SUMMARY
Adipose tissue in the mammary gland undergoes dramatic remodeling during reproduction. Adipocytes are replaced by mammary alveolar structures during pregnancy and lactation, then reappear upon weaning. The fate of the original adipocytes during lactation and the developmental origin of the re-appearing adipocyte post involution are unclear. Here, we reveal that adipocytes in the mammary gland de-differentiate into Pdgfrα+ preadipocyte- and fibroblast-like cells during pregnancy, and remain de-differentiated during lactation. Upon weaning, de-differentiated fibroblasts proliferate and re-differentiate into adipocytes. This cycle occurs over multiple pregnancies. These observations reveal the potential of terminally differentiated adipocytes to undergo repeated cycles of de-differentiation and re-differentiation in a physiological setting.
Keywords: adipose tissue, preadipocyte, de-differentiation, mammary gland, lactation
INTRODUCTION
Adipocytes comprise a large portion of the stromal compartment in the adult non-lactating mammary gland. Pregnancy triggers extensive and rapid proliferation of the ductal epithelium in the mammary gland to form the mammary alveolar structures, which further develop into milk-secreting alveoli during lactation (Rillema, 1994). Meanwhile, mammary adipocytes gradually disappear during pregnancy, freeing up room for the expanding mammary glands. Upon cessation of lactation at weaning, the mammary alveolar structures collapse through apoptosis (Li et al., 1997; Lund et al., 1996). At the same time, adipocytes regenerate rapidly and quickly fill up the space in the mammary gland (Cinti, 2007). Adipocytes are believed to serve an important endocrine function for the developing mammary gland during puberty (Hovey and Aimo, 2010; Hovey et al., 2001; Landskroner-Eiger et al., 2010; Pavlovich et al., 2010); however, the fate of adipocytes and their interaction with the mammary gland during pregnancy, lactation, and involution remains unresolved.
The origin of adipocytes in the mammary gland during reproduction is somewhat controversial and has been debated in the literature. Many studies have suggested that the mammary alveolar structures develop from specific mammary stem cells (Eirew et al., 2008; Rios et al., 2014; Shackleton et al., 2006; Tiede and Kang, 2011; Wang et al., 2015). However, Cinti and colleagues suggested that adipocytes trans-differentiate into mammary gland alveolar cells during pregnancy and lactation. After lactation, these mammary epithelial cells are postulated to undergo reverse trans-differentiation and become adipocytes (Giordano et al., 2014; Morroni et al., 2004). Utilizing our doxycycline-inducible mature adipocyte-specific tracking system – the “AdipoChaser mouse” (Wang et al., 2013), we find unambiguous evidence to support that adiponectin-expressing adipocytes undergo de-differentiation and remain as fibroblast-like cells between the mammary gland alveolar structures during pregnancy and lactation. Using the same technique, it is also clear that mammary gland milk-secreting alveolar cells do not arise from pre-existing adipocytes. The de-differentiated adipocytes/fibroblast-like cells initiate the expression of the adipogenic precursor (preadipocyte) marker Platelet-derived growth factor receptor (PDGFR) α, therefore undergoing a de-differentiation program rather than just depleting lipid droplets. Upon involution, these fibroblast-like cells effectively re-differentiate back into adipocytes. We conclude that adipocytes display a remarkable level of plasticity in terms of going back and forth from fully differentiated adipocytes to preadipocyte- and fibroblast-like cells.
RESULTS
Regenerated Mammary Adipocytes upon Weaning Arise from Pre-existing Adipocytes
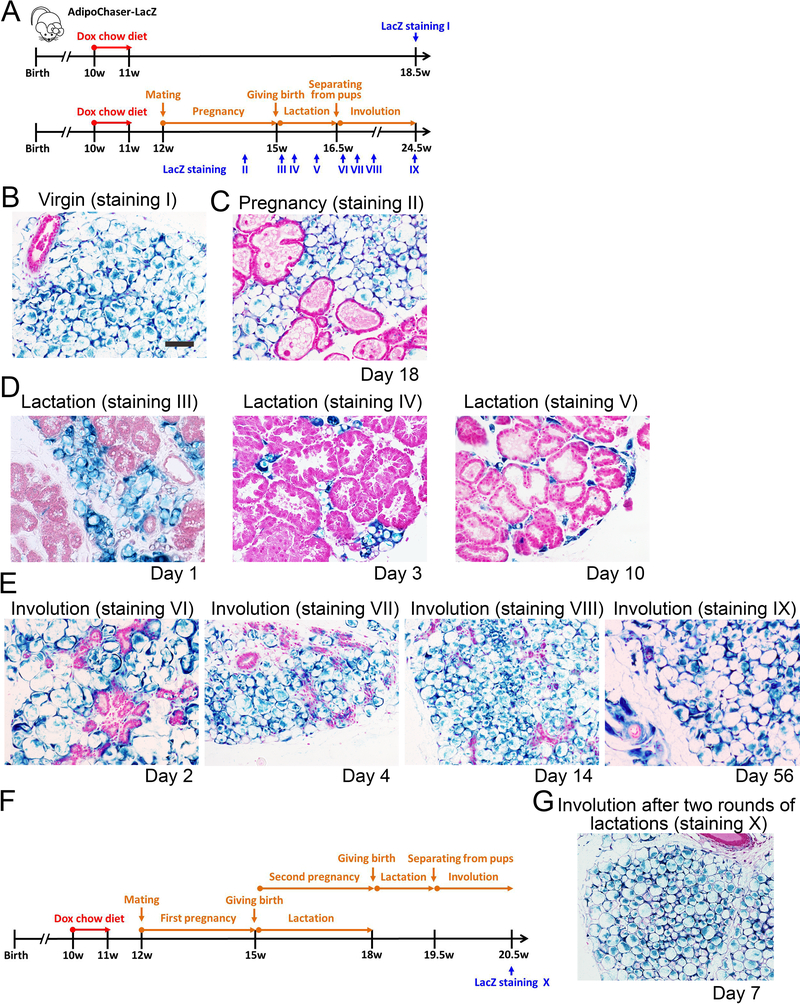
To better understand the dynamics of adipogenesis in vivo, we have previously developed the AdipoChaser mouse, a system for the inducible, permanent labeling of mature adipocytes (Wang et al., 2013). This doxycycline-based tet-responsive Cre-loxP system is derived from interbreeding three transgenic strains: 1) mice expressing the “tet-on” transcription factor rtTA under the control of the adiponectin gene promoter (Adn-rtTA); 2) mice expressing tet-responsive CRE (TRE-Cre) that can be activated with rtTA in the presence of doxycycline (dox); and 3) reporter mice expressing a LacZ reporter gene from the Rosa26 locus in a Cre-dependent manner (Rosa26-loxP-STOP-loxP-LacZ). Utilizing the AdipoChaser-LacZ system, we traced the fate of adiponectin-expressing adipocytes in the mammary gland during pregnancy, lactation and involution. 