Significance
Around the world insecticides are being deregistered and banned, as their environmental costs are deemed too great or their efficacy against pest insects is reduced through the evolution of insecticide resistance. With the introduction of replacement insecticides comes the responsibility to assess the way new insecticides perturb various levels of biological systems, from insect physiology to ecosystems. We used a systems genetics approach to identify genetic variants affecting survivorship of Drosophila melanogaster exposed to chlorantraniliprole. The study population was completely naïve to this insecticide chemistry and yet we find associations with variants in neuromuscular genes and coregulated detoxification genes. We predict that these variants will increase in populations of this “sentinel species” as these insecticides are applied in the environment.
Keywords: chlorantraniliprole, DGRP, Cap‘n’collar, Cyp12d1, Strn-Mlck
Abstract
Insecticide resistance is a paradigm of microevolution, and insecticides are responsible for the strongest cases of recent selection in the genome of Drosophila melanogaster. Here we use a naïve population and a novel insecticide class to examine the ab initio genetic architecture of a potential selective response. Genome-wide association studies (GWAS) of chlorantraniliprole susceptibility reveal variation in a gene of major effect, Stretchin Myosin light chain kinase (Strn-Mlck), which we validate with linkage mapping and transgenic manipulation of gene expression. We propose that allelic variation in Strn-Mlck alters sensitivity to the calcium depletion attributable to chlorantraniliprole’s mode of action. GWAS also reveal a network of genes involved in neuromuscular biology. In contrast, phenotype to transcriptome associations identify differences in constitutive levels of multiple transcripts regulated by cnc, the homolog of mammalian Nrf2. This suggests that genetic variation acts in trans to regulate multiple metabolic enzymes in this pathway. The most outstanding association is with the transcription level of Cyp12d1 which is also affected in cis by copy number variation. Transgenic overexpression of Cyp12d1 reduces susceptibility to both chlorantraniliprole and the closely related insecticide cyantraniliprole. This systems genetics study reveals multiple allelic variants segregating at intermediate frequency in a population that is completely naïve to this new insecticide chemistry and it foreshadows a selective response among natural populations to these chemicals.
An elaboration of the adage of Paracelsus (1493–1541) that “the dose makes the poison” is that there is a dose range of insecticides that kills some but not all insects in a population. By examining the genetic variation that contributes to survivorship on such discriminating doses, we can take a genetics approach to address a diverse set of questions relating to insecticide biology. Which genes have variants that affect survivorship, and how do they combine to provide the genetic architecture underpinning the trait? Do they provide insights into the mode of action of new insecticides? Do they suggest likely mechanisms by which insecticide resistance will arise? And what else do they tell us about past and future evolutionary responses to insecticides in pest and nontarget species?
Chlorantraniliprole (Rynaxapyr), is the first of the anthranilic diamides, a new class of insecticides. Unlike earlier insecticides that predominantly target neurotransmission, the anthranilic diamides are designed to target the ryanodine receptor, which is primarily involved in calcium homeostasis and muscle contraction (1, 2). Disruption of ryanodine receptor activity causes rapid incapacitation of the pest, leading to feeding cessation, lethargy, paralysis, and death (3, 4). Therefore, both the mode of action and the chemistry suggest that cross-resistance with older insecticides is unlikely.
Chlorantraniliprole was first sold in the Philippines in 2007, and worldwide soon after (5). Within years of introduction, resistance cases were reported in the diamondback moth Plutella xylostella (6, 7), and the tomato leafminer Tuta absoluta (8). While some of these cases can be attributed to mutations in the ryanodine receptor, the primary molecular target of these insecticides, there are others that suggest that resistance to this new insecticide class can arise through other means (9–12).
While Drosophila melanogaster is not a pest or a direct target of chlorantraniliprole applications, it is an organism of interest for two reasons. Firstly, D. melanogaster has long served as a model for insecticide resistance (13) and its status as a model organism more generally means that there is a wide variety of tools available to characterize genetic traits (14). Secondly, selective sweep analyses show that insecticides (particularly the organophosphates) have been major selective agents on D. melanogaster populations (15–18). These findings support the proposition that D. melanogaster can be used as a sentinel species for environmental pollutants, particularly insecticides (19, 20).
Like pest insects, D. melanogaster evolves insecticide resistance chiefly through target molecule insensitivity or detoxification enzyme adaptation, although other resistance mechanisms have been characterized (21). Resistance mutations in the target site genes typically diminish insecticide binding (e.g., ref. 22). Resistance mutations affecting detoxification enzymes can alter the protein sequence (e.g., ref. 23), but more generally increase transcriptional output through copy number variation (CNV) or cis-regulatory changes in the promoters of the resistance genes (e.g., refs. 24 and 25). Master regulatory genes that control, in trans, detoxification pathways have been reported in multiple arthropod species (26–33). Increased constitutive activation of these pathways has been shown to correlate with resistance in pest insects (32, 34) as well as in D. melanogaster (35), but so far natural variation underpinning such phenotypes has not been described at nucleotide resolution.
A powerful addition to the D. melanogaster toolkit is the Drosophila Genetic Reference Panel (DGRP) (36), which comprises 205 inbred lines derived from a single North American population. Each line in the DGRP has been bred for homozygosity and its genome sequenced, creating a “living library,” designed for genome-wide association studies (GWAS) that will associate genetic variants with phenotypes. The DGRP has been phenotyped for an extensive number of traits (37), including various insecticide phenotypes (16–18, 23, 38, 39) and “intermediate phenotypes” such as transcript abundance that enables eQTL to be mapped (40). Thus, the DGRP is becoming an important model for systems genetics of insects (41, 42), which, as we demonstrate here, enables deeper characterization of the genetic and regulatory mechanisms underpinning traits.
The DGRP lines were established from a collection of flies from the Farmers Market in Raleigh, NC in 2003 (36), before chlorantraniliprole became commercially available in 2007. Thus, they are naïve with respect to the completely novel class of chemistry of the group 28 insecticides. This provides a rare opportunity to examine the ab initio state of a potentially adaptive trait at unprecedented genetic resolution.
Here, we investigate the genetic architecture of chlorantraniliprole susceptibility across multiple doses. We interrogate associations between survivorship on food with varying concentrations of chlorantraniliprole and both genomic and transcriptomic variation. We find that an allele of large effect already segregates within this population, which is detectable only at higher levels of exposure. In addition, we show that phenotype to transcriptome associations reveal a completely different set of candidate genes, linked by a common trans-regulatory pathway.
