Abstract
Objective(s):
NOTCH signaling pathway is well known for its role in cell fate, cell survival, cell differentiation, and apoptosis. Some of the NOTCH signaling genes are critical for endometrial function and implantation in animals and appear to play a similar role in humans. The purpose of the current study was to investigate the potential roles of some main components of the NOTCH family in human endometrium during implantation period in common gynecological diseases.
Materials and Methods:
Endometrial NOTCH receptors NOTCH1, 3, 4 and ligand JAG1, 2 and survivin mRNA expression were investigated using the Q-PCR technique and the amount of the JAG1, 2 proteins was also determined by Western blot. Samples were obtained from 12 patients with endometriosis, 12 patients with repeated implantation failure (RIF), 12 patients with Polycystic Ovary Syndrome (PCOS) and 10 healthy fertile women as a control group. Data were analyzed using SPSS version 18. Group comparisons were performed by one-way ANOVA or Kruskal-Wallis.
Results:
All patient groups failed to show the expected mid-luteal increase in NOTCH1, JAG 1, 2, and survivin expression as documented in the control group. Moreover, a significant rise in NOTCH3 expression levels was found only in PCOS women. There was a direct correlation between gene expression and protein level for JAG 1, 2.
Conclusion:
Aberrant NOTCH signaling molecules expression suggests that altered development of the endometrium at the molecular level may be associated with the impaired decidualization and implantation failure in gynecological disorders such as endometriosis, PCOS, and RIF.
Key Words: Endometriosis, NOTCH signaling, PCOS, Repeated implantation-failure (RIF), Window of implantation
Introduction
Implantation is a key part of early pregnancy in which blastocyst adheres and penetrates into the wall of the receptive endometrium. A complex sequence of cellular and molecular changes should take place at the maternal endometrium in order to permit the embryo to invade the decidua (1). Until recently, the exact factors mediating the endometrial receptivity were not fully understood. Among these, NOTCH signaling genes are supposed to be important factors in the endometrial function (2).
NOTCH signaling is an evolutionarily conserved pathway, which regulates cell proliferation, cell fate determination, cell invasion, differentiation, and apoptosis (3, 4). Each of these processes absolutely exhibits an important role in endometrial remodeling. In mammals, NOTCH signaling family includes four NOTCH receptors (NOTCH1-4), five ligands, three Delta-like proteins (DLL1,3,4) and two Jagged proteins (JAG1,2). Both the NOTCH receptors and ligands are single-pass transmembrane proteins, hence, NOTCH signaling is activated upon cell-to-cell contact (5). Receptor/ligand interaction at the cell surface, activates proteolytic cleavage of the receptor, leading to the release of intracellular NOTCH receptor domain, which enters the cell nucleus and induces the NOTCH target genes expression (6, 7), including survivin (8).
There are only a few reports concerning the expression pattern and role of the NOTCH pathway in the human endometrium. In physiological conditions, significant increase in endometrial NOTCH1 and decrease in NOTCH4 during the luteal phase, suggests that NOTCH-4 likely contributes more to the controlling proliferation, while, NOTCH-1 more likely involves in the differentiation programming (2). In addition, an up-regulated expression of JAG1 and DLL4 as the NOTCH ligands localized in the endometrium have been reported during the mid-luteal phase, which implies their involvement in the endometrial receptivity(9).
It has been shown that endometrial remodeling, decidualization and embryo implantation could be modulated by the NOTCH signaling in mice. Afshar et. al. indicated a decrease in proliferation and up-regulation of apoptosis-associated genes via inhibition of NOTCH1 signaling during decidualization (10). PCOS (40%), endometriosis (50%), repeated implantation failure (10%) are three common causes of female infertility (11-13).
Women with PCOS, endometriosis, and RIF have been reported to exhibit an altered expression pattern of receptivity markers during the period of implantation (14-17). Therefore, the aim of this study was to evaluate the expression of the NOTCH receptors (NOTCH1, NOTCH2, NOTCH3), ligands (JAG1and JAG2) and target gene, and survivin, in the endometrium of patients with PCOS, endometriosis, repeated implantation failure (RIF) and healthy fertile women during the window of implantation.