10-week old female AdipoChaser-LacZ mice were first put on a doxycycline chow diet for 7 days to ensure uniform and permanent labeling of all mature adipocytes with active LacZ enzyme expression, followed by 7 days of chow diet to make sure that the doxycycline was fully washed out. Subsequently, these mice were bred with male mice of the same age; the mammary gland tissues were collected for LacZ staining at multiple time points during pregnancy, lactation and involution (Figure 1A). Doxycycline pre-labeled virgin female AdipoChaser-LacZ mice were used as positive controls, and these mice showed no new adipogenesis, judged by the fact that all fat depots displayed nearly 100% positive LacZ staining in adipocytes, even as long as 7 weeks post doxycycline treatment. Thus, extremely low levels of de novo adipogenesis were seen in subcutaneous fat pads in females (Figure 1B), which is consistent with the result we observed in males (Wang et al., 2013). When the mice were pregnant for 18 days, adipocytes in the subcutaneous fat pad still showed nearly 100% LacZ labeling, with no obvious morphological changes, while the expanding mammary gland took over 50% of the space in the mammary fat pad (Figure 1C). Notably, the developing mammary gland structures were completely free of LacZ positive cells, indicating that these cells are not generated from trans-differentiation of pre-existing adipocytes (Figure 1C). During lactation, while the mammary gland further expanded and finally established milk-producing alveolar structures, the LacZ positive adipocytes gradually morphed into “lipid droplet-free adipocytes”, morphologically resembling fibroblasts at the peak of lactation (day 10). At lactation day 10, these “empty adipocytes” totally lost their lipid droplets and reduced their size dramatically, yet were still intensely positive for LacZ activity (Figure 1D). We then removed the pups of AdipoChaser-LacZ mice at postnatal day 10, and performed LacZ staining on the mammary gland within either two days, four days, two weeks or 2 months after weaning (Figure 1E). The alveolar structures rapidly collapsed, and the adipocytes regenerated immediately during mammary gland involution. Surprisingly, these newly generated adipocytes were all positive for LacZ activity (Figure 1E). These observations clearly indicate that adipocytes do not trans-differentiate into milk-secreting alveolar cells, consistent with the studies that show that alveolar cells develop from their own mammary stem cells (Rios et al., 2014). Meanwhile, the adipocyte population regenerated during involution derives almost exclusively from pre-existing mature adipocytes. In an additional set of experiments, we pre-labeled all the mammary adipocytes and tested if the same phenomenon happens after two rounds of pregnancy (Figure 1F). Indeed, all the mammary adipocytes generated during the second round of involution are positive for LacZ activity (Figure 1G), indicating that adipocytes can undergo multiple rounds of de- and re-differentiation
Figure 1. Lineage tracing of adipocytes in the mammary gland during reproduction.
(A) Experimental design: 10-week old AdipoChaser-LacZ female mice were put on a doxycycline chow diet for 7 days to ensure that LacZ expression is turned on in all mature adipocytes. After doxycycline treatment, mice were switched back to regular chow diet for a week to ensure washout of doxycycline. Thereafter, mice were kept as virgins or bred with male mice to undergo pregnancy and lactation. LacZ staining is performed at the indicated time points. (B-E) Representative β-gal staining of the mammary gland from female AdipoChaser-LacZ mice. (B) Virgin female: 7 weeks after doxycycline diet treatment. (C) During pregnancy: female mice were pregnant for 18 days. (D) During lactation: female mice were lactating pups at day 1, day 3 and day 10. (E) Involution: pups were taken away from lactating female mice at L10 for 2 days, 4 days, two weeks or 2 months (56 days) (F) Experimental design: 10-week old AdipoChaser-LacZ female mice were treated with doxycycline chow diet to label all mature adipocytes as described in panel A. Mice were then bred with male mice to undergo pregnancy and lactation for two rounds. LacZ staining is performed 7 days after weaning. (G) Representative β-gal staining of the mammary gland from female AdipoChaser-LacZ mice, after two rounds of lactations. n = 3–5 mice per group. Scale bar: 100 μm. This experiment is representative of three independent experiments.