Results
Phenotype to Genome Associations.
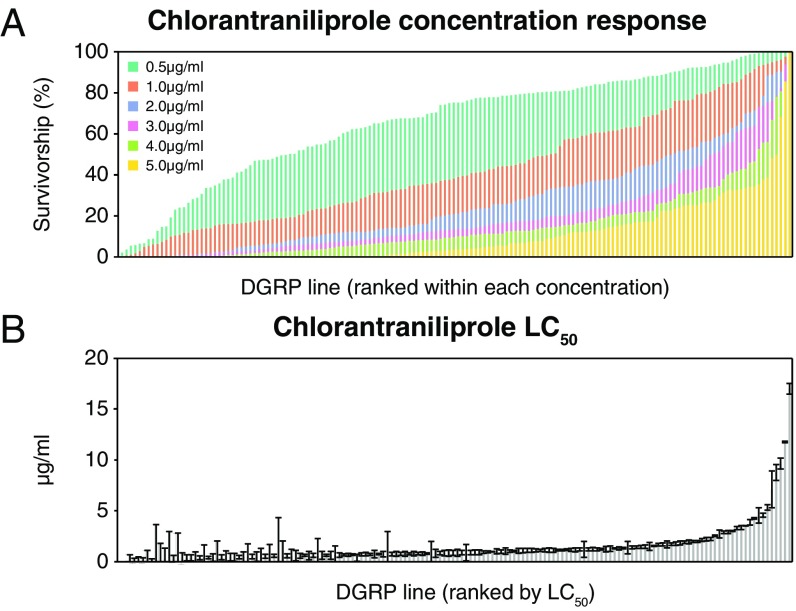
A total of 152 DGRP lines were scored for larval survivorship on six concentrations of chlorantraniliprole (Fig. 1A and SI Appendix, Fig. S1). The broad-sense heritability (H2) of the six concentrations ranged from 0.73 to 0.85, indicating a strong genetic component to chlorantraniliprole survivorship across the DGRP. GWAS on each of the concentrations identified 42–335 variants below the arbitrary genome-wide significance threshold (P < 1 × 10−5). The number of variants passing this threshold increased with concentration; however, this relationship failed to account for linkage disequilibrium (LD). Therefore, we examined the explanatory power of phenotype-associated DGRP variants for each chlorantraniliprole concentration using a multivariate genomic prediction model and found that more genes are required to explain the genetic architecture of the lowest concentration (0.5 μg/mL). For example, if the top 50 most-associated variants of the 0.5-μg/mL concentration are considered, they explain about the same amount of the phenotypic variation (R = 0.43) as the top five variants of the 5-μg/mL concentration (SI Appendix, Fig. S2).
Fig. 1.
Chlorantraniliprole phenotypes of 152 DGRP lines. (A) The survivorship of each line on the different concentrations (colored). The lines are arranged in ascending order for each concentration. (B) LC50, calculated from a minimum of six single-concentration phenotypes.
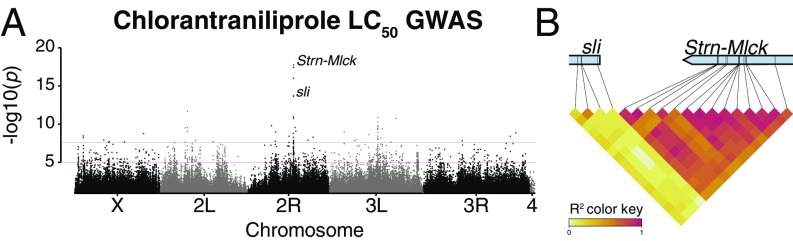
These data were supplemented with screening at additional concentrations for some DGRP lines to estimate the concentration of chlorantraniliprole required to kill 50% of individuals in each line (LC50). The mean LC50 was equal to 1.4 μg/mL (SD = 2.07 μg/mL) while the maximum LC50 was 17 μg/mL (Fig. 1B). A GWAS of the LC50 phenotype identified 931 associated variants below the arbitrary genome-wide significance threshold (P < 1 × 10−5; Fig. 2A). Ninety-six of these remained significant after a Bonferroni correction for multiple testing (2.65 × 10−8; SI Appendix, Supplementary File). The strongest association was to a single nucleotide polymorphism (SNP) in an intron of Stretchin myosin light chain kinase (Strn-Mlck) (2R:11853686; P = 2.03 × 10−17; Fig. 2), and variants annotated to this gene accounted for 15 of the 96 Bonferroni-significant GWAS variants.
Fig. 2.
(A) Top associations with genomic variants. Manhattan plot of DGRP chlorantraniliprole LC50 associations. Strn-Mlck is a standout candidate, with a minimum P value of 2.03 × 10−17. (B) LD at LC50 GWAS top candidates sli and Strn-Mlck.
To account for the fact that the strong effect of Strn-Mlck variation may be influencing other GWAS associations, we fitted the effect of Strn-Mlck to the chlorantraniliprole LC50 data and ran a GWAS on the residuals. The gene with the most highly associated variants after Strn-Mlck in the original LC50 GWAS, sli, was maintained in the Strn-Mlck–corrected GWAS. This demonstrates that despite its proximity to Strn-Mlck, associations with variants in sli are not artifacts of LD with variants in Strn-Mlck (Fig. 2B and SI Appendix, Fig. S3).
sli is like other genes harboring highly associated variants in that it is involved in axon guidance, sarcomere organization, intracellular signaling, and regulation of cell growth. The R spider software (43) was used to identify networks of genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) or Reactome pathways that were enriched for variants associated with chlorantraniprole phenotypes. The top 100 variants from LC50 genomic prediction both before and after correcting for the Strn-Mlck association were considered. The uncorrected LC50 network contained 12 genes, five with GWAS associations, out of a total of 54 recognized by R spider (Monte Carlo simulation P = 0.01; SI Appendix, Fig. S4); the Strn-Mlck–corrected LC50 network contained 10 genes, five with GWAS associations, out of a total of 53 genes with annotated function, of which seven could be mapped to a reference network previously described in KEGG (P = 0.005; SI Appendix, Fig. S4). Three GWAS-associated genes appeared in both networks: sli, robo3, and norpA. Although not connected to either network based on Reactome or KEGG databases, Strn-Mlck can be linked to the corrected LC50 network, as it has been shown to be one of the serine/threonine kinases that phosphorylates the transcription factor foxo (44).