Materials and Methods
Patients and tissue selection
This prospective study was performed, during a 12 months’ period from September 2015 to October 2016. Signed informed consent was taken from each participant and the study was approved by the local Ethics Committee at Iran University of Medical Sciences (IUMS, no 19950). Human endometrial tissue samples were obtained from PCOS women (n=12), endometriosis (n=12), RIF patients (n=12), and healthy fertile women in the secretory phase (on days 3–5 after ovulation, n=10). Patients with chronic oligo/anovulation (cycle length >35 days and < 6 months), hyperandrogenism, and polycystic ovaries were identified as PCOS (consistent with Rotterdam criteria). The timing of the LH surge, ultrasound scanning, and histological evaluation, confirmed that samples were harvested during the luteal phase. The mean age of the subjects was 26 years (range: 18–35). Women with any pathological uterine disorder and endometrial hyperplasia were not included. They also had not used hormonal medications or an intrauterine contraceptive device in the previous three months. Under sterile conditions, endometrial biopsies were collected using Pipelle catheters (Pipelle, Laboratoire CCD, France). Each endometrial sample was cut into two portions: for RNA and protein extraction, the first part was put inside a Cryo tube containing 1 ml RNA later (Ambion, Austin, TX, USA). Another part was fixed in formalin to perform histological dating.
RNA extraction, cDNA synthesis, and reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from endometrial tissue samples by homogenization in Trizol reagent (Sigma Pool, UK), following the manufacturer’s instructions. To remove genomic DNA contamination, RNA samples were purified with DNase I (Fermentas, Sankt Leon-Rot, Germany). RNA was reverse transcribed into cDNA using SuperScript First-Strand Synthesis System (200 U/μl, fermentase). Negative controls were prepared as non-reverse transcribed controls (RT controls). The RT-PCR was done using prepared cDNA, Platinum Blue PCR SuperMix (Invitrogen, Paisley, Scotland, UK), and the forward and reverse primers for NOTCH1-3, JAG1, JAG2 and survivin (Metabion, Martinsried, Bavaria, Germany). The used primer sequences are described in Table 1. The RT-PCR conditions for 40 cycles were as follows: 95 °C for 30 sec, 60 °C for 1 min, and 72 °C for 1 min. To separate PCR products, agarose gel (1.2%) electrophoresis and visualizing via an ultraviolet transillumination were carried out. The identity of the amplified product was confirmed by the sequencing of the RT-PCR products. β-actin was considered as a reference gene (18).
Table 1.
The primer sequences (5'-3') used in quantitative real-time polymerase chain reaction (qRT-PCR)
| Gene symbol | Forward primer | Reversed primer |
|---|---|---|
| β-actin | TGACCCAGATCATGTTTGAGACC | GGAGGAGCAATGATCTTGATCTTC |
| NOTCH1 | AGAAGATGCTCCAGCAACACA | GCAAGTCTCCTACAAAACACGG |
| NOTCH2 | AATGAGTGTCTGAGTGAACCCT | GACTCCATCAAATCCTGCCTG |
| NOTCH3 | CTCATGGTATCTGCACCAACCT | GGGTCACAGTCATTGATGTCCT |
| JAG1 | CTGTGGCTTGGATCTGTTGCT | CGTTGTTGGTGGTGTTGTCCT |
| JAG2 | CCTCTGCCTTGCTACAATGGTG | GCGATACCCGTTGATCTCATCC |
| Survivin | GATTTGATTCGCCCTCCTCCC | AACAGCCGAGATGACCTCCA |
Quantitative real-time polymerase chain reaction (qPCR)
All transcripts were quantified in triplicates using an ABI Prism 7300 apparatus (Applied Biosystems, Foster City, California, USA). The total reaction volume was 20 μl containing 250 ng cDNA, 5 pmol of each primer, and SYBR Green reagent (Applied Biosystems) with ROX dye as a passive control for signal intensity. Amplification was carried out at 95 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 30 sec. Melting curves of PCR reactions were used to check the formation of nonspecific products and primer-dimer. The efficiency of the primers was tested by the standard curves.