De-differentiated Mammary Adipocyte Can Proliferate and Differentiate into Adipocytes in Vitro
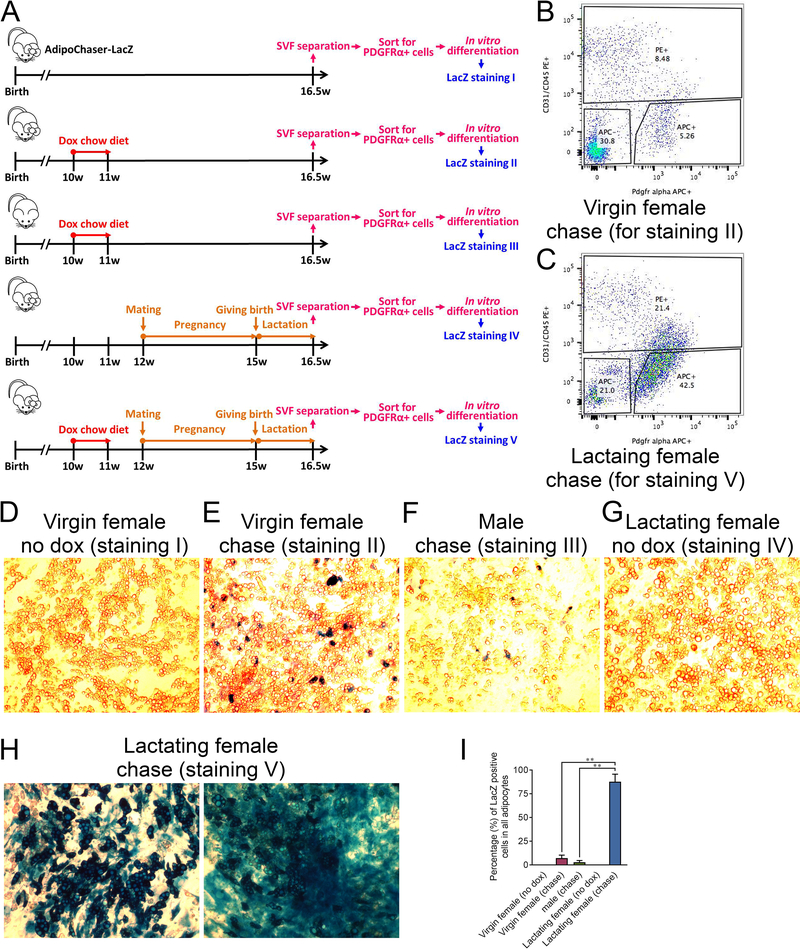
We aimed to determine whether adipocytes in the mammary gland completely de-differentiate or merely lose their lipid droplets during pregnancy and lactation. Most adipogenic cells within WAT are of non-endothelial and non-hematopoietic origin (CD31− and CD45−) (Berry et al., 2014), and express PDGFRα, a mesenchymal marker expressed in all described adipogenic preadipocyte populations (Berry and Rodeheffer, 2013; Hepler et al., 2017). 10-week old female or male AdipoChaser-LacZ mice were used in this study as indicated in Figure 2A. Initially, we labeled all mature adipocytes by exposing mice to a doxycycline-containing chow diet. We then allowed the female mice to undergo pregnancy and lactation. At the peak of lactation, we purified CD31−/CD45−/PDGFRα+ preadipocyte-enriched cells from the stromal vascular fraction (SVF) from the mammary gland through FACS, and used these isolated cells for in vitro differentiation assays (Figure 2A). As controls, we purified the corresponding population from virgin female AdipoChaser-LacZ littermates as well as male AdipoChaser-LacZ littermates, with or without doxycycline exposure (Figure 2A). FACS analysis reveals that the percentage of PDGFRα+ cells within the mammary gland of lactating female is much higher than that found in the control virgin littermates (Figure 2B, C). As expected, these CD31−/CD45−/PDGFRα+ cells isolated from the mammary glands can efficiently differentiate into adipocytes in a dish (Figure 2D–H). LacZ staining of these differentiated adipocytes from doxycycline pre-labeled mice reveals that there were very few lacZ positive adipocytes both in the virgin female (7%) and in male mice (3%), which reflects a low-level presence of these fibroblast-like cells that originate from mature adipocytes (Figure 2E, F, I). However, for the lactating females, 87% of adipocytes differentiated in vitro were positive for LacZ staining, indicating these cells arose from pre-existing adipocytes (Figure 2H, I). These results suggest that adipocytes in the mammary gland have not only lost their lipid content but actually regained preadipocyte features during pregnancy and lactation, as judged by their acquisition of PDGFRα expression, a protein not expressed in mature adipocytes. More importantly, these de-differentiated adipocytes can readily re-differentiate into mature adipocytes upon exposure to classical adipogenic stimuli in an in vitro setting.
Figure 2. In vitro isolation and differentiation of PDGFRα positive cells from the SVF of the lactating mammary gland.
(A) Experimental design: 5 cohorts of 10-week old AdipoChaser-LacZ female or male mice were collected, and 5 experimental conditions were performed as indicated. At the end of each experiment, the mammary glands (subcutaneous adipose tissues) were used for SVF separation. The separated SVF were then flow sorted for CD31−/CD45−/PDGFRα+ cells. The isolated CD31−/CD45−/PDGFRα+ cells were plated in dishes and differentiated into adipocyte for LacZ staining. (B-C) Flow sorting images for condition II and V. (D-H) β-gal staining on the differentiated adipocytes. (D) Adipocytes differentiated from the CD31−/CD45−/PDGFRα+ cells from the mammary gland SVF of virgin females without any doxycycline treatment. (E) Adipocytes differentiated from the CD31−/CD45−/PDGFRα+ cells from the mammary gland SVF of virgin females with doxycycline pre-labeling. (F) Adipocytes differentiated from the CD31−/CD45−/PDGFRα+ cells from the sWAT SVF of male mice with doxycycline pre-labeling. (G) Adipocytes differentiated from the CD31−/CD45−/PDGFRα+ cells from the mammary gland SVF of lactating female without doxycycline pre-labeling. (H) adipocytes differentiated from the CD31−/CD45−/PDGFRα+ cells from the mammary gland SVF of lactating female with doxycycline pre-labeling. (I) Quantification of the percentage of LacZ positive adipocytes in the total adipocytes. n = 3 images, Virgin female (no dox), Virgin female (chase), male (chase) and Lactating female (no dox); n = 7 images, Lactating female (chase). **, P < 0.01 compared to Virgin female (chase) or male (chase). This experiment is representative of three independent experiments.
