Abstract
Inbred mouse strains are a cornerstone of translational research but paradoxically many strains carry mild inborn errors of metabolism. For example, α-aminoadipic acidemia and branched-chain ketoacid dehydrogenase deficiency are known in C57BL/6J mice. Using RNA sequencing, we now reveal the causal variants in Dhtkd1 and Bckdhb, and the molecular mechanism underlying these metabolic defects. C57BL/6J mice have decreased Dhtkd1 mRNA expression due to a solitary long terminal repeat (LTR) in intron 4 of Dhtkd1. This LTR harbors an alternate splice donor site leading to a partial splicing defect and as a consequence decreased total and functional Dhtkd1 mRNA, decreased DHTKD1 protein and α-aminoadipic acidemia. Similarly, C57BL/6J mice have decreased Bckdhb mRNA expression due to an LTR retrotransposon in intron 1 of Bckdhb. This transposable element encodes an alternative exon 1 causing aberrant splicing, decreased total and functional Bckdhb mRNA and decreased BCKDHB protein. Using a targeted metabolomics screen, we also reveal elevated plasma C5-carnitine in 129 substrains. This biochemical phenotype resembles isovaleric acidemia and is caused by an exonic splice mutation in Ivd leading to partial skipping of exon 10 and IVD protein deficiency. In summary, this study identifies three causal variants underlying mild inborn errors of metabolism in commonly used inbred mouse strains.
Keywords: inbred mouse strains, recombinant inbred mouse strains, mouse genetics, splicing defects, inborn errors of metabolism
1. Introduction
Most animal studies using mice take advantage of introducing specific gene modifications by homologous recombination in embryonic stem cells or more recently by CRISPR-Cas9-based strategies in zygotes. Additionally, the long history of mice in research has led to a rich resource of natural genetic variation represented in numerous mouse strains. Some of these spontaneous mutant strains harbor a loss of function mutation equivalent to a gene knockout (KO) and may be used as models for human diseases (Table 1). Well studied examples in metabolic research include Leptin deficiency in the Ob/Ob mouse [1], nicotinamide nucleotide transhydrogenase deficiency in the C57BL/6J strain [2], short-chain acyl-CoA dehydrogenase deficiency in the BALB/cByJ strain [3] and primary carnitine deficiency in the juvenile visceral steatosis mouse [4].
Table 1.
List of selected (metabolic) defects due to spontaneous mutations in (commonly used) inbred mouse strains.
| Gene | Full name | Variant | Strain(s) | MIM | References |
|---|---|---|---|---|---|
| Acads | Short-chain acyl-CoA dehydrogenase (SCAD) | Deletion Chr5:115,110,929–115,111,207 |
BALB/cByJ | 201470 | [3] |
| Alpl | alkaline phosphatase | Unknown | C57BL/6J | 146300 241500 241510 |
[7] |
| Bckdhb | branched chain keto acid dehydrogenase E1 subunit beta | Insertion Chr9:83,942,547–83,949,878 |
C57BL/6J | 248600 | [8, 20] This paper |
| Cox7a2l | supercomplex assembly factor I | c.216_221delGCCCAT p.F72delinsLPI |
C57BL/6 substrains 129S5/SvEvBrd BALB/cJ |
Not reported | [20, 37] |
| D2hgdh | D-2-hydroxyglutarate dehydrogenase | Unknown | C57BL/6J | 600721 | [20] |
| Dhtkd1 | dehydrogenase E1 and transketolase domain containing 1 | Insertion chr2:5,924,595–5,925,150 |
C57BL/6J A/J AKR/J FVB/NJ NOD/LtJ NZO/HILtJ |
204750 245130 |
[8] This paper |
| Eci3 | enoyl-Coenzyme A delta isomerase 3 | Unknown | DBA/2J | Not applicable | [38] |
| Glyctk | glycerate kinase | c.196dupA p.R66KfsX143 rs243632237 |
129 substrains LP/J |
220120 | [39, 40] |
| Gpnmb | glycoprotein (transmembrane) nmb | c.448C>T p.R150X rs47598337 |
DBA/2J | Not reported | [41] |
| Ivd | isovaleryl-CoA dehydrogenase | c.1068G>A p.K356K rs27440099 |
129S1/SvImJ 129S2/SvPasCrl 129P2/OlaHsd 129S5/SvEvBrd BTBR T+ Itpr3tf/J BUB/BnJ LP/J |
243500 | This paper |
| Lep | Leptin (Obese) | c.313T>C p.R105X |
Ob/Ob | 614962 | [1, 42] |
| Nnt | nicotinamide nucleotide transhydrogenase | Deletion 17.8 kbp Chr13:119,375,448 |
C57BL/6J | 614736 | [2] |
| Slc22a5 | high-affinity sodium-dependent carnitine cotransporter (OCTN2) | c.1055T>G p.L352R |
juvenile visceral steatosis (Jvs) | 212140 | [4] |
| Tyr | tyrosinase (albino) | c.308G>C p.C103S rs31191169 |
A/J AKR/J BALB/cJ FVB/NJ NOD/LtJ |
203100 | [43] |
The table lists the official gene name of the affected gene, a commonly used full gene name and abbreviation, the causal variant, the strain in which the variant was first characterized, the MIM number (https://www.omim.org/) of the equivalent human genetic disorder and a selected reference.
Akin to the situation in humans, it has been hypothesized that the phenotypic diversity between mouse strains are complex traits caused by many genetic variants with different effect sizes on mRNA levels and/or protein function. Identification of the genetic architecture underlying these complex phenotypes has been facilitated by quantitative genetic approaches in populations of mice such as second progeny offspring (F2), outbred cohorts or recombinant inbred strains. Genome-wide genotyping of these mice allows quantitative trait locus (QTL) analysis, identification of key underlying loci and in some cases, resolution to the causative genes. Mapping complex phenotypes alongside complex traits such as transcript, protein and metabolite abundances can reveal the molecular mechanisms through which genes and variants act on phenotypic presentation [5, 6].
One such example is provided by the BXD recombinant inbred strains, a mouse panel derived from DBA/2J and C57BL/6J parents [7]. Detailed genomic, proteomic, metabolomic and phenomic analysis of BXD strains has revealed multiple novel QTLs with relatively large effect sizes reminiscent of autosomal recessive traits. A list of these traits including examples from other strains is provided in Table 1. Here, we followed up on our recent identification of defects in the α-ketoadipic acid dehydrogenase (KADH) complex and the branched-chain ketoacid dehydrogenase (BCKDH) complex in the BXD genetic reference population due to expression (e)QTLs for Dhtkd1 and Bckdhb, respectively [8].