To analysis qPCR results, the comparative CT method was utilized. ΔCT was defined as the difference between the number of cycles required for amplification of the candidate gene and the human β-actin (reference gene selected). Then, ΔΔCT was calculated as the difference between the groups and 2-ΔΔCT was reported as mRNA fold change (FC) (18).
Western blotting
A 50 µg protein aliquot from each sample was separated on SDS-polyacrylamide gel and then blotted onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad, USA). The membranes were blocked with 5% BSA and 0.1% Tween-20 (Sigma-Aldrich, St. Louis, MO, USA). Then, the membranes were incubated with the primary anti-JAG1 antibody (1:100; Abcam ab7771, Cambridge, UK), anti-JAG2 antibody (1:100; Abcam ab109627, Cambridge, UK) and β-actin (1:200; Sigma-Aldrich, St. Louis, MO, USA) overnight at 4 °C. The blots were then washed with TBST, incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:200; Abcam, ab7086) for 1 hr at room temperature. After washing with TBST, immunodetection was achieved with the chemiluminescent peroxidase substrate (ECL Advance, GE Healthcare, UK) and blots were exposed to x-ray films (GE, 28906835). Consequently, the films were scanned with a densitometer (GS-800, Bio-Rad, USA). The signal intensity of JAG1 and JAG2 relative to the density of β-actin band was computed using the Image G software. As a negative control, the blots were treated with non-immunized serum as the primary antibody.
Statistical analysis
Data were analyzed using SPSS version 18 and are described as mean ± SD. Distribution of data was checked with the Kolmogorov-Smirnov test. Between groups comparisons were performed by one-way ANOVA for the data with normal distribution and by the Kruskal-Wallis for non-parametric data. Statistical significance was set at P<0.05.
Results
Expression of NOTCH receptors in the endometrium of women with gynecological diseases compared with the healthy fertile women
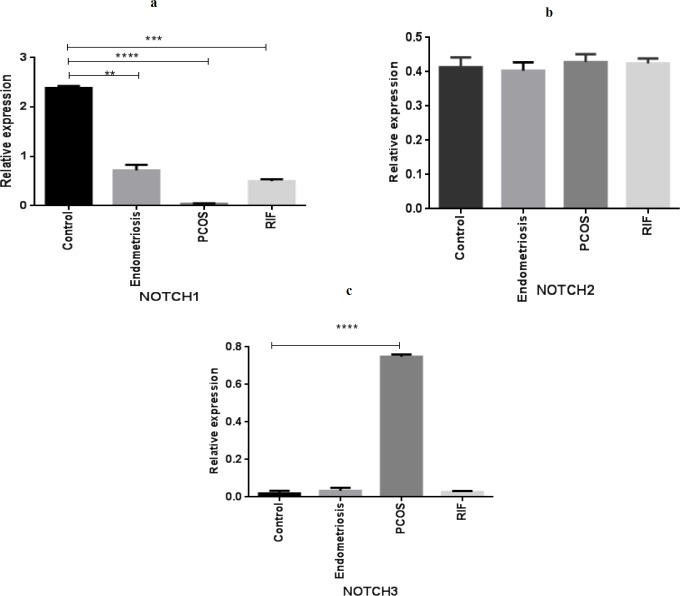
qPCR analysis indicated a significant decrease in NOTCH1 mRNA expression levels of women with PCOS, endometriosis and RIF groups compared with the healthy group during the implantation window (Figure 1). As shown in Figure 2, mRNA expression of NOTCH2 has no statistically significant difference between the women with different gynecological diseases and the control group (Figure 2). In addition, NOTCH3 was significantly up-regulated only in the PCOS group compared with the healthy fertile women (Figure 3).
Figure 1.