De-differentiated Mammary Adipocytes Proliferate In Vivo upon Weaning
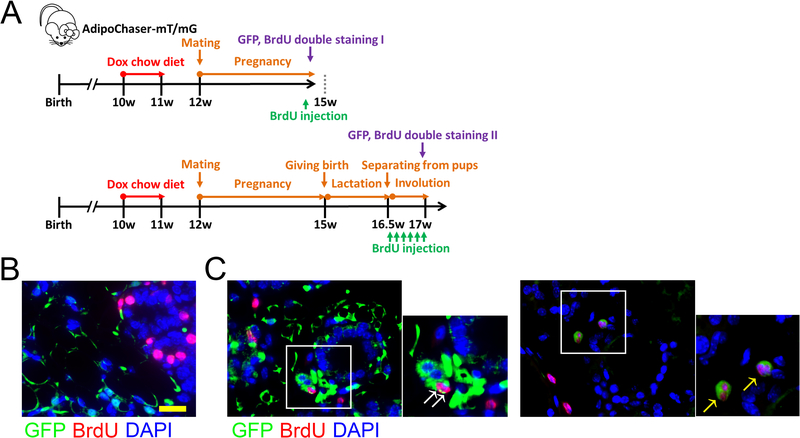
In order to determine whether these de-differentiated adipocytes can proliferate prior to re-differentiation in vivo, we switched to an AdipoChaser-mT/mG system we reported recently (Vishvanath et al., 2016; Ye et al., 2015) for immunofluorescence co-labeling. We first pre-labeled the mature adipocyte as GFP positive cells with doxycycline-containing chow diet, and thereafter examined these cells during pregnancy and lactation for proliferation by double staining the cells with GFP and BrdU (Figure 3A). During day 20 of pregnancy, as the mammary epithelium expands, BrdU positive cells were located in the developing alveolar structures (Figure 3B), and there was no overlap between BrdU positive mammary epithelial cells and GFP positive adipocytes. We then removed the pups after 10 days of lactation to initiate mammary gland involution. Three days after the separation of pups, BrdU positive cells were found in the regenerating adipocyte clusters, and these cells co-labeled with GFP (Figure 3C). Some of the co-labeled cells are captured as actively dividing cells, and adipocytes are not yet generated around these cells (Figure 3C). This result indicates that these proliferating cells correspond to the cellular pool of de-differentiated adipocytes. Thus, at least some of these de-differentiated adipocytes have the potential to proliferate prior to adipogenesis at this stage.
Figure 3. In vivo detection of proliferating de-differentiated adipocytes in the mammary gland upon weaning.
(A) Experimental design: 10-week old AdipoChaser-mT/mG female mice were put on a doxycycline chow diet for 7 days to ensure that Tomato expression is turned on in all mature adipocytes. After doxycycline treatment, mice were switched back to regular chow diet for a week to ensure washout of doxycycline. Thereafter, mice were bred with male mice to undergo pregnancy and lactation. BrdU injection was performed at the indicated time points. (B-C) Representative Immunofluorescence staining shows GFP (green), BrdU (red), and DAPI (blue) in the mammary gland during pregnancy (day 20) (B) and involution (pups removed from lactating female on lactation day 10, 3 days after weaning) (C). White arrow: co-localization of GFP and BrdU in the same cell. Yellow arrow: dividing cell with GFP and BrdU co-localization. Scale bar: 50 μm. n = 3 mice per group. This experiment is representative of 4 independent experiments.
De-differentiated Mammary Adipocytes Lose Mature Adipocyte Characteristics and Gain Preadipocyte Features
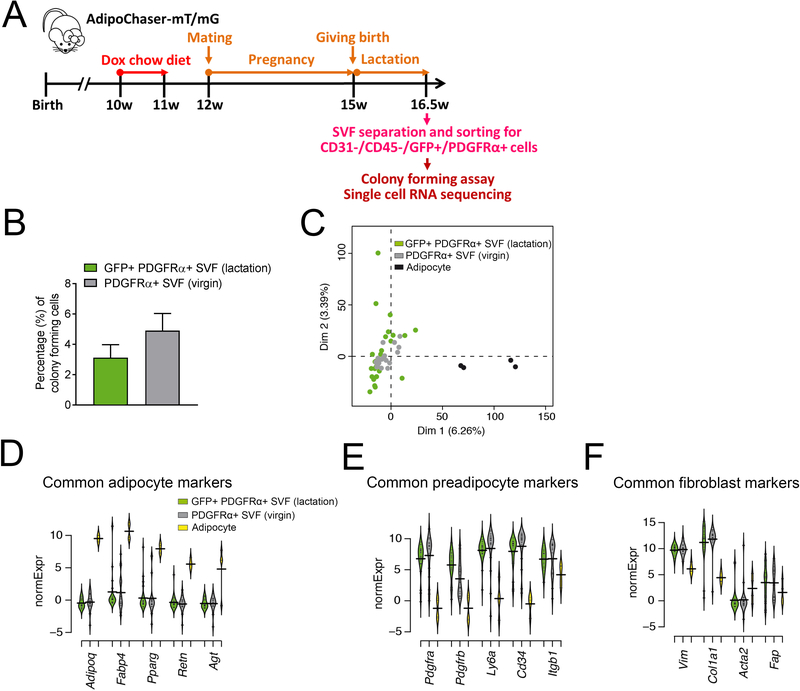
In order to determine the molecular signature of these de-differentiated adipocytes in the mammary gland, we compared these cells with classical adipocytes. Using the AdipoChaser-mT/mG system, we pre-labeled mature adipocytes with GFP expression to characterize the features of these de-differentiated adipocytes (Figure 4A), and then purified CD31−/CD45−/PDGFRα+/GFP− and CD31−/CD45−/PDGFRα+/GFP+ cells from the SVF of lactating mammary gland at the peak of lactation through FACS (Supplemental. Figure S1A). Colony forming assay showed that similar to the CD31−/CD45−/PDGFRα+ cells of WT virgin mice, which represent the classical preadipocyte pool, the CD31−/CD45−/PDGFRα+/GFP+ cells have the ability to form colonies (Figure 4B). Gene expression analyses showed the CD31−/CD45−/PDGFRα+/GFP+ cells were indeed enriched with GFP expression, while the CD31−/CD45−/PDGFRα+/Tomato+ cells were enriched with Tomato expression (Supplemental. Figure S1B, C). We then collected CD31−/CD45−/PDGFRα+/GFP+ cells as single-cells for subsequent single-cell RNA-sequencing analysis (Figure 4C–F, Supplemental. Figure S2A–G). After the flow sorting and single-cell RNA amplification, 26 CD31−/CD45−/PDGFRα+/GFP+ cells passed the quality control, and these cells were used for single-cell RNA-sequencing analysis. Adipocytes differentiated from the immortalized murine-derived brown preadipocyte cell line were used as mature adipocyte controls (Pradhan et al., 2017). It remains a major technical challenge to subject mature primary white adipocytes to flow cytometry and single cell sequencing. We therefore resorted to the slightly smaller, more manageable brown adipocyte cell line. While not perfect as a comparator, they serve as an excellent technical control for these experiments. Additionally, we also included population RNA-seq experiments, including three mature white adipocyte samples, two GFP positive, and six GFP negative ones. The RNA-sequencing data revealed that GFP+ de-differentiated adipocytes were transcriptionally highly distinct from the mature fat cells and highly similar to CD31−/CD45−/PDGFRα+ cells from the SVFs of WT virgin mice (termed as “GFP-”) (Figure 4C, Supplemental. Figure S2C, Table S1, 2). Strikingly, the GFP+ de-differentiated adipocytes have very low expression of the common adipocyte markers adiponectin (Adipoq), fatty acid binding protein 4 (Fabp4), peroxisome proliferator activated receptor gamma γ (Pparg), resistin (Retn) and angiotensinogen (Agt), similarly to GFP- cells (Figure 4D). Meanwhile, these GFP+ cells have regained expression of common preadipocyte markers, such as Pdgfra, Pdgfrb, lymphocyte antigen 6 complex, locus A (Ly6a), Cd34, and fibronectin receptor beta subunit (Itgb1) (Figure 4E) (Hepler et al., 2017; Wang and Seale, 2016). These de-differentiated adipocytes also increased expressions of some common fibroblast markers, such as Vimentin (Vim), Collagen Type I Alpha 1 Chain (Col1a1), Smooth Muscle α Actin (Acta2), and fibroblast activation protein (Fap), again, similarly to GFP- cells (Figure 4F). Based on the data shown in Supplemental Tables S1, 2, genes significantly lower expressed in GFP+ cells enriched for fat cell-related terms, such as “brown fat cell differentiation”, “triglyceride metabolic processes”, and “positive regulation of lipid storage”. The observed similarity between GFP+ and GFP- cells, in particular when it comes to low adipogenic and high pre-adipogenic gene expression were also confirmed in the population data, where comparisons to mature white adipocytes were performed (Figure S2C–G). These results indicate that these pre-existing mammary adipocytes have lost mature adipocyte features during de-differentiation, and they have regained preadipocyte features when they transform into the fibroblast-like cells.