Phenotype to Transcriptome Associations.
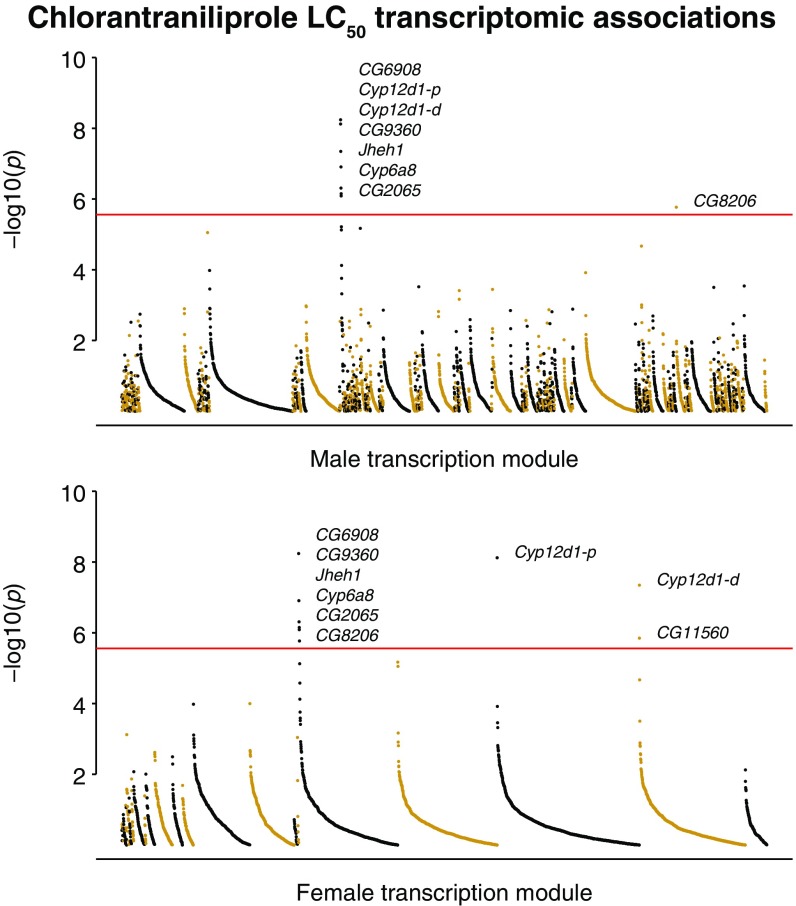
Transcript-level variation from the DGRP transcriptome dataset (mean for males and females) (40) was associated with chlorantraniliprole LC50 for nine genes below Bonferroni significance [generalized linear model (GLM)] (P < 2.76 × 10−6; Fig. 3). These top candidates are enriched for members of genetically correlated transcription modules (40) for both males (module 79; seven transcripts) and females (module 21; six transcripts). Furthermore, eight of the nine Bonferroni-significant transcripts have also been shown to be induced through ectopic expression of the transcription factor Cap‘n’collar (cnc) and following exposure to phenobarbital (Fig. 3) (45). One top candidate, Cyp12d1, exhibits CNV within the DGRP such that two copies (Cyp12d1-p and Cyp12d1-d) are observed in 24% of lines. The DGRP has been explicitly genotyped for duplication of Cyp12d1 (46, 47) and the duplication was found to be correlated with Cyp12d1 transcription levels (Cyp12d1-p: male r2 = 0.12, female r2 = 0.14 and Cyp12d1-d: male r2 = 0.38, female r2 = 0.27). Thus, two distinct contributions to Cyp12d1 transcript level can be identified: that which is attributable to cnc regulation and that which is attributable to CNV at the locus.
Fig. 3.
P values of association between the chlorantraniliprole LC50 phenotype and each DGRP transcript level, grouped into sex-specific genetically correlated transcriptional modules. The transcripts within each module are ordered by rank to give “opera house” plots. Eight of the nine Bonferroni-significant associations (red line; P < 2.76 × 10−6) are affected by ectopic cnc expression.
Validation of Strn-Mlck and Cyp12d1-P in Chlorantraniliprole Survivorship Traits.
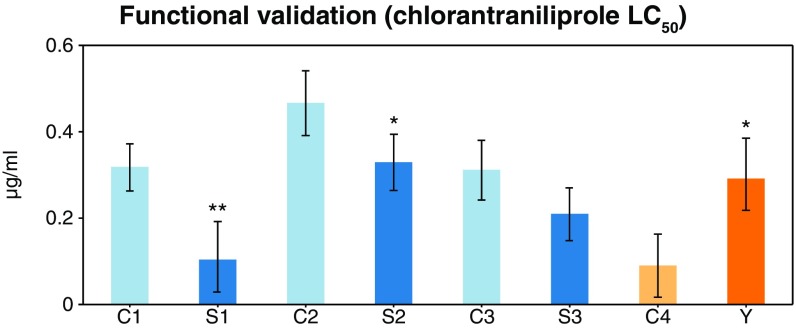
To support the involvement of Strn-Mlck in chlorantraniliprole survivorship, two DGRP lines (RAL-59 and RAL-399) that differ by both LC50 and Strn-Mlck variants, were crossed. Comparison of probit curves from parental lines and the F1 suggest that the more resistant allele is recessive (degree of dominance = −0.76 at LC50, −0.84 at LC90). A pair of restriction fragment length polymorphism (RFLP) assays showed that the Strn-Mlck haplotype identified in the GWAS was significantly enriched in chlorantraniliprole-screened F2 survivors over untreated controls (χ2 P = 9.97 × 10−7), indicating that this region is indeed associated with chlorantraniliprole survivorship. The importance of Strn-Mlck was also tested using three separate RNAi lines crossed to the Actin5c driver. In two of the three crosses, flies with Strn-Mlck knocked down showed significantly decreased LC50s relative to CyO siblings (Fig. 4).
Fig. 4.