DHTKD1 is the E1 subunit of the KADH complex, a mitochondrial enzyme that catalyzes the oxidative decarboxylation of α-ketoadipic acid into glutaryl-CoA in the lysine degradation pathway. Mutations in DHTKD1 cause α-ketoadipic and α-aminoadipic aciduria (MIM 204750 and 245130) [9–11], an autosomal recessive biochemical abnormality of questionable clinical significance [12]. Mice harboring the low expressing Dhtkd1 C57BL/6J allele (Dhtkd1B6) have higher plasma α-aminoadipic acid concentrations than mice with the high expressing DBA/2J allele (Dhtkd1D2) [8, 9]. The molecular mechanism underlying the decreased Dhtkd1 expression and the degree of enzymatic deficiency in C57BL/6J mice is currently unknown.
BCKDHB is a subunit of the BCKDH complex, which is a mitochondrial enzyme in the degradation pathway for branched-chain amino acids (BCAA). Together with BCKDHA, BCKDHB forms the E1 subunit of this complex, whereas DBT and DLD are the E2 and E3 subunits, respectively. A deficiency of the BCKDH complex in humans causes maple syrup urine disease (MSUD), a severe neurometabolic disorder diagnosed by the detection alloisoleucine in plasma (MIM 248600). The BCKDH complex defect in C57BL/6J is not severe enough to cause increased alloisoleucine levels, although an increase in the BCAA/alanine ratio was noted [8]. It is known that the defect is caused by decreased expression of the BckdhbB6 allele, but the causal variant has remained elusive.
We now report the identification of the causal variants and molecular mechanisms underlying the DHTKD1 and BCKDHB deficiencies in C57BL/6J mice. In addition, we describe an exonic splice mutation in Ivd encoding isovaleryl-CoA dehydrogenase as the underlying cause of elevated C5-carnitine in the 129 substrain thus mimicking isovaleric acidemia (IVA; MIM 243500).
2. Materials and Methods
2.1. Mouse genomic studies
The Dhtkd1 locus was genotyped using two different methods. The locus around the structural variant was amplified using the following forward and reverse primers; 5’-AAA ACA TTG CCA CGA AGG AC-3’ and 5’-AGG CAC CCA CAG AAT CCA TA-3’. This yields a product of 767 bp for Dhtkd1B6, and 211 bp for Dhtkd1129 and Dhtkd1D2. These products were Sanger sequenced to determine the size to the structural variant (GenBank: MH925964-MH925966). We also performed a restriction fragment length polymorphism (RFLP) assay for rs27092974 in exon 8 of Dhtkd1, which is approximately 6.7 kb downstream of the structural variant. For this we amplified 206bp of exon 8 containing rs27092974 using the following forward and reverse primers; 5’-ACC TAC GCA GAG CAC CTC AT-3’ and 5’-TCT AGA GGC ACC CCT GTG TC-3’. The SNP introduces an SphI site and the PCR product is cleaved for Dhtkd1129 and Dhtkd1D2, but not for Dhtkd1B6. The Bckdhb and Ivd loci were genotyped using RNA sequencing data covering rs13480313 (Bckdhb exon 11), and rs4905023 and rs47004510 (both in exon 9 of Ivd).
2.2. Animal experiments
All animal experiments were approved by the IACUC of the Icahn School of Medicine at Mount Sinai (# IACUC-2016–0490) or the local animal experimentation committee of the Canton de Vaud (Switzerland, license no. 2257.2), and comply with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). Six week old male 129S2/SvPasCrl, DBA/2J and C57BL/6J mice were ordered. At 8 weeks of age, blood for biochemical analyses was collected via a submandibular blood draw in the random fed state (4pm). Plasma was collected and stored at −20°C for future analyses. Mice were euthanized at 10 weeks of age exactly 2 hours after an intraperitoneal dose of L-lysine (100mg/kg). Immediately preceding necropsy, mice were anesthetized with pentobarbital (100mg/kg ip) and then exsanguinated via the vena cava inferior. Blood was collected for the preparation of EDTA plasma and organs were snap frozen in liquid nitrogen and stored at −80°C for future analyses.
The B6129PF2 cohort was generated using C57BL/6N and 129P2 parents. Mice were euthanized at 29–45 weeks of age (mean age is 37 weeks) after overnight food withdrawal. They were first anesthetized with pentobarbital (100mg/kg ip) and then exsanguinated via the vena cava inferior. Blood was collected for the preparation of EDTA plasma and organs were snap frozen in liquid nitrogen and stored at −80°C for further use.
Male A/J, CAST/EiJ, NZO/HILtJ and PWK/PhJ mice were euthanized at 9–11 weeks old (mean age is 10 weeks). They were first anesthetized with a ketamine/xylazine mixture after which blood was collected by cardiac puncture. Plasma was prepared in Li-heparin tubes.
For the amino acid challenge experiments, 5 to 6 week old male 129S2/SvPasCrl, DBA/2J and C57BL/6J mice were ordered. At 9 weeks of age, all mice received an intraperitoneal injection of 15mL/kg 0.9% saline. At 12 weeks of age, all mice received 500mg/kg BCAAs (V150, L221, I129). At 14 weeks of age, all mice received 500mg/kg L-lysine. At 15 weeks of age, the BCAA challenge was repeated in the 129S2/SvPasCrl and C57BL/6J mice. One hour after the ip injection, a blood sample was collected from the saphenous vein using a capillary blood tube (Microvette CB 300 K2E).
2.3. RNA sequencing analysis
RNA was isolated from liver using QIAzol lysis reagent and the RNeasy kit (Qiagen). RNA samples were submitted to the Genomics Core Facility at the Icahn Institute and Department of Genetics and Genomic Sciences. cDNA libraries were prepared using the Illumina TruSeq RNA Library Preparation kit (#RS-122–2001). Samples were run on Illumina HiSeq 2500, and 100 nucleotide, single end fragments were read at a depth of about 25 million reads per sample. Raw and processed data were returned, count files were generated by aligning the reads to the mouse genome mm10 (GRCm38.75) with STAR and counting overlaps with exons grouped at gene level by featureCounts [13]. The aligned reads and splice junctions were visualized using the Integrative Genomics Viewer (IGV) and its Sashimi plot feature [14, 15].
2.4. Immunoblotting
Total liver protein homogenates were separated on a Bolt™ 4–12% Bis-Tris Plus Gel and blotted onto nitrocellulose. DHTKD1 was detected using a polyclonal antibody against recombinant human DHTKD1 (GeneTex, GTX32561). BCKDHB was detected using a polyclonal antibody against recombinant human BCKDHB (Abcam, ab201225). IVD was detected using a polyclonal antibody against recombinant human IVD (ThermoFisher, PA5–59547). Citrate synthase was detected using a polyclonal antibody against recombinant human protein (GeneTex, GTX110624). Secondary antibodies goat anti-rabbit IRDye 800CW or 680RD were from LI-COR Biosciences (Lincoln, NE, USA) and immunoblot images were obtained using the Odyssey infrared imaging system (LI-COR Biosciences).