Figure 4. Characterization of de-differentiated adipocyte in the lactating mammary gland by single-cell RNA-seq.
(A) Experimental design: 10-week old AdipoChaser-mT/mG female mice were put on a doxycycline chow diet for 7 days to ensure that GFP expression is turned on in all mature mammary adipocytes. After doxycycline treatment, mice were switched back to regular chow diet for a week to ensure the washout of doxycycline. For chasing mature mammary adipocyte in lactating female, mice were bred with male mice to undergo pregnancy and lactation. When the mice had been lactating for 10 days, the mammary glands were used for SVF separation. The separated SVF was then flow-sorted for CD31−/CD45−/PDGFRα+/GFP+ cells for single-cell RNA sequencing analysis. (B) Quantification of colony-forming unit of FACS purified CD31−/CD45−/PDGFRα+/GFP+ from the mammary gland SVFs of lactating female, comparing to the classic preadipocyte population (CD31−/CD45−/PDGFRα+ cells) from the mammary gland SVFs of virgin female. (D-G) Gene expressions of these de-differentiated mammary adipocytes (CD31−/CD45−/PDGFRα+/GFP+ cells) from the mammary gland SVFs of lactating female were compared to the classic preadipocyte population (CD31−/CD45−/PDGFRα+ cells) from the mammary gland SVFs of virgin female and an adipocyte cell line through single-cell RNA-seq. (C) Overview of single-cell grouping by principal component analysis. (D-F) Beanplots showing the distribution of normalized expression values (normExpression) across cells that belong to one of these three categories of common adipocyte markers (D), common preadipocyte markers (E) and common fibroblast markers (F). n = 26 cells for CD31−/CD45−/PDGFRα+/GFP+ SVF cells from lactating female, n = 20 cells for CD31−/CD45−/PDGFRα+ SVF cells from virgin female, n = 4 cells for brown adipocyte cell line.
DISCUSSION
In summary, we describe a new phenomenon whereby adipocytes in the mammary gland undergo de-differentiation into fibroblast-like cells and reside between the alveolar structures during pregnancy and lactation. Upon cessation of lactation, these de-differentiated cells can proliferate and give rise to mature adipocytes. This can repeatedly happen with the same cells through at least two pregnancies. This is surprising in light of the current models that paint the adipocyte to be a terminally differentiated cell whose fate is continued survival as an adipocyte or the only alternative being necrotic or apoptotic cell death. The results presented here offer critical insights towards our understanding of how adipose tissue regresses and regenerates during a female reproductive cycle. It will be interesting to determine the underlying mechanisms that control this process of adipocyte de-differentiation and re-differentiation during this dramatic tissue remolding event. The hormones involved during pregnancy and lactation are highly likely the key regulators of this process, such as estrogen, progesterone and prolactin. Meanwhile, for the de-differentiated mammary adipocytes, besides their ability to undergo adipogenesis, it will be interesting to see whether they have gained pluripotency and can be converted to other cell types, such as osteoblasts or myocytes. It will also be interesting to determine whether this process is restricted to the mammary gland and lactation, or whether this occurs at a low frequency even under normal physiological conditions as well. The fact that we can find such fibroblastic cell types at very low levels even in the absence of pregnancy and lactation would hint at a broader significance of the phenomenon, suggesting that the differentiation/de-differentiation cycle may be part of an ongoing process at all times. A recent report shows that adiponectin-positive intradermal progenitors give rise to dermal myofibroblasts (Marangoni et al., 2015). We do not know whether metabolic challenges, such as high fat diet feeding, impose an impairment of adipose tissue de-differentiation and re-differentiation, and whether this impairment would have a critical impact on the function of the mammary gland and/or other fat pads. At the same time, we cannot formally rule out that there are some adipocytes that undergo apoptosis during pregnancy and lactation, since the number of LacZ positive “empty adipocytes” at rest seemed to be fewer compared to the number of LacZ positive adipocytes prior pregnancy. However, it is clear that all emerging adipocytes derive from previously adiponectin-positive cells. Importantly, we conclude that none of the adipocytes contribute towards the alveolar structures, as we failed to detect any lacZ label within the milk-producing lobules. Overall, this reflects a remarkable plasticity within the mature adipocyte population, events that may also take place in the skin during sclerosis as well as during the stromal infiltration of cancer cells into the fatty stroma of the mammary gland.