Transgenic manipulation of candidate genes. LC50 values of combined reciprocal crosses for RNAi knockdown of Strn-Mlck using three TRiP lines HMS02663 (S1), JF02171 (S2), and HMS01665 (S3) crossed to the Actin5c driver. In two out of three of the crosses, Strn-Mlck knockdown results in a significantly decreased chlorantraniliprole LC50 relative to CyO siblings (C1–3). Overexpression of Cyp12d1-p (Y) significantly increases chlorantraniliprole LC50 relative to control (C4). Error bars represent 95% confidence of probit fit at LC50. *P < 0.05, **P < 0.01.
The involvement of Cyp12d1-p was tested using the GAL4-UAS system and the 6g1HR-GAL4 driver (48); flies overexpressing Cyp12d1-p in key metabolic tissues were 2.2-fold more tolerant to chlorantraniliprole than controls (Fig. 4). This system was also employed to test cross-tolerance to the closely related insecticide, cyantraniliprole. At three concentrations, survivorship of flies overexpressing Cyp12d1-p was significantly increased relative to controls (P < 0.05, two-tailed t test assuming unequal variances), suggesting that the enzyme acts on chemical moieties that the two insecticides have in common.
Induction of Cyp12d1.
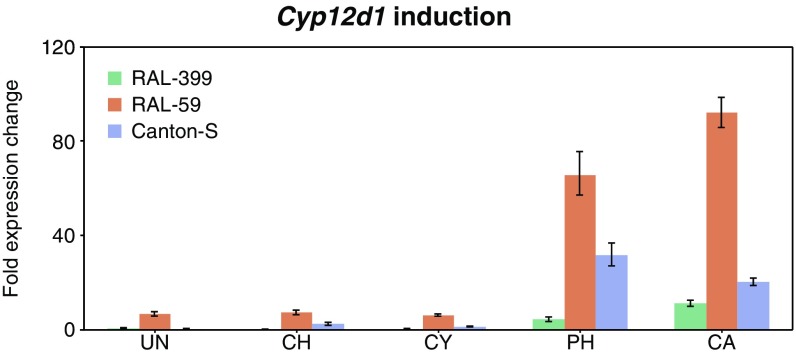
It has been previously demonstrated that both Cyp12d1-p and Cyp12d1-d are up-regulated in response to phenobarbital and caffeine (45). To test if chlorantraniliprole or cyantraniliprole induces Cyp12d1 expression, Cyp12d1 transcript levels (Cyp12d1-p and Cyp12d1-d were not distinguished) were quantified after larvae from a laboratory strain (Canton-S) and two DGRP lines (chlorantraniliprole-resistant RAL-59 and chlorantraniliprole-susceptible RAL-399) were exposed to chlorantraniliprole, cyantraniliprole, phenobarbital, and caffeine. In accordance with DGRP transcriptome data from Huang et al. (40), we found Cyp12d1 transcript abundance in unexposed larvae significantly higher in the resistant DGRP line RAL-59 relative to susceptible line RAL-399 (Fig. 5). In all three lines Cyp12d1 transcript levels were increased significantly following exposure to phenobarbital and caffeine, but not following exposure to chlorantraniliprole or cyantraniliprole (Fig. 5).
Fig. 5.
Variation and inducibility of Cyp12d1 expression. Cyp12d1 expression (both Cyp12d1-p and Cyp12d1-d detected; normalized to housekeeper CG11322) in third-instar larvae from three DGRP lines unexposed (UN) and after exposure to four xenobiotic compounds; chlorantraniliprole (CH), cyantraniliprole (CY), phenobarbital (PH), and caffeine (CA). Error bars represent SD of the first derivative.
Discussion
Genetic Architecture Changes with Insecticide Concentration.
Theoretical considerations have led to the prediction that the genetic architecture of insecticide resistance would become less polygenic with increasing concentration of an insecticide (49). The DGRP allows identification of polygenes affecting survivorship on different concentrations of an insecticide in an unprecedented way. Concurring with expectations, smaller effect sizes were observed in the lowest concentration GWAS. However, relatively few variants passed the significance threshold in the GWAS at this concentration compared with those performed at higher concentrations. This is a consequence of the increasingly nonnormal distribution of the survivorship among lines as concentration increases and reflects the statistical mechanics and distribution assumptions of GWAS, and the degree of LD around associated variants at lower frequencies. We addressed this by modeling the additive contribution of small-effect alleles using a Bayesian linear regression. At the three highest concentrations, and the concentration inferred to kill 50% of flies (LC50), many of the associated variants are within large stretches of LD around Strn-Mlck. When the LD is accounted for, the number of genes associated with survivorship on high concentrations was indeed lower, consistent with theoretical predictions. The genetic architecture of lower-concentration survivorship is far more polygenic, with very few of the top associations showing strong LD relationships. There is also a loss of explanatory power at 0.5 μg/mL, with R2 plateauing at 69%, making it obvious that the set of associated variants is not capturing all of the variation in this phenotype. In addition, the heritability at the lowest concentration is smaller, suggesting other environmental effects are more pronounced at this concentration, or that epistasis, which is likely to be more prevalent with a greater level of polygenicity, is playing a role (50).
Strn-Mlck: A Novel Gene of Major Effect.
The GWAS presented here identified 15 variants in the Strn-Mlck gene as marking an allele of major effect. This genome-wide and unbiased approach was confirmed by two methods: RNAi knockdown supports the potential of this gene to contribute to the trait, and linkage mapping indicates that a naturally occurring variant of major effect occurs in this vicinity. A member of the Titan family, Strn-Mlck is highly complex, with a total of 33 exons, three start sites, and three poly-A sites, leading to 16 predicted isoforms. Myosin light chain kinases (MLCKs) are serine/threonine kinases whose substrate is the myosin regulatory chain, a component of the thick filaments in muscles. Most MLCKs are calmodulin dependent, meaning they are proteins that are sensitive to calcium homeostasis. Strn-Mlck’s association with the muscle and its sensitivity to calcium, the intracellular release of which is perturbed with ryanodine insecticides, strengthen the case that Strn-Mlck is involved. The alleles that increase survivorship may act downstream of the insecticide binding to the ryanodine receptor, and simply ameliorate the effect of the insecticide. However, there is an alternative hypothesis. Recently several genome-wide analyses of insecticide phenotypes have identified the involvement of genes in neurogenic pathways (38, 51, 52). In this study, we found that GWAS-associated variants are enriched for a network of neurodevelopmental genes which can be linked to Strn-Mlck. Thus, the highly associated variants in Strn-Mlck act by influencing the development of the nervous system such that some flies are less sensitive to the insecticide.