2.5. Metabolite analysis
EDTA plasma (8 week old, random fed) was shipped to the Stable Isotope & Metabolomics Core of the Einstein-Mount Sinai Diabetes Research Center for a targeted metabolomics assay using the AbsoluteIDQ p180 kit (BIOCRATES Life Sciences AG, Innsbruck, Austria). This method quantifies ~180 metabolites in plasma including α-aminoadipic acid using liquid chromatography and flow injection analysis–mass spectrometry.
Plasma acylcarnitines, plasma amino acids and urine organic acids were measured by the Mount Sinai Biochemical Genetic Testing Lab (now Sema4). Plasma amino acids were analyzed as described [16]. Urine organic acids were quantified using a standard curve and pentadecanoic acid as internal standard.
2.6. Statistics
Data are displayed as the mean ± the standard deviation (SD) as indicated in the figure legends. Differences between groups of mice were evaluated using a two-sided t-test, one-way analysis of variance with Dunnett’s multiple comparison test, a Kruskal-Wallis test or a two-way analysis of variance as indicated. Significance is indicated in the figures. Allele effect sizes were determined using linear regression analysis. All analyses were performed in GraphPad Prism 6.
3. Results
3.1. Characterization of DHTKD1 deficiency in inbred mouse strains
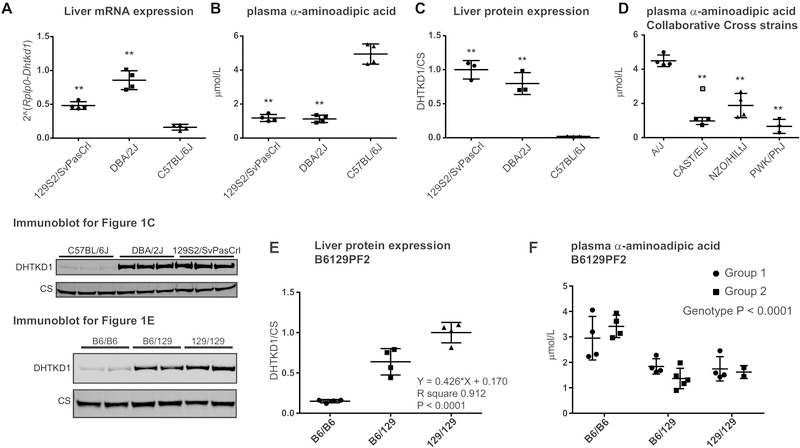
Our previous work has established that C57BL/6J mice are deficient in DHTKD1, but the degree of deficiency has remained unclear. We further characterized DHTKD1 function in C57BL/6J mice and compared these to mice with two other common inbred backgrounds; 129S2/SvPasCrl and DBA/2J. We focused on liver as this organ has the highest Dhtkd1 expression level. As expected based on previous studies [8, 17], C57BL/6J mice had the lowest Dhtkd1 mRNA in liver and the highest plasma α-aminoadipic acid levels (Figure 1A, B). We quantified DHTKD1 protein level in livers of these mice using immunoblot and found only 2% residual DHTKD1 protein in C57BL/6J (Figure 1C).
Figure 1.
Identification of the structural variant in Dhtkd1 causing α-aminoadipic acidemia in C57BL/6J mice. (A) Dhtkd1 mRNA expression in liver of 129S2/SvPasCrl, DBA/2J and C57BL/6J mice as measured by quantitative PCR. (B) Plasma α-aminoadipic acid concentrations in random fed mice as measured in the Biocrates panel. Samples were also analyzed by the Mount Sinai Biochemical Genetic Testing Lab with similar results. (C) Immunoblot analysis and quantification of DHTKD1 protein levels in liver. CS denotes citrate synthase. DHTKD1 expression was normalized using CS. The average DHTKD1/CS for 129S2/SvPasCrl was set to 1. Each group consisted of 4 male mice, but for the immunoblot only 3 mice per group were analyzed. Error bars indicate SD and ** P < 0.01 in a one-way ANOVA with Dunnett’s multiple comparisons test. (D) Plasma α-aminoadipic acid concentrations in 4 selected Collaborative Cross mouse strains (A/J, CAST/EiJ, NZO/HILtJ and PWK/PhJ) sacrificed at 1pm (random fed). Samples were analyzed by the Mount Sinai Biochemical Genetic Testing Lab. Each group consisted of 4 male mice, but one PWK/PhJ mouse died before the sample collection and one CAST/EiJ sample was deemed to be an outlier (grey square in the figure). Error bars indicate SD and ** P < 0.01 in a one-way ANOVA with Dunnett’s multiple comparisons test. (E) Immunoblot analysis and quantification of DHTKD1 protein levels in liver of the B6129PF2 cohort. The effect of the Dhtkd1B6 allele was estimated using linear regression. (F) Concentration of α-aminoadipic acid in plasma of two groups in the B6129PF2 cohort. Group 1 was a control group, whereas group 2 was treated with a compound (L-aminocarnitine). The inheritance of the elevated α-aminoadipic acid concentration is recessive and we did not perform linear regression analysis. A two-way ANOVA revealed a highly significant genotype effect.
DHTKD1 deficiency is not confined to the C57BL/6J strain. In the diversity outbred mouse model, which is derived from the 8 founder laboratory strains of the Collaborative Cross, the low expressing Dhtkd1 allele was shared between the A/J, C57BL/6J, NOD/LtJ and NZO/HILtJ founder strains, while the high expressing allele was found in 129S1/SvImJ, CAST/EiJ, PWK/PhJ and WSB/EiJ strains [17]. We now measured plasma α-aminoadipic acid concentration in mice from 2 low (A/J and NZO/HILtJ) and 2 high (CAST/EiJ and PWK/PhJ) DHTKD1 expressing Collaborative Cross strains. A/J mice had the highest α-aminoadipic acid concentration, NZO/HILtJ mice had intermediate values, and the lowest values were observed in CAST/EiJ and PWK/PhJ mice (Figure 1D), which is consistent with the DHTKD1 expression levels in these strains [17].