LIMITATIONS OF STUDY
There are some limitations to this study. We have characterized the basic gene expression of the de-differentiated adipocytes and clearly seen a characteristic mRNA fingerprint of preadipocytes. However, we do not know whether these precursors would have the potential to differentiate into other cell types if given an appropriate stimulus. Other critical gaps are the unknown nature of the signal(s) that drive the de-differentiation process initially, as well as the signal(s) active during involution that lead to a rapid recruitment of the precursor cells. How are these precursors selected preferentially over other “virgin” mural cell that have never undergone a previous differentiation process? Future studies will need to address these questions.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the Lead Contact, Philipp E. Scherer (Philipp.Scherer@utsouthwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were maintained in a 12 h dark/light cycle and housed in groups of three to five with unlimited access to water and food (chow, number 5058, lab diet; doxycycline chow diet (600 mg kg−1), S4107, Bio-Serv). The Institutional Animal Care and Use Committees of the University of Texas Southwestern Medical Center, Dallas, and City of Hope, Duarte, have approved all animal experiments. All mice were on C57BL/6J background. Adn-rtTA mouse was generated as previously described (Sun et al., 2012). TRE-Cre, Rosa26-loxP-STOP-loxP-LacZ and Rosa26-loxP-STOP-loxP-mT/mG mouse lines were obtained from the Jackson Laboratories. For lineage tracing studies, Adn-rtTA mice were intercrossed to the TRE-Cre and Rosa26-loxP-STOP-loxP-LacZ mice, or Rosa26-loxP-STOP-loxP-mT/mG mice, we refer to these triple transgenic mice as AdipoChaser-LacZ or AdipoChaser-mT/mG mice. Female AdipoChaser-LacZ or AdipoChaser-mT/mG mice were used in all experiments; male AdipoChaser-LacZ mice were used for SVF isolation as a control in Figure 2. All experiments were started when mice are 10 weeks old, mice were kept in room temperature at all time (24 °C).
SVF Culture and Adipocyte Differentiation
SVF fractions from the mammary gland were obtained from female mice. Briefly, dissected mammary glands were digested for 1 hour at 37 degrees in PBS containing 10 mM CaCl2, 2.4 units/mL Dispase II (Roche Diagnostics Corporation, Indianapolis, IN) and 1.5 units/mL collagenase D (Roche Diagnostics Corporation, Indianapolis, IN). Digested cell/tissue mixture was filtered through a 100 μm cell strainer to remove undigested tissues. The flow-through cells were centrifuged for 5 minutes at 600xg at 4ºC. The cell pellets were resuspended in complete SVF culture medium (DMEM/F12 Invitrogen, Carlsbad, CA; plus Glutamax, Pen/Strep, and 10% FBS) and then filtered through a 40 μM cell strainer to remove clumps and large adipocytes. The flow-through was centrifuged for 5 minutes at 600 g at 4ºC, SVF pellet was then resuspended in complete SVF culture medium and plated onto a 6-well tissue culture plate, maintained at 37 °C humidified atmosphere with 10% CO2. For adipocyte differentiation, once confluence was reached, cells were exposed to the adipogenic cocktail containing dexamethasone (1 μM), insulin (5 μg/ml), isobutylmethylxanthine (0.5 mM) (DMI) and rosiglitazone (1 μM) in complete SVF culture medium. Forty-eight hours after induction, cells were maintained in SVF culture medium containing insulin (5 μg/ml) and rosiglitazone (1 μM) until LacZ staining.
METHOD DETAILS
β-gal staining of adipose tissue or in vitro differentiated adipocytes
For adipose tissue, mice were anesthetized and perfused with 0.2% glutaraldehyde in PBS. Fat depots of interest were removed and chopped into small pieces. The tissue pieces were washed with rinse buffer (100 mM sodium phosphate, 2 mM MgCl2, 0.01% Sodium deoxycholate and 0.02% NP-40) for three times and then soaked in β-gal staining buffer (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide and 1 mg ml−1 β-gal substrate in rinse buffer) for 48 hours at room temperature with mild shaking. Upon developing the β-gal stain, the tissue pieces were moved into 10% formalin overnight for post-fixation. The post-fixed tissue was processed with a standard paraffin tissue embedding protocol to produce sections. Following paraffin embedding and sectioning, tissues were counterstained with nuclear fast red. For in vitro differentiated adipocytes, cells were fixed by 0.2% glutaraldehyde in PBS for 2 hours, followed by three washings by the rinse buffer. Then cells were incubated with β-gal staining buffer for 24 hours before taking images. For both adipose tissue and in vitro differentiated adipocytes, the staining was imaged using a Keyence microscope.
Immunofluorescence staining
Formalin-fixed, paraffin-embedded sections from the mammary gland were dewaxed in xylene and hydrated in a graded series of ethanol to ddH2O. Slides were placed in 10mM sodium citrate buffer and boiled for 30 minutes, then blocked in PBST with 5% BSA. Primary antibody used was BrdU 1:200 (Novus) and GFP 1:500 (Abcam); secondary antibodies (1:200 dilution) used were Cy3 Goat Anti-Rat IgG H&L 1:200 (Abcam) and Alexa Fluor 488 Goat anti-Chicken IgY H&L 1:200 (Invitrogen). Slides were counterstained with DAPI and mounted with prolong Anti-Fade mounting medium (Invitrogen). Images were acquired using a Keyence microscope.
Flow cytometry
Samples of SVF cells from the mammary glands (subcutaneous adipose tissue) were first incubated on ice for 20 minutes in 200 uL of 2% FBS/PBS containing anti-mouse CD16/CD32 Fc Block (clone 2.4G2, BD) (1:200). Cells were then incubated with primary antibody (anti-CD31, 1:200, anti-CD45, 1:200, anti-CD140a, 1:100, all from BioLegend) and were incubated rotating at 4°C for 30 minutes. They were then washed three times with 2% FBS/PBS, and either analyzed using a FACSCantoIITM flow cytometer or sorted by a FACSAriaTM flow cytometer (UT Southwestern Medical Center Flow Cytometry Core Facility). For sorting, cells were initially selected by size, on the basis of forward scatter (FSC) and side scatter (SSC). Live cells were then gated on both SSC and FSC Width singlets, ensuring that individual cells were analyzed. SVF cells isolated from wild-type mice, along with fluorescent-minus-one (FMO) controls, were used to determine background fluorescence levels. Cells were sorted into FBS and then cultured in a 24-well dish. All flow cytometry antibodies were rat-derived and conjugated to APC or PE.