Gene expression pattern of NOTCH receptors (NOTCH1, NOTCH2, NOTCH3) in women with polycystic ovary syndrome(PCOS), endometriosis, and repeated implantation failure(RIF) groups compared with the control group (mean± SEM)
** represents significance at P<0.01, *** represents significance at P<0.001
****represents significance at P<0.0001
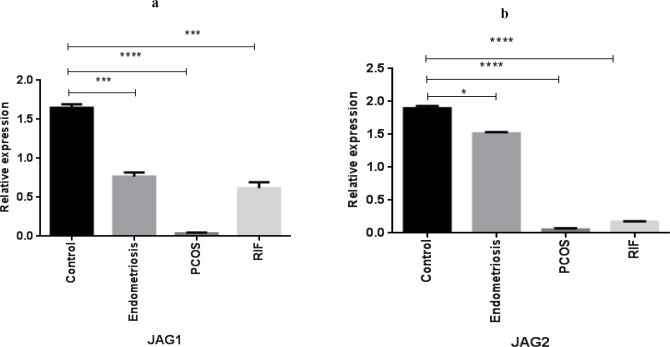
Figure 2.
Gene expression pattern of NOTCH ligands (JAG1, JAG2) in women with polycystic ovary syndrome (PCOS), endometriosis, and repeated implantation failure (RIF) groups compared with the control group (mean± SEM)
* represents significance at P<0.05, *** represents significance at P<0.001
****represents significance at P<0.0001
Figure 3.
Gene expression pattern of NOTCH target gene survivin in women with polycystic ovary syndrome (PCOS), endometriosis and repeated implantation failure (RIF) groups compared with the control group (mean±SEM)
** represents significance at P<0.01
Expression of NOTCH ligands in the endometrium of women with gynecological diseases compared with the healthy fertile women
A significantly lower expression of JAG1 was observed in PCOS (P<0.0001), endometriosis (P<0.001), and RIF (P<0.001) groups compared with the healthy fertile women (Figure 2, a). Furthermore, JAG2 gene expression was significantly down-regulated in the endometrium of PCOS (P<0.0001), endometriosis (P<0.05), and RIF (P<0.0001) groups compared with control subjects (Figure 2, b).
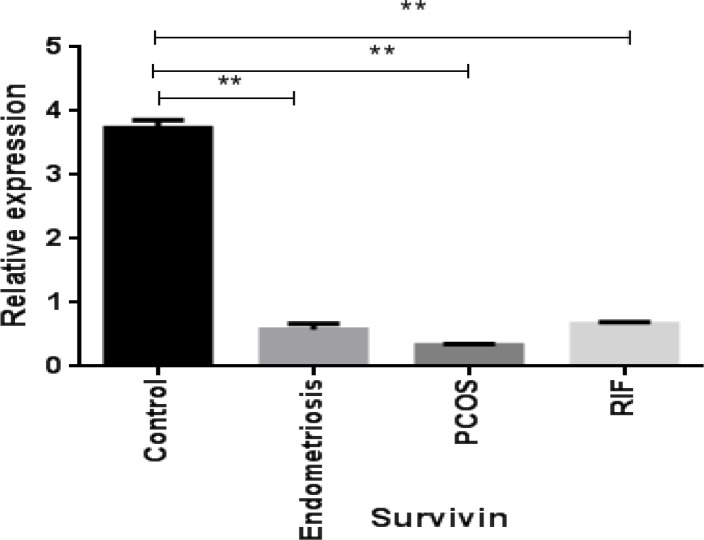
Expression of the NOTCH target gene, survivin, in the endometrium of women with gynecological diseases compared with the healthy fertile women
As shown in Figure 3, survivin gene expression was decreased in the endometrial samples of PCOS women, and endometriosis and RIF groups compared with healthy women.
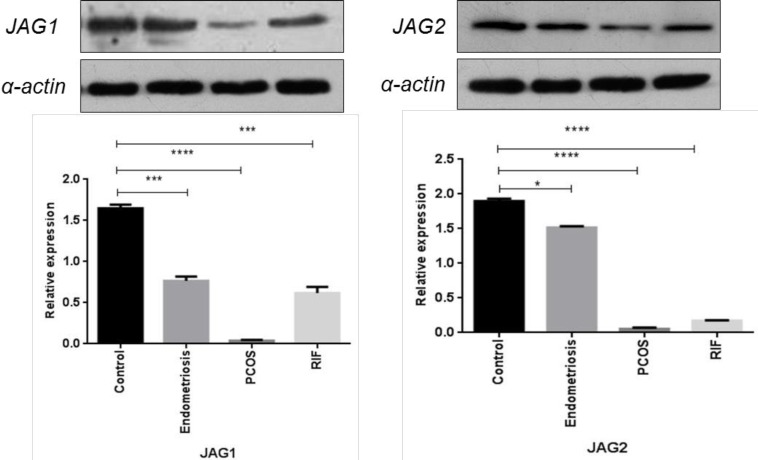
Immunoblotting analysis of NOTCH ligands in the endometrium of women with gynecological diseases compared with the healthy fertile women
Western blot analysis showed a significantly lower JAG1 and JAG2 protein expression in the endometrial samples of PCOS patients, and endometriosis and RIF groups compared with the healthy fertile ones (Figure 4).