3.2. Identification and characterization of the Dhtkd1 variant causing α-aminoadipic acidemia in mice
The Mouse Genomes Project has sequenced the genomes of key laboratory mouse strains and identified SNPs, insertions/deletions and structural variants [18, 19]. We focused on an insertion of 556 base pair (bp) (chr2:5,924,595–5,925,150), because it was identified in all low Dhtkd1 expressing strains [8, 17]. This insertion in intron 4 is a solitary long terminal repeat (LTR). Using reverse transcriptase (RT) PCR on liver mRNA, we demonstrate that this insertion introduced an alternate splice donor site (chr2:5,924,677). Using RNA sequencing data, we furthermore observed that this insertion leads to; (1) aberrant transcriptional activity in intron 4, (2) read-through from exon 4 into intron 4, (3) decreased joining of exon 4 to exon 5, and (4) joining of intron 4 to exon 5 at the alternate splice donor site at position chr2:5,924,677. These observations indicate that the alternate splice donor in intron 4 competes with the splice donor at exon 4. None of these abnormalities were observed in mice without the insertion. A detailed description of all experiments describing the identification of the causal variant, characterization of the molecular mechanism underlying the DHTKD1 defect, estimation of the effect size of the Dhtkd1B6 allele, and the confirmation of the RNA sequencing data using quantitative RT PCR can be found in the online supplement (Supplemental Figures 1 and 2).
3.3. Experimental confirmation of the biochemical defect associated with the Dhtkd1B6 variant
We next used a B6129PF2 cohort of mice to confirm the biochemical defect associated with the Dhtkd1B6 variant. The animals in the B6129PF2 cohort carry recombined genomes from their C57BL/6N and 129P2 parents, which are DHTKD1 deficient and wild type, respectively. We determined the Dhtkd1 genotype and DHTKD1 protein level in these mice (Figure 1E). Consistent with the data from the inbred strains, Dhtkd1129/129 animals have the highest DHTKD1 protein levels. Using linear regression analysis, we estimated the effect size of the mutation and found that for each Dhtkd1B6 allele, DHTKD1 protein levels decrease by 43%. Lastly, we measured α-aminoadipic acid in plasma of two groups of B6129PF2 mice (Figure 1F). The concentration of α-aminoadipic acid was significantly higher in Dhtkd1B6/B6 mice when compared to Dhtkd1B6/129 and Dhtkd1129/129 animals, which is consistent with the established autosomal recessive inheritance of the biochemical phenotype in humans.
3.4. Identification of the causal variant for the branched-chain ketoacid dehydrogenase defect in C57BL/6J mice
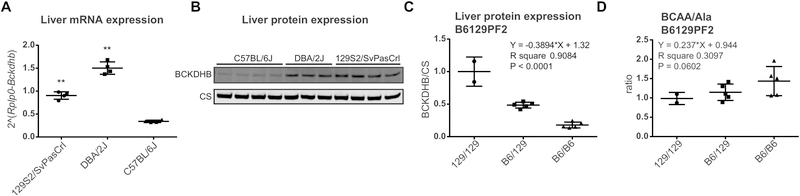
We next set out to identify the molecular mechanism and causal variant underlying BCKDHB deficiency in C57BL/6J mice [8, 20]. We first compared expression of Bckdhb between livers of 129S2/SvPasCrl, DBA/2J and C57BL/6J mice and confirmed that expression of Bckdhb is lowest in C57BL/6J (Figure 2A). The Mouse Genomes Project lists one large structural variant that is common to 129, DBA/2J and all other tested strains, a deletion of 7332 bp in intron 1 (Chr9:83,942,547–83,949,878). This region is an LTR retrotransposon (IAPLTR1_Mm, Supplemental Figure 3A) and is therefore an insertion in the C57BL/6J reference genome. Two murine Bckdhb transcripts are described; the canonical variant (NM_001305935.1) and a variant containing an alternate exon 1 encoding a protein that uses a potential downstream startcodon (Met69, NM_199195.1). This alternate exon 1 is part of the LTR retrotransposon in intron 1 and the encoded variant protein lacks the mitochondrial targeting sequence; therefore, it is unlikely to be a functional part of the BCKDH complex.
Figure 2.
Identification of the structural variant in Bckdhb causing BCKDHB deficiency in C57BL/6J mice. (A) Bckdhb mRNA expression in liver of 129S2/SvPasCrl, DBA/2J and C57BL/6J mice as measured by quantitative PCR. Error bars indicate SD and ** P < 0.01 in a one-way ANOVA with Dunnett’s multiple comparisons test. (B) Immunoblot analysis of BCKDHB protein levels in livers of 129S2/SvPasCrl, DBA/2J and C57BL/6J mice. (C) Quantification of BCKDHB protein levels in liver of mice from the B6129PF2 cohort. The effect size of each mutant Bckdhb allele was estimated using linear regression. (D) Ratio of the plasma concentration of BCAA over alanine (Ala).
We used RNA sequencing data in order to study the molecular mechanism underlying the decreased Bckdhb expression from the BckdhbB6 allele. We visualized reads and splice junctions focusing on the first exons. In mice with one or two BckdhbB6 alleles, we observed; (1) usage of alternate exon 1 and joining with exon 2, (2) decreased joining of exon 1 with exon 2, (3) skipping of exons 2 and 3, (4) various other abnormal splicing events, and (5) aberrant transcriptional activity in intron 1 (Supplemental Figure 3A). There was a clear gene dosage effect for all these events. None of these abnormalities were observed in mice with the Bckdhb129/129 genotype. A detailed description of all experiments describing the molecular analysis and estimation of the effect size of the BckdhbB6 allele can be found in the online supplement (Supplemental Figure 3). Using immunoblotting, we estimated that the residual BCKDHB protein expression is 18% in C57BL/6J (compared to 129S2/SvPasCrl) and 18% in BckdhbB6/B6 mice of the B6129PF2 cohort (Figure 2B, C). The BCAA/Ala was not affected by the number of BckdhbB6 alleles (Figure 2D).
3.5. Isovaleryl-CoA dehydrogenase deficiency in 129 mice
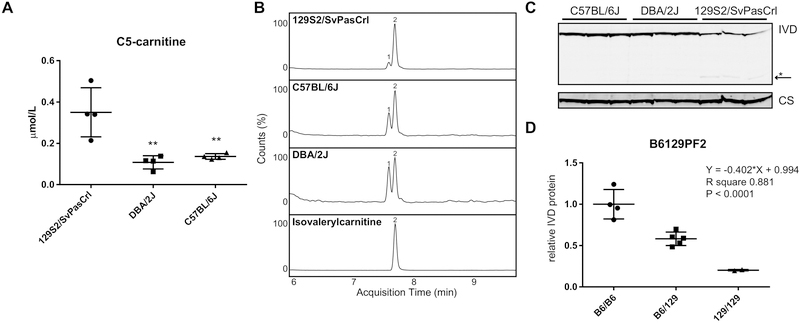
We noted elevated C5-carnitine concentrations in 129S2/SvPasCrl mice upon analysis of acylcarnitines in plasma of different mouse strains (Figure 3A). A similar observation was recently reported for 129S1/SvImJ and 129S6/SvEvTac [21]. Elevated C5-carnitine is found in IVA and 2-methylbutyrylglycinuria caused by IVD and ACADSB mutations, respectively. IVA is caused by a defect in isovaleryl-CoA dehydrogenase, an enzyme in the leucine degradation pathway. ACADSB encodes the short/branched chain acyl-CoA dehydrogenase, which is an enzyme in the isoleucine degradation pathway. The accumulating C5-carnitine isomer in the former disorder is isovalerylcarnitine, whereas it is 2-methylbutyrylcarnitine in the latter. UPLC-MS/MS analysis demonstrated that in plasma of C57BL/6J and DBA/2J mice the C5-carnitine is a mixture of 2 isomers with isovalerylcarnitine a bit more abundant than 2-methylbutyrylcarnitine (69% and 60% respectively). In contrast 88% of the C5-carnitine in 129S2/SvPasCrl mice is isovalerylcarnitine (Figure 3B), which makes a defect in Ivd most likely.