Transcriptomic Associations Identify a Known Detoxification Pathway.
Underscoring the power of the systems approach that is afforded by the DGRP, associations between chlorantraniliprole tolerance phenotypes and gene expression levels (40) implicated a completely different set of genes from GWAS candidates. A strong association was observed between chlorantraniliprole LC50 and the expression of a group of genes known to be coregulated in response to xenobiotics (45). These genes have previously been found to be regulated by cnc (45), a transcription factor well conserved across both vertebrates and invertebrates (53). cnc and its mammalian homolog Nrf2 are CNC-bZIP transcription factors that play a well-conserved role in oxidative stress response (54). While most studies characterize this pathway in terms of its induction in response to oxidative stress, the transcript-level variation identified in this study comes from unexposed flies, suggesting variation in the constitutive expression level of cnc-regulated transcripts among DGRP lines, a phenomenon that has previously been linked to insecticide resistance (34, 35, 54).
The constitutively up-regulated cnc-regulated transcripts identified in the DGRP include potential detoxification enzymes, including two cytochrome P450s (Cyp12d1 and Cyp6a8), a juvenile hormone epoxide hydrolase (Jheh1), two oxidoreductases (CG2065 and CG9360), and an ecdysone kinase-like enzyme (CG6908). Guio et al. (55) demonstrated that the insertion of a Bari1 transposable element upstream of Jheh1 introduces additional cnc binding motifs. This results in increased inducibility of Jheh1 and its tandem paralog, Jheh2, in response to oxidative stress, conferring increased resistance to the organophosphate insecticide malathion.
Cyp12d1 induction has been associated with exposure to multiple xenobiotics, including DDT, caffeine (56), pyrethrum (57), atrazine (58), and piperonyl butoxide (PBO) (60), and its overexpression increases survivorship to DDT, dicyclanil, and malathion (17, 60). Due to its polymorphic status, it has been assumed that the Cyp12d1 duplication is a recent event (61). Duplication frequency in the DGRP is correlated with higher expression levels in adults, a pattern that is also seen in other outbred populations (62). While the DGRP transcriptome data were measured in adults, our qPCR results suggest that this difference is also observed in larvae.
As the transcription output of a gene can be affected by numerous variants, including rare variants, genes that are missed in the GWAS may be identified by phenotype to transcriptome associations. Given that the transcriptome analysis implicated Cyp12d1, we examined its position in all of the datasets more carefully. We identified two cis-eQTL of Cyp12d1 (2R:6994376 and 2R:7007339) (40), associated with survivorship at 4-μg/mL and 1-μg/mL concentrations, respectively. Both these variants are in LD with the duplicated state of Cyp12d1 (R2 = 0.46 and 0.72, respectively), suggesting the effect of the Cyp12d1 duplication was indirectly detected in the GWAS. Huang et al. (40) also report a trans-eQTL for Cyp12d1 occurring in sli, the gene that ranks second in the LC50 GWAS after Strn-Mlck. Thus, trans-regulatory variation may be combining with cis-regulatory variation to affect the transcriptional output of Cyp12d1 and chlorantraniliprole survivorship.
The observation that an eQTL for a cnc-regulated gene maps to a region strongly implicated in the GWAS is intriguing. This study raises the possibility that there is a link between muscle function as implicated by the GWAS and the oxidative stress pathway as implicated by the transcriptome associations. There is some support for this in the literature: mutations in LamC, a LC50 GWAS candidate, trigger cellular redox imbalance, an enrichment of cnc in the cytosol of larval muscle genes, and an increase in the baseline expression of genes shown to be up-regulated by ectopic cnc expression (35, 63).
Conclusions.
Here we set out to explore the genetic architecture underlying a completely novel insecticide chemistry for which there was no expected adaptive precedence because the population sample was collected before chlorantraniliprole was deployed in the field, and is therefore completely naïve to this insecticide and indeed all of the insecticides in the new anthranilic diamide class. This contrasts to several recent studies where insecticide resistance loci have been found to feature genes of major effect that have been built through a series of adaptive substitutions (25, 64, 65). We explored the genetic architecture of chlorantraniliprole survivorship at multiple concentrations and surprisingly found that at higher concentrations there was clearly a gene of major effect. Standing variation in the neuromuscular gene Strn-Mlck increased the LC50 by ∼3 μg/mL. This is not the first time that molecular changes underpinning insecticide tolerance can be considered as exaptations (e.g., ref. 66); however this case unambiguously demonstrates that standing variation in a population is relevant to a future selective agent.
D. melanogaster is not a pest insect but it is common in orchards where these new insecticides are being applied. It may therefore be thought of a sentinel species (19), such that if, in the future, there is evidence for selection at Strn-Mlck or the loci we identified here, then this may be attributed to the use of these new insecticides. Given the precedent for the involvement of the cnc pathway in insecticide resistance in pest species (35), perhaps the factors which cause the constitutive activation of this regulatory hub present the greatest concern.
Materials and Methods
Fly Lines.
All DGRP lines and Transgenic RNAi Project (TRiP) lines (67) were obtained from the Bloomington Drosophila Stock Center. UAS-Cyp12d1-p and 6g1HR-GAL4 were obtained from the Batterham Lab at the University of Melbourne (48). Lines were maintained on cornmeal-yeast-agar media and were kept at 25 °C at constant light for at least one generation before use.
Chlorantraniliprole Phenotyping.
Altacor (350 g/kg chlorantraniliprole/Rynaxypyr) was obtained from DuPont Australia. Wettable granules were dispersed in water to make a stock solution of 100 μg/mL chlorantraniliprole. Chlorantraniliprole was administered through cornmeal-yeast-agar fly media. Insecticide was added to the media once it had cooled below 55 °C. Food dye was added in conjunction with the insecticide to ensure even dispersal through the food. Fifty first-instar larvae were then placed into fly vials, each containing 10 mL of chlorantraniliprole-laced media. This was done in triplicate for each line on each concentration.
A total of six concentrations for each DGRP line were used in the initial screens (0.5, 1, 2, 3, 4, and 5 μg/mL), with a number of lines having to be rescreened on higher concentrations (6, 8, and 12 μg/mL) to accurately determine the LC50. Survivorship was scored strictly 11 d after picking, with survivors being defined as any fly that successfully eclosed.