Single cell colony assay
The colony-forming unit assay was performed as previously described (Vishvanath et al., 2016). Briefly, sorted cells (450 cells/10cm plate) were seeded in growth media containing 2% FBS for 2–3 weeks at 10% CO2. Cells were fixed for 5 minutes in 10% Formalin, washed twice with PBS and then stained with 0.05% Crystal Violet for 30 minutes. After staining, the cells were washed with ddH2O three times and air dried. The total number of colonies was counted under light microscopy. Four replicate plates were quantified for each population.
Single-cell and population RNA-seq
The single-cell RNA-seq was performed using the Smart-seq2 method (Picelli et al., 2013), with several modifications (Chen et al., 2017). Briefly, single cells were sorted by FACS into 96-well PCR plate with lysis buffer (2 ul of 0.2% TritonX-100, 1 ul of dNTP (10 uM each), 1 ul of 10 uM biotin-oligo-dT30VN oligo, 0.1 ul of 40U/ul RNase Inhibitor (Clontech), 0.1 ul of ERCC RNA Spike-In Mix (1:106 diluted, ThermoFisher)). The single-cell lysates were then incubated at 72 °C for 3 min and put on ice immediately. RT mix (0.06 ul of 1M MgCl2, 2 ul of 2M Betaine, 0.1 ul of Biotin-TSO-LNA oligo, 0.25 ul of RNase Inhibitor, 2 ul of 5* Maxima H- RT buffer, 0.1 ul of Maxima H Minus Reverse Transcriptase (ThermoFisher), 1.39 ul of H2O) was added to each well, and the samples were subjected to reverse transcription. The cDNAs were subsequently amplified with PCR mix (12.5 ul 2* KAPA HiFi HotStart ReadyMix (KAPA), 0.25 ul of 10 uM Biotin-ISPCR oligo, 2.25 ul of H2O). The amplified cDNAs were then purified, tagmentated by in-house Tn5 (Picelli et al., 2014) and subsequently amplified using Nextera i5 and i7 Adapters (Illumina). The yielding libraries are purified, pooled and subjected to sequencing in Nextseq-500 (Illumina).
All the oligos are synthesized and HPLC purified in IDT, with sequence listed below:
biotin-oligo-dT30VN: /5Biosg/AAGCAGTGGTATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN
biotin-TSO-LNA:
/5Biosg/AAGCAGTGGTATCAACGCAGAGTACATrGrG+G
biotin-ISPCR:
/5Biosg/AAGCAGTGGTATCAACGCAGAGT
One biological replicate contained single GFP+ cells and IBA cells as controls, the second biological replicate contained GFP+ cells, GFP- cells and IBA cells as controls. Negative controls with no cells were included in both replicates, and the samples with cDNA yield lower than or equal to negative controls were filtered.
The population RNA-seq samples were processed in an analogous way, but with a higher number of cells per well. Each GFP+ and GFP- sample, respectively, contained 50 cells, while the control white adipose tissue samples contained 1 ng purified total RNA.
Single-cell and population RNA-seq data analysis
FastQ files containing paired-end sequenced tags (reads) each cell were trimmed and filtered using prinseq 0.20.3 (Schmieder and Edwards, 2011) with the parameters “-custom_params ‘A 70%;T 70%;G 70%;C 70%’ -trim_tail_left 36 -trim_tail_right 36 -lc_method dust -lc_threshold 45 -min_gc 1 -out_format 3” and cutadapt 1.5 (Martin, 2011) with the parameters “-q 15 --stringency 3 -e 0.05 --length 36”. The retained tags were evaluated using FastQC v0.11.2 and aligned to the Ensembl 84 gene annotation of the NCBI38/mm10 mouse genome using STAR 2.4.0g (Dobin et al., 2013) with the parameters “--runThreadN 4 --outReadsUnmapped Fastx --runMode alignReads --genomeDir $genomedir --outFilterScoreMinOverLread 0.20 --outFilterMatchNminOverLread 0.20 --outFilterMultimapNmax 1 --outSAMtype BAM Unsorted SortedByCoordinate”. Number of tags per gene were calculated using htseq-count 0.6.0 (Anders et al., 2015) with the parameters “htseq-count -m intersection-nonempty -s no -a 10 -t exon -i gene_id”. For each gene, expression estimates per gene were expressed as log-transformed counts per million (log_cpm), by dividing total tags per gene by the total number of gene-aligned reads per cell and taking the log(x+1) of this value. Genes with a count per million (cpm) greater than 2 at least 2 samples were retained and raw counts were then normalized using mean-variance modeling at the observational level, as implemented in the voom function in limma_3.30.4 (Ritchie et al., 2015), and additionally ComBat to correct for batch effects between the two biological replicates in the single-cell samples (Johnson et al., 2007). Differential expression was computed on the normalized values using the limma_3.30.4 pipeline at an FDR of 0.05 for the single-cell experiments and 0.1 for the population ones, as well as a fold-change cutoff of 2. Heatmaps displaying row-normalized (z-score transformation) expression (log_cpm) values were generated using pheatmap_1.0.8 and the parameters “clustering_distance_rows = “correlation”, clustering_distance_cols = “correlation”, clustering_method = “average”, scale = “row”. Hierarchical clustering was performed using pvclust 2.0 and the parameters method.hclust=“ward.D2”, method.dist=“correlation”, nboot=100.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as means ± s.e.m.. Differences were analyzed by unpaired two-tailed Student’s t-test between two groups using Excel. No method was used to determine whether the data met assumptions of Student’s t-test. For single-cell and population RNA-seq data analysis, statistical analysis can be found in the STAR METHODS. The statistical parameters (i.e. the exact n numbers, p values, and numbers of biological repeats) can be found in the Figure Legends.
DATA AND SOFTWARE AVAILABILITY
The entire RNA-sequencing raw data, including single-cell analysis and population analysis are provided as supplemental tables. The data set is also uploaded to ArrayExpress (E-MTAB-6818) (https://www.ebi.ac.uk/arrayexpress/).