Figure 3.
Isovaleric acidemia in 129 mice. (A) Plasma C5-carnitine concentrations in random fed 129S2/SvPasCrl, DBA/2J and C57BL/6J mice as measured in the Biocrates panel. Samples were also analyzed by the Mount Sinai Biochemical Genetic Testing Lab with similar results. Error bars indicate SD and ** P < 0.01 in a one-way ANOVA with Dunnett’s multiple comparisons test. (B) UPLC-MS/MS analysis of C5-carnitine in plasma of 129S2/SvPasCrl, C57BL/6J and DBA/2J mice. Peak 2 was identified as isovalerylcarnitine based on a reference standard. Peak 1 is most likely 2-methylbutyrylcarnitine. Counts of the highest peak were normalized to 100%. (C) Immunoblot analysis of IVD protein levels in livers of 129S2/SvPasCrl, DBA/2J and C57BL/6J mice. The arrow indicates the position of an IVD proteolytic fragment. (D) Quantification of IVD protein levels in liver of mice from the B6129PF2 cohort.
In order to identify a cause for the elevated isovalerylcarnitine, we studied expression of Ivd in the RNA sequencing data of the liver samples of B6129PF2 mice and observed partial skipping of exon 10 in mice with IvdB6/129 and Ivd129/129 genotypes (Supplemental Figure 4A). The variant most likely responsible for the decreased splicing efficiency is a synonymous SNP at the last nucleotide of exon 10 (rs27440099) that changes the conserved G of a splice donor site into an A. This variant can therefore be classified as an exonic splice mutation. The detailed molecular characterization of the Ivd129 can be found in the online supplement (Supplemental Figure 4).
The canonical Ivd mRNA encodes a 424 amino acid protein with a predicted molecular weight of 46.3 kDa for the mitochondrial precursor and ~43 kDa after processing [22]. Ivd mRNA with an in-frame deletion in exon 10 encodes a protein that is 35 amino acids shorter, 4 kDa smaller, and lacks part of the substrate and FAD binding sites (p.Leu322_Lys356del, XP_006499988.1). Upon immunoblotting, we detected two immunoreactive bands in liver of 129S2/SvPasCrl mice. One band migrated at the same molecular weight as IVD in C57BL/6J and DBA/2J, a second band migrated just below the 28 kDa marker and was only detected in 129S2/SvPasCrl mice (Figure 4C). The levels of the full length IVD protein were decreased, which is consistent with partial exon skipping. Similar results were obtained in our cohort of B6129PF2 mice (Figure 4D). The residual IVD protein levels were 18% in 129S2/SvPasCrl and 20% in the Ivd129/129 mice of the B6129PF2 cohort. Our data are in line with proteome analysis of the diversity outbred mouse model, which listed a cis pQTL for IVD in the supplemental data [17]. Due to its autosomal recessive inheritance pattern, we did not have sufficient samples to evaluate the effect of the Ivd129/129 genotype on C5-carnitine in the B6129PF2 mice. Nonetheless, our combined data show that mice of the 129 substrains have a mild, but biochemically detectable defect in IVD.
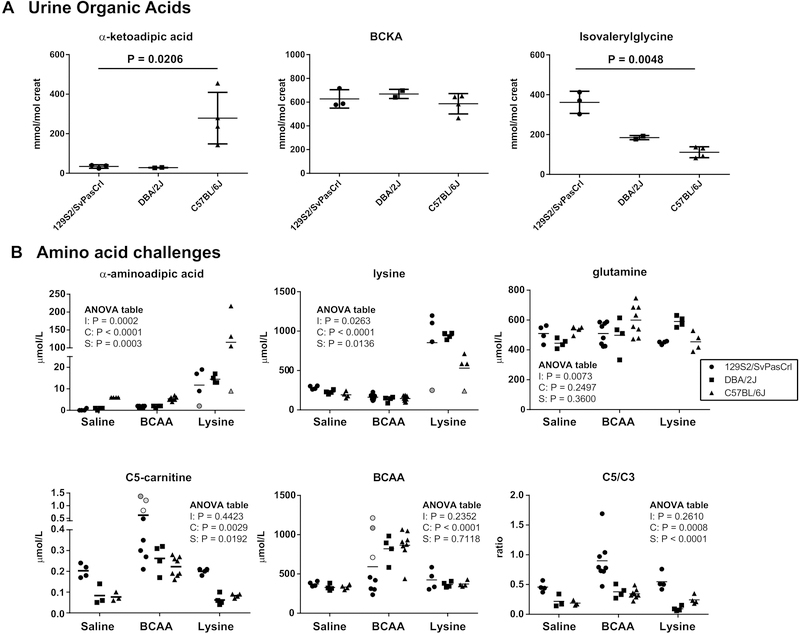
Figure 4.
Functional assessment of the identified variants in 129S2/SvPasCrl, DBA/2J and C57BL/6J mice. (A) Quantification of α-ketoadipic acid, BCKA (sum of 2-ketoisovaleric, 2-keto-3-methylvaleric and 2-ketoisocaproic acids) and isovalerylglycine in urine samples from 129S2/SvPasCrl (n = 3 of which one pooled sample), DBA/2J (n = 2 pooled samples) and C57BL/6J (n = 4 of which one pooled sample) mice. P value indicates the result of a Kruskal-Wallis test. (B) Mice (129S2/SvPasCrl, DBA/2J and C57BL/6J) were challenged with an ip injection of saline, 500mg/kg BCAA (V150, L221, I129) or 500mg/kg lysine. Plasma amino acids and acylcarnitines were analyzed one hour after the injection. In the lysine challenge one 129S2/SvPasCrl mouse and one C57BL/6J mouse were non-responders. These samples are indicated with a grey circle and grey triangle respectively. For unknown reasons, many of the 129S2/SvPasCrl mice did not respond the BCAA challenge. The samples with the largest increase in C5-carnitine were also the samples in which the BCAA concentration increased to similar levels as in the DBA/2J and C57BL/6J strain. These samples are indicated in circles with different scales of grey. A two-way analysis of variance was performed and the results are displayed in a table for each graph. I denotes the interaction term, C denotes the challenge effect and S denotes the strain effect.