Figure 4.
Protein expression pattern of NOTCH ligands (JAG1, JAG2) in women with polycystic ovary syndrome (PCOS), endometriosis, and repeated implantation failure(RIF) groups compared with the control group (mean±SEM)
* represents significance at P<0.05, *** represents significance at P<0.001
****represents significance at P<0.0001
Discussion
In the current study, the expression pattern of NOTCH signaling molecules as potential biomarkers was assessed in the endometrial samples taken during the mid-luteal phase of women with endometriosis, PCOS, and RIF. Our results showed that NOTCH pathway molecules including NOTCH1, 3, JAG1, 2, and survivin are differently expressed in the patient groups compared with healthy controls. A large body of evidence suggests that aberrant endometrial receptivity may attribute to the undesired reproductive outcomes in common gynecological disorders including PCOS, endometriosis, and RIF (19-22). Various biological molecules have been proposed to contribute to the impairment of decidualization and endometrial receptivity (23).
NOTCH pathway mediates cell differentiation, cell progression, and cell death and regulates angiogenesis (24-26). All of these processes are critical for successful implantation (1, 27). Su et. al. indicated that endometrial NOTCH molecules attenuate in patients with endometriosis. They suggested that the decreased NOTCH signaling contributes to the decidualization defects and therefore insufficient uterine receptivity (28).
Inconsistently, the current study demonstrated that NOTCH1, JAG-1, JAG-2, and survivin, as a down target molecule of NOTCH pathway, significantly decrease in women with PCOS, endometriosis, and RIF. It is documented that NOTCH1 is involved in decidual angiogenesis and endometrial differentiation, while NOTCH3 and 4 control the proliferation (2, 29). Animal studies have shown that NOTCH1 silencing leads to the decrease in IL-11 and IGFBP1 as two main human decidualization biomarkers (10). Moreover, NOTCH1 deficiency results in the reduction of bone morphogenetic protein2 (bmp2) and wnt4 in the mouse uterus, which are necessary for the decidualization (10). A recent study has revealed that NOTCH1 and NOTCH ligands including JAG1 and DLL1 are down-regulated in the endometrium of women with unexplained infertility during the implantation window compared with the fertile subjects (30). DLL1 changes integrin α6 and integrin β1 expression, which have essential roles in the embryo implantation (31). The expression of JAG1 was increased from proliferative into secretory phase, which proposes a responsibility for JAG1 in the endometrial receptivity (2, 30), and the endometrial expression of JAG2 was decreased in endometriosis, which is contributed to the decidualization failure (28).
Interestingly, we found NOTCH-3 increased in PCOS exclusively. Previous studies revealed that the NOTCH system plays a pivotal role in maintaining balance between cell proliferation and cell death (32). It has been shown that NOTCH3 increases endometrial cell proliferation, and NOTCH1 promotes differentiation (2, 29). It seems both underexpression of NOTCH1, as well as overexpression of NOTCH3, may have detrimental effects on uterine receptivity in PCOS patients. Also, increased NOTCH3 expression may be related to the elevated endometrial thickness and hyperplasia that is seen in women with PCOS.
Conclusion
In summary, our findings illustrate that dysregulated NOTCH signaling molecules during the window of implantation may be associated with implantation problems and consequently poor outcomes observed in endometriosis, PCOS, and RIF. However, our knowledge is still limited due to the complexity of this pathway. Hence, much remains to be discovered about the role of the NOTCH signaling pathway and its changes during the mid-luteal phase in normal and disease conditions.