For calculation of LC50s for both DGRP and transgenic lines, linear models were fitted to concentration-mortality data on a log-probit scale using “glm” in the R statistical package and scripts from Johnson et al. (68). LC50 values and 95% confidence intervals were calculated using Fieller’s method from fitted linear models (69).
GWAS.
GWAS were performed on six single-concentration phenotypes and the LC50 for 152 DGRP lines using the DGRP webtool (70) (http://dgrp2.gnets.ncsu.edu). This involved uploading each of the seven phenotype files and for each running a pipeline that fits a linear model between the phenotype and each site variant among DGRP genomes. The linear model incorporates as covariates Wolbachia pipientis infection status, and five common chromosomal inversion genotypes that vary among the 205 DGRP lines, as described in ref. 70. A total of 1,887,900 variants were tested in each GWAS. To correct for the effect of the most associated variant on the LC50 phenotype, the genotype of this variant was fitted as a fixed effect in a linear model. The residuals were then extracted and submitted to the DGRP pipeline.
Genomic Prediction.
For genomic prediction analysis across the top variants associated with each concentration phenotype and the LC50, a Bayesian linear regression coupled with LASSO was implemented using the Bayesian linear regression package (71) in R, with the phenotype acting as the data vector and the genotypes, the incidence matrix for βL. Up to 500 of the top variants associated with each concentration phenotype (as ranked by P value) were used to explain the phenotype. Each model was implemented with 5,500 iterations, with a burn in of 5,000 iterations and a thinning interval of 50.
Network Based Analysis.
To examine annotated interactions between different GWAS candidates, R spider software (43) (http://www.bioprofiling.de/R_spider.html) was used to map GWAS candidates to KEGG and Reactome pathways.
Phenotype to Transcriptome Associations.
Transcriptome data for 1- to 3-d-old adult flies from 185 DGRP lines were recovered from the DGRP website (http://dgrp.gnets.ncsu.edu/data.html) (40). Mean transcription level was calculated for each gene in each sex from two biological replicates, to give a mean level for each of the 18,140 transcripts measured by Huang et al. (40) in each DGRP line, for both males and females. The mean of male and female transcript levels was then calculated. A linear model was fit between mean transcription level of each gene measured by Huang et al. (40) and chlorantraniliprole ;LC50.
Strn-Mlck RFLP Mapping Crosses.
To examine the dominance of chlorantraniliprole tolerance and to confirm the association of GWAS-associated Strn-Mlck variants with survivorship, crosses were set up between DGRP lines RAL-399 and RAL-59. These lines have contrasting chlorantraniliprole phenotypes, are free of any of the major cosmopolitan inversions (70), and carry different states at the top GWAS variants in Strn-Mlck. Reciprocal crosses were performed to obtain two classes of F1 progeny to check for maternal and X-linked effects. Offspring from the reciprocal crosses were combined in equal numbers to establish an F2 population.
For the cross-typing, single-fly DNA extractions were performed. Two sets of primers for RFLPs were designed to distinguish “resistant” and “susceptible” Strn-Mlck haplotypes: EcoRV: 5′ TCAGTTCGTTGGTGTTCAGG 3′, 5′ ACTTCACGGTCAACCTGTCC 3′ and HpaII: 5′ GTTCAGATTCATCGCCATCC 3′, 5′ ACCTGCACTACCACGTACCC 3′. A total of 190 flies were scored on both assays. For each sample, 10 μL PCRs were set up using 5 μL of GoTaq Green (9PIM712, Promega), 1 μL each of forward and reverse primers (5 μL), and 3 μL of sterile water.
Knockdown of Strn-Mlck.
RNAi knockdown of different transcripts of Strn-Mlck was carried out using three different UAS lines from the TRiP Library (67), as no single line targets all of the predicted isoforms of Strn-Mlck. The lines, HMS01665, HMS02663, and JF02171 target exons 1, 4, and 30, respectively, and were reciprocally crossed to the ubiquitous actin5c driver. The presence of the CyO balancer chromosome in the Actin5c driver line meant that not all offspring of each cross would inherit the UAS-RNAi construct. For these crosses, the CyO-carrying offspring were used as controls for the UAS siblings.
Overexpression of Cyp12d1-P.
Cyp12d1-p was overexpressed using the GAL4/UAS system (72) and the 6g1HR-GAL4 driver described by Chung et al. (48). The 6g1HR-GAL4 virgin females, in which GAL4 is regulated by Cyp6g1 upstream sequence originating from Hikone-R line flies, were crossed to males carrying an additional copy of Cyp12d1-p under control of a UAS promoter (60). The w1118 line was used as a control for UAS-Cyp12d1-p.
Cyp12d1 qRT-PCR Induction Assays.
For each replicate of each line, 50 third-instar larvae were added to plates containing cornmeal-yeast-agar media and either 2.5 μg/mL chlorantraniliprole, 0.5 μg/mL cyantraniliprole, 10 mM phenobarbital (dissolved in ETOH), or 1.5 mg/mL caffeine (dissolved in 80 °C water). Control plates were untreated. Larvae were allowed to feed for 4 h before being suspended in TRIsure (Bioline) and snap frozen in liquid nitrogen for storage at −70 °C. RNA was later extracted following the standard TRIsure protocol. cDNA was synthesized using M-MuLV reverse transcriptase (NEB) and random nonamer primers following the standard NEB reverse transcriptase protocol. qRT-PCR was performed on the Roche Light Cycler 480 using the following primers: Cyp12d1 (Cyp12d1-p or Cyp12d1-d): 5′ GGGGAAAACTACGATCAGCC 3′, 5′ CGGATTTCCTTAATGCGCTCT 3′ and CG11322 (housekeeper): 5′ TGGGCAGTGCCTTCTACATTT 3′, 5′ CGTACGCACCTCGCTTGTT 3′.
Supplementary Material
Acknowledgments
We thank Trudy Mackay for advice and wisdom; Stephen Wilcox for his assistance with molecular biology analyses; Phil Batterham for UAS lines and discussion; Josefa González for discussion; and John McKenzie for comments on the manuscript. This work was partially funded by the Australia Research Council Grant DP0985013.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821713116/-/DCSupplemental.