3.6. α-ketoadipic aciduria in C57BL/6J and isovaleryl glycinuria in 129S2/SvPasCrl
We have provided evidence that C57BL/6J and 129S2/SvPasCrl carry specific mild inborn errors of metabolism. These biochemical abnormalities are not only diagnosed through analyses of plasma acylcarnitines and amino acids, but also of urine organic acids. We therefore analyzed the urine organic acid profile in randomly collected urine samples from C57BL/6J, 129S2/SvPasCrl and DBA/2J mice. Consistent with DHTKD1 deficiency, the urine of C57BL/6J mice had elevated α-ketoadipic acid levels when compared to the two other strains (Figure 4A). The urinary organic acid profile of MSUD patients is characterized by elevated branched-chain 2-keto (and 2-hydroxy) acids. Mouse urine contained relatively high basal levels of these branched-chain ketoacids, but there was no apparent increase in urine from C57BL/6J mice (Figure 4A). This result, in combination with the absence of plasma alloisoleucine, indicates that the Bckdhb variant in C57BL/6J is likely of limited functional relevance. IVA is characterized by elevated urine isovalerylglycine and 3-hydroxyisovaleric acid. Isovalerylglycine was elevated in urine of 129S2/SvPasCrl mice, but 3-hydroxyisovaleric acid was not detected (Figure 4A). These results are consistent with IVD deficiency in the 129S2/SvPasCrl strain.
3.7. Amino acid challenges provide additional evidence for the functional significance of the DHTKD1 and IVD defects
An intraperitoneal (ip) challenge with 20mg lysine in C57BL/6JUnib mice leads to a dramatic increase in plasma and tissue α-aminoadipic acid levels [23, 24]. In order to establish that this increase is caused by the DHTKD1 defect in C57BL/6 mice, we challenged C57BL/6J, 129S2/SvPasCrl and DBA/2J with lysine and analyzed plasma amino acids one hour after the ip injection. The lysine challenge increased plasma lysine levels in all strains, although this increase was somewhat less pronounced in C57BL/6J mice (Figure 4B). Plasma α-aminoadipic acid concentrations increased after the ip challenge in all strains, but C57BL/6J mice reached values >100μmol/L, while concentration in 129S2/SvPasCrl and DBA/2J remained < 20μmol/L (Figure 4B). In one 129S2/SvPasCrl and one C57BL/6J mouse both plasma lysine and α-aminoadipic acid concentrations did not increase after the lysine challenge, which is likely caused by a technical failure of the ip injection (Figure 4B).
Under basal conditions, the BCKDHB defect in C57BL/6J does not lead to detectable alloisoleucine levels. We tested whether a BCAA challenge could unmask this MSUD biochemical phenotype. One hour after the BCAA challenge, the concentration of plasma BCAA was increased (Figure 4B), but alloisoleucine remained virtually undetectable in all strains (not shown).
Leucine is one of the three BCAAs, and therefore the BCAA challenge also allowed us to further study the IVD defect in 129S2/SvPasCrl mice. Although we had some technical difficulties obtaining a reproducible increase in plasma BCAA levels in 129S2/SvPasCrl mice, the mice that did respond to the challenge also displayed the most striking elevation of C5-carnitine (Figure 4B). The results of the amino acid challenges further question the significance of the BCKDHB defect in C57BL/6J mice, but illustrate the functional consequences of the DHTKD1 and IVD defects in C57BL/6J and 129S2/SvPasCrl mice, respectively.
4. Discussion
Inbred mouse strains remain a cornerstone of translational biomedical research. Here we report our characterization of the causal variants and molecular mechanisms underlying three mild inborn errors of metabolism in commonly used inbred mouse strains. We found that α-aminoadipic acidemia in C57BL/6J mice is caused by a structural variant in intron 4 of Dhtkd1 that introduces an alternative splice donor site leading to a partial splicing defect. As a result, functional Dhtkd1 mRNA and DHTKD1 protein levels are decreased, and plasma α-aminoadipic acid concentration is increased. Similarly, BCKDHB deficiency in C57BL/6J mice is caused by a structural variant in intron 1 that introduces an alternative exon 1 and leads to aberrant splicing, apparently without an obvious biochemical consequence. Finally, elevated C5-carnitine in 129 mice is caused by an exonic splice site mutation in Ivd that leads to skipping of exon 10, a deficiency in IVD protein and a biochemical phenotype mimicking IVA.
The C57BL/6J inbred strain has been used to generate the mouse reference genome. The Mouse Genomes Projects lists the causal structural variants in Dhtkd1 and Bckdhb as deletions in the non C57BL/6J strains. Given that these deletions abolish aberrant splicing in both cases, it is fair to assume that the allele with the deletion is in fact the wild type allele and the structural variant is the result of an insertion. Indeed, both variants are the result of transposable element (TE) activity. The causal variant in Dhtkd1 is annotated as a repeat region called MT2-Mm and this sequence is 86% identical to the LTR from the mouse endogenous retroviral (ERV) sequence MuERV-L (Y12713.1), a class III endogenous retrovirus [25, 26]. This is a solitary LTR, likely the result of a recombination between 5’ and 3’ LTRs that deleted the gag, pol and dUTPase genes [25, 26]. The C57BL/6J genome carries 37,000–38,000 MT2-Mm copies [27]. The causal variant in Bckdhb is a so-called intracisternal A-particle (IAP) mobile element, a class II ERV. The C57BL/6J genome carries ~10,000 IAP LTR elements [28]. Combined, ERVs comprise 8–10% of the mouse genome and, due to their ongoing activity, they account for the majority of spontaneous mouse mutants [25, 26]. Indeed more than 5000 TE variants were identified between DBA/2J and C57BL/6J strains [29], and over 100,000 TE variants between 13 classical laboratory and 4 wild-derived mouse inbred strains [30]. Of the TE variants with a single annotation, 52% were LTRs, 43% were long interspersed nuclear elements (LINEs) and 5% were short interspersed nuclear elements (SINEs) [29]. On an evolutionary scale, TEs are thought to have a significant impact via the generation of new exons and alternative splicing [31, 32].