Acknowledgment
The authors would like to thank the Shahid Akbarabadi Clinical Research Development Unit (Sh A CRDU), Iran University of Medical Sciences (IUMS), Tehran, Iran for sample collection. This study was financed by Iran University of Medical Science (grant no. 26493).
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- 1.Boeddeker SJ, Hess AP. The role of apoptosis in human embryo implantation. J Reprod Immunol. 2015;108:114–22. doi: 10.1016/j.jri.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Cobellis L, Caprio F, Trabucco E, Mastrogiacomo A, Coppola G, Manente L, et al. The pattern of expression of notch protein members in normal and pathological endometrium. J ANAT. 2008;213:464–72. doi: 10.1111/j.1469-7580.2008.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohishi K, Katayama N, Shiku H, Varnum-Finney B, Bernstein I, editors Notch signalling in hematopoiesis. Semin Cell Dev Biol. 2003;14:111–158. doi: 10.1016/s1084-9521(02)00183-0. [DOI] [PubMed] [Google Scholar]
- 4.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, et al. Activation of notch-1 signaling maintains the neoplastic phenotype in human ras-transformed cells. NAT MED. 2002;8:979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C-P, Yang J-L, Zhang J, Li L, Huang L, Ji S-Y, et al. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology. 2011;152:2437–47. doi: 10.1210/en.2010-1182. [DOI] [PubMed] [Google Scholar]
- 6.Sasnauskienė A, Jonušienė V, Krikštaponienė A, Butkytė S, Dabkevičienė D, Kanopienė D, et al. NOTCH1, NOTCH3, NOTCH4, and JAG2 protein levels in human endometrial cancer. Medicina. 2014;50:14–8. doi: 10.1016/j.medici.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Maillard I, Pear WS. Notch and cancer: best to avoid the ups and downs. Cancer cell . 2003;3:203–5. doi: 10.1016/s1535-6108(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 8.Altieri DC. New wirings in the survivin networks. Oncogene. 2008;27:6276–84. doi: 10.1038/onc.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsuhashi Y, Horiuchi A, Miyamoto T, Kashima H, Suzuki A, Shiozawa T. Prognostic significance of notch signalling molecules and their involvement in the invasiveness of endometrial carcinoma cells. Histopathology. 2012;60:826–37. doi: 10.1111/j.1365-2559.2011.04158.x. [DOI] [PubMed] [Google Scholar]
- 10.Afshar Y, Jeong J-W, Roqueiro D, DeMayo F, Lydon J, Radtke F, et al. Notch1 mediates uterine stromal differentiation and is critical for complete decidualization in the mouse. FASEB J. 2012;26:282–94. doi: 10.1096/fj.11-184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunselman G, Vermeulen N, Becker C, Calhaz-Jorge C, D’Hooghe T, De Bie B, et al. Eshre guideline: management of women with endometriosis. Hum Reprod. 2014;29:400–12. doi: 10.1093/humrep/det457. [DOI] [PubMed] [Google Scholar]
- 12.Baqai Z, Khanam M, Parveen S. Prevalence of pcos in infertile patients. Med Channel. 2010;16:609. [Google Scholar]
- 13.Somigliana E, Vigano P, Busnelli A, Paffoni A, Vegetti W, Vercellini P. Repeated implantation failure at the crossroad between statistics, clinics and over-diagnosis. Reprod Biomed Online. 2017;36:32–38. doi: 10.1016/j.rbmo.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Shi C, Han HJ, Fan LJ, Guan J, Zheng XB, Chen X, et al. Diverse endometrial mRNA signatures during the window of implantation in patients with repeated implantation failure. HUM FERTIL . 2017;21:183–194. doi: 10.1080/14647273.2017.1324180. [DOI] [PubMed] [Google Scholar]