References
- 1.Nauen R. (2006) Insecticide mode of action: Return of the ryanodine receptor. Pest Manag Sci 62:690–692. [DOI] [PubMed] [Google Scholar]
- 2.Sparks TC, Nauen R (2015) IRAC: Mode of action classification and insecticide resistance management. Pestic Biochem Physiol 121:122–128. [DOI] [PubMed] [Google Scholar]
- 3.Cordova D, et al. (2006) Anthranilic diamides: A new class of insecticides with a novel mode of action, ryanodine receptor activation. Pestic Biochem Physiol 84:196–214. [Google Scholar]
- 4.Lahm GP, et al. (2007) Rynaxypyr: A new insecticidal anthranilic diamide that acts as a potent and selective ryanodine receptor activator. Bioorg Med Chem Lett 17:6274–6279. [DOI] [PubMed] [Google Scholar]
- 5.Lahm GP, Cordova D, Barry JD (2009) New and selective ryanodine receptor activators for insect control. Bioorg Med Chem 17:4127–4133. [DOI] [PubMed] [Google Scholar]
- 6.Troczka B, et al. (2012) Resistance to diamide insecticides in diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) is associated with a mutation in the membrane-spanning domain of the ryanodine receptor. Insect Biochem Mol Biol 42:873–880. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Wu S, Yang Y, Wu Y (2012) Molecular cloning, characterization and mRNA expression of a ryanodine receptor gene from diamondback moth, Plutella xylostella. Pestic Biochem Physiol 102:204–212. [Google Scholar]
- 8.Steinbach D, et al. (2015) Geographic spread, genetics and functional characteristics of ryanodine receptor based target-site resistance to diamide insecticides in diamondback moth, Plutella xylostella. Insect Biochem Mol Biol 63:14–22. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, et al. (2014) Identification of a novel cytochrome P450 gene, CYP321E1 from the diamondback moth, Plutella xylostella (L.) and RNA interference to evaluate its role in chlorantraniliprole resistance. Bull Entomol Res 104:716–723. [DOI] [PubMed] [Google Scholar]
- 10.Nauen R, Steinbach D (2016) Resistance to diamide insecticides in lepidopteran pests. Advances in Insect Control and Resistance Management (Springer, Cham, Switzerland: ), pp 219–240. [Google Scholar]
- 11.Kim AY, Kwon DH, Jeong IH, Koh YH (2018) An investigation of the molecular and biochemical basis underlying chlorantraniliprole-resistant Drosophila strains and their cross-resistance to other insecticides. Arch Insect Biochem Physiol 99:e21514. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. (2018) Molecular identification of four novel cytochrome P450 genes related to the development of resistance of Spodoptera exigua (Lepidoptera: Noctuidae) to chlorantraniliprole. Pest Manag Sci 74:1938–1952. [DOI] [PubMed] [Google Scholar]
- 13.Bartlett BRJ. (1952) A study of insecticide resistance in strains of Drosophila melanogaster Meig. Can Entomol 84:189–205. [Google Scholar]
- 14.Perry T, Batterham P (2018) Harnessing model organisms to study insecticide resistance. Curr Opin Insect Sci 27:61–67. [DOI] [PubMed] [Google Scholar]
- 15.Garud NR, Messer PW, Buzbas EO, Petrov DAJ (2015) Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet 11:e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battlay P, Schmidt JM, Fournier-Level A, Robin C (2016) Genomic and transcriptomic associations identify a new insecticide resistance phenotype for the selective sweep at the Cyp6g1 locus of Drosophila melanogaster. G3 (Bethesda) 6:2573–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battlay P, et al. (2018) Structural variants and selective sweep foci contribute to insecticide resistance in the Drosophila genetic reference panel. G3 (Bethesda) 8:3489–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duneau D, et al. (2018) Signatures of insecticide selection in the genome of Drosophila melanogaster. G3 (Bethesda) 8:3469–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson TG. (2005) Drosophila: Sentinels of environmental toxicants. Integr Comp Biol 45:127–136. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann AA, Daborn PJ (2007) Towards genetic markers in animal populations as biomonitors for human-induced environmental change. Ecol Lett 10:63–76. [DOI] [PubMed] [Google Scholar]
- 21.Strycharz JP, et al. (2013) Resistance in the highly DDT-resistant 91-R strain of Drosophila melanogaster involves decreased penetration, increased metabolism, and direct excretion. Pestic Biochem Physiol 107:207–217. [Google Scholar]
- 22.Mutero A, Pralavorio M, Bride JM, Fournier D (1994) Resistance-associated point mutations in insecticide-insensitive acetylcholinesterase. Proc Natl Acad Sci USA 91:5922–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt JM, et al. (2017) Insights into DDT resistance from the Drosophila melanogaster genetic reference panel. Genetics 207:1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daborn P, Boundy S, Yen J, Pittendrigh B, ffrench-Constant R (2001) DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol Genet Genomics 266:556–563. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt JM, et al. (2010) Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet 6:e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plapp FW., Jr (1984) The genetic basis of insecticide resistance in the house fly: Evidence that a single locus plays a major role in metabolic resistance to insecticides. Pestic Biochem Physiol 22:194–201. [Google Scholar]
- 27.Liu N, Scott JG (1997) Inheritance of CYP6D1-mediated pyrethroid resistance in house fly (Diptera: Muscidae). J Econ Entomol 90:1478–1481. [DOI] [PubMed] [Google Scholar]
- 28.Peng T, et al. (2016) Cytochrome P450 CYP6DA2 regulated by cap ‘n’collar isoform C (CncC) is associated with gossypol tolerance in Aphis gossypii Glover. Insect Mol Biol 25:450–459. [DOI] [PubMed] [Google Scholar]
- 29.Ingham VA, Pignatelli P, Moore JD, Wagstaff S, Ranson H (2017) The transcription factor Maf-S regulates metabolic resistance to insecticides in the malaria vector Anopheles gambiae. BMC Genomics 18:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalsi M, Palli SR (2017) Cap n collar transcription factor regulates multiple genes coding for proteins involved in insecticide detoxification in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol 90:43–52. [DOI] [PubMed] [Google Scholar]
- 31.Kalsi M, Palli SR (2017) Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem Mol Biol 83:1–12. [DOI] [PubMed] [Google Scholar]
- 32.Shi L, et al. (2017) The expression of P450 genes mediating fenpropathrin resistance is regulated by CncC and Maf in Tetranychus cinnabarinus (Boisduval). Comp Biochem Physiol C Toxicol Pharmacol 198:28–36. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, et al. (2018) Cloning and functional analysis of CncC and Keap1 genes in silkworm. J Agric Food Chem 66:2630–2636. [DOI] [PubMed] [Google Scholar]