DHTKD1 deficiency causing α-ketoadipic and α-aminoadipic aciduria is currently considered a biochemical abnormality of questionable clinical significance [12]. This means that this disorder can be diagnosed through biochemical and genetic methods, but is not considered harmful. Any previous associations of α-ketoadipic and α-aminoadipic aciduria with disease phenotypes are therefore due to ascertainment bias [12]. However, given the rarity of DHTKD1 deficiency, conclusive evidence for this is difficult to provide. We have established that the residual DHTKD1 protein levels were ~2% in C57BL/6J liver relative to 129S2/SvPasCrl and DBA/2J liver, and ~15% in liver of B6129PF2 mice with the Dhtkd1B6/B6 genotype. The Dhtkd1B6/B6 genotype renders the KADH complex rate-limiting in lysine degradation as evidenced by increased plasma α-aminoadipic acid concentration, increased urine α-ketoadipic acid levels, and a marked further increase in plasma α-aminoadipic acid concentration upon a lysine challenge. Therefore the mutant Dhtkd1 allele identified in C57BL/6J mice could be used to study the consequences of DHTDK1 deficiency. In addition to this natural variant, a targeted Dhtkd1 KO mouse model was recently generated using the TALEN approach [33].
Mutations in BCKDHB lead to a defect in the BCKDH complex and are a cause of MSUD in humans. We estimated that the residual BCKDHB protein expression is ~18% in mice with the structural variant. This is within the range of residual BCKDH complex activity reported in patients with milder (variant) MSUD [34]. However, the absence of pathognomonic alloisoleucine in plasma even after a BCAA load and the absence of elevated BCKA in urine suggest that the BCKDHB defect in C57BL/6J is of limited physiological significance.
IVA due to mutations in IVD is an inborn error of leucine degradation. IVA is a harmful and potentially life-threatening disorder, but early diagnosis through newborn screening and dietary intervention improves patient outcomes [35]. There is currently no mouse model available to study the pathophysiology of IVA. Our work suggests that 129 mice have IVA and thus the Ivd129 allele may be useful to generate a mouse model for IVA. Moreover, all early transgenic mouse models were generated using ES cells derived from 129 substrains, and although these models are now often backcrossed to other inbred strains, many mouse models are still on a full or partial 129 background, and therefore carry the hypomorphic Ivd129 allele.
The genetic standardization resulting from the use of inbred mouse strains has improved the reproducibility of experiments by decreasing a major source of variation between individuals. At the same time, studying one individual mouse inbred line is not unlike human case reports, which have marginal validity on the population level. Therefore, the use of single inbred lines may underpin the fact that data cannot easily be generalized to other strains; most importantly, this may contribute to the limited translation of findings in mouse to humans. In addition, we have shown that any chosen inbred strain can harbor impactful genetic variants (Table 1). A particular variant can make a strain less suitable to study a specific process due to gene-gene or gene-environment interactions [36]. Our study reinforces the notion that using diverse populations of mice such as BXD or collaborative cross recombinant inbred strains will avoid many of the pitfalls of studying mice on a single genetic background.
Supplementary Material
Acknowledgements
We acknowledge the help of the shared resource facilities at the Icahn School of Medicine at Mount Sinai (Colony Management, Real Time Polymerase Chain Reaction (qPCR), Mouse Genetics and Gene Targeting, Scientific Computing and the Genomics Core) and the Stable Isotope and Metabolomics Core Facility of the Diabetes Research and Training Center (DRTC) of the Albert Einstein College of Medicine (Supported by NIH/NCI grant P60DK020541). We thank Dr. Hongjie Chen and Purvika Patel for their expert assistance with the analysis of the urine organic acids.
Funding
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number R03HD092878. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. JA is supported by grants from the École Polytechnique Fédérale de Lausanne, the Swiss National Science Foundation (31003A-140780), the AgingX program of the Swiss Initiative for Systems Biology (51RTP0-151019), and the NIH (R01AG043930).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ingalls AM, Dickie MM, Snell GD, Obese, a new mutation in the house mouse J Hered 41 (1950) 317–318. [DOI] [PubMed] [Google Scholar]
- [2].Huang TT, Naeemuddin M, Elchuri S, Yamaguchi M, Kozy HM, Carlson EJ, Epstein CJ, Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase Hum Mol Genet 15 (2006) 1187–1194. [DOI] [PubMed] [Google Scholar]
- [3].Hinsdale ME, Kelly CL, Wood PA, Null allele at Bcd-1 locus in BALB/cByJ mice is due to a deletion in the short-chain acyl-CoA dehydrogenase gene and results in missplicing of mRNA Genomics 16 (1993) 605–611. [DOI] [PubMed] [Google Scholar]
- [4].Nezu J, Tamai I, Oku A, Ohashi R, Yabuuchi H, Hashimoto N, Nikaido H, Sai Y, Koizumi A, Shoji Y, Takada G, Matsuishi T, Yoshino M, Kato H, Ohura T, Tsujimoto G, Hayakawa J, Shimane M, Tsuji A, Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion-dependent carnitine transporter Nat. Genet 21 (1999) 91–94. [DOI] [PubMed] [Google Scholar]
- [5].Argmann CA, Chambon P, Auwerx J, Mouse phenogenomics: the fast track to “systems metabolism” Cell Metab 2 (2005) 349–360. [DOI] [PubMed] [Google Scholar]
- [6].Williams EG, Auwerx J, The Convergence of Systems and Reductionist Approaches in Complex Trait Analysis Cell 162 (2015) 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, Schoonjans K, Williams RW, Auwerx J, Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits Cell 150 (2012) 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, Zamboni N, Auwerx J, Aebersold R, Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population Cell 158 (2014) 1415–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hagen J, te Brinke H, Wanders RJ, Knegt AC, Oussoren E, Hoogeboom AJ, Ruijter GJ, Becker D, Schwab KO, Franke I, Duran M, Waterham HR, Sass JO, Houten SM, Genetic basis of alpha-aminoadipic and alpha-ketoadipic aciduria Journal of inherited metabolic disease 38 (2015) 873–879. [DOI] [PubMed] [Google Scholar]
- [10].Stiles AR, Venturoni L, Mucci G, Elbalalesy N, Woontner M, Goodman S, Abdenur JE, New Cases of DHTKD1 Mutations in Patients with 2-Ketoadipic Aciduria JIMD reports (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Danhauser K, Sauer SW, Haack TB, Wieland T, Staufner C, Graf E, Zschocke J, Strom TM, Traub T, Okun JG, Meitinger T, Hoffmann GF, Prokisch H, Kolker S, DHTKD1 Mutations Cause 2-Aminoadipic and 2-Oxoadipic Aciduria Am J Hum Genet 91 (2012) 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Goodman SI, Duran M, Biochemical phenotypes of questionable clinical significance, in: Blau N, Duran M, Gibson KM, Dionisi-Vici C (Eds.), Physician’s guide to the diagnosis, treatment, and follow-up of inherited metabolic diseases, Springer Verlag, Heidelberg, 2014, pp. 691–705. [Google Scholar]
