. Oncotarget 7:61069–61080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez D, et al. (2017) Exogenous IL-33 restores dendritic cell activation and maturation in established cancer. J Immunol 198:1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S, et al. (2015) Host miR155 promotes tumor growth through a myeloid-derived suppressor cell-dependent mechanism. Cancer Res 75:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.