- 34.Bottino-Rojas V, et al. (2018) The redox-sensing gene Nrf2 affects intestinal homeostasis, insecticide resistance, and Zika virus susceptibility in the mosquito Aedes aegypti. J Biol Chem 293:9053–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra JR, Lam G, Thummel CS (2013) Constitutive activation of the Nrf2/Keap1 pathway in insecticide-resistant strains of Drosophila. Insect Biochem Mol Biol 43:1116–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackay TF, et al. (2012) The Drosophila melanogaster genetic reference panel. Nature 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anholt RRH, Mackay TFC (2018) The road less traveled: From genotype to phenotype in flies and humans. Mamm Genome 29:5–23. [DOI] [PubMed] [Google Scholar]
- 38.Denecke S, et al. (2017) Multiple P450s and variation in neuronal genes underpins the response to the insecticide imidacloprid in a population of Drosophila melanogaster. Sci Rep 7:11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Najarro MA, Hackett JL, Macdonald SJ (2017) Loci contributing to boric acid toxicity in two reference populations of Drosophila melanogaster. G3 (Bethesda) 7:1631–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, et al. (2015) Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc Natl Acad Sci USA 112:E6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayroles JF, et al. (2009) Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robin C, Battlay P, Fournier-Level A (2019) What can genetic association panels tell us about evolutionary processes in insects? Curr Opin Insect Sci 31:99–105. [DOI] [PubMed] [Google Scholar]
- 43.Antonov AV, Schmidt EE, Dietmann S, Krestyaninova M, Hermjakob H (2010) R spider: A network-based analysis of gene lists by combining signaling and metabolic pathways from reactome and KEGG databases. Nucleic Acids Res 38:W78–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattila J, Kallijärvi J, Puig O (2008) RNAi screening for kinases and phosphatases identifies FoxO regulators. Proc Natl Acad Sci USA 105:14873–14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misra JR, Horner MA, Lam G, Thummel CS (2011) Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev 25:1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Good RT, et al. (2014) The molecular evolution of cytochrome P450 genes within and between Drosophila species. Genome Biol Evol 6:1118–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Najarro MA, et al. (2015) Identifying loci contributing to natural variation in xenobiotic resistance in Drosophila. PLoS Genet 11:e1005663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung H, et al. (2007) Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175:1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenzie JA, Batterham P (1994) The genetic, molecular and phenotypic consequences of selection for insecticide resistance. Trends Ecol Evol 9:166–169. [DOI] [PubMed] [Google Scholar]
- 50.Howard R, Carriquiry AL, Beavis WD (2014) Parametric and nonparametric statistical methods for genomic selection of traits with additive and epistatic genetic architectures. G3 (Bethesda) 4:1027–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seong KM, Coates BS, Sun W, Clark JM, Pittendrigh BR (2017) Changes in neuronal signaling and cell stress response pathways are associated with a multigenic response of Drosophila melanogaster to DDT selection. Genome Biol Evol 9:3356–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fournier-Level A, et al. (2019) The spread of resistance to imidacloprid is restricted by thermotolerance in natural populations of Drosophila melanogaster. Nat Ecol Evol 3:647–656. [DOI] [PubMed] [Google Scholar]
- 53.Li L, et al. (2008) Molecular evolution of Keap1. Two Keap1 molecules with distinctive intervening region structures are conserved among fish. J Biol Chem 283:3248–3255. [DOI] [PubMed] [Google Scholar]
- 54.Wilding CS. (2018) Regulating resistance: CncC:Maf, antioxidant response elements and the overexpression of detoxification genes in insecticide resistance. Curr Opin Insect Sci 27:89–96. [DOI] [PubMed] [Google Scholar]
- 55.Guio L, Barrón MG, González J (2014) The transposable element Bari-Jheh mediates oxidative stress response in Drosophila. Mol Ecol 23:2020–2030. [DOI] [PubMed] [Google Scholar]
- 56.Willoughby L, et al. (2006) A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem Mol Biol 36:934–942. [DOI] [PubMed] [Google Scholar]
- 57.Jensen HR, Scott IM, Sims S, Trudeau VL, Arnason JT (2006) Gene expression profiles of Drosophila melanogaster exposed to an insecticidal extract of Piper nigrum. J Agric Food Chem 54:1289–1295. [DOI] [PubMed] [Google Scholar]
- 58.Le Goff G, et al. (2006) Xenobiotic response in Drosophila melanogaster: Sex dependence of P450 and GST gene induction. Insect Biochem Mol Biol 36:674–682. [DOI] [PubMed] [Google Scholar]
- 59.Willoughby L, Batterham P, Daborn PJ (2007) Piperonyl butoxide induces the expression of cytochrome P450 and glutathione S-transferase genes in Drosophila melanogaster. Pest Manag Sci 63:803–808. [DOI] [PubMed] [Google Scholar]
- 60.Daborn PJ, et al. (2007) Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol 37:512–519. [DOI] [PubMed] [Google Scholar]
- 61.McDonnell CM, et al. (2012) Evolutionary toxicogenomics: Diversification of the Cyp12d1 and Cyp12d3 genes in Drosophila species. J Mol Evol 74:281–296. [DOI] [PubMed] [Google Scholar]
- 62.Schrider DR, Hahn MW, Begun DJ (2016) Parallel evolution of copy-number variation across continents in Drosophila melanogaster. Mol Biol Evol 33:1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dialynas G, et al. (2015) Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet 11:e1005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karasov T, Messer PW, Petrov DA (2010) Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet 6:e1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM (2011) Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet 7:e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zimmer CT, et al. (2018) Neofunctionalization of duplicated P450 genes drives the evolution of insecticide resistance in the brown planthopper. Curr Biol 28:268–274.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ni JQ, et al. (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8:405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson RM, Dahlgren L, Siegfried BD, Ellis MD (2013) Acaricide, fungicide and drug interactions in honey bees (Apis mellifera). PLoS One 8:e54092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finney D. (1971) Probit Analysis (Cambridge Univ Press, New York: ), p 32. [Google Scholar]
- 70.Huang W, et al. (2014) Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res 24:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de los Campos G, et al. (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.