- [13].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner Bioinformatics 29 (2013) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, Integrative genomics viewer Nature biotechnology 29 (2011) 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thorvaldsdottir H, Robinson JT, Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration Brief Bioinform 14 (2013) 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Le A, Ng A, Kwan T, Cusmano-Ozog K, Cowan TM , A rapid, sensitive method for quantitative analysis of underivatized amino acids by liquid chromatography-tandem mass spectrometry (LC-MS/MS) J Chromatogr B Analyt Technol Biomed Life Sci 944 (2014) 166–174. [DOI] [PubMed] [Google Scholar]
- [17].Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP, Defining the consequences of genetic variation on a proteome-wide scale Nature 534 (2016) 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellaker C, Goodstadt L, Nicod J, Bhomra A, Hernandez-Pliego P, Whitley H, Cleak J, Dutton R, Janowitz D, Mott R, Adams DJ, Flint J, Sequence-based characterization of structural variation in the mouse genome Nature 477 (2011) 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ, Mouse genomic variation and its effect on phenotypes and gene regulation Nature 477 (2011) 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, Aebersold R, Auwerx J, Systems proteomics of liver mitochondria function Science 352 (2016) aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fujisaka S, Avila-Pacheco J, Soto M, Kostic A, Dreyfuss JM, Pan H, Ussar S, Altindis E, Li N, Bry L, Clish CB, Kahn CR, Diet, Genetics, and the Gut Microbiome Drive Dynamic Changes in Plasma Metabolites Cell reports 22 (2018) 3072–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ikeda Y, Keese SM, Tanaka K, Molecular heterogeneity of variant isovaleryl-CoA dehydrogenase from cultured isovaleric acidemia fibroblasts Proc Natl Acad Sci U S A 82 (1985) 7081–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pena IA, Marques LA, Laranjeira AB, Yunes JA, Eberlin MN, Arruda P, Simultaneous detection of lysine metabolites by a single LC-MS/MS method: monitoring lysine degradation in mouse plasma Springerplus 5 (2016) 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pena IA, Marques LA, Laranjeira AB, Yunes JA, Eberlin MN, MacKenzie A, Arruda P, Mouse lysine catabolism to aminoadipate occurs primarily through the saccharopine pathway; implications for pyridoxine dependent epilepsy (PDE) Biochim Biophys Acta 1863 (2017) 121–128. [DOI] [PubMed] [Google Scholar]
- [25].Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL , Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line PLoS Genet 2 (2006) e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stocking C, Kozak CA, Murine endogenous retroviruses Cellular and molecular life sciences : CMLS 65 (2008) 3383–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schoorlemmer J, Perez-Palacios R, Climent M, Guallar D, Muniesa P, Regulation of Mouse Retroelement MuERV-L/MERVL Expression by REX1 and Epigenetic Control of Stem Cell Potency Front Oncol 4 (2014) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Faulk C, Barks A, Dolinoy DC, Phylogenetic and DNA methylation analysis reveal novel regions of variable methylation in the mouse IAP class of transposons BMC genomics 14 (2013) 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, Hurles ME, Mell JC, Hall IM, Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome Genome research 20 (2010) 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nellaker C, Keane TM, Yalcin B, Wong K, Agam A, Belgard TG, Flint J, Adams DJ, Frankel WN, Ponting CP, The genomic landscape shaped by selection on transposable elements across 18 mouse strains Genome Biol 13 (2012) R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kazazian HH Jr., Mobile elements: drivers of genome evolution Science 303 (2004) 1626–1632. [DOI] [PubMed] [Google Scholar]
- [32].Kim DS, Huh JW, Kim YH, Park SJ, Kim HS, Chang KT, Bioinformatic analysis of TE-spliced new exons within human, mouse and zebrafish genomes Genomics 96 (2010) 266–271. [DOI] [PubMed] [Google Scholar]
- [33].Biagosch C, Ediga RD, Hensler SV, Faerberboeck M, Kuehn R, Wurst W, Meitinger T, Kolker S, Sauer S, Prokisch H, Elevated glutaric acid levels in Dhtkd1-/Gcdh-double knockout mice challenge our current understanding of lysine metabolism Biochim Biophys Acta 1863 (2017) 2220–2228. [DOI] [PubMed] [Google Scholar]
- [34].Simon E, Flaschker N, Schadewaldt P, Langenbeck U, Wendel U, Variant maple syrup urine disease (MSUD)--the entire spectrum Journal of inherited metabolic disease 29 (2006) 716–724. [DOI] [PubMed] [Google Scholar]
- [35].Vockley J, Ensenauer R, Isovaleric acidemia: new aspects of genetic and phenotypic heterogeneity American journal of medical genetics. Part C, Seminars in medical genetics 142C (2006) 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Enríquez JA, Mind your mouse strain Nat. Metab 1 (2019) 5–7. [DOI] [PubMed] [Google Scholar]
- [37].Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, Navas P, Cruz R, Carracedo A, Lopez-Otin C, Perez-Martos A, Fernandez-Silva P, Fernandez-Vizarra E, Enriquez JA, Supercomplex assembly determines electron flux in the mitochondrial electron transport chain Science 340 (2013) 1567–1570. [DOI] [PubMed] [Google Scholar]
- [38].van Weeghel M, Ofman R, Argmann CA, Ruiter JP, Claessen N, Oussoren SV, Wanders RJ, Aten J, Houten SM, Identification and characterization of Eci3, a murine kidney-specific Delta3,Delta2-enoyl-CoA isomerase FASEB J 28 (2014) 1365–1374. [DOI] [PubMed] [Google Scholar]
- [39].Cazier JB, Kaisaki PJ, Argoud K, Blaise BJ, Veselkov K, Ebbels TM, Tsang T, Wang Y, Bihoreau MT, Mitchell SC, Holmes EC, Lindon JC, Scott J, Nicholson JK, Dumas ME, Gauguier D, Untargeted metabolome quantitative trait locus mapping associates variation in urine glycerate to mutant glycerate kinase Journal of proteome research 11 (2012) 631–642. [DOI] [PubMed] [Google Scholar]
- [40].Qiao Q, Li T, Sun J, Liu X, Ren J, Fei J, Metabolomic analysis of normal (C57BL/6J, 129S1/SvImJ) mice by gas chromatography-mass spectrometry: detection of strain and gender differences Talanta 85 (2011) 718–724. [DOI] [PubMed] [Google Scholar]
- [41].Anderson MG, Smith RS, Hawes NL, Zabaleta A, Chang B, Wiggs JL, John SW, Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice Nat Genet 30 (2002) 81–85. [DOI] [PubMed] [Google Scholar]
- [42].Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM, Positional cloning of the mouse obese gene and its human homologue Nature 372 (1994) 425–432. [DOI] [PubMed] [Google Scholar]
- [43].Yokoyama T, Silversides DW, Waymire KG, Kwon BS, Takeuchi T, Overbeek PA, Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice Nucleic Acids Res 18 (1990) 7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.