- 15.Amjadi F, Aflatoonian R, Javanmard SH, Saifi B, Ashrafi M, Mehdizadeh M. Apolipoprotein A1 as a novel anti-implantation biomarker in polycystic ovary syndrome: a case-control study. J RES MED SCI . 2015;20:1039–1045. doi: 10.4103/1735-1995.172813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimitriadis E, Stoikos C, Stafford-Bell M, Clark I, Paiva P, Kovacs G, et al. Interleukin-11, IL-11 receptorα and leukemia inhibitory factor are dysregulated in endometrium of infertile women with endometriosis during the implantation window. J REPROD IMMUNOL. 2006;69:53–64. doi: 10.1016/j.jri.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Amjadi F, Mehdizadeh M, Ashrafi M, Nasrabadi D, Taleahmad S, Mirzaei M, et al. Distinct changes in the proteome profile of endometrial tissues in polycystic ovary syndrome compared with healthy fertile women. Reprod Biomed Online . 2018;37:184–200. doi: 10.1016/j.rbmo.2018.04.043. [DOI] [PubMed] [Google Scholar]
- 18.Taghavi SA, Ashrafi M, Mehdizadeh M, Karimian L, Joghataie MT, Aflatoonian R. Toll-like receptors expression in follicular cells of patients with poor ovarian response. Int J Fertil Steril . 2014;8:183–92. [PMC free article] [PubMed] [Google Scholar]
- 19.Giudice LC. Endometrium in pcos: implantation and predisposition to endocrine CA. BEST PRACT RES CL EN Journal . 2006;20:235–44. doi: 10.1016/j.beem.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Timeva T, Shterev A, Kyurkchiev S. Recurrent implantation failure: the role of the endometrium. J Reprod Infertil . 2014;15:173–183. [PMC free article] [PubMed] [Google Scholar]
- 21.Selam B, Arici A. Implantation defect in endometriosis: endometrium or peritoneal fluid. J Reprod Fertil Suppl. 2000;55:121–8. [PubMed] [Google Scholar]
- 22.Matteo M, Cicinelli E, Neri M, Carrubba R, Carpagnano F, Romeo F, et al. Pro-inflammatory M1/Th1 type immune network and increased expression of TSG-6 in the eutopic endometrium from women with endometriosis. EUR J OBSTET GYN R B. 2017;218:99–105. doi: 10.1016/j.ejogrb.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Cavagna M, Mantese J. Biomarkers of endometrial receptivity—a review. Placenta . 2003;24:S39–S47. doi: 10.1016/s0143-4004(03)00184-x. [DOI] [PubMed] [Google Scholar]
- 24.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 25.Leong KG, Karsan A. Recent insights into the role of notch signaling in tumorigenesis. Blood . 2006;107:2223–33. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 26.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, et al. Notch signaling is essential for vascular morphogenesis in mice. GENE DEV Journal . 2000;14:1343–52. [PMC free article] [PubMed] [Google Scholar]
- 27.Torry DS, Leavenworth J, Chang M, Maheshwari V, Groesch K, Ball ER, et al. Angiogenesis in implantation. J ASSIST REPROD GEN . 2007;24:303–15. doi: 10.1007/s10815-007-9152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su R-W, Strug MR, Joshi NR, Jeong J-W, Miele L, Lessey BA, et al. Decreased notch pathway signaling in the endometrium of women with endometriosis impairs decidualization. J CLIN ENDOCR METAB. 2015;100:E433–E42. doi: 10.1210/jc.2014-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, et al. Notch3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer research. 2008;68:1881–8. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- 30.Van Sinderen M, Cuman C, Gamage T, Rainczuk K, Osianlis T, Rombauts L, et al. Localisation of the notch family in the human endometrium of fertile and infertile women. J MOL HISTOL . 2014;45:697–706. doi: 10.1007/s10735-014-9587-y. [DOI] [PubMed] [Google Scholar]
- 31.Estrach S, Cordes R, Hozumi K, Gossler A, Watt FM. Role of the notch ligand Delta1 in embryonic and adult mouse epidermis. J INVEST DERMATOL . 2008;128:825–32. doi: 10.1038/sj.jid.5701113. [DOI] [PubMed] [Google Scholar]
- 32.Liu N, Zhang J, Ji C. The emerging roles of notch signaling in leukemia and stem cells. Biomarker research . 2013;1:23. doi: 10.1186/2050-7771-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]