Supplementary Material
ACKNOWLEDGEMENTS
The authors were supported by US National Institutes of Health grants R01DK55758, R01DK099110, P01DK088761 and P01AG051459 (P.E.S.) and K01DK107788, R03HD095414 (Q.A.W.) as well as by Cancer Prevention Research Institute of Texas grant RP140412 (P.E.S.). PES was also supported by an unrestricted grant from the Novo Nordisk Research Foundation.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information includes two Supplemental Figures, and two Supplemental Tables, can be found with this article online.
Supplemental Figure 1. FACS sorting of the de-differentiated mammary adipocytes. Related to Figure 4.
Supplemental Figure 2. Sample clustering of the GFP positive single-cells for RNA-sequencing analysis. Related to Figure 4.
Table S1. Single-cell RNA-seq raw data. Related to Figure 4
Table S2. Population RNA-seq raw data. Related to Figure 4
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R, Jeffery E, and Rodeheffer, Matthew S (2014). Weighing in on Adipocyte Precursors. Cell Metabolism 19, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R, and Rodeheffer MS (2013). Characterization of the adipocyte cellular lineage in vivo. Nature cell biology 15, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gardeux V, Meireles-Filho A, and Deplancke B (2017). Profiling of Single-Cell Transcriptomes. Current protocols in mouse biology 7, 145–175. [DOI] [PubMed] [Google Scholar]
- Cinti S (2007). The adipose organ In Adipose tissue and adipokines in health and disease (Springer; ), pp. 3–19. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, and Eaves CJ (2008). A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med 14, 1384–1389. [DOI] [PubMed] [Google Scholar]
- Giordano A, Smorlesi A, Frontini A, Barbatelli G, and Cinti S (2014). MECHANISMS IN ENDOCRINOLOGY: White, brown and pink adipocytes: the extraordinary plasticity of the adipose organ. European Journal of Endocrinology 170, R159–R171. [DOI] [PubMed] [Google Scholar]
- Hepler C, Vishvanath L, and Gupta RK (2017). Sorting out adipocyte precursors and their role in physiology and disease. Genes & development 31, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovey RC, and Aimo L (2010). Diverse and active roles for adipocytes during mammary gland growth and function. Journal of mammary gland biology and neoplasia 15, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovey RC, Goldhar AS, Baffi J, and Vonderhaar BK (2001). Transcriptional regulation of vascular endothelial growth factor expression in epithelial and stromal cells during mouse mammary gland development. Molecular Endocrinology 15, 819–831. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, and Rabinovic A (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 8, 118–127. [DOI] [PubMed] [Google Scholar]
- Landskroner-Eiger S, Park J, Israel D, Pollard JW, and Scherer PE (2010). Morphogenesis of the developing mammary gland: stage-dependent impact of adipocytes. Developmental biology 344, 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liu X, Robinson G, Bar-Peled U, Wagner KU, Young WS, Hennighausen L, and Furth PA (1997). Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution. Proc Natl Acad Sci U S A 94, 3425–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund LR, Romer J, Thomasset N, Solberg H, Pyke C, Bissell MJ, Dano K, and Werb Z (1996). Two distinct phases of apoptosis in mammary gland involution: proteinase-independent and -dependent pathways. Development 122, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni RG, Korman BD, Wei J, Wood TA, Graham LV, Whitfield ML, Scherer PE, Tourtellotte WG, and Varga J (2015). Myofibroblasts in murine cutaneous fibrosis originate from adiponectin-positive intradermal progenitors. Arthritis & rheumatology (Hoboken, N.J.) 67, 1062–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal 17, pp. 10–12. [Google Scholar]
- Morroni M, Giordano A, Zingaretti MC, Boiani R, De Matteis R, Kahn BB, Nisoli E, Tonello C, Pisoschi C, and Luchetti MM (2004). Reversible transdifferentiation of secretory epithelial cells into adipocytes in the mammary gland. Proceedings of the National Academy of Sciences of the United States of America 101, 16801–16806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovich AL, Manivannan S, and Nelson CM (2010). Adipose stroma induces branching morphogenesis of engineered epithelial tubules. Tissue Engineering Part A 16, 3719–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, and Sandberg R (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Meth 10, 1096–1098. [DOI] [PubMed] [Google Scholar]
- Picelli S, Björklund ÅK, Reinius B, Sagasser S, Winberg G, and Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Research 24, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan RN, Bues JJ, Gardeux V, Schwalie PC, Alpern D, Chen W, Russeil J, Raghav SK, and Deplancke B (2017). Dissecting the brown adipogenic regulatory network using integrative genomics. Scientific reports 7, 42130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillema JA (1994). Development of the mammary gland and lactation. Trends in Endocrinology & Metabolism 5, 149–154. [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, and Visvader JE (2014). In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322–327. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, and Edwards R (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics (Oxford, England) 27, 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M-L, Wu L, Lindeman GJ, and Visvader JE (2006). Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88. [DOI] [PubMed] [Google Scholar]
- Sun K, Asterholm IW, Kusminski CM, Bueno AC, Wang ZV, Pollard JW, Brekken RA, and Scherer PE (2012). Dichotomous effects of VEGF-A on adipose tissue dysfunction. Proceedings of the National Academy of Sciences 109, 5874–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede B, and Kang Y (2011). From milk to malignancy: the role of mammary stem cells in development, pregnancy and breast cancer. Cell research 21, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishvanath L, MacPherson KA, Hepler C, Wang QA, Shao M, Spurgin SB, Wang MY, Kusminski CM, Morley TS, and Gupta RK (2016). Pdgfrbeta+ Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell Metab 23, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang X-O, Yang L, and Zeng YA (2015). Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517, 81–84. [DOI] [PubMed] [Google Scholar]
- Wang QA, Tao C, Gupta RK, and Scherer PE (2013). Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med 19, 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, and Seale P (2016). Control of brown and beige fat development. Nat Rev Mol Cell Biol 17, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Wang QA, Tao C, Vishvanath L, Shao M, McDonald JG, Gupta RK, and Scherer PE (2015). Impact of tamoxifen on adipocyte lineage tracing: Inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Molecular metabolism 4, 771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The entire RNA-sequencing raw data, including single-cell analysis and population analysis are provided as supplemental tables. The data set is also uploaded to ArrayExpress (E-MTAB-6818) (https://www.ebi.ac.uk/arrayexpress/).